Abstract
Although cigarette smoke (CS) is the primary risk factor for chronic obstructive pulmonary disease (COPD), the underlying molecular mechanisms for the significant variability in developing COPD in response to CS are incompletely understood. We performed lung gene expression profiling of two different wild-type murine strains (C57BL/6 and NZW/LacJ) and two genetic models with mutations in COPD genome-wide association study genes (HHIP and FAM13A) after 6 months of chronic CS exposure and compared the results to human COPD lung tissues. We identified gene expression patterns that correlate with severity of emphysema in murine and human lungs. Xenobiotic metabolism and nuclear erythroid 2-related factor 2-mediated oxidative stress response were commonly regulated molecular response patterns in C57BL/6, Hhip+/−, and Fam13a−/− murine strains exposed chronically to CS. The CS-resistant Fam13a−/− mouse and NZW/LacJ strain revealed gene expression response pattern differences. The Fam13a−/− strain diverged in gene expression compared with C57BL/6 control only after CS exposure. However, the NZW/LacJ strain had a unique baseline expression pattern, enriched for nuclear erythroid 2-related factor 2-mediated oxidative stress response and xenobiotic metabolism, and converged to a gene expression pattern similar to the more susceptible wild-type C57BL/6 after CS exposure. These results suggest that distinct molecular pathways may account for resistance to emphysema. Surprisingly, there were few genes commonly modulated in mice and humans. Our study suggests that gene expression responses to CS may be largely species and model dependent, yet shared pathways could provide biologically significant insights underlying individual susceptibility to CS.
Keywords: gene expression, mouse model, chronic obstructive pulmonary disease, cigarette smoking
Clinical Relevance
Mouse emphysema models are an important tool to study potential mechanisms of resistance and susceptibility to develop chronic obstructive pulmonary disease (COPD) in human smokers. We found more differences than similarities comparing lung gene expression profiles across several wild-type and genetically targeted murine models, and also comparing these results to expression profiling in human COPD. Understanding these differences is important for translation of therapeutic targets for COPD from mouse to human studies.
Chronic obstructive pulmonary disease (COPD) is a disease characterized by persistent, usually progressive airflow limitation. It is the fourth leading cause of death worldwide, and the number of affected individuals is growing (1). Although cigarette smoke (CS) is the primary risk factor for COPD, not all smokers develop COPD (2, 3). Genetic factors likely play a role in determining disease susceptibility (4, 5). Genome-wide association studies (GWAS) have identified genetic variants associated with susceptibility or resistance, but the underlying molecular mechanisms are poorly understood.
Murine models have provided valuable insights into COPD pathogenesis (6). Genetic models with targeted disruption of genes identified in GWAS show variation of susceptibility/resistance to emphysema (7, 8). Mice with Hhip haploinsufficiency are more susceptible to CS-induced airspace enlargement, whereas Fam13a knockout mice are protected. On the other hand, murine strains with different genetic backgrounds show significant variability in airspace enlargement after chronic exposure to CS (9, 10). Whether the underlying molecular mechanism of susceptibility/resistance to CS is shared among the genetic models, different background strains, and, ultimately, human pathobiology has not been fully determined. For instance, many murine models for COPD susceptibility and resistance are based on short-term CS exposure, making it difficult to compare with lung specimens from human subjects with COPD, who are usually long-term smokers. Studying both susceptible and resistant models will be important to identify mechanisms of both predisposition to and protection from the harmful effects of CS.
Microarray gene expression profiling of tissues provides a genome-wide assessment of molecular state and responses associated with the disease. Herein, we present the results of transcriptomic analysis on lung samples from two different strains of wild-type (WT) mice and two different lines of gene-targeted mice that are either susceptible or resistant to emphysema development after the animals had been exposed chronically to air or CS. We assessed the relationship between gene expression and phenotypic emphysema, and compared the results with gene expression in human COPD lungs to gain understanding of shared and distinct pathways between human and murine models. We hypothesized, based on our transcriptomic analysis, that there would be shared biological pathways between a susceptible WT strain and a susceptible gene-targeted line of mice, between a resistant WT strain and a resistant gene-targeted line of mice, and also between susceptible murine strains or lines and human lung samples.
Materials and Methods
Mouse Models of Emphysema
All animal studies were approved by the Harvard Medical School (Boston, MA) Institutional Animal Care and Use Committee.
Fam13a−/− and Hhip+/− mice were generated in C57BL/6 background as described previously (7, 8). Female, 10-week-old Fam13a−/− (n = 13) and Hhip+/− (n = 19) mice, both in a pure C57BL/6 background, as well as C57BL/6 WT littermate controls (n = 13 and 17, respectively) and NZW/LacJ mice (Jackson Laboratory, Bar Harbor, ME, n = 8), were exposed to mixed main-stream and side-stream CS from 3R4F Kentucky Research cigarettes (University of Kentucky, Lexington, KY) for 6 d/wk in a whole-body chamber (Teague TE 10z, TSP 100–200 mg/m3, CO2 levels ∼6 ppm; Woodland, CA) for 6 months, as described previously (8). Control groups were exposed to filtered air for the same time period (Fam13a−/−, n = 7; Hhip+/−, n = 14; NZW/LacJ, n = 9; C57BL/6, n = 26; see Figure E1 in the online supplement).
For airspace enlargement measurements, randomly captured lung section images were analyzed for mean chord length (MCL) to assess for CS-induced airspace enlargement using Scion Image software (Scion Corporation, Frederick, MD) (11).
Human Lung Tissues
Human lung samples were obtained from patients undergoing thoracic surgery at Brigham and Women’s Hospital (Boston, MA), St. Elizabeth’s Hospital (Brighton, MA), and Temple University Hospital (Philadelphia, PA), as described previously (12). All patients were former smokers with severe COPD (Global Initiative for Chronic Obstructive Lung Disease spirometry stages 3–4), whereas control smokers had normal spirometry. Institutional Review Board approval was obtained at each hospital, and all subjects provided written informed consent.
Microarray Gene Expression Profiling and Data Analysis
Detailed microarray processing methods are available in the online supplement.
Murine lung RNA samples were hybridized onto a Sentrix MouseRef-8 v2.0 Expression BeadChip Array (Illumina, San Diego, CA). Human lung samples were hybridized onto HumanHT-12 BeadChips (Illumina). After quality control, 151 human samples and 109 murine samples were available for analysis. Expression data have been deposited to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession nos. GSE87292 [murine] and GSE76925 [human]).
Batch effects were explored using classical multidimensional scaling plots. The Hhip+/− smoke exposure experiment did not show significant batch effects, so no batch correction was performed. Fam13a−/− mice were compared with WT C57BL/6 littermate controls (n = 4) and WT C57BL/6 (n = 6) controls from the NZW/LacJ experiment, so Combat batch correction was used (13). Combat was also applied to correct for batch effects in the NZW/LacJ experiment. Differential expression analyses were performed using the R package “limma” (14). P values were adjusted for multiple comparisons using the Benjamini and Hochberg false discovery rate (15).
For functional enrichment tests of the candidate genes, WebGestalt (16) was used for Gene Ontology term analysis. Pathway and network analyses were generated using Ingenuity Pathway Analysis software (IPA; Qiagen, Redwood City, CA; www.qiagen.com/ingenuity). Analyses were performed in murine experiments and human samples, which were then compared.
Comparison of human and mouse gene expression data was performed by mapping murine genes to orthologous human genes (www.ensembl.org/biomart). We performed the Fisher’s exact test to compare the distribution of genes that show the same direction of expression between the human and murine models.
To identify genes associated with the MCL, we fitted a linear regression model, adjusted for treatment (air or CS) and genotype (WT, Fam13a−/−, or Hhip+/−).
