Abstract
Diatoms have attracted considerable attention due to their success in diverse environmental conditions, which probably is a consequence of their complex origins. Studies of their metabolism will provide insight into their adaptation capacity and are a prerequisite for metabolic engineering. Several years of investigation have led to the development of the genome engineering tools required for such studies, and a profusion of appropriate tools is now available for exploring and exploiting the metabolism of these organisms. Diatoms are highly prized in industrial biotechnology, due to both their richness in natural lipids and carotenoids and their ability to produce recombinant proteins, of considerable value in diverse markets. This review provides an overview of recent advances in genetic engineering methods for diatoms, from the development of gene expression cassettes and gene delivery methods, to cutting-edge genome-editing technologies. It also highlights the contributions of these rapid developments to both basic and applied research: they have improved our understanding of key physiological processes; and they have made it possible to modify the natural metabolism to favour the production of specific compounds or to produce new compounds for green chemistry and pharmaceutical applications.
This article is part of the themed issue ‘The peculiar carbon metabolism in diatoms’.
Keywords: genetic engineering, metabolic engineering, biotechnology
1. Introduction
Diatoms are the most prevalent microalgae worldwide due to their ability to grow in a wide range of environments, from lakes to oceans. They account for about 40% of marine primary production and are, alone, responsible for the production of one-fifth of the oxygen we breathe [1]. In addition to their ecological importance, the complex evolutionary history and outstanding physiological properties of diatoms have attracted considerable scientific interest, both because of their fantastic potential and because of their possible uses in biotechnological applications [2–4]. The identification or development of models appears to be essential for such studies. But how can we best define a scientific model organism? In our view, the most appropriate definition is that given by Rachel A. Ankeny in 2011: ‘Model organisms are non-human species that are extensively studied in order to understand a range of biological phenomena, with the hope that data and theories generated through use of the model will be applicable to other organisms, particularly those that are in some way more complex than the original model’ [5, p. 313]. Based on these criteria, several diatom species can be considered appropriate models. These species include Thalassiosira pseudonana for silica biomineralization, Phaeodactylum tricornutum for carbohydrate metabolism, the xanthophyll cycle and lipid metabolism, Cylindrotheca fusiformis for ecophysiological and silicification research, Pseudonitzschia for both sexual reproduction and harmful algal blooms, Chaetoceros for harmful algal blooms and Fragilariopsis cylindrus for algal adaptation to polar marine conditions. However, an organism can be considered a model organism only if it can be manipulated experimentally.
In recent decades, systems biology research and genetic engineering approaches have been developed to improve our understanding of complex biological systems and for the manipulation of such systems. Systems biology combines high-throughput experimentation with large-scale ‘-omics’ technologies (genomics, transcriptomics, proteomics, metabolomics and fluxomics), quantitative analysis and modelling, to provide insight into processes contributing to the organization and dynamics of cells. Genetic approaches can be used to identify and confirm gene functions and to design model organisms, and such approaches have been extremely successful in recent years. This review focuses on systems and approaches developed for the genetic manipulation of diatoms for both basic research, through functional analyses of diatom genomes; and applied research, by making it possible to improve strains for biotechnological applications.
2. Molecular genetic approaches to understand and manipulate diatom model species
The tremendous accumulation of genomic and transcriptomic data (ensemblgenomes.org; genomeportal.jgi.doe.gov) over the last decade and the development of screening methods for the production of specific molecules have provided valuable information about the regulation of diatom metabolism. Genetic engineering techniques have opened up possibilities for manipulating the genetic heritage of diatom cells by modifying, deleting and introducing genomic sequences. Such manipulations are essential for investigations of the function of genes and for improving diatom metabolic potential.
(a). Creation of a gene expression toolbox
Genetic engineering requires procedures for introducing DNA into diatom cells and for ensuring its integration into the host genome for gene expression. This requires (i) the assembly of a vector carrying all the structural elements controlling transgene expression (e.g. promoters and terminators of transcription, 5′UTR, 3′UTR), (ii) the identification of selectable markers for the isolation of transformed cells, and (iii) the development of efficient methods for stable DNA delivery.
(i). Regulatory elements driving efficient transgene expression
Expression vectors are entities designed to carry the gene of interest plus the regulatory sequences (promoters and terminators) driving gene expression. Here, we describe some constitutive and inducible endogenous promoters that have been used to drive transgene expression in diatoms. Endogenous promoters have been obtained from genes encoding a chlorophyll a/c-binding light-harvesting complex protein (Lhcf, formerly called fcp; about 15 Lhcf genes are known, Lhcf1-15 [6]), a histone gene (h4) and the elongation factor 2 (ef2) genes. All these promoters are constitutive, driving constant, high levels of transgene expression. The Lhcf promoters were initially identified in P. tricornutum, and promoters of this type have proved effective in various diatom species including both pennates and centrics [7–12]. These promoters are widely used, but their dependence on light renders them unsuitable for studies of transgenes in conditions of darkness [13]. The h4 promoter is one possible alternative; it drives light-independent expression, but at levels lower than those that can be achieved with Lhcf promoters [14]. This promoter, which was first used in P. tricornutum, has been successfully identified and used in other diatom species (table 1). A constitutive promoter derived from the elongation factor 2 (ef2) gene has recently been used to drive expression to levels greater than those achieved with Lhcf2 promoters (at least 1.2 times higher) in P. tricornutum [17]. Another promoter (Lhcr5), derived from the red algal-like light-harvesting complex protein (Lhcr, also constituting a multigene family), has also been used as a constitutive promoter in Chaetoceros gracilis [11]. Diverse promoters are thus available, but the terminators used to date have been limited to those of the Lhcf1, Lhcf9, nr (nitrate reductase), rbcL (rubisco small subunit) and Lhcr14 genes [7,8,10–12,26,27]. Most expression vectors used can express one gene of interest. However, new versions of expression vectors able to express two genes simultaneously have recently been developed. These vectors are valuable for co-localization studies [32] and for work with metabolic pathways controlled by more than one gene [33].
Table 1.
Toolbox for diatom transformation including selectable markers, reporters and promoters used. ACCase, acetyl-CoA carboxylase gene; ca1, beta-carbonic anhydrase 1, cat, chloramphenicol acetyltransferase, conferring resistance to chloramphenicol; CaMV, cauliflower mosaic virus 35 S; CMV, cytomegalovirus; RSV, Rous sarcoma virus; ClP1, Chaetoceros lorenzianus-infecting DNA virus; Cf-Lhcf1, Lhcf1 promoter from Cylindrotheca fusiformis; ef2, elongation factor 2; fbp1, ferrichrome-binding protein 1; fld, flavodoxin; GFP, green fluorescent protein; YFP, yellow fluorescent protein; CFP, cyan fluorescent protein; GUS, beta-glucuronidase; Isi1, iron starvation-induced protein 1; LUC, luciferase; nat, nourseothricin acetyl transferase, conferring resistance to nourseothricin; nptII, neomycin phosphotransferase II, conferring resistance to neomycin; psba, D1 polypeptide of the photosystem II; rbcL, rubisco large subunit gene; sat, streptothricin acetyl transferase, conferring resistance to nourseothricin; sh ble, conferring resistance to zeocin or phleomycin.
