Abstract
Endothelial cell (EC) activation underlies many vascular diseases, including pulmonary arterial hypertension (PAH). Several members of the E–twenty six (ETS) family of transcription factors are important regulators of the gene network governing endothelial homeostasis, and their aberrant expression is associated with pathological angiogenesis. The goal of this study was to determine whether deficiencies of the ETS family member, Friend leukemia integration 1 transcription factor (FLI1), and its closest homolog, ETS-related gene (ERG), are associated with PAH. We found that endothelial ERG was significantly reduced in the lung samples from patients with PAH, as well as in chronically hypoxic mice. Functional studies revealed that depletion of ERG or FLI1 in human pulmonary ECs led to increased expression of inflammatory genes, including IFN genes, whereas genes regulating endothelial homeostasis and cell–cell adhesion were down-regulated. Simultaneous knockdown of both ERG and FLI1 had synergistic or additive effects on the expression of these genes, suggesting that ERG and FLI1 coregulate at least a subset of their target genes. Functionally, knockdown of ERG and FLI1 induced cell monolayer permeability with a potency similar to that of vascular endothelial growth factor. Notably, stimulation of ECs with Toll-like receptor 3 ligand poly(I:C) suppressed ERG expression and induced ERG dissociation from the IFNB1 promoter, while promoting signal transducers and activators of transcription 1 (STAT1) recruitment. Consistent with the up-regulation of inflammatory genes seen in vitro, Erg and Fli1 double-heterozygote mice showed increased immune cell infiltration and expression of cytokines in the lung. In conclusion, loss of ERG and FLI1 might contribute to the pathogenesis of vascular lung complications through the induction of inflammation.
Keywords: systemic sclerosis, pulmonary arterial hypertension, E–twenty six–related gene, Friend leukemia integration 1 transcription factor, IFN
Clinical Relevance
Immune aspects of pulmonary arterial hypertension (PAH) are incompletely understood. This work shows that two closely related ETS family transcription factors, ERG and FLI1, are key regulators of interferon-related genes in pulmonary vasculature. ERG and FLI1 are down-regulated in pulmonary vasculature of scleroderma- and idiopathic-PAH patients, and their loss leads to de-repression of IFN-β1 gene and marked up-regulation of interferon-responsive pro-inflammatory genes. Additionally, partial ablation of Erg and Fli1 in a murine model leads to spontaneous pulmonary inflammation. Targeted therapies toward normalizing ERG and FLI1 levels could be beneficial for patients with PAH.
Pulmonary arterial hypertension (PAH) is a devastating cardiopulmonary disease characterized by perivascular inflammation and increased vasoconstriction in lung. Endothelial dysfunction, together with abnormal vascular remodeling, leads to elevated pulmonary arterial pressure, followed by right-heart hypertrophy, which, if left untreated, can lead to heart failure (1). PAH is also the most serious organ complication in systemic sclerosis (SSc), an autoimmune disease characterized by vasculopathy and fibrosis (2). Systemic vasculopathy and chronic inflammation are likely factors contributing to the more severe PAH disease manifestation in those patients (3). Structural changes in the pulmonary vasculature are driven by perturbations in a number of physiological processes, with growing evidence suggesting inflammation as a central mediator (4). Inflammation triggered by oxidative stress, leading to endothelial injury and the switch from the quiescent to activated state in endothelial cells (ECs), is partially mediated by transcriptional regulation. E–twenty six (ETS) transcription factors consist of a large family of proteins, and many of them are essential for hematopoietic and EC development, as well as the maintenance of endothelial homeostasis in adults (5). Our previous work demonstrated that the ETS family member, Friend leukemia integration 1 (FLI1), is an essential repressor of profibrotic genes (6, 7), and plays a pivotal role in maintaining vascular integrity by promoting expression of cell adhesion and proangiogenic molecules (8). FLI1 is expressed in skin microvascular endothelium of healthy adults; however, FLI1 is depleted from the endothelium of patients with SSc, which might, at least in part, explain the observed increased vascular permeability, perivascular inflammation, and degeneration of small capillaries (9).
ETS-related gene (ERG) is the closest FLI1 homolog and the most highly expressed ETS member in resting ECs. ERG is important in maintaining quiescence of the endothelium by regulating EC homeostasis, survival, and differentiation (10). We previously demonstrated that endothelial ERG is essential in mouse embryogenesis by regulating vascular integrity (11). ERG is also important in repressing proinflammatory NF-κB and IL-8, thereby keeping the endothelium from activation under normal physiological conditions. Interestingly, animal studies show that proinflammatory stimuli, such as LPS and TNF-α treatments, down-regulate endothelial ERG expression, confirming that ERG plays an important role in inhibiting EC activation in vivo (12, 13).
In this study, we investigate the association between ERG and FLI1 loss and vascular inflammation in lungs of patients with PAH. We show that simultaneous loss of FLI1 and ERG in ECs causes a dramatic proinflammatory response and IFN pathway activation. In vivo, partial depletion of both Erg and Fli1, but not single depletion, leads to increased lung inflammation, demonstrating the antiinflammatory role of those two factors.
Materials and Methods
Human Samples
Lung samples were obtained from patients with SSc-PAH and patients with idiopathic PAH (iPAH) who underwent lung transplantation and control subjects at the University of Pittsburgh Medical Center (Pittsburgh, PA), under a protocol approved by the Institutional Review Board. Detailed patient information is listed in Figure E1 in the online supplement.
Mice
Erg+/−Fli1+/− mice used for experiments were 6–8 weeks old, both male and female, on a C57BL/6 genetic background. The derivation of Erg+/− and Fli1+/− mice has been described previously (11, 14, 15).
All of the experiments were performed under the guidelines of and were approved by the Boston University (Boston, MA) Institutional Animal Care and Use Committee (protocol AN-15037). For hypoxic conditions, 6-week-old male C57BL/6 mice were placed in a hypoxic chamber (10% oxygen; BioSpherix, Parish, NY) for 3 weeks. The left lung was fixed by tracheal perfusion either with 4% paraformaldehyde or 50% optimal cutting temperature (OCT) compound (Thermo Fisher Scientific, Waltham, MA) in PBS, and the right lung used for RNA isolation.
Immunohistochemistry and Immunofluorescence
For immunohistochemistry and immunofluorescent staining, slides were processed and stained as described previously (16). Primary antibodies used in immunohistochemistry were: ERG (Biocare Medical, Concord, CA), van Willebrand factor (Dako, Carpinteria, CA), and MAC3 and CD45 (BD Biosciences, San Jose, CA). Primary antibodies used in immunofluorescence were: ERG (clone EPR3863; Abcam, Cambridge, MA) and CD31 (clone SZ31; Dianova, Hamburg, Germany).