Results
Mouse Models Show Varying Degrees of Airspace Enlargement after CS Exposure
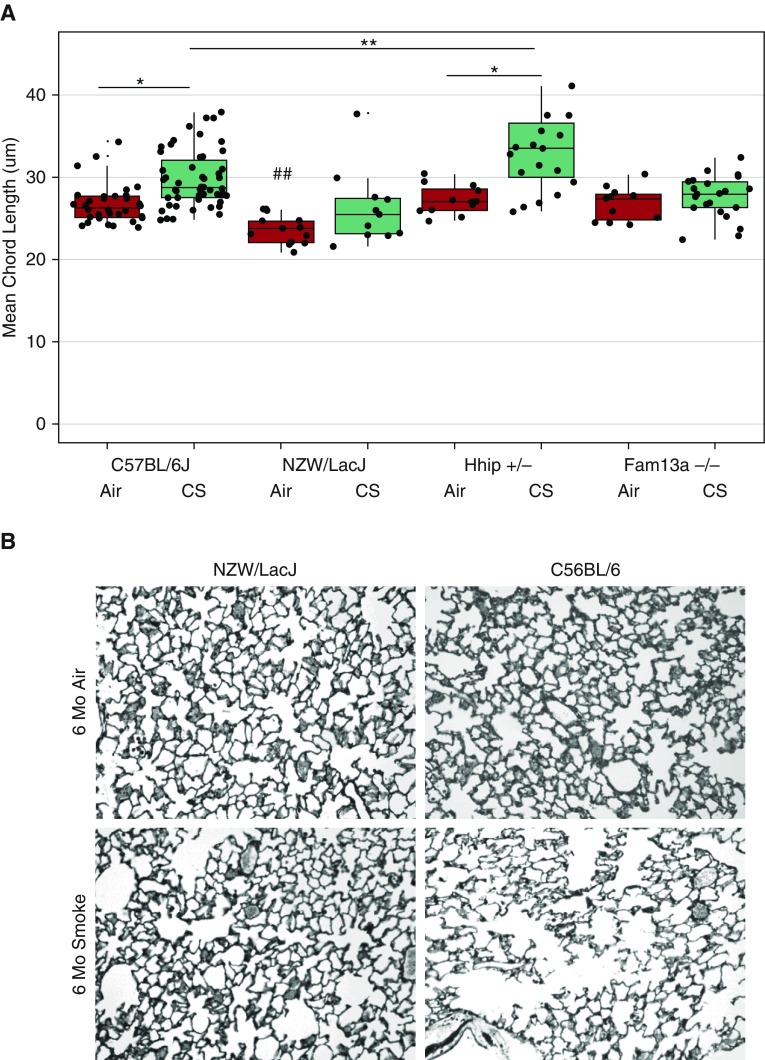
Four different murine models were exposed to filtered air or CS for 6 months. We have previously reported the phenotypes of Hhip+/− and Fam13a−/− mice exposed to CS for 6 months, and used microarray data from these studies (7, 8). Air-exposed WT C57BL/6, Hhip+/−, Fam13a−/−, and NZW/LacJ mice had normal lung architecture at 2 months of age, as determined by histologic analysis (Figure 1). MCL, a morphometric measure of alveolar space enlargement, was significantly lower in NZW/LacJ air-exposed mice compared with the other groups (ANOVA, P < 0.001).
Figure 1.
Different strains and genetic models display varying degrees of emphysema upon 6 months of cigarette smoke (CS) exposure. (A) Mean chord length (MCL) varies among strain and targeted genetic models after 6 months of CS exposure. Baseline controls (exposed to filtered air) of C57BL6/J, Hhip+/−, and Fam13a−/− do not show difference in MCL, but NZWLac/J is significantly lower (ANOVA, Tukey’s honest significant difference post hoc test, ##P < 0.001). NZWLac/J and Fam13a−/−, strain and genetic models resistant to CS-induced emphysema, show no difference in MCL between baseline and CS groups (P > 0.05, t test). CS and baseline groups of C57BL6/J and Hhip+/− differ in MCL (*P < 0.01, t test), and the CS groups of C57BL6/J and Hhip+/− differ in MCL as well (**P < 0.001, t test). (B) Lung histology sections from NZW/LacJ and C57BL/6J mice exposed to room air or CS. There is minimal airspace enlargement in NZW/LacJ mice compared with C57BL/6J mice. Original magnification: ×200.
When compared with air-exposed mice, C57BL/6 and Hhip+/− mice had significantly higher MCL after 6 month of CS exposure. NZW/LacJ and Fam13a−/− did not have significantly different MCL compared with air controls. The extent of alveolar space enlargement after CS exposure was markedly higher in Hhip+/− mice than in C57BL/6 mice, as reported previously (7) (Figure 1).
CS Response Genes Are Involved in Oxidative Stress and Xenobiotic Metabolism
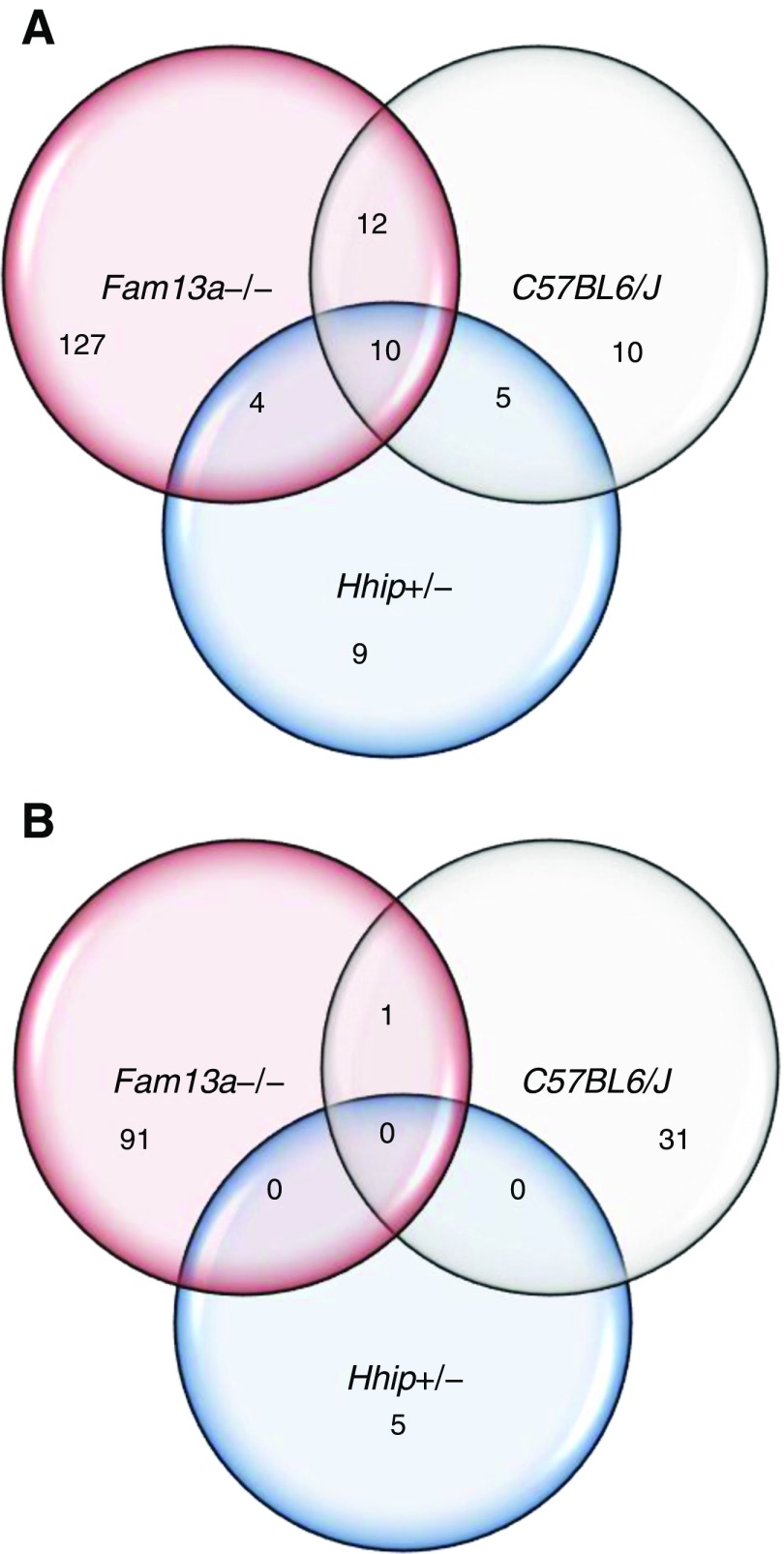
Genes that were differentially expressed (at false discovery rate < 0.05) in CS-exposed lungs from Fam13a−/−, C57BL/6, and Hhip+/− mice compared with air-exposed genotype-matched controls were termed CS-response genes (Figure 2). In each comparison, there were more genes that were up-regulated than down-regulated in response to CS. In Fam13a−/− mice, 153 genes were up-regulated and 92 genes were down-regulated in response to CS. In C57BL/6 mice, 37 genes were up-regulated and 32 genes were down-regulated, and, in Hhip+/− mice, 28 genes were up-regulated and 5 genes were down-regulated. Comparing the three models, there was more overlap for up-regulated than down-regulated CS-response genes (Figure 2). Table 1 lists the 10 up-regulated genes that had shared responses among Fam13a−/−, C57BL/6, and Hhip+/− mice. Aldh3a1 and Gstp1 genes are involved in aryl hydrocarbon receptor signaling and xenobiotic metabolism, whereas Gpx2 and Gstp1 genes are involved in the nuclear erythroid 2-related factor 2 (Nrf2)-mediated oxidative stress response, as expected with CS exposure. The number of up-regulated genes involved in xenobiotic metabolism and Nrf2-mediated oxidative stress response was highest in Fam13a−/− mice and lowest in Hhip+/− mice, inversely correlating with severity of airspace enlargement (Table 2).