| diatom species | transformation methods (B) biolistic (E) electroporation (C) conjugation |
selectable gene or reporter | promoters developed (C) constitutive (I) inducible (H) heterologous |
|
|---|---|---|---|---|
| pennates | Phaeodactylum tricornutum | B |
sh ble [7,8,12], nat [12], nptII [12], cat [7,12], sat [12] LUC [8], GUS [12], GFP [12], YFP [15], CFP [16] |
(C): Lhcf 1,2,3,5,6 [7,8,12], h4 [14], ef2 [17] (I): nr [18], fbp1 [19], fld [19], Isi1 [19], ca1 [20], (H): Cf-Lhcf1 [21], CMV [22], RSV [22], CaMV [22], ClP1 [21] |
| E | cat [23], sh ble [24,25], GUS [24,25], GFP [24,25] | (C): Lhcf1-2 [24,25], (I): nr [23] |
||
| C |
sh ble [26], GFP [26], YFP [26], CFP [26] |
(C): Lhcf2 [26] (I): nr [26] |
||
| chloroplast via E [27] and B [28] |
cat [27], GFP [27] psba [28] |
(C): rbcL [27] | ||
| Cylindrotheca fusiformis | B | sh ble [10], GFP [10] | (I): nr [10] | |
| Navicula saprophila | B | nptII [29] | (C): ACCase [29] | |
| Fistulifera sp. | B | nptII [9], GFP [9] | (C): Lhcf2 [9] (C) h4 [9], (H): Pt Lhcf2 [9], RSV [9], CaMV [9] |
|
| Pseudonitzschia multistriata | B | sh ble [30] | (C): h4 [30] | |
| Pseudonitzschia arenysensis | GUS [30], GFP [30] | |||
| centrics | Cyclotella cryptica | B | nptII [29] | (C): ACCase [29] |
| Thalassiosira weissflogii | B | GUS [8] | (C): Lhcf2 [8] | |
| Thalassiosira pseudonana | B |
sh ble [31], nat [31], GFP [31] |
(C): Lhcf9 [31] (I): nr [31] |
|
| C | nat [26], YFP [26] | (C): Lhcf9 [26] | ||
| Chaetoceros gracilis | E | nat [11], GFP [11], LUC [11] | (C): Lhcr5 [11] (I): nr [11] |
Inducible promoters that can be used to switch transgene expression on and off in an efficient and reversible manner have been found. These promoters are particularly useful for producing toxic compounds for the pharmaceutical or green chemistry industries. One of the most widely used inducible promoters in diatoms is the nitrate reductase (nr) promoter. This promoter is induced by the presence of nitrate as the sole nitrogen source in the medium, and is inactivated in the presence of ammonium ions [10]. The nr promoter, which was first investigated in Cylindrotheca fusiformis, has since been identified and used in T. pseudonana, P. tricornutum and Chaetoceros [10,11,18,31]. However, a recent study revealed that the promoter nr shows residual activity in the absence of nitrate, leading to a low level of gene expression [34]. Promoters induced by iron starvation and derived from the iron starvation-induced protein 1 (Isi1), ferrichrome-binding protein 1 (fbp1) and flavodoxin (fld) genes [19] have recently been described, providing additional possibilities for the modulation of gene expression in diatoms. A CO2-responsive promoter derived from the plastid carbonic anhydrase gene (ca1) of P. tricornutum has also been identified [20].
Heterologous or synthetic promoters are required to drive transgene expression without a negative effect on endogenous regulatory networks. Examples of such promoters are the Lhcf2 promoter from P. tricornutum, which is used in Fistulifera species [9], and the Lhcf1 promoter from C. fusiformis, which is used in P. tricornutum [21]. In addition, viral promoters have been reported to be functional in P. tricornutum species. Thus, the CaMV (cauliflower mosaic virus) promoter, the CMV (cytomegalovirus) promoter and the RSV promoter (Rous sarcoma virus) drive reporter gene expression [22]. Similarly, the ClP1 promoter from diatom-infecting viruses can drive stable expression and does so to levels higher than those observed with endogenous diatom promoters and other viral promoters (CaMV, CMV and RSV) [21]. Overall, the diversity of diatom promoters now available opens up new opportunities to control gene expression at specific times during culture and to fine-tune enzyme levels, thereby facilitating metabolic pathway engineering.
(ii). Genetic reporter systems and selectable markers
The genetic reporter systems widely used to identify and characterize promoters in diatoms (see above) have also greatly contributed to studies of gene expression and regulation, and the spatial localization of proteins. Reporter genes for which expression can be monitored on the basis of enzymatic activity, notably the bacterial β-glucuronidase gene (GUS, also known as uidA) [12] and the firefly luciferase (LUC) gene [8], can be used to assess promoter activity. Fluorescent proteins such as the green fluorescent protein (GFP) [12,26], the cyan fluorescent protein (CFP) [26] and the yellow fluorescent protein (YFP) [15,26] have been used as reporters for monitoring protein localization in vivo. Selectable markers conferring resistance to antibiotics have been developed for diatoms to facilitate the isolation of genetic transformants. The most commonly used selectable markers in diatoms are nourseothricin (nat), neomycin (nptII) and phleomycin/zeocin (sh ble) (table 1), and the appropriate concentrations differ between species (table 2).
Table 2.
Antibiotic concentrations used to select resistant transformants.
| selectable markers | antibiotics | species | concentration |
|---|---|---|---|
| sh ble | zeocin | Phaeodactylum tricornutum | 100 µg ml−1 [7] |
| Cylindrotheca fusiformis | 1 mg ml−1 [10] | ||
| Thalassiosira pseudonana | 1 µg ml−1 [31] | ||
| Pseudonitzschia multistriata | 50 µg ml−1 [30] | ||
| Pseudonitzschia arenysensis | 50 µg ml−1 [30] | ||
| sh ble | phleomycin | Phaeodactylum tricornutum | 20 or 100 µg ml−1 [8,26] |
| nat | nourseothricin | Phaeodactylum tricornutum | 300 µg ml−1 [35] |
| Chaetoceros gracilis | 400 µg mg ml−1 [11] | ||
| Thalassiosira pseudonana | 100 µg ml−1 [31] | ||
| nptII | neomycin | Phaeodactylum tricornutum | 100 µg ml−1 [12] |
| Navicula saprophila | 25 µg ml−1 [29] | ||
| Fistulifera sp. | 500 µg ml−1 [9] | ||
| Cyclotella cryptica | 50 or 100 µg ml−1 [29] |
(iii). Gene delivery methods for the nuclear and plastid genomes
Three techniques for nuclear gene transfer are commonly used in diatoms: biolistics, electroporation and bacterial conjugation. In biolistics, also known as ‘particle bombardment’ or the ‘gene gun technique’, fine particles (microgold or tungsten) coated with DNA are used to deliver transgenes directly into diatom cells. This method has the advantage of being suitable for use with several diatom species. Both pennate and centric diatoms have been successfully transformed with this technique (table 1). Transformation efficiency is from 10−7 to 10−8 diatom cells µg−1 DNA [8,21,35,36]. Biolistic guns can be used with diverse species, and to introduce multiple transgenes simultaneously [35], such that it is feasible to construct a genetic circuit for synthetic biology. This method is efficient but expensive due to the equipment required and running costs involved. Consequently, another cheaper and simpler method has been developed: electroporation. This technique, based on the application of a strong electrical field to enhance pore formation in the cell membrane, has been successfully used to transform C. gracilis [11] and P. tricornutum [23–25]. Various protocols, differing in pulse length, the type and duration of the electrical field and the topology of the DNA delivered, have been used in this second species [23–25]. Several studies have shown that markedly higher transformation efficiencies can be achieved with a linear than with a circular plasmid. Transformation frequency varies from 2000 to 4000 transgenic colonies per 108 cells, an efficiency 10–100 times higher than that for biolistic approaches [24,25]. However, the success of this technique seems to depend on the laboratory and the apparatus used. The third technique used is an episome delivery method involving bacterial conjugation. It is effective for both DNA delivery (with a transformation efficiency 1000 times higher than that for biolistics) and long-term protein production [26]. This strategy circumvents the major issue of uncontrolled genetic modifications due to the random integration into the chromosome of plasmids delivered by biolistic and electroporation methods. The use of this method is likely to increase over the next few years.