Cell Culture, Transient Transfections
Human pulmonary artery ECs (HPAECs) and human pulmonary microvascular ECs (HPMECs) were purchased from Lonza (Basel, Switzerland). Poly(I:C) was from Invivogen (San Diego, CA); IFN-β1a was from PBL Assay Science (Piscataway, NJ); B18R was from eBioscience (San Diego, CA); and the Toll-like receptor (TLR) 3/complex inhibitor was from EMD Millipore (Billerica, MA). Cells were transfected with small interfering RNA (siRNA) specific to human ERG, FLI1, or scrambled siRNA (ON-TARGETplus SMART pool) using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Endothelial Permeability Assay
HPMECs were treated with siRNA specific to human ERG, FLI1, or scrambled siRNA for 48 hours and a permeability assay was performed as described previously (17).
RNA Isolation, Quantitative RT-PCR
Total RNA was isolated using TRIzol reagent (MRC, Inc., Cincinnati, OH) for cell culture samples or by using the RNeasy Fibrous Tissue Mini Kit (Qiagen, Valencia, CA) for tissue samples. Real-time PCR was performed using the StepOnePlus Real-Time PCR System using SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA). The primer sequences used are available upon request.
Microarray Analysis
Microarray analysis was performed at the Boston University Microarray Core Facility.
Affymetrix CEL files were normalized to produce gene-level expression values using the implementation of the Robust Multiarray Average (RMA) (18) in the affy package (version 1.36.1; Affymetrix, Santa Clara, CA) (19) included within in the Bioconductor software suite (version 2.12; https://www.bioconductor.org/help/faq/) (20) and an Entrez Gene-specific probeset mapping from BrainArray (version 16.0.0; brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/CDF_download.asp) (21). RLE and NUSE quality metrics were computed using the affyPLM Bioconductor package (version 1.34.0; www.bioconductor.org) (22). All microarray analyses were performed using the R environment for statistical computing (version 2.15.1; www.r-project.org).
The private link to the Gene Expression Omnibus–deposited data is: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=idkrsmsqrlsrzqxandacc=GSE83641.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as previously described (23) using ChIP-grade ERG antibody (clone EPR3863; Abcam) and signal transducers and activators of transcription (STAT) 1 antibody (clone C-24; Santa Cruz Biotechnology, Santa Cruz, CA). Real-time quantitative PCR was performed in triplicate using specific primers for the human IFNB1 gene promoter, as shown in Figure 6E. The amount of immunoprecipitated DNA (% recovery) was calculated from a standard curve generated with the serial dilutions of input chromatin.
Figure 6.
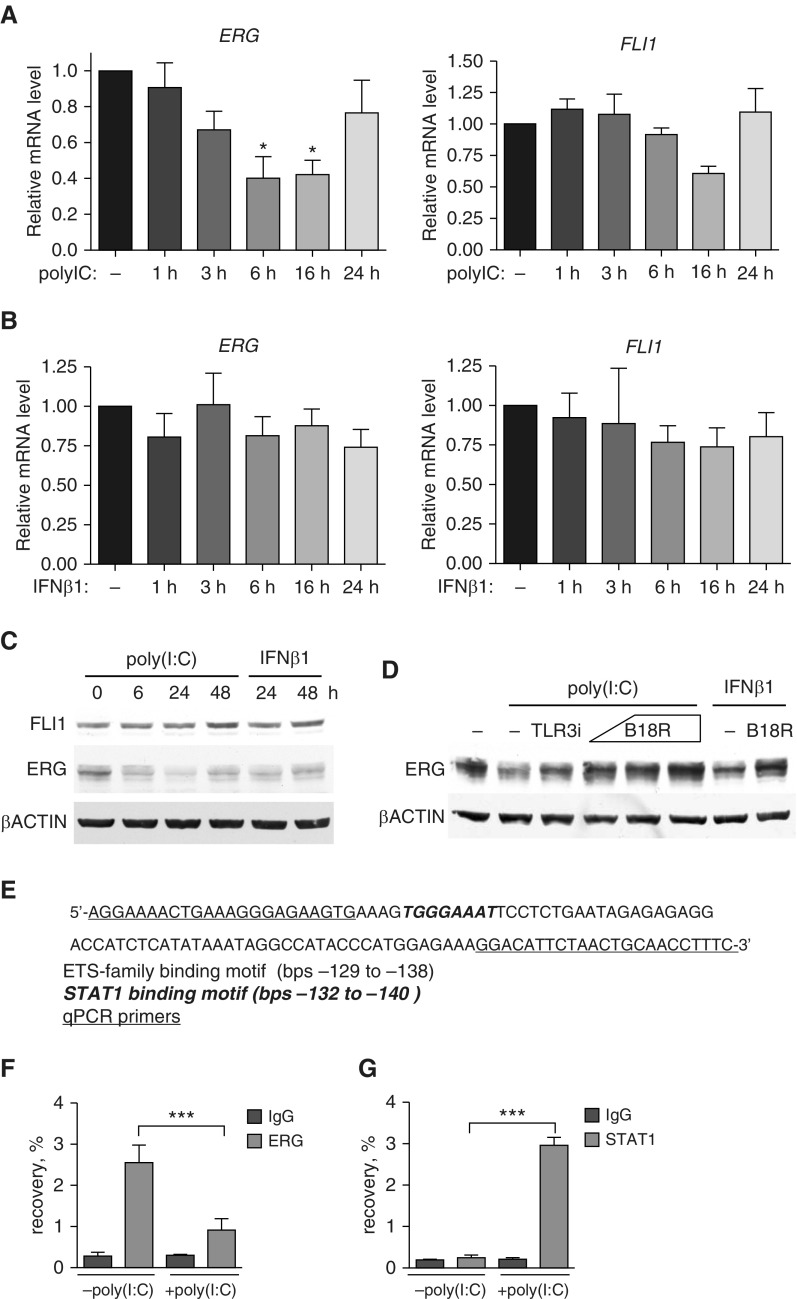
Poly(I:C) and IFN-β1 treatments down-regulate ERG, but not FLI1. HPAECs were treated with 2.5 μg/ml poly(I:C) (Toll-like receptor [TLR] 3) agonist or and IFN-β1 for indicated periods of time. (A and B) Relative mRNA expression was determined by quantitative RT-PCR. (C) Representative Western blot of HPAEC lysates treated with 2.5 μg/ml poly(I:C) or 1,000 U/ml IFN-β1 showing levels of FLI1 and ERG (n = 3 independent experiments). (D) HPEACs were pretreated with a TLR3/complex inhibitor (TLR3i; 50 μM) or IFN inhibitor, B18R (0.1 and 0.2 μg/ml) for 1 hour and then incubated with poly(I:C) or IFN-β1 for 24 hours. Chromatin immunoprecipitation followed by quantitative RT-PCR with primers targeting IFN-β1 promoter region (E) was performed on HPAECs treated for 6 hours with 2.5 μg/ml poly(I:C). (F) ERG or (G) signal transducers and activators of transcription (STAT) 1 antibodies were used. Data are presented as a percent of input DNA recovery. IgG, isotype control (n = 3 independent experiments). All data shown as mean (±SD). One-way ANOVA followed by Tukey’s multiple comparison were used for statistical analysis. *P < 0.05, ***P < 0.001.