Figure 2.
Transcriptional response upon CS exposure. Venn diagrams show number of genes commonly or uniquely regulated in different mouse models. (A) Number of up-regulated genes (false discovery rate [FDR] < 0.05). (B) Number of down-regulated genes (FDR < 0.05).
Table 1.
Common Cigarette Smoke Response Genes among Fam13a−/−, Hhip+/−, and C57BL/6J Mice
| Symbol | Entrez Gene Name | LogFC (Fam13a−/−) | LogFC (C57BL/6J) | LogFC (Hhip+/−) |
|---|---|---|---|---|
| Acox2 | Acyl-CoA oxidase 2, branched chain | 0.743 | 0.604 | 0.558 |
| Agrp | Agouti related neuropeptide | 0.81 | 0.505 | 0.51 |
| Akr1b8 | Aldo-keto reductase family 1, member B8 (aldose reductase) | 1.311 | 1.335 | 1.175 |
| Aldh3a1 | Aldehyde dehydrogenase 3 family, member A1 | 1.737 | 1.992 | 1.554 |
| Cbr3 | Carbonyl reductase 3 | 1.128 | 1.617 | 0.884 |
| Ces1f | Carboxylesterase 1F | 1.014 | 0.752 | 0.769 |
| Cxcl17* | Chemokine (C-X-C motif) ligand 17 | 1.397 | 0.724 | 0.665 |
| Gpx2 | Glutathione peroxidase 2 | 1.321 | 1.088 | 1.007 |
| Gstp1* | Glutathione S-transferase pi 1 | 1.077 | 0.76 | 0.837 |
| Ugt1a10 | UDP glucuronosyltransferase 1 family, polypeptide A10 | 1.07 | 0.569 | 0.714 |
Definition of abbreviation: FC, fold change.
Genes that had false discovery rate less than 0.05 in less than all three models are shown.
Multiple probes corresponding to the gene; probes with the greatest variance are shown.
Table 2.
Expression of Xenobiotic Metabolism and Nuclear Erythroid 2-Related Factor 2-Mediated Oxidative Stress Response Genes in Hhip+/−, C57BL/6, Fam13a−/− Mice in Response to 6 Months of Cigarette Smoke Exposure
| Pathways |
Hhip+/− |
C57BL/6 |
Fam13a−/− |
|||
|---|---|---|---|---|---|---|
| Gene | Log FC | Gene | LogFC | Gene | LogFC | |
| Xenobiotic metabolism signaling | Aldh3a1 | 1.554 | Aldh3a1 | 1.992 | Aldh3a1 | 1.737 |
| Cyp1a1 | 0.171 | Cyp1a1 | 0.633 | |||
| Cyp1b1 | 1.211 | Cyp1b1 | 2.257 | Cyp1b1 | 0.957 | |
| Gsta3 | 0.339 | |||||
| Gstp1 | 0.837 | Gstp1 | 0.76 | Gstp1 | 1.077 | |
| Gsto1 | 0.495 | Gsto1 | 0.948 | |||
| Nqo1 | 1.215 | Nqo1 | 1.131 | Nqo1 | 1.24 | |
| Nqo2 | 0.339 | |||||
| Ugt1a6 | 0.609 | Ugt1a6 | 0.473 | Ugt1a6 | 1.02 | |
| Ugt1a7 | 0.714 | Ugt1a7 | 0.569 | Ugt1a7 | 1.07 | |
| Pik3r3 | 0.213 | Ahrr | 0.207 | Ces1 g | 0.099 | |
| Mapk13 | 0.476 | |||||
| Mgst1 | 0.484 | |||||
| Mgst2 | 0.553 | |||||
| Smox | 0.343 | |||||
| Sra1 | 0.253 | |||||
| Sult1d1 | 0.839 | |||||
| Nrf2-mediated oxidative stress response | Gpx2 | 1.007 | Gpx2 | 1.088 | Gpx2 | 1.321 |
| Gstp1 | 0.837 | Gstp1 | 0.76 | Gstp1 | 1.077 | |
| Nqo1 | 1.215 | Nqo1 | 1.131 | Nqo1 | 1.24 | |
| Pik3r3 | 0.213 | Nqo2 | 0.339 | |||
| Gsto1 | 0.495 | Gsto1 | 0.948 | |||
| Cbr1 | 0.456 | Cbr1 | 0.637 | |||
| Gclm | 0.881 | Actc1 | 0.735 | |||
| Akr7a2 | 0.212 | |||||
| Cdc34 | 0.215 | |||||
| Gsta3 | 0.339 | |||||
| Mgst1 | 0.484 | |||||
| Mgst2 | 0.553 | |||||
Definition of abbreviations: FC, fold change; Nrf2, nuclear erythroid 2-related factor 2.
Pathway analysis was performed using ingenuity pathway analysis.
Comparisons of Differentially Expressed Genes between Models Suggest Candidates Involved in Susceptibility
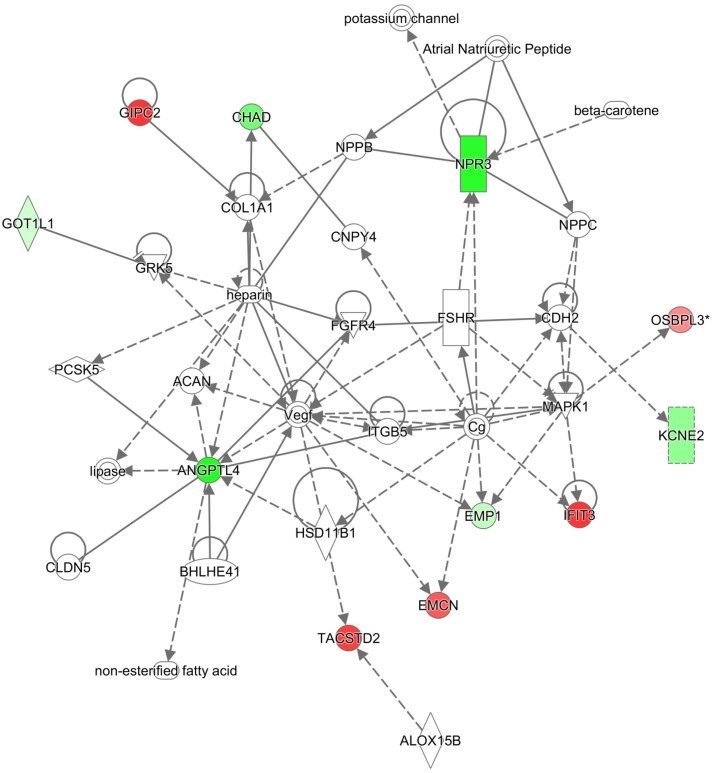
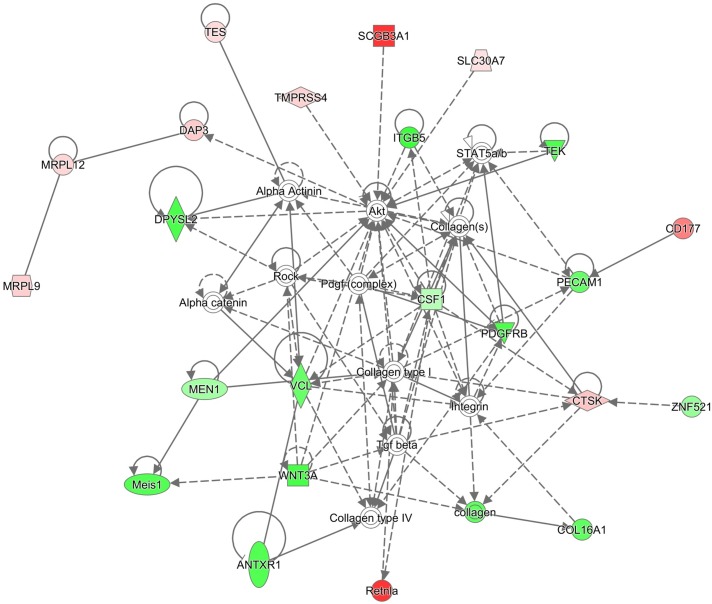
Based on the varying susceptibility of murine models with different genetic backgrounds to CS-induced emphysema (Figure 1), genes that are differentially expressed only in susceptible or resistant models might pinpoint potential mechanisms underlying the variable susceptibility of mice and/or humans to CS-induced emphysema (17). We assessed common biological pathways or networks enriched in the differentially expressed genes found only in susceptible or resistant strains. Among the 14 genes that were differentially expressed only in CS-susceptible Hhip+/− mice, 11 genes are involved in connective tissue development and function network (Figure 3). The 218 genes that were differentially expressed only in CS-resistant Fam13a−/− mice were involved in multiple functional networks, including carbohydrate and lipid metabolism, and molecular transport (Figure 4).