Genetic manipulation of the chloroplast genome has also received considerable attention due to the importance of this organelle for fundamental biological processes, such as photosynthesis, and its capacity to serve as a cellular biofactory. The chloroplast has all the essential characteristics for stable and efficient transgene expression: (i) a high homologous recombination frequency facilitating gene insertion and preventing position effects, (ii) the organization of plastid genes into operons, making it possible to insert multiple genes, and (iii) an absence of epigenetic marks ensuring high levels of stable gene expression [37]. Plastids are ideal subcellular compartments for the production of large amounts of protein without interfering with central metabolism [38]. Algal chloroplast transformation was first achieved in 1988, in the model green alga Chlamydomonas reinhardtii, but it took another 20 years for chloroplast transformation to be demonstrated in diatoms [27,28]. This was largely due to the complexity of diatom plastids, which are surrounded by four membranes because of secondary endosymbiosis rather than the two membranes of primary plastids of land plants and green and red algae. Two studies have reported plastid transformation by homologous recombination in P. tricornutum [27,28]. However, as plastid transformation techniques are not well established in the diatom community, alternative strategies are frequently used. For example, the gene of interest fused to a sequence encoding a plastid-targeting signal sequence can be integrated into the nuclear genome, such that the protein is produced and transported into plastids. Plastid-targeting signal sequences consist of a signal peptide followed by a transit peptide-like sequence. These bipartite presequences are sufficient for plastid import [16,39]. However, plastid import requires the presence of the conserved ‘ASAFAP’-motif between the signal peptide and the transit peptide-like domains [16]. The functionality of plastid-targeting signal sequences has been demonstrated by fusion to the N-terminus of GFP [16,39,40].
(b). Genetic engineering approaches for the manipulation of diatom genomes
Two approaches are widely used to study gene function: (i) classical genetic approaches based on random insertional mutagenesis with a vector-encoded selectable marker gene and high-throughput screening for a specific phenotype and (ii) reverse genetic approaches involving the disruption or modification of a specific gene. The diatom community has tended to focus on reverse genetics rather than on classical genetics. The reasons for this include a desire to link physiological processes to gene functions, and the difficulties of gene inactivation in diploids like diatoms (random insertion is vanishingly unlikely to provide inactivation of both alleles of a gene). In reverse genetics, the goal is to investigate the consequences of changes to a particular gene and thereby to infer its function. Two approaches are used: (i) the deregulation of gene expression through the overexpression of endogenous or heterologous genes and (ii) the down-regulation of endogenous genes and genome-editing technologies based on the use of double-strand-break (DSB) mechanisms efficiently providing targeted modifications.
(i). Modulation of gene expression
The development of expression vectors and delivery methods has made it possible to increase the expression of a specific gene and thus (i) to study the localization of the protein concerned through fusion to a fluorescent marker, (ii) to assess the consequences of this overexpression for metabolism, (iii) to complement loss of function in a mutant strain, (iv) to investigate gene functions across species barriers, by exploring functional conservation in heterologous expression experiments, and (v) to create new functions through the introduction of foreign genes controlling a metabolic pathway, the starting point for synthetic biology (see §3 on Metabolic engineering). Today, only the first two of these approaches have been used in diatoms but we expect a substantial increase over the next few years. Methods based on the knock-down of gene expression have been developed for diatoms, making it possible to link genes to particular functions: RNA interference (RNAi) [14,41,42] and more recently the artificial miRNAs [43]. The proof of concept of RNAi was in 2009 with the GUS reporter gene [14]. Constructs expressing either anti-sense or inverted repeat sequences were introduced into a P. tricornutum GUS transgenic strain and repressed GUS expression. RNAi is an extremely powerful tool for modulating gene expression in diatoms: it has already been exploited to confirm the functions of key enzymes in metabolic networks, and to increase the production of valuable compounds in both P. tricornutum and T. pseudonana [14,41,42,44,45].
(ii). Targeted genome engineering with site-specific nucleases
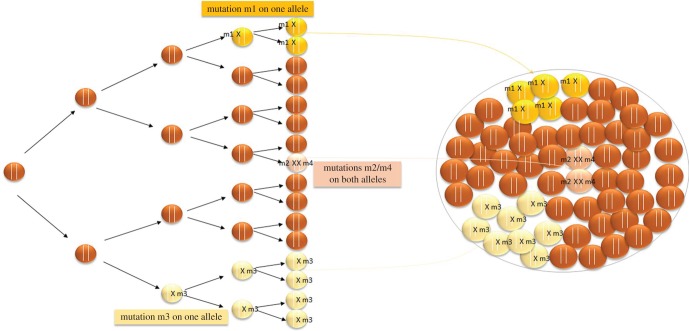
The use of double-strand DNA break (DSB) repair mechanisms has recently emerged as a revolutionary method for highly efficient targeted genome modifications. Targeted genome engineering has several advantages over conventional methods based on the modulation of gene expression. First, it involves transgene integration at a specific locus. This avoids problems of genome instability and the absence of transgene expression sometimes encountered with classical methods based on random vector integration. Second, it can be used to generate deletions or to introduce mutations into specific target genes, a particularly difficult task in diploid organisms, but essential for both basic and applied research. The frequency of homologous recombination in microalgae is very low (less than 10−6), but molecular scissors able to induce DSBs at a specific locus increases this frequency by at least three orders of magnitude [35,46]. Three classes of sequence-specific nucleases have been successfully used to induce targeted modifications of diatom genomes (meganucleases, TALENs, CRISPR/Cas9). All of these nucleases are derived from natural molecules, the basic functions of which have been modified to create powerful tools for genome engineering. Meganucleases (MNs) are derived from the homing endonucleases involved in the lateral transfer of introns or inteins in yeasts [47,48]. TALENs are derived from the transcriptional activator-like effectors produced by the plant pathogenic bacterium Xanthomonas, which activate the transcription of a specific plant gene promoting bacterial infection [49,50]. Finally, the CRISPR/Cas9 system is derived from an RNA-guided DNA cleavage defence system present in bacteria and archaea and conferring a degree of ‘acquired immunity’ against bacteriophages through storage of the foreign DNA in their genome memory, at the CRISPR locus [51,52]. Two peculiarities in diatoms complicate genome editing: first the diatoms are diplonts, which means that a mutagenic event must occur in both alleles to inactivate the targeted gene; and second, most DSBs are repaired faithfully. Therefore, the initially transformed cell is not necessarily subject to the mutagenesis and each resulting colony is a mixed population of cells with or without mutations and with mutations of different types (mosaicism). Consequently, a subsequent additional subcloning step is required (figure 1). Such mosaicism has been described for the yeast transformed with the I-CreI meganuclease [53].
Figure 1.