Western Blot
Cells were lysed and processed as described previosuly (24). Antibodies used were: ERG (Biocare Medical); FLI1 (custom made); β-actin (Thermo Fisher Scientific); IRF3 (Santa Cruz Biotechnology); and IRF7 (Cell Signaling, Danvers, MA).
Statistical Analysis
Prism 5 (GraphPad Software, Inc., La Jolla, CA) was used for all analyses.
Results
Endothelial ERG Is Reduced in Lungs of Patients with PAH and in Lungs of Chronic Hypoxic Mice
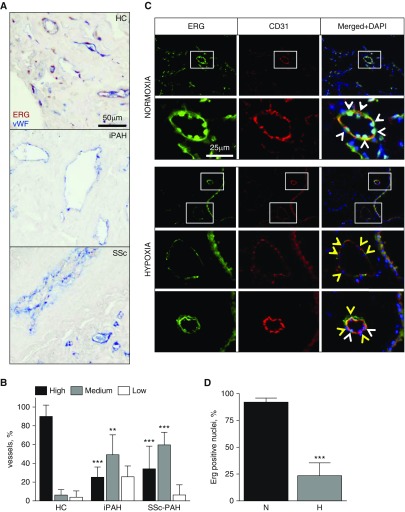
Down-regulation of endothelial ERG has been previously shown in injured vasculature (12, 13, 25). To evaluate the levels of endothelial ERG in diseased pulmonary vasculature in humans, lung sections from iPAH, SSc-PAH, and healthy control (HC) were immunostained with endothelial marker, van Willebrand Factor, and ERG. Additional patient information is listed in Table E1. As shown in Figure 1, the endothelium in HC lung had high levels of nuclear ERG as compared with significantly lower levels or even complete loss of ERG in the endothelium of patients with iPAH and SSc-PAH. When quantified, the differences in vessel numbers with high and medium ERG levels between HC and PAH were statistically significant (Figure 1B). To demonstrate that loss of endothelial ERG is a general phenomenon occurring upon vascular injury, we assessed ERG levels in lungs of chronically hypoxic mice. Chronic hypoxia is a known factor that induces pulmonary vascular defects, including endothelial dysfunction and perivascular inflammation (26). Immunostaining shown in Figure 1C demonstrates ERG nuclear localization in ECs (CD31 positive) under normoxic conditions; however, under hypoxic conditions, nuclear ERG is visibly depleted from ECs. The statistical significance of this observation was verified by quantification of ERG-positive and -negative EC nuclei in normoxic and hypoxic mice (Figure 1D).
Figure 1.
Endothelial E–twenty six (ETS)–related gene (ERG) is down-regulated in lungs of patients with pulmonary arterial hypertension (PAH) and in chronically hypoxic mice. (A) Paraffin-embedded lung sections from healthy control (HC), idiopathic PAH (iPAH), and systemic sclerosis (SSc)–associated PAH were immunostained with van Willebrand factor (vWF) and ERG antibodies. Scale bar: 50 μm. (B) Vessels from five fields of view were categorized, based on endothelial nuclear ERG signal, as high, medium, or low (n = 3 for HC, n = 5 for iPAH/SSc). (C) C57BL/6 mice were kept in hypoxia (10% O2) or normoxia (21% O2) for 21 days. Frozen lung sections were immunostained with CD31 and ERG antibodies. Scale bar: 25 μm. (D) Endothelial nuclei from five fields of view were counted and categorized as ERG positive (white arrowheads) or ERG negative (yellow arrowheads) (n = 4 mice per group). Data shown as mean (±SD). One-way ANOVA followed by Tukey’s multiple comparison and a Student t test were used for statistical analysis. **P < 0.01, ***P < 0.001. DAPI, 4′,6-diamidino-2-phenylindole; H, hypoxia; N, normoxia.
In conclusion, endothelial ERG is down-regulated in lungs of human subjects with systemic and pulmonary vascular disorders, as well as in a hypoxic mice model.
FLI1 and ERG Synergistically Induce IFN-Related Gene Expression in HPAECs
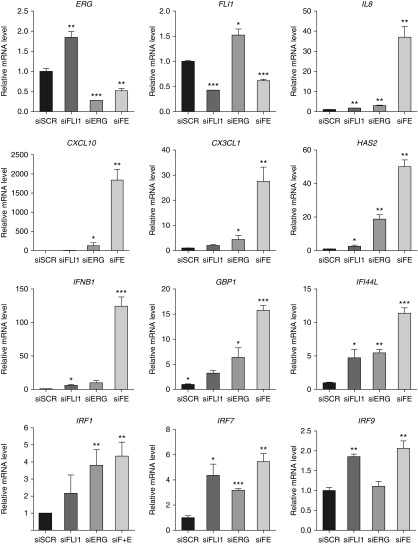
FLI1 is the closest homolog of ERG and, similar to ERG, is known to regulate genes that sustain endothelial homeostasis. We therefore asked if there is a transcriptional synergy between those two factors. We addressed this question by performing FLI1/ERG double-siRNA–mediated knockdown in HPAECs followed by gene expression array. The top 20 up-regulated genes are listed in Table 1 and the top 20 down-regulated genes are listed in Table 2. As predicted, the list of down-regulated targets was enriched with genes that regulate cell adhesion and endothelium homeostasis, including fatty acid binding protein 4 (FABP4) (27), lymphatic vessel endothelial hyaluronan receptor 1 (LYVE-1) (28) and apelin (APLN) (29) (Table 2). Strikingly, ERG and FLI1 knockdown induced remarkable overexpression of proinflammatory (CXCL10, HAS2, IL1A, CCL5, CXCL11, and VCAM1) and IFN-responsive genes (GBP4, IFIT2, OASL, and IRF6) (Table 1). Importantly, the quantitative PCR verification of C-X-C motif chemokine 10 (CXCL10) and hyaluronan synthase 2 (HAS2), the targets found in the array, showed a synergistic effect of ERG and FLI1 loss in inducing their gene expression (Figure 2).
Table 1.