Figure 3.
Genes uniquely modulated in Hhip+/− mice are involved in the connective tissue development and function network. Red color indicates up-regulation; green indicates down-regulation. Network diagram was generated using the Ingenuity Pathway Analysis software.
Figure 4.
Genes uniquely modulated in Fam13a−/− mice are involved in carbohydrate, lipid metabolism, and molecular transport network. Red color indicates up-regulation; green indicates down-regulation. Network diagram was generated using the Ingenuity Pathway Analysis software.
Furthermore, we tested whether the relative changes in expression levels of genes between the air- and CS-exposed mice from the three strains correlated with the degree of emphysema susceptibility: Fam13a−/− (resistant), C57BL/6 (moderately susceptible), and Hhip+/− (susceptible). We found seven genes, the expression levels of which were negatively correlated with susceptibility (Table 3). Acox2 and Gpx2 are involved in oxidoreductase activity, whereas Abcb6 and Ugt1a6a are Nrf2-regulated genes.
Table 3.
Resistance Candidate Genes
| Symbol | Entrez Gene Name | LogFC (Fam13a−/−) | LogFC (C57BL/6J) | LogFC (Hhip+/−) |
|---|---|---|---|---|
| Abcb6 | ATP-binding cassette, subfamily B | 0.474 | 0.359 | 0.337 |
| Acox2 | Acyl-coenzyme A oxidase2, branched chain | 0.743 | 0.604 | 0.558 |
| Av249152 | Mus musculus expressed sequence Av249152 | 0.775 | 0.290 | 0.225 |
| Bc024561 | Mus musuculus cDNA sequence Bc024561 | 1.382 | 0.724 | 0.665 |
| Bc048546 | Mus musuculus cDNA sequence Bc048546 | 1.427 | 0.672 | 0.563 |
| Bc054509 | Mus musuculus cDNA sequence Bc054509 | 0.564 | 0.307 | 0.262 |
| Cxcl17 | Chemokine (C-X-C motif) ligand 17 | 1.397 | 0.684 | 0.581 |
| Gpx2 | Glutathione peroxidase 2 | 1.321 | 1.088 | 1.007 |
For definition of abbreviation, see Table 1.
Genes that are negatively correlated with emphysema susceptibility in the three different models (Fam13a−/−, resistant; C57BL/6J, moderate; Hhip+/−, susceptible). Genes with false discovery rate less than 0.2 in all three models are shown.
Gene Expression in Resistant Models Reveals Different Mechanisms of Resistance to CS-Induced Emphysema
The NZW/LacJ strain did not have any genes that were significantly differentially expressed after CS exposure. However, the gene expression profile of air-exposed mice is different when comparing NZW/LacJ to C57BL/6 mice (Table 4). This result suggests that baseline gene expression differences, rather than the response pattern to CS exposure, may account for the phenotypic resistance of the NZW/LacJ strain. There were 99 up-regulated genes and 141 down-regulated genes between air-exposed NZW/LacJ mice and air-exposed C57BL/6 mice. The up-regulated genes were involved in pathways including RhoA signaling and glutathione-mediated detoxification, and the down-regulated genes were involved in cholesterol biosynthesis, ketolysis, and systemic lupus erythematous signaling (Table 4, Table E1).
Table 4.
Gene Expression Pattern in Resistant Phenotype NZW/LacJ and Fam13a−/− Mice
| Group | Air |
CS |
||
|---|---|---|---|---|
| Up-Regulated | Down-Regulated | Up-Regulated | Down-Regulated | |
| NZW/LacJ versus C57BL/6 | 99 | 141 | 0 | 0 |
| RhoA signaling | Superpathway of cholesterol biosynthesis | |||
| PCP pathway | Ketolysis | |||
| LPS/IL-1–mediated inhibition of RXR function | Systemic lupus erythematosus signaling | |||
| Autophagy | Ketogenesis | |||
| Hypoxia signaling in the cardiovascular system | ATM signaling | |||
| Fam13a−/− versus C57BL/6 | 0 | 0 | 24 | 42 |
| Melatonin degradation I | Ephrin receptor signaling | |||
| LPS/IL-1–mediated inhibition of RXR function | Bladder cancer signaling | |||
| Superpathway of melatonin degradation | VEGF family ligand-receptor interactions | |||
| S-adenosyl-L-methionine biosynthesis | VEGF signaling | |||
| Embryonic stem cell differentiation into cardiac lineages | Leukotriene biosynthesis | |||
Definition of abbreviations: ATM, ataxia telangiectasis mutated protein; CS, cigarette smoke; PCP, planar cell polarity; RXR, retinoid X receptor; VEGF, vascular endothelial growth factor.
The number of differentially expressed genes and the top five pathways in CS-exposed NZW/LacJ mice and Fam13a−/− mice compared to C57BL/6 are shown (false discovery rate < 0.2). Pathway analysis was performed using Ingenuity Pathway Analysis software. For complete list of differentially expressed genes, please refer to Tables E1 and E2 in the online supplement.
On the other hand, Fam13a−/− mice, which are also resistant to CS-induced airspace enlargement (Figure 1), did not show any differences in gene expression pattern when compared with C57BL/6 WT mice after air exposure, but differed from C57BL/6 WT mice in lung gene expression after CS exposure. Fam13a−/− murine lungs had 24 up-regulated genes and 42 down-regulated genes that are involved in melatonin degradation, LPS/IL-1–mediated inhibition of retinoic acid receptor function, and ephrin receptor signaling, which might contribute to their resistance to CS-induced lung injury (Table 4, Table E2).
Comparison of Human and Murine Lung Gene Expression Profiles
To compare patterns of resistance/susceptibility upon CS exposure, we compared the murine gene expression results to the human lung transcriptomic profiles from previously reported studies including: (1) patients with severe COPD and control smokers (18); and (2) smokers and nonsmokers (19). We reasoned that Hhip+/− or WT C57BL/6 murine models with airspace enlargement after CS may share similar gene expression pathways as patients with COPD, whereas resistant NZW/LacJ or Fam13a−/− may share similar molecular mechanisms with smokers without COPD. Therefore, we compared differentially expressed genes between CS-exposed mice with differentially expressed genes from subjects with severe COPD and smoking control subjects, which we termed COPD genes. Compared with differentially expressed genes in human subjects with COPD, there were 5 genes that were shared with C57BL/6 compared with Fam13a−/− (CS exposed), 14 genes with C57BL/6 compared with Hhip+/− (CS exposed), and 36 genes with Fam13a−/− compared with Hhip+/− (CS exposed) (Table 5). Although there were few genes overlapping between human and mouse, several shared genes have been previously reported to be associated with COPD, including VEGFA and HDAC5. The purinergic receptor, P2Y14, was a common differentially expressed gene in all three comparisons (Table E3).
Table 5.
Shared Genes among Human Chronic Obstructive Pulmonary Disease, Emphysema, and Murine Models
| Comparison Group (No. of Genes) | COPD Genes (1,003) | Emphysema |
|
|---|---|---|---|
| Perc15 (1,433) | LAA950 (1,740) | ||
| WT CS versus Hhip+/− CS (343) | 14 | 23 | 32 |
| WT CS versus Fam13a−/− CS (66) | 5 | 5 | 5 |
| Fam13a−/− versus Hhip+/− CS (1,010) | 36 | 55 | 74 |
Definition of abbreviations: COPD, chronic obstructive pulmonary disease; CS, cigarette smoke; LAA950, low attenuation areas at −950 Hounsfield units on chest computed tomography scans; Perc15, 15th percentile of the lung density histogram on chest computed tomography scans; WT, wild-type C57BL/6J mice.
Number of shared orthologous genes between mice models and human subjects, at false discovery rate less than 0.2. See Table E3 for the individual gene lists.