Illustration of the mosaicism concept within a colony obtained from transformation with an engineered nuclease. Phaeodactylum tricornutum cells were transformed with engineered nuclease designed to recognize a specific sequence. The majority of the double-strand breaks induced by the nucleases are repaired by faithful re-ligation such that there is no mutation. However, some double-strand breaks are repaired by the non-homologous end-joining (NHEJ) mechanism, in which the broken chromosomes are rejoined, often imprecisely, thereby introducing nucleotide changes at the break site. Consequently, each colony is a mixed population of cells with or without mutations and with mutations of different types (the mutation m1, m2, m3 and m4 can be different). The inactivation of one gene requires a mutagenic event in both alleles, which is not the most frequent case observed within a colony. In addition, the simultaneous introduction of nuclease and a DNA template with sequences displaying similarity to the targeted sequence leads to the formation of colonies harbouring a mixed population of cells with or without integrating DNA matrix and with or without mutation induced by NHEJ.
The first proof-of-concept for targeted genome modification in diatoms was provided in 2013; a high frequency of both targeted mutagenesis and homologous recombination was achieved in P. tricornutum using two types of designer nucleases: MNs and TALENs [35,54,55]. The proof-of-concept for metabolic engineering in diatoms was provided a few months later, with the creation of a strain producing large amounts of lipids paving the way for a new field of investigation. This was done by inactivating a key gene involved in the storage of energy as sugars. Several groups have since produced and/or used TALEN technology in P. tricornutum [35,56,57] and in T. pseudonana [54]. The CRISPR/Cas9 system is very efficient and easy to use, and can produce multiple gene modifications in several organisms [58]. Consequently, several groups have embarked on the evaluation of the CRISPR/Cas9 system in diatoms [36]. Two groups have successfully used the system in P. tricornutum and T. pseudonana [36,59]. For genome editing in P. tricornutum, a codon-optimized Cas9 was placed under the control of an Lhcf2 promoter and the guide RNA was placed under the control of the P. tricornutum U6 snRNA promoter [36]. In T. pseudonana, a human codon-optimized Cas9 and a U6 small nuclear RNA from T. pseudonana were used [59].
3. Metabolic engineering by genetic engineering approaches
(a). A greater knowledge of physiological mechanisms
Genome engineering methods for expressing, silencing or deleting the gene of interest greatly facilitate studies of fundamental questions regarding the physiology, the evolutionary ecology and the metabolism of diatoms. This section will provide a brief overview of how genome engineering has already led to progress. We will present specific examples illustrating the three key areas considered above rather than providing an exhaustive description of the use of genome engineering in diatom research.
(i). Elucidating the capacity of diatoms to tolerate intense light
Diatoms must cope with rapid changes in light intensity and periodic exposure to very high light intensity; these microalgae are extremely tolerant to intense light [60,61]. Diatoms have various strategies for dissipating the excess light as heat, through so-called non-photochemical quenching (NPQ). RNAi and overexpression approaches have been used to analyse the mechanisms underlying NPQ. These studies led to the identification of the key enzyme (de-epoxidase) catalysing the conversion of diatoxanthin into diadinoxanthin, and demonstrated the role of LCHX (a protein from the light-harvesting protein family) in high-light stress responses [62,63]. The RNAi experiment also revealed that the photoreceptors (blue light photoreceptors cryptochrome PtCPF1 and aureochromes), which can perceive light, may play a much more important role in photoacclimation than those of higher plants [14,18]. Combined genetic, physiology and biophysics approaches have shown that diatoms optimize their photosynthesis through extensive energy exchanges between plastids and mitochondria [64].
(ii). Improving our knowledge of carbon fixation
The CO2-concentrating mechanism (CCM) enables diatoms to play an important role in the global carbon cycle by avoiding CO2 limitation in seawater. However, little is known on the carbon fixation pathway in diatoms. Over the last two decades, several groups have highlighted the existence of a biophysical C3 CCM in both P. tricornutum and T. pseudonana, by RNAi methods and subcellular localization of the putative carbonic anhydrases and bicarbonate transporters [65,66]. In addition to the C3 pathway, it has been suggested that diatoms might also make use of the C4 CCM pathway, like higher plants [67]. However, the absence of a plastid decarboxylase in P. tricornutum and the lack of a significant effect on inorganic carbon acquisition after silencing of the pyruvate-orthophosphate dikinase involved in the C4 pathway [68] have raised debates about the possible presence of a conventional CCM pathway in diatoms [66].
(iii). Identifying enzymes controlling carbohydrate metabolism pathways
The biochemical pathway for the synthesis of the principal storage carbohydrate of diatoms, chrysolaminarin, has been studied. Chrysolaminarin is a β-1,3-glucan chain with β-1,6-branches, stored in diatom vacuoles. By contrast, plants and green algae store carbohydrates (starch consisting of α-1,4-glucans) in chloroplasts. This vacuolar localization implies that the degradative enzymes are also localized there. Several studies have been undertaken to elucidate this particularity. One of the UDP-glucose pyrophosphorylases (UGP1) has been shown to be involved in chrysolaminarin synthesis: knocking the expression of the UGP1 gene out or down significantly decreases the amount of chrysolaminarin in P. tricornutum [35,69]. Two of the three β-1,6-transglycosylases of diatoms [70] catalysing the branching of β-1,3-glucan with β-1,6-glucan units have been found in vacuoles and characterized by complementing a yeast mutant [70]. The key enzyme forming the β-1,3-glucan bonds of chrysolaminarin, β-1,3-glucan synthase, has been identified and characterized by Huang and colleagues (data to be published in a forthcoming paper).
These three examples illustrate the power of the genome engineering techniques already available in diatoms to dissect complex and sophisticated mechanisms. They will undoubtedly contribute to work on molecular mechanisms underlying the ecological success of diatoms, to increasing our knowledge of diatom physiology particularly cell wall biogenesis and regulation of the life cycles, and to discovering enzymes controlling metabolism.
(b). Redesigning the metabolic potential of cells for biotechnological applications
A goal of industrial biotechnology is to develop new bio-based technologies to convert renewable raw materials into chemicals, useful materials and bioenergy. One particularly challenging domain is metabolic engineering, or the redesign of natural metabolic pathways to increase the production of desired metabolites and to produce new compounds (for cost-effective fuel, or synthesis of valuable chemicals or drugs) [71,72]. Diatoms have substantial biotechnological potential: they contain an abundance of marketable bioproducts (lipids, pigments and nanomaterials); production can be enhanced and new compounds created by genetic engineering; industrial processes involving diatoms are cost-effective; diatoms perform well in large-scale culture; and they are robust when faced with harsh environmental conditions [73]. P. tricornutum is the only diatom species that has been demonstrated to be robust in industrial processes, as demonstrated by its industrial uses in aquaculture and eicosapentaenoic acid production [74–76].