Top 20 Genes Up-Regulated in Human Pulmonary Artery Endothelial Cells after Simultaneous E–Twenty Six–Related Gene and Friend Leukemia Integration 1 Transcription Factor Knockdown
| Gene Symbol | Gene Name | Fold Change |
|---|---|---|
| CXCL10 | Chemokine (C-X-C motif) ligand 10 | 116.73 |
| TNFSF13B | TNF (ligand) superfamily, member 13b/BAFF | 28.37 |
| HAS2 | Hyaluronan synthase 2 | 24.49 |
| IL1A | IL-1α | 19.72 |
| CCL5 | Chemokine (C-C motif) ligand 5 | 17.97 |
| IDO1 | Indoleamine 2,3-dioxygenase 1 | 17.90 |
| GBP4 | Guanylate binding protein 4 | 17.38 |
| IFIT2 | IFN-induced protein with tetratricopeptide repeats 2 | 14.59 |
| OASL | 2′-5′-oligoadenylate synthetase-like | 12.78 |
| HERC5 | HECT and RLD domain containing E3 ubiquitin protein ligase 5 | 12.18 |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | 11.95 |
| VCAM1 | Vascular cell adhesion molecule 1 | 11.82 |
| RSAD2 | Radical S-adenosyl methionine domain containing 2 | 11.31 |
| LGALS3BP | Lectin, galactoside-binding, soluble, 3 binding protein | 11.15 |
| SECTM1 | Secreted and transmembrane 1 | 11.04 |
| RTP4 | Receptor (chemosensory) transporter protein 4 | 10.62 |
| IRF6 | IFN regulatory factor 6 | 9.83 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 9.69 |
| SEMA3C | Sema domain, Ig domain, short basic domain, secreted, (semaphorin) 3C | 8.91 |
| TNFAIP3 | TNF-α–induced protein 3 | 8.84 |
Definition of abbreviations: BAFF, B-cell activating factor; HECT, homologous to the E6-AP carboxyl terminus; RLD, regulator of chromosome condensation 1.
Table 2.
Top 20 Genes Down-Regulated in Human Pulmonary Artery Endothelial Cells after E–Twenty Six–Related Gene and Friend Leukemia Integration 1 Transcription Factor Knockdown
| Gene Symbol | Gene Name | Fold Change |
|---|---|---|
| FABP4 | Fatty acid binding protein 4, adipocyte | −42.42 |
| LYVE1 | Lymphatic vessel endothelial hyaluronan receptor 1 | −18.04 |
| RGS7BP | Regulator of G-protein signaling 7 binding protein | −15.94 |
| DLL4 | Delta-like 4 (Drosophila) | −12.06 |
| CHST1 | Carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | −11.52 |
| KIT | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | −11.09 |
| TSPAN18 | Tetraspanin 18 | −10.47 |
| ACE | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | −9.65 |
| C15orf54 | Chromosome 15 open reading frame 54 | −9.62 |
| RASSF2 | Ras association (RalGDS/AF-6) domain family member 2 | −9.02 |
| APLN | Apelin | −8.80 |
| CXorf36 | Chromosome X open reading frame 36 | −8.48 |
| PLEKHG1 | Pleckstrin homology domain containing, family G (with rhogef domain) member 1 | −8.37 |
| CYTL1 | Cytokine-like 1 | −8.37 |
| GDPD5 | Glycerophosphodiester phosphodiesterase domain containing 5 | −8.14 |
| PLXNA4 | Plexin A4 | −7.85 |
| CPNE5 | Copine V | −7.28 |
| GPR116 | G protein–coupled receptor 116 | −7.25 |
| TRAC | T cell receptor α constant | −7.07 |
| ADAMTS18 | ADAM metallopeptidase with thrombospondin type 1 motif, 18 | −6.88 |
Definition of abbreviation: ADAM, a disintegrin and metalloprotease.
Figure 2.
Simultaneous loss of ERG and Friend leukemia integration 1 transcription factor (FLI1) in human pulmonary artery endothelial cells (HPAECs) synergistically induces the expression of inflammatory and IFN genes. HPAECs were subjected to small interfering RNA (siRNA)-mediated silencing of FLI1 and/or ERG for 48 hours. siFE indicates double FLI1 and ERG siRNA treatment. Scrambled siRNA (siSCR) was used as a control. Relative mRNA expression was determined by quantitative RT-PCR (n = 3 independent experiments). Data shown as mean (±SD). One-way ANOVA followed by Tukey’s multiple comparison were used for statistical analysis. *P < 0.05, **P < 0.01, ***P < 0.001. CXCL 10, chemokine (C-X-C motif) ligand 10; CX3CL1, chemokine (C-X3-C motif) ligand 1; GBP1, guanylate-binding protein 1; HAS2, hyaluronan synthase 2; IFI44L, IFN-induced protein 44-like; IFNB1, interferon β 1; IRF, IFN-regulatory factors.
In addition to the genes shown in Table 2, we measured the expression of other IFN pathway genes, including IFN-induced guanylate-binding protein 1 (GBP1), IFN-β (IFNB1), IFN-induced protein 44-like (IFI44L), and IFN-regulatory factors 1, 7, and 9 (IRF1, -7, and -9), as well as proinflammatory genes, IL8 and chemokine (C-X3-C motif) ligand 1 (CX3CL1) (Figure 2). The response of those genes to ERG and FLI1 silencing showed either synergistic or additive effects in gene up-regulation upon depletion of these two factors. In summary, simultaneous knockdown of ERG and FLI1 in HPAECs leads to up-regulation of proinflammatory and IFN pathway–related genes in magnitudes exceeding that from knockdown of ERG or FLI1 alone.
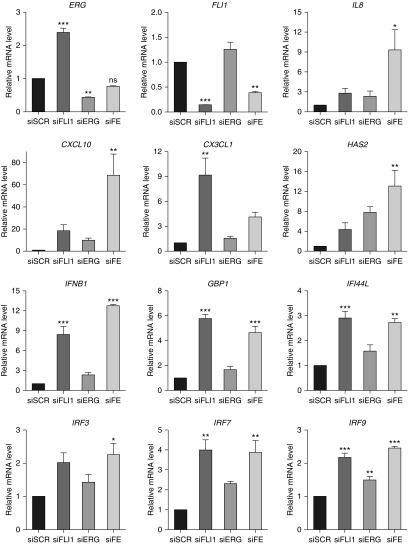
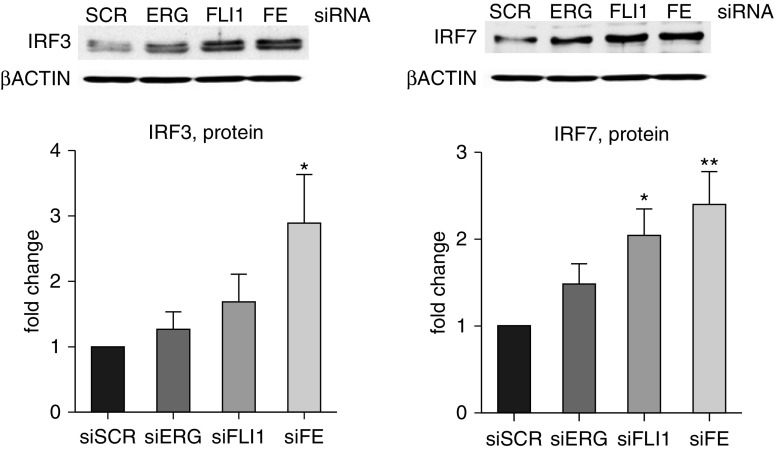
We also investigated the transcriptional response to ERG and FLI1 loss in HPMECs, which are more relevant to the early pathological changes in PAH (Figure 3, Figure E2). Similar to HPAECs, there was an additive effect in expression of several genes, including IL-8 and CXCL10; however, overall magnitude of response was smaller in HPMECs compared with HPAECs. It also appeared that expression of selected genes (e.g., CX3CL1, GBP1, and IFI44L) was affected more by the depletion of FLI1 in HPMECs, which was not observed in HPAECs. The effects of ERG and FLI1 depletion on the protein levels of two of the target genes, IRF3 and IRF7, was consistent with the effects on mRNA levels (Figure 4).