Second, we compared differentially expressed genes between air- and CS-exposed mice with smoking signature genes reported previously in human lungs (19). In Hhip+/− mice, 14 genes were shared, and, in Fam13a −/− mice, 157 genes were shared with the human smoking signature genes. Not surprisingly, genes commonly shared among humans and different murine models are involved in CS related metabolism and inflammation, including AGRP, ALDH3A1, CXCL17, CYP1B1, GPX2, and NQO1.
Concordance between Human and Mouse Gene Expression Differs between Model Systems
To most closely resemble the mouse Hhip+/− or Fam13a−/− versus WT genetic models, we stratified the human subjects according to gene expression levels of HHIP or FAM13A above or below the median expression level, and performed differential expression analysis for the rest of the genes. We then examined the directionality of differentially expressed genes between species. The overlap of the differentially expressed genes between human high versus low expressers of HHIP and C57BL/6 compared with Hhip+/− mice upon CS exposure was significantly higher than what would be expected by chance (Fisher’s exact test, P = 0.016; Table 6, Table E4). The up-regulated genes are involved in the inflammatory response and connective tissue disorders (Table 7), whereas the down-regulated genes are involved in cell death and survival pathways (Table 8). In contrast, far fewer genes were shared among human high versus low expressers of FAM13A and C57BL/6 compared with Fam13a−/− mice exposed to CS (Table E5).
Table 6.
Concordance between Hhip+/− Mice and Human Lung Gene Expression
| Number of Genes | Human HHIP High versus Low |
Human HHIP High versus Low |
|---|---|---|
| Up | Down | |
| Hhip+/− up | 56 | 19 |
| Hhip+/− down | 75 | 56 |
Contingency table comparing human HHIP high versus low expressers and Hhip+/− differentially expressed genes (compared to wild-type C57BL/6J cigarette smoke).
See Table E4 for the list of individual genes.
Table 7.
Up-Regulated Genes and Pathways between Hhip+/− Mice and Human Lung
| Canonical Pathways | Genes | P Value |
|---|---|---|
| Gαq signaling | GNA14, HTR2B, PLCB2, PTK2B | 1.38 × 10−3 |
| IL-15 production | PTK2B, STAT1 | 3.11 × 10−3 |
| Chemokine signaling | PLCB2, PTK2B | 1.86 × 10−2 |
Top 3 pathways and genes are shown from 56 shared up-regulated genes from Table 6 (false discovery rate <0.2).
Table 8.
Down-Regulated Genes and Pathways between Hhip+/− Mice and Human Lung
| Canonical Pathways | Genes | P Value |
|---|---|---|
| Cholecystokinin/gastrin-mediated signaling | CREM, RHOJ, PRKCA | 3.03 × 10−2 |
| Superpathway of inositol phosphate compounds | DUSP8, MINPP1, PAWR, PPP4R1 | 1.85 × 10−2 |
| Protein kinase A signaling | CREM, DHH, DUSP8, PED4D, PRKCA | 1.34 × 10−2 |
Top 3 pathways and genes are shown from 56 shared down-regulated genes from Table 6 (false discovery rate < 0.2).
Comparison of Emphysema Genes between Human and Mouse
The expression profiles of CS-exposed murine lungs also may contain information related to the extent of airspace enlargement. Exploiting the known phenotypic variability even within the same strain of mouse (20), we used linear regression models to identify genes associated with MCL. Overall, 25 genes were associated with MCL, after adjusting for treatment (CS) and genotype, which were enriched for genes involved in the inflammatory response and cell-to-cell signaling and interactions (Table 9). Few genes from the MCL gene set overlapped with human genes associated with COPD and emphysema. When murine MCL genes were compared with genes associated with quantitative computed tomography (CT) scan emphysema measurements in humans (18), only two genes overlapped: MEFV and TNFSF14. Similarly, only one gene (KAT14 [lysine acetyltransferase14, Csrp2bp]) was up-regulated in both the human COPD and murine MCL gene datasets.
Table 9.
Genes Associated with Mean Chord Length in Mouse Model
| Symbol | Entrez Gene Name | Adjusted P Value |
|---|---|---|
|
Scrg1 |
Scrapie responsive gene 1 |
0.001 |
|
Prok2 |
Prokinetin 2 |
0.004 |
|
Stfa2 |
Stefin A2 |
0.010 |
|
Celsr3 |
Cadherin, EGF LAG seven-pass G-type receptor 3 |
0.010 |
|
Prtn3 |
Proteinase 3 |
0.010 |
|
Hrh2 |
Histamine receptor H2 |
0.011 |
|
Mefv |
Mediterranean fever |
0.051 |
|
Mrgpra2 |
MAS-related GPR, member A2B |
0.070 |
|
Foxd4 |
Forkhead box d4 |
0.089 |
|
Itih5 |
Inter-alpha (globulin) inhibitor H5 |
0.090 |
|
Ndn |
Necdin |
0.095 |
|
Otop3 |
Otopetrin3 |
0.095 |
|
Clec4e |
C-type lectin domain family 4, member e |
0.115 |
|
Csrp2bp |
Cysteine and glycine-rich protein 2 binding protein |
0.115 |
|
Tpd52 |
Tumor protein D52 |
0.121 |
|
Soat2 |
Sterol O-acyltransferase |
0.129 |
|
Tlr6 |
Toll like receptor 6 |
0.147 |
|
V1rh9 |
Vomeronasal 1 receptor 208 |
0.171 |
|
Cxcl2 |
Chemokine (C-X-C motif) ligand 2 |
0.181 |
|
Slc2a3 |
Solute carrier family 2 member 3 |
0.181 |
|
Rdh12 |
Retinol dehydrogenase 12 |
0.181 |
|
Il21r |
Interleukin 21 receptor |
0.181 |
|
Gm249 |
Protease, serine 38 |
0.192 |
| Tnfsf14 | Tumor necrosis factor superfamily member 14 | 0.192 |
Definition of abbreviations: EGF, epidermal growth factor; GPR, G protein–coupled receptor; LAG, laminin G.
Linear regression model was fitted to identify genes associated with mean chord length in wild-type C57BL/6J, Hhip+/−, and Fam13a−/− mice. Adjusted P value for each of the genes from the linear regression model is shown.
Discussion
We compared the gene expression profiles of different murine models of CS-induced emphysema to address variable susceptibility in developing COPD. We sought to understand the relevance of these murine models by comparing them with human lung transcriptomic data. Although there are many studies investigating gene expression changes with acute CS exposure in murine models (17, 21–26), few gene expression studies have been performed in the chronic CS model in which emphysematous changes take place (27–31). Our study is the first to compare global gene expression patterns of murine lungs after chronic CS exposure with those of human COPD lungs. By incorporating susceptible versus resistant strains of WT mice (C57BL/6 and NZW/LacJ, respectively) and susceptible versus resistant genetic models (Hhip+/− and Fam13a−/− mice, respectively), we investigated both common and model-specific mechanisms underlying COPD. We found that induction of antioxidant and detoxification genes were a common response to CS exposure, which was inversely correlated with emphysema susceptibility in C57BL/6, Hhip+/−, and Fam13a−/− mice. The resistant mouse models, NZW/LacJ and Fam13a−/−, showed distinct patterns of gene expression after CS exposure. Furthermore, comparison with murine and human gene expression demonstrated that no single murine model could account for the complete gene response pattern in human emphysema, underscoring the heterogeneous nature of COPD.
Shared Transcriptional Signature in Murine Models of CS-Induced Emphysema
The current understanding of COPD pathogenesis involves multiple mechanisms, including oxidative stress, inflammation, extracellular matrix destruction, cellular senescence, and apoptosis. Oxidants generated by CS enhance inflammation, tissue destruction, and apoptosis (32), and antioxidant capacity is one of the mechanisms underlying differential sensitivity to CS (33, 34). We found that antioxidant and detoxification genes were up-regulated in response to CS exposure across C57BL/6, Fam13a−/−, and Hhip+/− mice. Among the 10 genes that were shared in the 3 models, the genes Acox2, Aldh3a1, Gpx2, and Gstp1 are involved in xenobiotic metabolism and the Nrf2-mediated oxidative stress response. These antioxidant (Gpx3, Cyp1b1) and detoxification (Nqo1, Aldh3a1) genes have also been reported to be up-regulated in C57BL/6, ICR, and DBA/2 mice exposed to 1 month of CS (17), as well as in 6-month CS exposure in A/J mice (Nqo1, Cyp1b1) (27). We also observed that induction of antioxidant and detoxification genes was a shared response between human smoking and murine CS exposure models, demonstrating a conserved response to CS exposure.