(i). Redesigning endogenous metabolic pathways to increase yield
The ability of diatoms to produce large amounts of lipids (30% of dry weight, and up to 46% in conditions of nitrate starvation) has excited considerable interest in their possible use as a diesel fuel feedstock, or food or feed [73,77]. Twenty years ago, the Aquatic Species Program selected from the 3000 microalgae strains screened, a list of the 50 most promising microalgal strains for biofuel production: 65% are diatoms [73,78]. One of the major biotechnological challenges is overcoming the dependence of this lipid production on stress, which results in lower biomass productivity when the lipid metabolism pathway is engineered. Several approaches have been used to increase lipid content and storage in triacylglycerol (TAG) form or to modify the characteristics of the lipids generated. In general, shorter carbon chain lengths in biofuels are desirable for optimal cold-flow properties [73,79] and saturated fatty acids are more desirable because they confer good ignition quality (cetane number) on the fuel. By contrast, omega-3 (ω-3) fatty acids are long chain polyunsaturated fatty acids (C20:5, C22:4) with significant health benefits as human food. The lipid metabolism in three diatoms has been manipulated: P. tricornutum, T. pseudonana and F. solaris. Lipid content was increased by (i) overproducing enzymes involved in fatty-acid biosynthesis [80], (ii) overexpressing genes involved in lipid storage in the TAG form [81–83], (iii) silencing or deleting genes controlling competing pathways, such as the carbohydrate pathway [35,69], (iv) silencing genes involved in lipid catabolism [42,84] and (v) knocking down expression of the nitrate reductase gene [85]. Desaturases and thioesterases have also been overexpressed to modify fatty-acid profiles [75,86–88]. Specific modifications of the diatom genome have resulted in the production of several high-lipid producers of potential interest for industrial purposes.
Another class of compounds, carotenoids, has been studied in detail in diatoms. These molecules are important in light capture, protecting cells against the damaging effects of free radicals (they are also precursors in the synthesis of hormones in humans and are used as food-colouring agents and in animal feed supplements) [89]. Carotenoid production in wild-type microalgae has been commercially successful. The key genes involved in the complex synthetic pathway have been identified, including phytoene synthase [43,90–92], zeaxanthin epoxidase [93] and violaxanthin epoxidase [63], a prerequisite for the production of carotenoids for industrial applications.
(ii). Production of new compounds
It would be useful to be able to introduce heterologous genes controlling a metabolic pathway absent from wild-type strains into algae and diatoms. Manipulations of this type can be used to create new products for industry and to reduce costs through the production of valuable co-products. The first proof-of-concept that diatoms can serve as efficient cell factories for the production of new compounds was provided in 2011. A human IgG antibody against the hepatitis B virus surface protein was produced in P. tricornutum (HBsAg was 9% of total soluble protein) [94,95]. A bioplastic poly-3-hydroxybutyrate has similarly been produced in P. tricornutum (10.6% by cellular dry weight) following overexpression of the three genes controlling the metabolic pathway [33]. These studies constitute a major advance and modifications to the N-glycosylation pathway are currently being made to develop an efficient platform for the production of pharmaceutical compounds [96,97].
(iii). Nanocarriers for therapeutic applications
A recent study suggested that diatoms can be considered as nanoporous silica-based materials for drug delivery in anticancer treatment rather than as molecule producers [98]. Thalasiossira pseudonona was manipulated so as to serve as therapeutic biosilica nanoparticles: antibodies were attached to the surface for the specific recognition of cancer cells, and the same diatom silica particle was loaded with drugs to destroy the tumour. This exciting progress should generate further innovative ideas in the field.
4. Conclusion
We are at the dawn of a new era in diatom biology. The first brick has been laid and the building of the pyramid has just begun. The accumulation of genomic and transcriptomic data associated with the development of cheap and easy-to-use genome-editing technologies will facilitate continued rapid progress in diatom biology. In this review, we summarize a range of recent technical advances from transformation methodologies to genome engineering strategies, and the consequences for basic and applied research. Henceforth, new fields can be addressed, for example exploring the extraordinary capacity of diatoms to adapt to environmental conditions, the molecular basis of biofilm interactions, diatom morphological versatility and the control of sexual reproduction in diatoms. The development of genetic tools has paved the way for metabolic pathway engineering for biotechnological applications, as illustrated by the generation of high-lipid and carotenoid producers. In the next few years, yields should improve, due in part to genomic modifications made possible by the development of the CRISPR/Cas9 system. The extension of these tools to an increasing number of diatom species will be of great value for both basic and applied research.
Acknowledgment
We thank Dr Thiyagarajan Gnanasekaran for proofreading and discussion.
Data accessibility
This article has no additional data.
Authors' contribution
F.D. conceived and designed the format of the manuscript. F.D. and W.H. contributed equally in writing the manuscript.
Competing interests
We declare we have no competing interests.
Funding
The postdoctoral grant for Weichao Huang was funded by the Région Midi-Pyrénées 15058490. This work was supported by the grant 15058490 from Région Midi-Pyrénées (financial support for ‘Accueil d'Equipes d'Excellence’).
References
- 1.Falkowski PG, Barber RT, Smetacek V. 1998. Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–207. ( 10.1126/science.281.5374.200) [DOI] [PubMed] [Google Scholar]
- 2.Kroth P. 2007. Molecular biology and the biotechnological potential of diatoms. Adv. Exp. Med. Biol. 616, 23–33. ( 10.1007/978-0-387-75532-8_3) [DOI] [PubMed] [Google Scholar]
- 3.Kröger N. 2007. Prescribing diatom morphology: toward genetic engineering of biological nanomaterials. Curr. Opin. Chem. Biol. 11, 662–669. ( 10.1016/j.cbpa.2007.10.009) [DOI] [PubMed] [Google Scholar]
- 4.Bozarth A, Maier U-G, Zauner S. 2009. Diatoms in biotechnology: modern tools and applications. Appl. Microbiol. Biotechnol. 82, 195–201. ( 10.1007/s00253-008-1804-8) [DOI] [PubMed] [Google Scholar]
- 5.Ankeny RA, Leonelli S. 2011. What's so special about model organisms? Stud. Hist. Philos. Sci. Part A 42, 313–323. ( 10.1016/j.shpsa.2010.11.039) [DOI] [Google Scholar]