Figure 3.
Loss of ERG and FLI1 induces the expression of inflammatory and IFN genes in HPMECs. Human pulmonary microvascular ECs (HPMECs) were subjected to siRNA-mediated silencing of FLI1 and/or ERG for 48 hours. Scrambled siRNA (siRNA) was used as a control. Relative mRNA expression was determined by quantitative RT-PCR (n = 3 independent experiments). Data shown as mean (±SD). One-way ANOVA followed by Tukey’s multiple comparison were used for statistical analysis. *P < 0.05, **P < 0.01, ***P < 0.001. ns, not significant.
Figure 4.
Loss of ERG and FLI1 induces the expression of IRF proteins in HPMECs. HPMECs were subjected to siRNA-mediated silencing of FLI1 and/or ERG for 48 hours. siSCR was used as a control. Representative immunoblots and bar graphs with the densitometric analysis are shown (n = 3 independent experiments). Data shown as mean (±SD). One-way ANOVA followed by Tukey’s multiple comparison were used for statistical analysis. *P < 0.05, **P < 0.01.
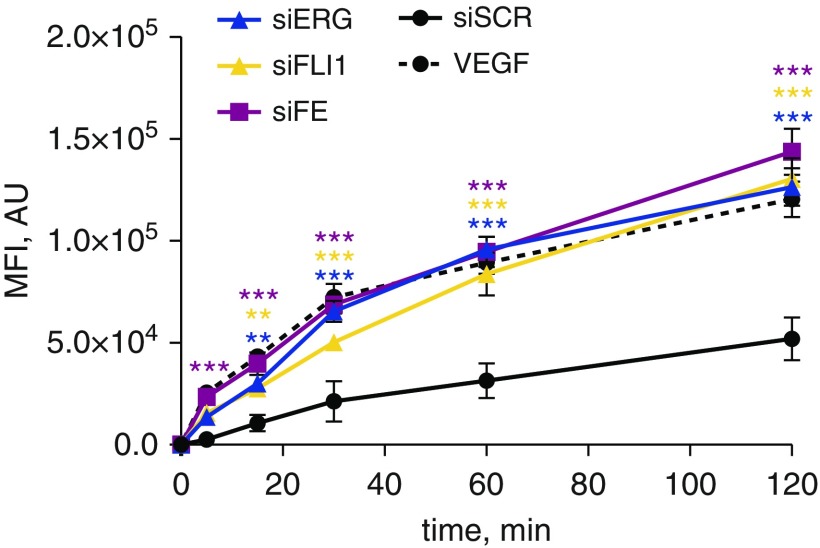
To test the physiological importance of ERG and FLI1 knockdown in endothelial barrier integrity, we performed a permeability assay in HPMEC monolayers treated with FLI1 and/or ERG siRNA. In Figure 5, we demonstrate that ERG and/or FLI1 knockdown led to the significantly increased monolayer permeability, which was similar in magnitude to vascular endothelial growth factor treatment, a known permeability-inducing agent. We did not observe a synergistic or additive effect in FLI1 and ERG loss in this assay, which might indicate that the integrity of the endothelial monolayer in this assay cannot be well quantified, and any barrier perturbation results in similar output measurement.
Figure 5.
Down-regulation of ERG and FLI1 in HPMECs induces increased cell monolayer permeability. HPMECs were subjected to siRNA-mediated silencing of FLI1 and/or ERG for 48 hours. siSCR was used as a control. Fluorescence of 3 kD dextran sulfate–FITC that penetrated the endothelial layer grown on transwell inserts was measured. Vascular endothelial growth factor (VEGF; 100 ng/ml) treatment was used as a positive control. The experiment was done in triplicate. Data shown as mean (±SD). One-way ANOVA followed by Tukey’s multiple comparison were used for statistical analysis. **P < 0.01, ***P < 0.001. AU, arbitrary units; MFI, mean fluorescent intensity.
Poly(I:C) and IFN-β1 Treatments Down-Regulate Expression of ERG, but Not FLI1
The IFN pathway plays an essential role in the innate immunity response to various pathogens, and its activation is mediated via IFN receptors and TLR. In our study, we observed activation of IFN-β, which is specifically stimulated by TLR3. To test if there is a potential feedback mechanism between IFN-β pathway and ERG or FLI1, we treated HPAECs with the TLR3 agonist, poly(I:C), and the IFN receptor ligand, IFN-β1. Treatment with 2.5 μg/ml poly(I:C) down-regulated ERG gene and protein expression over the course of time ranging from 1 to 48 hours, but had no effect on FLI1 (Figures 6A and 6C). IFN-β1 treatment (1,000 U/ml) down-regulated ERG protein levels, but, surprisingly, did not significantly alter ERG gene expression (Figures 6B and 6C). Similar to poly(I:C), INFβ1 had no effect on FLI1 protein or gene expression (Figures 6B and 6C). These distinct effects of poly(I:C) or IFN-β1 treatment on ERG and FLI1 expression are consistent with the in vivo observations from patients with PAH who showed primarily reduced protein levels of ERG, but not FLI1, in the lung vasculature.
We next sought to verify whether the observed poly(I:C) and INFβ1 suppression of ERG protein is dependent on TLR3 and IFN receptor signaling by treating cells with 50 μM of a TLR3/complex inhibitor (TLR3i) or 0.1 μg/ml of a type I IFN-binding protein (B18R) before and during a 24-hour poly(I:C) or INFβ1 stimulation. Inhibition of TLR3 resulted in partial suppression of poly(I:C)-mediated ERG depletion, whereas inhibition of IFN type I (B18R treatment) lead to a dose-dependent full suppression of poly(I:C)-mediated ERG depletion (Figure 6D). B18R also blocked IFN-β1-mediated ERG down-regulation, demonstrating that B18R is a potent inhibitor of type I IFN (Figure 6D). These results show that activation of IFN signaling through TLR3 significantly reduced ERG, but not FLI1 expression, at both mRNA and protein levels.
In addition, to demonstrate that ERG and FLI1 serve as transcriptional repressors of IFN genes in HPAECs, we overexpressed ERG and/or FLI1, followed by a 3-hour poly(I:C) treatment. Adenoviral overexpression of ERG or FLI1, or ERG and FLI1 together, significantly reduced poly(I:C)-induced IFNB1 overexpression (Figure E3). Adenoviral overexpression of ERG and FLI1 together also reduced poly(I:C)-induced GBP1 overexpression (Figure E3). These results strongly suggest that ERG and FLI1 serve as transcriptional repressors of IFNB1.