Although antioxidant genes were consistently increased with CS exposure, the specific up-regulated genes were variable across murine models. The number of up-regulated genes involved in xenobiotic metabolism and Nrf2-mediated oxidative stress response was highest in Fam13a−/− mice and lowest in Hhip+/− mice, inversely correlated with severity of airspace enlargement. Similar to our murine study findings, the expression of a Nrf2-modulated genes is negatively associated with COPD in humans (35), indicating that emphysema is associated with decreased expression of Nrf2-regulated antioxidant and detoxification genes. Nrf2 is a key transcription factor regulating multiple antioxidant and detoxification genes, such as heme oxygenase-1, glutathione reductase, and glutathione peroxidase, and is critical for protection against CS-induced lung injury (33). Induction of Nrf2-regulated antioxidant genes is seen in short-term CS exposure murine models and in the lungs of human smokers (36). Conversely, A/J WT mice and ApoE−/− mice, which are both susceptible to developing emphysema after chronic CS exposure, show significantly reduced expression or no change in expression of Nrf2-regulated genes (27, 37), whereas CS-resistant ICR mice showed induction of Nrf2-regulated genes (33). These results suggests that antioxidant genes are a shared defense mechanism in response to CS exposure, and the number of xenobiotic-related genes induced by CS could explain the differences in emphysema susceptibility.
Distinct Transcriptional Signatures in Murine Models of CS-Induced Emphysema
NZW/LacJ mice showed no differentially expressed genes after CS exposure compared with air-exposed mice or compared with CS-exposed C57BL/6 mice, but had differentially expressed genes at baseline when compared with C57BL/6 air-exposed mice. Up-regulated genes were involved in RhoA signaling, xenobiotic metabolism, and Nrf2-mediated oxidative stress response, converging to a gene expression pattern similar to more susceptible WT C57BL/6 after CS exposure, and down-regulated genes were associated with inflammatory pathways. Although xenobiotic metabolism and Nrf2-mediated oxidative stress response were activated, the directionality of RhoA pathway was less clear, as the genes enriched in RhoA pathway included inhibitors (RhoGTPase activating proteins [RhoGAPs]: Arhgap12, Dlc1) as well as an activator (Rapgef6), and a target of RhoA pathway (binder of RhoGAPs, Cdc42ep2). Activated RhoA signaling was shown to inhibit clearance of apoptotic cells (38), leading to increased inflammation under oxidative stress induced by CS (39). Increased apoptosis is observed in human COPD lungs (40, 41), which suggests that imbalance between apoptosis, proliferation (42, 43), and apoptotic cell clearance (39) may contribute to the pathogenesis of COPD. Experimental validation would be required to test whether up-regulated RhoGAP plays a role in protection against emphysema. Similarly, genes in the glutathione-mediated detoxification pathway were both up-regulated and down-regulated; however, it is notable that Gstp1, a major detoxification enzyme, had higher expression in air-exposed NZW/LacJ compared with C57BL/6 air-exposed mice. Up-regulation of detoxification pathways and down-regulation of inflammatory pathways at baseline may explain the resistance to CS exposure in NZW/LacJ mice.
In contrast, although Fam13a−/− mice showed phenotypically similar resistance against CS with NZW/LacJ mice, the baseline gene expression in Fam13a−/− mice was similar compared with air-exposed C57BL/6 mice, and only diverged after CS exposure. The genes that were differentially expressed compared with C57BL/6 CS-exposed mice provide insight into potential mechanisms of resistance. The most significant up-regulated genes were Cyp2a6 and Sult1d1 and down-regulated genes were Vegfa and Vegfd. Cyp2a6 is involved in nicotine degradation, and Sult1d1 is involved in xenobiotic metabolism, in line with the heightened protective response against CS compared with C57BL/6 WT mice. Down-regulation of Vegfa and Vegfd likely contribute to emphysema, as inactivation of the pro-survival growth factor, vascular endothelial growth factor, has been shown to induce apoptosis and subsequent emphysema in animal models (43). These results suggest that related processes, such as apoptosis, cellular maintenance, and metabolism, could have different expression patterns in response to CS, even in phenotypically similar models. Although differences in antioxidant defense capacity correlate with emphysema susceptibility and resistance, the gene expression patterns and additional pathways were distinct in each model. These findings suggest that diverse mechanisms underlie susceptibility versus resistance of different murine models to the development of CS-induced emphysema.
Comparison of Murine Models to Human COPD
Because of the heterogeneous nature of COPD, it is possible that the murine models represent subsets of human COPD. To this end, we compared the differentially expressed genes from the murine models to human lung transcriptome of smokers and patients with severe COPD, as well as high versus low expressors of HHIP and FAM13A. We also compared genes associated with human emphysema to genes that correlate with murine MCL measurements.
As expected, different murine strains overlap with only a portion of the differentially expressed genes found in the human lung. Two genes were perturbed in all murine COPD models as well as the human COPD case–control study: P2Y14 and CHN2. Their expression was also associated with human quantitative chest CT scan emphysema measurements. The direction of differential regulation in the mouse models did not concur with what was observed in human lungs. P2Y14 gene expression was decreased in patients with COPD, but was up-regulated in the susceptible Hhip+/− mice exposed to CS, and was down-regulated in the resistant Fam13a−/− mice upon CS exposure. Because the purinergic receptor, P2Y14, is expressed in both airway epithelial cells and immune cells (44, 45), and is involved in sensing cellular stresses, such as radiation and aging, as well as inhibiting cellular senescence (46), the murine models may represent earlier responses of lung tissue damage compared with the patients with severe COPD. On the other hand, human patients with COPD had a higher level of CHN2 expression, but Fam13a−/− mice also displayed high expression of Chn2. CHN2 gene encodes β-chimerin, and GWAS have identified polymorphisms in this gene that are associated with addiction vulnerability and with cigarette smoking in a candidate gene study (47, 48). It is possible that differences in smoking status between the human former smokers and the murine CS exposure model may explain the differences in gene expression.
Given the spectrum of airspace enlargement across the murine models, we combined all experimental animals together and modeled MCL as a quantitative trait to identify emphysema susceptibility genes. When the 25 MCL-associated genes were compared with human quantitative CT scan–defined emphysema genes, MEFV and TNFSF14 were the only overlapping genes. TNFSF14 promotes CD8+ T cell activation (49), which has been linked to emphysema development in mice (50).
There are several factors that might have contributed to the apparent low level of overlap between the murine models and human COPD lung, beyond the heterogeneity of both human COPD and the mouse models. First, incomplete orthology between human and murine genes may have limited the number of genes that can be compared. In addition, differences in human and mouse lung development and anatomy, differences between the experimental smoking protocol and human cigarette smoking (6, 20), and differences in the morphometric analysis performed in mouse strains and human patients (20) might have contributed to the low concordance. Moreover, despite the fact that the chronic CS exposure model serves as the gold standard murine model for emphysema development, it is a poor replicator of chronic bronchitis and mucus hypersecretion, features of human COPD. Thus, the CS-induced emphysema mouse captures only part of the phenotypic spectrum of COPD. Furthermore, human lung gene expression profiling data are heterogeneous in themselves, due to difference in populations, case definition, smoking status, and phenotypic variability (51). Finally, as these gene expression data are obtained from homogenized lung tissues, they represent the averaged expression from mixtures of cell types. Inflammation and structural destruction from the COPD process can alter cellular proportions, and there may be a clearer signal if comparison of individual cell types were possible.
Although our study included several WT and genetic models exposed to CS, we did not include all the reported murine smoking models and other emphysema models, such as elastase, nor did we include all possible human COPD expression datasets. The human subjects had severe COPD, so information on mild COPD could have been lost. There are multiple possible methods for network and pathway analysis. Unsupervised analysis, such as weighted gene coexpression network analysis or gene set variation analysis, would be difficult to interpret, given the difference in genetic background and exposure conditions, with relatively small numbers in each group. However, the method we used for pathway analysis did show relationships to known mechanisms of COPD.