- 6.Lepetit B, Volke D, Gilbert M, Wilhelm C, Goss R. 2010. Evidence for the existence of one antenna-associated, lipid-dissolved and two protein-bound pools of diadinoxanthin cycle pigments in diatoms. Plant Physiol. 154, 1905–1920. ( 10.1104/pp.110.166454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apt KE, Kroth-Pancic PG, Grossman AR. 1996. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 252, 572–579. [DOI] [PubMed] [Google Scholar]
- 8.Falciatore A, Casotti C, Leblanc C, Abrescia C, Bowler C. 1999. Transformation of nonselectable reporter genes in marine diatoms. Mar. Biotechnol. 239–251. ( 10.1007/PL00011773) [DOI] [PubMed] [Google Scholar]
- 9.Muto M, Fukuda Y, Nemoto M, Yoshino T, Matsunaga T, Tanaka T. 2013. Establishment of a genetic transformation system for the marine pennate diatom Fistulifera sp. strain JPCC DA0580--a high triglyceride producer. Mar. Biotechnol. 15, 48–55. ( 10.1007/s10126-012-9457-0) [DOI] [PubMed] [Google Scholar]
- 10.Poulsen N, Kröger N. 2005. A new molecular tool for transgenic diatoms: control of mRNA and protein biosynthesis by an inducible promoter-terminator cassette. FEBS J. 272, 3413–3423. ( 10.1111/j.1742-4658.2005.04760.x) [DOI] [PubMed] [Google Scholar]
- 11.Ifuku K, Yan D, Miyahara M, Inoue-Kashino N, Yamamoto YY, Kashino Y. 2015. A stable and efficient nuclear transformation system for the diatom Chaetoceros gracilis. Photosynth. Res. 123, 203–211. ( 10.1007/s11120-014-0048-y) [DOI] [PubMed] [Google Scholar]
- 12.Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossman AR, Apt KE. 2000. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J. Phycol. 36, 379–386. ( 10.1046/j.1529-8817.2000.99164.x) [DOI] [Google Scholar]
- 13.Nymark M, Valle KC, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM, Brembu T. 2013. Molecular and photosynthetic responses to prolonged darkness and subsequent acclimation to re-illumination in the diatom Phaeodactylum tricornutum. PLoS ONE 8, e58722 ( 10.1371/journal.pone.0058722) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riso V De, Raniello R, Maumus F, Rogato A, Bowler C, Falciatore A. 2009. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 37, e96 ( 10.1093/nar/gkp448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huysman MJJ, Tanaka A, Bowler C, Vyverman W, De Veylder L. 2015. Functional characterization of the diatom cyclin-dependent kinase A2 as a mitotic regulator reveals plant-like properties in a non-green lineage. BMC Plant Biol. 15, 86 ( 10.1186/s12870-015-0469-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilian O, Kroth PG. 2005. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. Cell Mol. Biol. 41, 175–183. ( 10.1111/j.1365-313X.2004.02294.x) [DOI] [PubMed] [Google Scholar]
- 17.Seo S, Jeon H, Hwang S, Jin E, Chang KS. 2015. Development of a new constitutive expression system for the transformation of the diatom Phaeodactylum tricornutum. Algal Res. 11, 50–54. ( 10.1016/j.algal.2015.05.012) [DOI] [Google Scholar]
- 18.Schellenberger Costa B, Sachse M, Jungandreas A, Bartulos CR, Gruber A, Jakob T, Kroth PG, Wilhelm C. 2013. Aureochrome 1a is involved in the photoacclimation of the diatom Phaeodactylum tricornutum. PLoS ONE 8, e74451 ( 10.1371/journal.pone.0074451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshinaga R, Niwa-Kubota M, Matsui H, Matsuda Y. 2014. Characterization of iron-responsive promoters in the marine diatom Phaeodactylum tricornutum. Mar. Genomics 16, 55–62. ( 10.1016/j.margen.2014.01.005) [DOI] [PubMed] [Google Scholar]
- 20.Harada H, Nakatsuma D, Ishida M, Matsuda Y. 2005. Regulation of the expression of intracellular beta-carbonic anhydrase in response to CO2 and light in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 139, 1041–1050. ( 10.1104/pp.105.065185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadono T, et al. 2015. Characterization of marine diatom-infecting virus promoters in the model diatom Phaeodactylum tricornutum. Sci. Rep. 5, 18708 ( 10.1038/srep18708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakaue K, Harada H, Matsuda Y. 2008. Development of gene expression system in a marine diatom using viral promoters of a wide variety of origin. Physiol. Plant. 133, 59–67. ( 10.1111/j.1399-3054.2008.01089.x) [DOI] [PubMed] [Google Scholar]
- 23.Niu Y-F, Yang Z-K, Zhang M-H, Zhu C-C, Yang W-D, Liu J-S, Li H-Y. 2012. Transformation of diatom Phaeodactylum tricornutum by electroporation and establishment of inducible selection marker. BioTechniques 52, 1–3. ( 10.2144/000113881) [DOI] [PubMed] [Google Scholar]
- 24.Miyahara M, Aoi M, Inoue-Kashino N, Kashino Y, Ifuku K. 2013. Highly efficient transformation of the diatom Phaeodactylum tricornutum by multi-pulse electroporation. Biosci. Biotechnol. Biochem. 77, 874–876. ( 10.1271/bbb.120936) [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, Hu H. 2014. High-efficiency nuclear transformation of the diatom Phaeodactylum tricornutum by electroporation. Mar. Genomics 16, 63–66. ( 10.1016/j.margen.2013.10.003) [DOI] [PubMed] [Google Scholar]
- 26.Karas BJ, et al. 2015. Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 6, 6925 ( 10.1038/ncomms7925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie W-H, Zhu C-C, Zhang N-S, Li D-W, Yang W-D, Liu J-S, Sathishkumar R, Li H-Y. 2014. Construction of novel chloroplast expression vector and development of an efficient transformation system for the diatom Phaeodactylum tricornutum. Mar. Biotechnol. 16, 538–546. ( 10.1007/s10126-014-9570-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Materna AC, Sturm S, Kroth PG, Lavaud J. 2009. First induced plastid genome mutations in an alga with secondary plastids: psba mutations in the diatom Phaeodactylum tricornutum (Bacillariophyceae) reveal consequences on the regulation of photosynthesis. J. Phycol. 45, 838–846. ( 10.1111/j.1529-8817.2009.00711.x) [DOI] [PubMed] [Google Scholar]
- 29.Dunahay TG, Jarvis EE, Roessler PG. 1995. Genetic transformation of the diatoms Cyclotella cryptica and Navicula saprophila. J. Phycol. 31, 1004–1012. ( 10.1111/j.0022-3646.1995.01004.x) [DOI] [Google Scholar]
- 30.Sabatino V, Russo MT, Patil S, d'Ippolito G, Fontana A, Ferrante MI. 2015. Establishment of genetic transformation in the sexually reproducing diatoms pseudo-nitzschia multistriata and Pseudo-nitzschia arenysensis and inheritance of the transgene. Mar. Biotechnol. 17, 452–462. ( 10.1007/s10126-015-9633-0) [DOI] [PubMed] [Google Scholar]
- 31.Poulsen N, Chesley PM, Kröger N. 2006. Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae)1. J. Phycol. 42, 1059–1065. ( 10.1111/j.1529-8817.2006.00269.x) [DOI] [Google Scholar]
- 32.Liu X, Hempel F, Stork S, Bolte K, Moog D, Heimerl T, Maier UG, Zauner S. 2016. Addressing various compartments of the diatom model organism Phaeodactylum tricornutum via sub-cellular marker proteins. Algal Res. 20, 249–257. ( 10.1016/j.algal.2016.10.018) [DOI] [Google Scholar]
- 33.Hempel F, Bozarth AS, Lindenkamp N, Klingl A, Zauner S, Linne U, Steinbüchel A, Maier UG. 2011. Microalgae as bioreactors for bioplastic production. Microb. Cell Factories 10, 81 ( 10.1186/1475-2859-10-81) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu L, Ewe D, Río Bártulos C, Kroth PG, Gruber A. 2016. Rapid induction of GFP expression by the nitrate reductase promoter in the diatom Phaeodactylum tricornutum. PeerJ 4, e2344 ( 10.7717/peerj.2344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daboussi F, et al. 2014. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun. 5, 3831 ( 10.1038/ncomms4831) [DOI] [PubMed] [Google Scholar]