Poly(I:C) Induces ERG Dissociation and STAT1 Recruitment to the IFNB1 Promoter
To our knowledge, direct regulation of INFB1 gene by ERG or FLI1 has not been previously demonstrated. We therefore performed ChIP with an ERG antibody followed by quantitative PCR including the -129 to -138 fragment of INFB1 promoter (Figures 6E and 6F), which contains two putative ETS binding motifs. We show not only that ERG bound to IFNB1 promoter region, but also that this binding is abolished by 6 h of poly(I:C) treatment (Figure 6F). Notably, this INFB1 promoter region also contains a putative binding site for activating transcription factor STAT1 (Figure 6E). This interaction could explain robust INFB1 activation upon de-repression induced by loss of ERG binding. To verify that possibility we performed another ChIP using anti-STAT1 antibody followed by quantitative PCR (Figure 6G). As hypothesized, STAT1 binding to INFB1 promoter was induced by poly(I:C) treatment (Figure 6G). This result demonstrates a dynamic remodeling of the transcription factor complex on the IFNB1 promoter in vivo in response to poly(I:C).
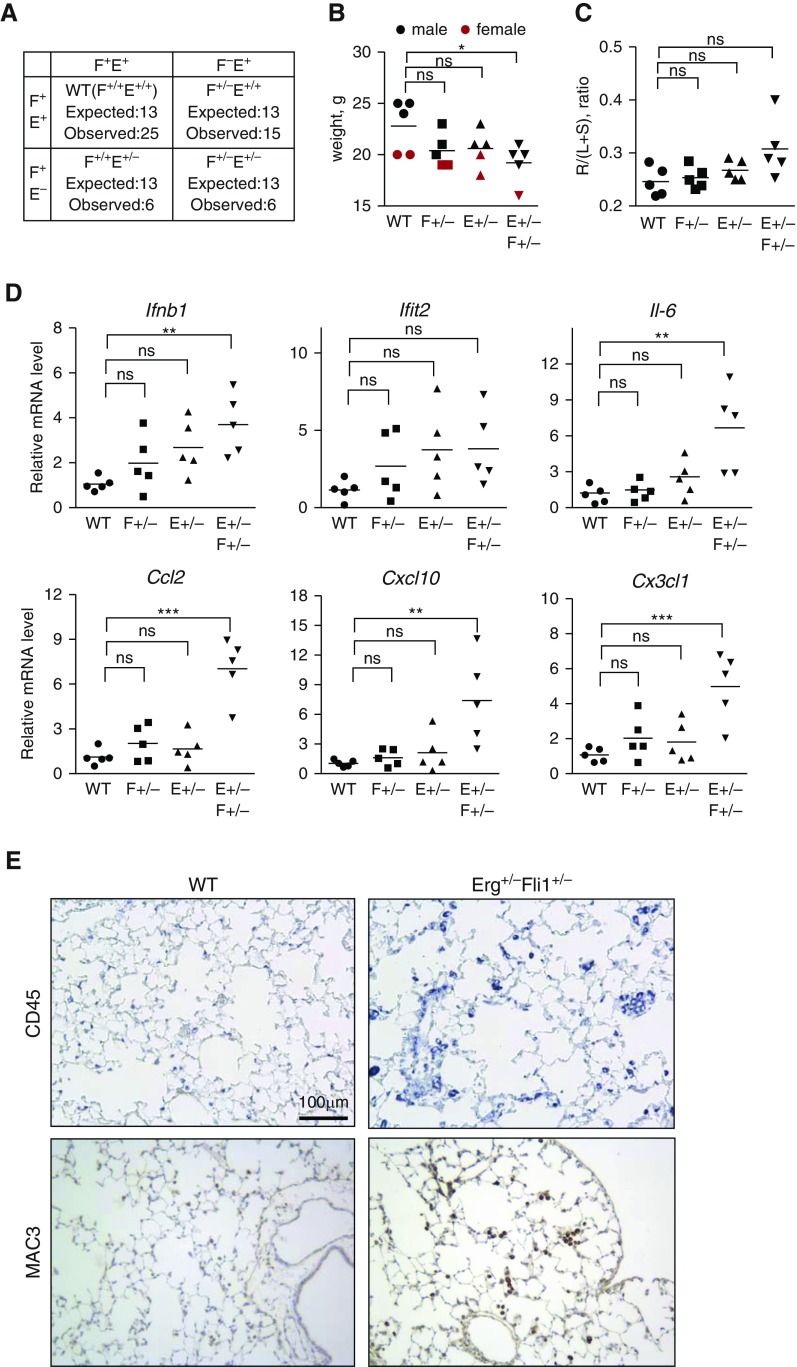
Mice Heterozygous for Erg and Fli1 have Elevated Inflammation in Lungs
To confirm important, synergistic role of ERG and FLI1 loss in vivo, we generated double heterozygous mice (Erg+/−Fli1+/−). As shown in Figure 7A, adult mice (4 weeks old) were not present in Mendelian distribution, with Erg+/− being a skewing factor. 8-week old Erg+/−Fli1+/− mice, but not single heterozygotes, were also significantly smaller than wild type mice (Figure 7B). To investigate the possibility that Erg+/−Fli1+/− mice are susceptible to develop pulmonary hypertension we measured the weight of right heart ventricle as compared with left ventricle with the septum (Figure 7C). Although, a trend toward increased right ventricle was observed in double heterozygous mice as compared with WT mice, the differences did not reach statistical significance (Figure 7C). Consistent with cell culture results, Erg+/−Fli1+/− mouse lungs, and not single heterozygotes, had significantly increased proinflammatory and IFN-related gene expression (Figure 7D). Immunostaining with the lymphocyte marker CD45 and macrophage marker MAC3 revealed higher immune cell infiltrate in double heterozygotes as compared with wild type littermates (Figure 7E). Due to the abnormally small litters and difficulties in breeding, flow cytometry analysis or inflammatory challenge of animals was not feasible. In conclusion, simultaneous partial depletion of Erg and Fli1 in mice resulted in decreased survival and increased inflammation in lungs.
Figure 7.
Mice heterozygous for both Erg and Fli1 have elevated inflammation in lungs. (A) Distribution of genotypes of adult (4 wk old) mice as a result of breeding Erg+/− with Fli1+/− mice. “Expected,” calculated from Mendelian distribution; “observed,” number of pups that survived to adulthood. (B) Body weight of 6- to 8-week-old mice. (C) Right heart hypertrophy was accessed by measuring weight of heart dissected into right ventricle (R) and left ventricle plus septum (L + S). (D) Gene expression of proinflammatory genes in whole lung of 6- to 8-week-old mice as measured by quantitative RT-PCR. (E) Paraffin-embedded lung sections were immunostained with CD45 and MAC3 antibodies to visualize total leukocytes and macrophages, respectively. One-way ANOVA followed by Tukey’s multiple comparison were used for statistical analysis. *P < 0.05, **P < 0.01, ***P < 0.001. Ifit2, interferon induced protein with tetratricopeptide repeats 2; WT, wild type.