Conclusions
By using a spectrum of CS-exposed murine models, we show that differential expression in key pathway genes in response to CS may underlie differences in emphysema susceptibility. We have identified distinct gene expression patterns in resistant phenotypes. Given the heterogeneity of COPD, no single murine model can recapitulate human disease. Correlation with animal models may be improved by better disease subtyping and phenotypic characterization. We found commonly regulated genes that offer novel insights into the pathogenesis and discovery of new therapeutic targets for COPD. As for future directions, the molecular signatures represent the ensemble of distinct tissue compartments and cell types. It will be important to compare and contrast the expression response in whole-lung samples to that in different cell types in the lung that play key roles in COPD development. Recent advances in approaches to decipher the molecular status of tissues in near–single-cell resolution could give further insight in to the mechanisms of susceptibility and resistance to CS exposure. Although we focused this study on the emphysema phenotype, future studies could examine differences in inflammation between different CS-induced murine models and human subjects with COPD to provide a greater understanding of COPD susceptibility.
Footnotes
This work was supported by National Institutes of Health (NIH) grants R01HL094635 (C.P.H.), R01HL125583 (C.P.H.), R01HL130512 (C.P.H.), R01HL112176 (X.Z.), R33HL114305 (X.Z.), P01HL105339 (E.K.S.), and R01HL089856 (E.K.S.).
Author Contributions: Conception and design—J.H.Y., X.Z., and C.P.H.; data acquisition—T.L., Z.J., and X.Z.; data analysis—J.H.Y., J.M., W.Q., and K.G.; data interpretation—J.H.Y., J.M., C.A.O., M.A.P., E.K.S., X.Z., and C.P.H.; manuscript writing—J.H.Y. and C.P.H.; manuscript revising/editing—J.H.Y., C.A.O., M.A.P., E.K.S., X.Z., and C.P.H.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2016-0328OC on March 1, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Mannino DM, Buist AS. Global burden of COPD: risk factors, prevalence, and future trends. Lancet. 2007;370:765–773. doi: 10.1016/S0140-6736(07)61380-4. [DOI] [PubMed] [Google Scholar]
- 2.Mannino D. The natural history of chronic obstructive pulmonary disease. Pneumonol Alergol Pol. 2011;79:139–143. [PubMed] [Google Scholar]
- 3.Lundbäck B, Lindberg A, Lindström M, Rönmark E, Jonsson AC, Jönsson E, Larsson LG, Andersson S, Sandström T, Larsson K Obstructive Lung Disease in Northern Sweden Studies. Not 15 but 50% of smokers develop COPD?—Report from the Obstructive Lung Disease in Northern Sweden Studies. Respir Med. 2003;97:115–122. doi: 10.1053/rmed.2003.1446. [DOI] [PubMed] [Google Scholar]
- 4.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O’Donnell WJ, Reilly JJ, Ginns L, Mentzer S, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease: risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med. 1998;157:1770–1778. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- 5.Berndt A, Leme AS, Shapiro SD. Emerging genetics of COPD. EMBO Mol Med. 2012;4:1144–1155. doi: 10.1002/emmm.201100627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leberl M, Kratzer A, Taraseviciene-Stewart L. Tobacco smoke induced COPD/emphysema in the animal model—are we all on the same page? Front Physiol. 2013;4:91. doi: 10.3389/fphys.2013.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lao T, Glass K, Qiu W, Polverino F, Gupta K, Morrow J, Mancini JD, Vuong L, Perrella MA, Hersh CP, et al. Haploinsufficiency of Hedgehog interacting protein causes increased emphysema induced by cigarette smoke through network rewiring. Genome Med. 2015;7:12. doi: 10.1186/s13073-015-0137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Z, Lao T, Qiu W, Polverino F, Gupta K, Guo F, Mancini JD, Naing ZZC, Cho MH, Castaldi PJ, et al. A chronic obstructive pulmonary disease susceptibility gene, FAM13A, regulates protein stability of β-catenin. Am J Respir Crit Care Med. 2016;194:185–197. doi: 10.1164/rccm.201505-0999OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerassimov A, Hoshino Y, Takubo Y, Turcotte A, Yamamoto M, Ghezzo H, Triantafillopoulos A, Whittaker K, Hoidal JR, Cosio MG. The development of emphysema in cigarette smoke–exposed mice is strain dependent. Am J Respir Crit Care Med. 2004;170:974–980. doi: 10.1164/rccm.200309-1270OC. [DOI] [PubMed] [Google Scholar]
- 10.Foronjy RF, Mercer BA, Maxfield MW, Powell CA, D’Armiento J, Okada Y. Structural emphysema does not correlate with lung compliance: lessons from the mouse smoking model. Exp Lung Res. 2005;31:547–562. doi: 10.1080/019021490951522. [DOI] [PubMed] [Google Scholar]
- 11.Laucho-Contreras ME, Taylor KL, Mahadeva R, Boukedes SS, Owen CA. Automated measurement of pulmonary emphysema and small airway remodeling in cigarette smoke–exposed mice. J Vis Exp. 2015;(95):52236. doi: 10.3791/52236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrow JD, Cho MH, Hersh CP, Pinto-Plata V, Celli B, Marchetti N, Criner G, Bueno R, Washko G, Glass K, et al. DNA methylation profiling in human lung tissue identifies genes associated with COPD. Epigenetics. 2016;11:1–10. doi: 10.1080/15592294.2016.1226451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 14.Ritchie MD, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 16.Wang J, Duncan D, Shi Z, Zhang B.WEB-based gene set analysis toolkit (WebGestalt): update 2013 Nucleic Acids Res 201341web server issue):W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavarra E, Fardin P, Fineschi S, Ricciardi A, De Cunto G, Sallustio F, Zorzetto M, Luisetti M, Pfeffer U, Lungarella G, et al. Early response of gene clusters is associated with mouse lung resistance or sensitivity to cigarette smoke. Am J Physiol Lung Cell Mol Physiol. 2009;296:L418–L429. doi: 10.1152/ajplung.90382.2008. [DOI] [PubMed] [Google Scholar]
- 18.Morrow JD, Zhou X, Lao T, Jiang Z, DeMeo DL, Cho MH, Qiu W, Cloonan S, Pinto-Plata V, Celli B, et al. Functional interactors of three genome-wide association study genes are differentially expressed in severe chronic obstructive pulmonary disease lung tissue. Sci Rep. 2017;7:44232. doi: 10.1038/srep44232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bossé Y, Postma DS, Sin DD, Lamontagne M, Couture C, Gaudreault N, Joubert P, Wong V, Elliott M, van den Berge M, et al. Molecular signature of smoking in human lung tissues. Cancer Res. 2012;72:3753–3763. doi: 10.1158/0008-5472.CAN-12-1160. [DOI] [PubMed] [Google Scholar]
- 20.Wright JL, Cosio M, Churg A. Animal models of chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol. 2008;295:L1–L15. doi: 10.1152/ajplung.90200.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izzotti A, Cartiglia C, Longobardi M, Bagnasco M, Merello A, You M, Lubet RA, De Flora S. Gene expression in the lung of p53 mutant mice exposed to cigarette smoke. Cancer Res. 2004;64:8566–8572. doi: 10.1158/0008-5472.CAN-04-1420. [DOI] [PubMed] [Google Scholar]
- 22.Izzotti A, Cartiglia C, Longobardi M, Balansky RM, D’Agostini F, Lubet RA, De Flora S. Alterations of gene expression in skin and lung of mice exposed to light and cigarette smoke. FASEB J. 2004;18:1559–1561. doi: 10.1096/fj.04-1877fje. [DOI] [PubMed] [Google Scholar]
- 23.Meng QR, Gideon KM, Harbo SJ, Renne RA, Lee MK, Brys AM, Jones R. Gene expression profiling in lung tissues from mice exposed to cigarette smoke, lipopolysaccharide, or smoke plus lipopolysaccharide by inhalation. Inhal Toxicol. 2006;18:555–568. doi: 10.1080/08958370600686226. [DOI] [PubMed] [Google Scholar]
- 24.Halappanavar S, Russell M, Stampfli MR, Williams A, Yauk CL. Induction of the interleukin 6/signal transducer and activator of transcription pathway in the lungs of mice sub-chronically exposed to mainstream tobacco smoke. BMC Med Genomics. 2009;2:56. doi: 10.1186/1755-8794-2-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agarwal AR, Zhao L, Sancheti H, Sundar IK, Rahman I, Cadenas E. Short-term cigarette smoke exposure induces reversible changes in energy metabolism and cellular redox status independent of inflammatory responses in mouse lungs. Am J Physiol Lung Cell Mol Physiol. 2012;303:L889–L898. doi: 10.1152/ajplung.00219.2012. [DOI] [PubMed] [Google Scholar]