- 36.Nymark M, Sharma AK, Sparstad T, Bones AM, Winge P. 2016. A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 6, 24951 ( 10.1038/srep24951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purton S, Szaub JB, Wannathong T, Young R, Economou CK. 2013. Genetic engineering of algal chloroplasts: progress and prospects. Russ. J. Plant Physiol. 60, 491–499. ( 10.1134/S1021443713040146) [DOI] [Google Scholar]
- 38.Bogorad L. 2000. Engineering chloroplasts: an alternative site for foreign genes, proteins, reactions and products. Trends Biotechnol. 18, 257–263. ( 10.1016/S0167-7799(00)01444-X) [DOI] [PubMed] [Google Scholar]
- 39.Apt KE, Zaslavkaia L, Lippmeier JC, Lang M, Kilian O, Wetherbee R, Grossman AR, Kroth PG. 2002. In vivo characterization of diatom multipartite plastid targeting signals. J. Cell Sci. 115, 4061–4069. ( 10.1242/jcs.00092) [DOI] [PubMed] [Google Scholar]
- 40.Gruber A, Vugrinec S, Hempel F, Gould SB, Maier U-G, Kroth PG. 2007. Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol. Biol. 64, 519–530. ( 10.1007/s11103-007-9171-x) [DOI] [PubMed] [Google Scholar]
- 41.Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, Allen A, Manfredonia A, Falciatore A, Bowler C. 2007. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406, 23–35. ( 10.1016/j.gene.2007.05.022) [DOI] [PubMed] [Google Scholar]
- 42.Trentacoste EM, Shrestha RP, Smith SR, Glé C, Hartmann AC, Hildebrand M, Gerwick WH. 2013. Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc. Natl Acad. Sci. USA 110, 19 748–19 753. ( 10.1073/pnas.1309299110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaur S, Spillane C. 2015. Reduction in carotenoid levels in the marine diatom Phaeodactylum tricornutum by artificial microRNAs targeted against the endogenous phytoene synthase gene. Mar. Biotechnol. 17, 1–7. ( 10.1007/s10126-014-9593-9) [DOI] [PubMed] [Google Scholar]
- 44.Bailleul B, Rogato A, de Martino A, Coesel S, Cardol P, Bowler C, Falciatore A, Finazzi G. 2010. An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proc. Natl Acad. Sci. USA 107, 18 214–18 219. ( 10.1073/pnas.1007703107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huysman MJJ, et al. 2013. AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum). Plant Cell 25, 215–228. ( 10.1105/tpc.112.106377) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rouet P, Smih F, Jasin M. 1994. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA 91, 6064–6068. ( 10.1073/pnas.91.13.6064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, Pâques F. 2011. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr. Gene Ther. 11, 11–27. ( 10.2174/156652311794520111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Epinat J-C, et al. 2003. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 31, 2952–2962. ( 10.1093/nar/gkg375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogdanove AJ, Schornack S, Lahaye T. 2010. TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13, 394–401. ( 10.1016/j.pbi.2010.04.010) [DOI] [PubMed] [Google Scholar]
- 50.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761. ( 10.1534/genetics.110.120717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. ( 10.1038/nature09886) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. ( 10.1126/science.1225829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL. 2002. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 30, 3870–3879. ( 10.1093/nar/gkf495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duchateau P, Daboussi F. 2013. Method for targeted modification of algae genomes. Patent no. WO2014076571.
- 55.Duchateau P, Daboussi F, Sourdive D, Epinat J. 2014. Modified diatoms for biofuel production. Patent no. WO2014207043.
- 56.Fortunato AE, et al. 2016. Diatom phytochromes reveal the existence of far-red-light-based sensing in the Ocean. Plant Cell 28, 616–628. ( 10.1105/tpc.15.00928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weyman PD, Beeri K, Lefebvre SC, Rivera J, McCarthy JK, Heuberger AL, Peers G, Allen AE, Dupont CL. 2015. Inactivation of Phaeodactylum tricornutum urease gene using transcription activator-like effector nuclease-based targeted mutagenesis. Plant Biotechnol. J. 13, 460–470. ( 10.1111/pbi.12254) [DOI] [PubMed] [Google Scholar]
- 58.Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. ( 10.1016/j.cell.2014.05.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hopes A, Nekrasov V, Kamoun S, Mock T. 2016. Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Methods 12, 49 ( 10.1186/s13007-016-0148-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goss R, Lepetit B. 2015. Biodiversity of NPQ. J. Plant Physiol. 172, 13–32. ( 10.1016/j.jplph.2014.03.004) [DOI] [PubMed] [Google Scholar]
- 61.Wilhelm C, Jungandreas A, Jakob T, Goss R. 2014. Light acclimation in diatoms: from phenomenology to mechanisms. Mar. Genomics 16, 5–15. ( 10.1016/j.margen.2013.12.003) [DOI] [PubMed] [Google Scholar]
- 62.Taddei L, et al. 2016. Multisignal control of expression of the LHCX protein family in the marine diatom Phaeodactylum tricornutum. J. Exp. Bot. 67, 3939–3951. ( 10.1093/jxb/erw198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lavaud J, Materna AC, Sturm S, Vugrinec S, Kroth PG. 2012. Silencing of the violaxanthin de-epoxidase gene in the diatom Phaeodactylum tricornutum reduces diatoxanthin synthesis and non-photochemical quenching. PLoS ONE 7, e36806 ( 10.1371/journal.pone.0036806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bailleul B, et al. 2015. Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369. ( 10.1038/nature14599) [DOI] [PubMed] [Google Scholar]
- 65.Kikutani S, Nakajima K, Nagasato C, Tsuji Y, Miyatake A, Matsuda Y. 2016. Thylakoid luminal θ-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc. Natl Acad. Sci. USA 113, 9828–9833. ( 10.1073/pnas.1603112113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hopkinson BM, Dupont CL, Matsuda Y. 2016. The physiology and genetics of CO2 concentrating mechanisms in model diatoms. Curr. Opin. Plant Biol. 31, 51–57. ( 10.1016/j.pbi.2016.03.013) [DOI] [PubMed] [Google Scholar]
- 67.Reinfelder JR, Kraepiel AM, Morel FM. 2000. Unicellular C4 photosynthesis in a marine diatom. Nature 407, 996–999. ( 10.1038/35039612) [DOI] [PubMed] [Google Scholar]
- 68.Haimovich-Dayan M, Garfinkel N, Ewe D, Marcus Y, Gruber A, Wagner H, Kroth PG, Kaplan A. 2013. The role of C4 metabolism in the marine diatom Phaeodactylum tricornutum. New Phytol. 197, 177–185. ( 10.1111/j.1469-8137.2012.04375.x) [DOI] [PubMed] [Google Scholar]
- 69.Zhu B-H, Shi H-P, Yang G-P, Lv N-N, Yang M, Pan K-H. 2016. Silencing UDP-glucose pyrophosphorylase gene in Phaeodactylum tricornutum affects carbon allocation. New Biotechnol. 33, 237–244. ( 10.1016/j.nbt.2015.06.003) [DOI] [PubMed] [Google Scholar]
- 70.Huang W, Río Bártulos C, Kroth PG. 2016. Diatom vacuolar 1,6-β-transglycosylases can functionally complement the respective yeast mutants. J. Eukaryot. Microbiol. 63, 536–546. ( 10.1111/jeu.12298) [DOI] [PubMed] [Google Scholar]