Discussion
FLI1 is down-regulated in the skin endothelium of patients with SSc, but it was not known whether FLI1 is similarly down-regulated in the endothelium of diseased pulmonary vasculature. We found low levels of FLI1 in the pulmonary endothelium in lung specimens from healthy subjects and a low degree of FLI1 down-regulation in PAH. Interestingly, we found high expression of ERG, the closest homolog of FLI1, in the endothelium of healthy lungs in human and mouse. We also show, for the first time, that diseased lung endothelium in PAH, as well as in chronically hypoxic mice, exhibit loss of ERG. This result is in accordance with previous observations of endothelial ERG down-regulation in human coronary disease and animal models treated with TNF-α (12). It illustrates a universal mechanism of ERG suppression in injured vasculature. In our experiments, the perivascular ERG staining observed in examined mouse lung samples was likely an artifact of cryopreserved sections, because primary nuclear localization of ERG is consistent with previously described ERG protein expression pattern in adult mammalian tissues (30).
Loss of ERG and FLI1 after siRNA-mediated gene knockdown in HPAECs and HPMECs led to up-regulation of a number of cytokines and IFN pathway genes, suggesting that FLI1 and ERG loss derepresses proinflammatory genes. Both types of cells responded in a similar manner, although the magnitude of response was much greater in HPAECs. Double ERG and FLI1 knockdown, as compared with single knockdowns, caused synergistic or additive target gene up-regulation, suggesting that FLI1 and ERG transcriptionally coregulate a subset of proinflammatory genes. The interaction of FLI1 and ERG proteins has previously been demonstrated (31), and could potentially indicate the cooperative transcriptional action of these two factors. Further studies identifying the mediators involved in this process are of great importance to understanding the regulatory mechanism of endothelium homeostasis.
It was previously demonstrated that the interaction of FLI1 and transforming growth factor (TGF)-β signaling involves feedback mechanism, where FLI1 represses TGF-β target genes and TGF-β down-regulates activity of FLI1 (6, 32). By analogy, we sought a similar mechanism in FLI1- and ERG-mediated regulation of IFN type I. Overabundance of IFN type I signaling has been demonstrated in SSc-PAH, and is thought to have an important pathological role in disease development and progression (33, 34). The source of the stimulatory signal is not known, but it has been speculated that viral infections and double-stranded RNA might be contributing to TLR-mediated activation of the innate immunity response. Our results clearly show that activation of IFN signaling through TLR3 significantly reduced ERG, but not FLI1, expression at both the mRNA and protein levels. In addition, we show that IFNB1 gene is directly regulated by ERG binding the IFNB1 promoter region, and that interaction is lost upon TLR3 activation, which correspond with the decrease of ERG protein level in the same experimental conditions. Derepression of IFNB1 promoter caused by loss of ERG binding is concomitant with binding of activating STAT1 transcription factor to the same promoter region.
Lastly, we confirmed the importance of the ERG and FLI1 synergistic antiinflammatory effects in vivo by generating double-heterozygous mouse Erg+/−Fli1+/−. Heterozygosity for Erg seemed to be a factor decreasing mouse health and survival, because distribution of Erg+/− and Erg+/−Fli1+/− adult mice was non-Mendelian, and those mice had significantly reduced body mass. Erg+/− mice, however, did not have significantly elevated lung inflammation, which might be due to health issues in those animals being related to earlier developmental problems (35). However, only lungs of Erg+/−Fli1+/− double heterozygotes had significantly increased baseline gene expression of cytokines and IFN genes, as well as increased immune cell infiltration. As mentioned previously, breeding difficulties and small litters made it unfeasible to proceed with more elaborate experiments.
In conclusion, endothelial ERG and FLI1 play essential roles in regulating vascular homeostasis, including suppression of proinflammatory genes. Alterations of those proteins might, therefore, be important contributors to the mechanism of vasculopathy in PAH and SSc.
Acknowledgments
Acknowledgments
The authors thank Carol Feghali-Bostwick (Medical University of South Carolina, Charleston, SC) for providing patient and healthy control lung samples.
Footnotes
This work was supported by National Institutes of Health grant RO1 AR42334, grant P30 AR061271, and CTSA grant UL1-TR000157 for micro-array analysis.
Author Contributions: Conception and design—A.P.L., R.H., and M.T.; performing experiments and collecting data—A.P.L., R.H., L.S., and G.M.; generating mice—R.H. and M.I.; analysis and interpretation—A.P.L. and R.H.; drafting the manuscript for important intellectual content—A.P.L., R.H., and M.T.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2016-0200OC on March 1, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Farber HW, Loscalzo J. Pulmonary arterial hypertension. N Engl J Med. 2004;351:1655–1665. doi: 10.1056/NEJMra035488. [DOI] [PubMed] [Google Scholar]
- 2.McLaughlin V, Humbert M, Coghlan G, Nash P, Steen V. Pulmonary arterial hypertension: the most devastating vascular complication of systemic sclerosis. Rheumatology (Oxford) 2009;48(suppl 3):iii25–iii31. doi: 10.1093/rheumatology/kep107. [DOI] [PubMed] [Google Scholar]
- 3.Le Pavec J, Humbert M, Mouthon L, Hassoun PM. Systemic sclerosis–associated pulmonary arterial hypertension. Am J Respir Crit Care Med. 2010;181:1285–1293. doi: 10.1164/rccm.200909-1331PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassoun PM, Mouthon L, Barberà JA, Eddahibi S, Flores SC, Grimminger F, Jones PL, Maitland ML, Michelakis ED, Morrell NW, et al. Inflammation, growth factors, and pulmonary vascular remodeling J Am Coll Cardiol 2009541 suppl 1)S10–S19. [DOI] [PubMed] [Google Scholar]
- 5.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 6.Nakerakanti SS, Kapanadze B, Yamasaki M, Markiewicz M, Trojanowska M. Fli1 and Ets1 have distinct roles in connective tissue growth factor/CCN2 gene regulation and induction of the profibrotic gene program. J Biol Chem. 2006;281:25259–25269. doi: 10.1074/jbc.M600466200. [DOI] [PubMed] [Google Scholar]
- 7.Czuwara-Ladykowska J, Shirasaki F, Jackers P, Watson DK, Trojanowska M. Fli-1 inhibits collagen type I production in dermal fibroblasts via an Sp1-dependent pathway. J Biol Chem. 2001;276:20839–20848. doi: 10.1074/jbc.M010133200. [DOI] [PubMed] [Google Scholar]