- 26.Morissette MC, Lamontagne M, Bérubé JC, Gaschler G, Williams A, Yauk C, Couture C, Laviolette M, Hogg JC, Timens W, et al. Impact of cigarette smoke on the human and mouse lungs: a gene-expression comparison study. PLoS One. 2014;9:1–11. doi: 10.1371/journal.pone.0092498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rangasamy T, Misra V, Zhen L, Tankersley CG, Tuder RM, Biswal S. Cigarette smoke–induced emphysema in A/J mice is associated with pulmonary oxidative stress, apoptosis of lung cells, and global alterations in gene expression. Am J Physiol Lung Cell Mol Physiol. 2009;296:L888–L900. doi: 10.1152/ajplung.90369.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gebel S, Diehl S, Pype J, Friedrichs B, Weiler H, Schüller J, Xu H, Taguchi K, Yamamoto M, Müller T. The transcriptome of Nrf2−/− mice provides evidence for impaired cell cycle progression in the development of cigarette smoke–induced emphysematous changes. Toxicol Sci. 2010;115:238–252. doi: 10.1093/toxsci/kfq039. [DOI] [PubMed] [Google Scholar]
- 29.Luettich K, Xiang Y, Iskandar A, Sewer A, Martin F, Talikka M, Vanscheeuwijck P, Berges A, Veljkovic E. Systems toxicology approaches enable mechanistic comparison of spontaneous and cigarette smoke–related lung tumor development in the A/J mouse model. Interdiscip Toxicol. 2014;7:73–84. doi: 10.2478/intox-2014-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabanski M, Fields B, Boue S, Boukharov N, DeLeon H, Dror N, Geertz M, Guedj E, Iskandar A, Kogel U, et al. Transcriptional profiling and targeted proteomics reveals common molecular changes associated with cigarette smoke–induced lung emphysema development in five susceptible mouse strains. Inflamm Res. 2015;64:471–486. doi: 10.1007/s00011-015-0820-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cloonan SM, Glass K, Laucho-Contreras ME, Bhashyam AR, Cervo M, Pabón MA, Konrad C, Polverino F, Siempos II, Perez E, et al. Mitochondrial iron chelation ameliorates cigarette smoke–induced bronchitis and emphysema in mice. Nat Med. 2016;22:163–174. doi: 10.1038/nm.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida T, Tuder RM. Pathobiology of cigarette smoke–induced chronic obstructive pulmonary disease. Physiol Rev. 2007;87:1047–1082. doi: 10.1152/physrev.00048.2006. [DOI] [PubMed] [Google Scholar]
- 33.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke–induced emphysema in mice. J Clin Invest. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao H, Rahman I. Current concepts on oxidative/carbonyl stress, inflammation and epigenetics in pathogenesis of chronic obstructive pulmonary disease. Toxicol Appl Pharmacol. 2011;254:72–85. doi: 10.1016/j.taap.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh A, Ling G, Suhasini AN, Zhang P, Yamamoto M, Navas-Acien A, Cosgrove G, Tuder RM, Kensler TW, Watson WH, et al. Nrf2-dependent sulfiredoxin-1 expression protects against cigarette smoke-induced oxidative stress in lungs. Free Radic Biol Med. 2009;46:376–386. doi: 10.1016/j.freeradbiomed.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morissette MC, Lamontagne M, Bérubé J-C, Gaschler G, Williams A, Yauk C, Couture C, Laviolette M, Hogg JC, Timens W, et al. Impact of cigarette smoke on the human and mouse lungs: a gene-expression comparison study. PLoS One. 2014;9:e92498. doi: 10.1371/journal.pone.0092498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boué S, De León H, Schlage WK, Peck MJ, Weiler H, Berges A, Vuillaume G, Martin F, Friedrichs B, Lebrun S, et al. Cigarette smoke induces molecular responses in respiratory tissues of ApoE−/− mice that are progressively deactivated upon cessation. Toxicology. 2013;314:112–124. doi: 10.1016/j.tox.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Tosello-Trampont A-C, Nakada-Tsukui K, Ravichandran KS. Engulfment of apoptotic cells is negatively regulated by Rho-mediated signaling. J Biol Chem. 2003;278:49911–49919. doi: 10.1074/jbc.M306079200. [DOI] [PubMed] [Google Scholar]
- 39.Richens TR, Linderman DJ, Horstmann SA, Lambert C, Xiao Y-Q, Keith RL, Boé DM, Morimoto K, Bowler RP, Day BJ, et al. Cigarette smoke impairs clearance of apoptotic cells through oxidant-dependent activation of RhoA. Am J Respir Crit Care Med. 2009;179:1011–1021. doi: 10.1164/rccm.200807-1148OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokohori N, Aoshiba K, Nagai A Respiratory Failure Research Group in Japan. Increased levels of cell death and proliferation in alveolar wall cells in patients with pulmonary emphysema. Chest. 2004;125:626–632. doi: 10.1378/chest.125.2.626. [DOI] [PubMed] [Google Scholar]
- 41.Imai K, Mercer BA, Schulman LL, Sonett JR, D’Armiento JM. Correlation of lung surface area to apoptosis and proliferation in human emphysema. Eur Respir J. 2005;25:250–258. doi: 10.1183/09031936.05.00023704. [DOI] [PubMed] [Google Scholar]
- 42.Kasahara Y, Tuder RM, Taraseviciene-Stewart L, Le Cras TD, Abman S, Hirth PK, Waltenberger J, Voelkel NF. Inhibition of VEGF receptors causes lung cell apoptosis and emphysema. J Clin Invest. 2000;106:1311–1319. doi: 10.1172/JCI10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang K, Rossiter HB, Wagner PD, Breen EC.Lung-targeted VEGF inactivation leads to an emphysema phenotype in mice J Appl Physiol (1985) 2004971559–1566.[Discussion, p. 1549.] [DOI] [PubMed] [Google Scholar]
- 44.Müller T, Bayer H, Myrtek D, Ferrari D, Sorichter S, Ziegenhagen MW, Zissel G, Virchow JC, Jr, Luttmann W, Norgauer J, et al. The P2Y14 receptor of airway epithelial cells: coupling to intracellular Ca2+ and IL-8 secretion. Am J Respir Cell Mol Biol. 2005;33:601–609. doi: 10.1165/rcmb.2005-0181OC. [DOI] [PubMed] [Google Scholar]
- 45.Moore DJ, Murdock PR, Watson JM, Faull RLM, Waldvogel HJ, Szekeres PG, Wilson S, Freeman KB, Emson PC. GPR105, a novel Gi/o-coupled UDP-glucose receptor expressed on brain glia and peripheral immune cells, is regulated by immunologic challenge: possible role in neuroimmune function. Brain Res Mol Brain Res. 2003;118:10–23. doi: 10.1016/s0169-328x(03)00330-9. [DOI] [PubMed] [Google Scholar]
- 46.Cho J, Yusuf R, Kook S, Attar E, Lee D, Park B, Cheng T, Scadden DT, Lee BC. Purinergic P2Y14 receptor modulates stress-induced hematopoietic stem/progenitor cell senescence. J Clin Invest. 2014;124:3159–3171. doi: 10.1172/JCI61636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrio-Real L, Barrueco M, González-Sarmiento R, Caloca MJ. Association of a novel polymorphism of the β2-chimaerin gene (CHN2) with smoking. J Investig Med. 2013;61:1129–1131. doi: 10.2310/JIM.0b013e3182a32ff9. [DOI] [PubMed] [Google Scholar]
- 48.Liu Q-R, Drgon T, Johnson C, Walther D, Hess J, Uhl GR. Addiction molecular genetics: 639,401 SNP whole genome association identifies many “cell adhesion” genes. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:918–925. doi: 10.1002/ajmg.b.30436. [DOI] [PubMed] [Google Scholar]
- 49.Scheu S, Alferink J, Pötzel T, Barchet W, Kalinke U, Pfeffer K. Targeted disruption of LIGHT causes defects in costimulatory T cell activation and reveals cooperation with lymphotoxin beta in mesenteric lymph node genesis. J Exp Med. 2002;195:1613–1624. doi: 10.1084/jem.20020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maeno T, Houghton AM, Quintero PA, Grumelli S, Owen CA, Shapiro SD. CD8+ T cells are required for inflammation and destruction in cigarette smoke–induced emphysema in mice. J Immunol. 2007;178:8090–8096. doi: 10.4049/jimmunol.178.12.8090. [DOI] [PubMed] [Google Scholar]
- 51.Zeskind JE, Lenburg ME, Spira A. Translating the COPD transcriptome: insights into pathogenesis and tools for clinical management. Proc Am Thorac Soc. 2008;5:834–841. doi: 10.1513/pats.200807-074TH. [DOI] [PMC free article] [PubMed] [Google Scholar]