- 71.Cho C, Choi SY, Luo ZW, Lee SY. 2015. Recent advances in microbial production of fuels and chemicals using tools and strategies of systems metabolic engineering. Biotechnol. Adv. 33, 1455–1466. ( 10.1016/j.biotechadv.2014.11.006) [DOI] [PubMed] [Google Scholar]
- 72.Kung Y, Runguphan W, Keasling JD. 2012. From fields to fuels: recent advances in the microbial production of biofuels. ACS Synth. Biol. 1, 498–513. ( 10.1021/sb300074k) [DOI] [PubMed] [Google Scholar]
- 73.Hildebrand M, Davis AK, Smith SR, Traller JC, Abbriano R. 2012. The place of diatoms in the biofuels industry. Biofuels 3, 221–240. ( 10.4155/bfs.11.157) [DOI] [Google Scholar]
- 74.Hamilton ML, Powers S, Napier JA, Sayanova O. 2016. Heterotrophic production of omega-3 long-chain polyunsaturated fatty acids by trophically converted marine diatom Phaeodactylum tricornutum. Mar. Drugs 14, 53 ( 10.3390/md14030053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hamilton ML, Haslam RP, Napier JA, Sayanova O. 2014. Metabolic engineering of Phaeodactylum tricornutum for the enhanced accumulation of omega-3 long chain polyunsaturated fatty acids. Metab. Eng. 22, 3–9. ( 10.1016/j.ymben.2013.12.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamilton ML, Warwick J, Terry A, Allen MJ, Napier JA, Sayanova O. 2015. Towards the industrial production of omega-3 long chain polyunsaturated fatty acids from a genetically modified diatom Phaeodactylum tricornutum. PLoS ONE 10, e0144054 ( 10.1371/journal.pone.0144054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barra L, Chandrasekaran R, Corato F, Brunet C. 2014. The challenge of ecophysiological biodiversity for biotechnological applications of marine microalgae. Mar. Drugs 12, 1641–1675. ( 10.3390/md12031641) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.John Sheehan TDJ. 1998. A look back at the U.S. Department of Energy's Aquatic Species Program: Biodiesel from Algae. Technical Report, Golden, CO: National Renewable Energy Lab. [Google Scholar]
- 79.Radakovits R, Jinkerson RE, Darzins A, Posewitz MC. 2010. Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 9, 486–501. ( 10.1128/EC.00364-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xue J, Niu Y-F, Huang T, Yang W-D, Liu J-S, Li H-Y. 2015. Genetic improvement of the microalga Phaeodactylum tricornutum for boosting neutral lipid accumulation. Metab. Eng. 27, 1–9. ( 10.1016/j.ymben.2014.10.002) [DOI] [PubMed] [Google Scholar]
- 81.Niu Y-F, Zhang M-H, Li D-W, Yang W-D, Liu J-S, Bai W-B, Li H-Y. 2013. Improvement of neutral lipid and polyunsaturated fatty acid biosynthesis by overexpressing a type 2 diacylglycerol acyltransferase in marine diatom Phaeodactylum tricornutum. Mar. Drugs 11, 4558–4569. ( 10.3390/md11114558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guihéneuf F, Leu S, Zarka A, Khozin-Goldberg I, Khalilov I, Boussiba S. 2011. Cloning and molecular characterization of a novel acyl-CoA:diacylglycerol acyltransferase 1-like gene (PtDGAT1) from the diatom Phaeodactylum tricornutum. FEBS J. 278, 3651–3666. ( 10.1111/j.1742-4658.2011.08284.x) [DOI] [PubMed] [Google Scholar]
- 83.Gong Y, Zhang J, Guo X, Wan X, Liang Z, Hu CJ, Jiang M. 2013. Identification and characterization of PtDGAT2B, an acyltransferase of the DGAT2 acyl-coenzyme A: diacylglycerol acyltransferase family in the diatom Phaeodactylum tricornutum. FEBS Lett. 587, 481–487. ( 10.1016/j.febslet.2013.01.015) [DOI] [PubMed] [Google Scholar]
- 84.Barka F, Angstenberger M, Ahrendt T, Lorenzen W, Bode HB, Büchel C. 2016. Identification of a triacylglycerol lipase in the diatom Phaeodactylum tricornutum. Biochim. Biophys. Acta 1861, 239–248. ( 10.1016/j.bbalip.2015.12.023) [DOI] [PubMed] [Google Scholar]
- 85.Levitan O, Dinamarca J, Zelzion E, Gorbunov MY, Falkowski PG. 2015. An RNA interference knock-down of nitrate reductase enhances lipid biosynthesis in the diatom Phaeodactylum tricornutum. Plant J. Cell Mol. Biol. 84, 963–973. ( 10.1111/tpj.13052) [DOI] [PubMed] [Google Scholar]
- 86.Radakovits R, Eduafo PM, Posewitz MC. 2011. Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab. Eng. 13, 89–95. ( 10.1016/j.ymben.2010.10.003) [DOI] [PubMed] [Google Scholar]
- 87.Peng K-T, Zheng C-N, Xue J, Chen X-Y, Yang W-D, Liu J-S, Bai W, Li H-Y. 2014. Delta 5 fatty acid desaturase upregulates the synthesis of polyunsaturated fatty acids in the marine diatom Phaeodactylum tricornutum. J. Agric. Food Chem. 62, 8773–8776. ( 10.1021/jf5031086) [DOI] [PubMed] [Google Scholar]
- 88.Dolch L-J, Maréchal E. 2015. Inventory of fatty acid desaturases in the pennate diatom Phaeodactylum tricornutum. Mar. Drugs 13, 1317–1339. ( 10.3390/md13031317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takaichi S. 2011. Carotenoids in algae: distributions, biosyntheses and functions. Mar. Drugs 9, 1101–1118. ( 10.3390/md9061101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kadono T, et al. 2015. Effect of an introduced phytoene synthase gene expression on carotenoid biosynthesis in the marine diatom Phaeodactylum tricornutum. Mar. Drugs 13, 5334–5357. ( 10.3390/md13085334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eilers U, Bikoulis A, Breitenbach J, Büchel C, Sandmann G. 2016. Limitations in the biosynthesis of fucoxanthin as targets for genetic engineering in Phaeodactylum tricornutum. J. Appl. Phycol. 128, 123–129. ( 10.1007/s10811-015-0583-8) [DOI] [Google Scholar]
- 92.Dambek M, Eilers U, Breitenbach J, Steiger S, Büchel C, Sandmann G. 2012. Biosynthesis of fucoxanthin and diadinoxanthin and function of initial pathway genes in Phaeodactylum tricornutum. J. Exp. Bot. 63, 5607–5612. ( 10.1093/jxb/ers211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eilers U, Dietzel L, Breitenbach J, Büchel C, Sandmann G. 2016. Identification of genes coding for functional zeaxanthin epoxidases in the diatom Phaeodactylum tricornutum. J. Plant Physiol. 192, 64–70. ( 10.1016/j.jplph.2016.01.006) [DOI] [PubMed] [Google Scholar]
- 94.Hempel F, Lau J, Klingl A, Maier UG. 2011. Algae as protein factories: expression of a human antibody and the respective antigen in the diatom Phaeodactylum tricornutum. PLoS ONE 6, e28424 ( 10.1371/journal.pone.0028424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hempel F, Maier UG. 2012. An engineered diatom acting like a plasma cell secreting human IgG antibodies with high efficiency. Microb. Cell Factories 11, 126 ( 10.1186/1475-2859-11-126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mathieu-Rivet E, Kiefer-Meyer M-C, Vanier G, Ovide C, Burel C, Lerouge P, Bardor M. 2014. Protein N-glycosylation in eukaryotic microalgae and its impact on the production of nuclear expressed biopharmaceuticals. Front. Plant Sci. 5, 359 ( 10.3389/fpls.2014.00359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vanier G, Hempel F, Chan P, Rodamer M, Vaudry D, Maier UG, Lerouge P, Bardor M. 2015. Biochemical characterization of human anti-hepatitis B monoclonal antibody produced in the microalgae Phaeodactylum tricornutum. PLoS ONE 10, e0139282 ( 10.1371/journal.pone.0139282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Delalat B, et al. 2015. Targeted drug delivery using genetically engineered diatom biosilica. Nat. Commun. 6, 8791 ( 10.1038/ncomms9791) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.