- 8.Asano Y, Stawski L, Hant F, Highland K, Silver R, Szalai G, Watson DK, Trojanowska M. Endothelial Fli1 deficiency impairs vascular homeostasis: a role in scleroderma vasculopathy. Am J Pathol. 2010;176:1983–1998. doi: 10.2353/ajpath.2010.090593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubo M, Czuwara-Ladykowska J, Moussa O, Markiewicz M, Smith E, Silver RM, Jablonska S, Blaszczyk M, Watson DK, Trojanowska M. Persistent down-regulation of Fli1, a suppressor of collagen transcription, in fibrotic scleroderma skin. Am J Pathol. 2003;163:571–581. doi: 10.1016/S0002-9440(10)63685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah AV, Birdsey GM, Randi AM. Regulation of endothelial homeostasis, vascular development and angiogenesis by the transcription factor ERG. Vascul Pharmacol. 2016;86:3–13. doi: 10.1016/j.vph.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han R, Pacifici M, Iwamoto M, Trojanowska M. Endothelial Erg expression is required for embryogenesis and vascular integrity. Organogenesis. 2015;11:75–86. doi: 10.1080/15476278.2015.1031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sperone A, Dryden NH, Birdsey GM, Madden L, Johns M, Evans PC, Mason JC, Haskard DO, Boyle JJ, Paleolog EM, et al. The transcription factor Erg inhibits vascular inflammation by repressing NF-κB activation and proinflammatory gene expression in endothelial cells. Arterioscler Thromb Vasc Biol. 2011;31:142–150. doi: 10.1161/ATVBAHA.110.216473. [DOI] [PubMed] [Google Scholar]
- 13.Yuan L, Nikolova-Krstevski V, Zhan Y, Kondo M, Bhasin M, Varghese L, Yano K, Carman CV, Aird WC, Oettgen P. Antiinflammatory effects of the ETS factor ERG in endothelial cells are mediated through transcriptional repression of the interleukin-8 gene. Circ Res. 2009;104:1049–1057. doi: 10.1161/CIRCRESAHA.108.190751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, Watson DK. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol. 2000;20:5643–5652. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohta Y, Okabe T, Larmour C, Di Rocco A, Maijenburg MW, Phillips A, Speck NA, Wakitani S, Nakamura T, Yamada Y, et al. Articular cartilage endurance and resistance to osteoarthritic changes require transcription factor Erg. Arthritis Rheumatol. 2015;67:2679–2690. doi: 10.1002/art.39243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stawski L, Han R, Bujor AM, Trojanowska M. Angiotensin II induces skin fibrosis: a novel mouse model of dermal fibrosis. Arthritis Res Ther. 2012;14:R194. doi: 10.1186/ar4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martins Green M, Petreaca M, Yao M.Chapter 8an assay system for in vitro detection of permeability in human “endothelium.” Cheresh DA, editor. Angiogenesis—In Vitro systemsSan Diego, CA: Academic Press & Elsevier2008137–153. [DOI] [PubMed] [Google Scholar]
- 18.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 19.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai L, Toor N, Olson R, Keeping A, Zimmerly S. Database for mobile group II introns. Nucleic Acids Res. 2003;31:424–426. doi: 10.1093/nar/gkg049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brettschneider J, Collin F, Bolstad BM, Speed TP. Quality assessment for short oligonucleotide microarray data. Technometrics. 2008;50:241–264. [Google Scholar]
- 23.Shirasaki F, Makhluf HA, LeRoy C, Watson DK, Trojanowska M. Ets transcription factors cooperate with Sp1 to activate the human tenascin-C promoter. Oncogene. 1999;18:7755–7764. doi: 10.1038/sj.onc.1203360. [DOI] [PubMed] [Google Scholar]
- 24.Pannu J, Gardner H, Shearstone JR, Smith E, Trojanowska M. Increased levels of transforming growth factor β receptor type I and up-regulation of matrix gene program: a model of scleroderma. Arthritis Rheum. 2006;54:3011–3021. doi: 10.1002/art.22063. [DOI] [PubMed] [Google Scholar]
- 25.Yuan L, Le Bras A, Sacharidou A, Itagaki K, Zhan Y, Kondo M, Carman CV, Davis GE, Aird WC, Oettgen P. ETS-related gene (ERG) controls endothelial cell permeability via transcriptional regulation of the claudin 5 (CLDN5) gene. J Biol Chem. 2012;287:6582–6591. doi: 10.1074/jbc.M111.300236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voelkel NF, Tuder RM. Hypoxia-induced pulmonary vascular remodeling: a model for what human disease? J Clin Invest. 2000;106:733–738. doi: 10.1172/JCI11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghelfi E, Yu CW, Elmasri H, Terwelp M, Lee CG, Bhandari V, Comhair SA, Erzurum SC, Hotamisligil GS, Elias JA, et al. Fatty acid binding protein 4 regulates VEGF-induced airway angiogenesis and inflammation in a transgenic mouse model: implications for asthma. Am J Pathol. 2013;182:1425–1433. doi: 10.1016/j.ajpath.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arimoto J, Ikura Y, Suekane T, Nakagawa M, Kitabayashi C, Iwasa Y, Sugioka K, Naruko T, Arakawa T, Ueda M. Expression of LYVE-1 in sinusoidal endothelium is reduced in chronically inflamed human livers. J Gastroenterol. 2010;45:317–325. doi: 10.1007/s00535-009-0152-5. [DOI] [PubMed] [Google Scholar]
- 29.Kasai A, Shintani N, Oda M, Kakuda M, Hashimoto H, Matsuda T, Hinuma S, Baba A. Apelin is a novel angiogenic factor in retinal endothelial cells. Biochem Biophys Res Commun. 2004;325:395–400. doi: 10.1016/j.bbrc.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 30.Mohamed AA, Tan SH, Mikhalkevich N, Ponniah S, Vasioukhin V, Bieberich CJ, Sesterhenn IA, Dobi A, Srivastava S, Sreenath TL. Ets family protein, erg expression in developing and adult mouse tissues by a highly specific monoclonal antibody. J Cancer. 2010;1:197–208. doi: 10.7150/jca.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrère S, Verger A, Flourens A, Stehelin D, Duterque-Coquillaud M. Erg proteins, transcription factors of the Ets family, form homo, heterodimers and ternary complexes via two distinct domains. Oncogene. 1998;16:3261–3268. doi: 10.1038/sj.onc.1201868. [DOI] [PubMed] [Google Scholar]
- 32.Asano Y, Czuwara J, Trojanowska M. Transforming growth factor-β regulates DNA binding activity of transcription factor Fli1 by p300/CREB-binding protein-associated factor-dependent acetylation. J Biol Chem. 2007;282:34672–34683. doi: 10.1074/jbc.M703907200. [DOI] [PubMed] [Google Scholar]
- 33.Tan FK, Zhou X, Mayes MD, Gourh P, Guo X, Marcum C, Jin L, Arnett FC., Jr Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology (Oxford) 2006;45:694–702. doi: 10.1093/rheumatology/kei244. [DOI] [PubMed] [Google Scholar]
- 34.Christmann RB, Hayes E, Pendergrass S, Padilla C, Farina G, Affandi AJ, Whitfield ML, Farber HW, Lafyatis R. Interferon and alternative activation of monocyte/macrophages in systemic sclerosis-associated pulmonary arterial hypertension. Arthritis Rheum. 2011;63:1718–1728. doi: 10.1002/art.30318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, Henley KJ, Ellis S, Voss AK, Metcalf D, et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol. 2008;9:810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]