Abstract
The heritability of chronic obstructive pulmonary disease (COPD) cannot be fully explained by recognized genetic risk factors identified as achieving genome-wide significance. In addition, the combined contribution of genetic variation to COPD risk has not been fully explored. We sought to determine: (1) whether studies of variants from previous studies of COPD or lung function in a larger sample could identify additional associated variants, particularly for severe COPD; and (2) the impact of genetic risk scores on COPD. We genotyped 3,346 single-nucleotide polymorphisms (SNPs) in 2,588 cases (1,803 severe COPD) and 1,782 control subjects from four cohorts, and performed association testing with COPD, combining these results with existing genotyping data from 6,633 cases (3,497 severe COPD) and 5,704 control subjects. In addition, we developed genetic risk scores from SNPs associated with lung function and COPD and tested their discriminatory power for COPD-related measures. We identified significant associations between SNPs near PPIC (P = 1.28 × 10−8) and PPP4R4/SERPINA1 (P = 1.01 × 10−8) and severe COPD; the latter association may be driven by recognized variants in SERPINA1. Genetic risk scores based on SNPs previously associated with COPD and lung function had a modest ability to discriminate COPD (area under the curve, ∼0.6), and accounted for a mean 0.9–1.9% lower forced expiratory volume in 1 second percent predicted for each additional risk allele. In a large genetic association analysis, we identified associations with severe COPD near PPIC and SERPINA1. A risk score based on combining genetic variants had modest, but significant, effects on risk of COPD and lung function.
Keywords: chronic obstructive pulmonary disease, genetic epidemiology, genetic risk factors, alpha-1 antitrypsin, genetic risk score
Clinical Relevance
In meta-analyses of a chronic obstructive pulmonary disease (COPD) genome-wide association study involving over 16,000 subjects, we found two single-nucleotide polymorphisms (SNPs; rs112458284 and rs6860095) not previously described in genome-wide studies that were associated with Global Initiative for Chronic Obstructive Lung Disease spirometric stage III–IV COPD at genome-wide significance levels. One of these likely tags the SERPINA1 Z allele based on its linkage disequilibrium pattern and conditional analysis results. In addition, we describe two genetic risk scores based on COPD- and lung function–associated SNPs and show their applications in explaining COPD severity and COPD affection status risk.
Chronic obstructive pulmonary disease (COPD), a progressive lung disease characterized by irreversible airflow obstruction, is a leading cause of morbidity and mortality worldwide (1, 2). Although cigarette smoking is the major determinant of COPD susceptibility in the industrialized world (3–5), pulmonary response to cigarette smoking is highly variable (6). Genetic factors contribute to variability in response to smoking, and multiple studies have identified genetic variants associated with increased COPD susceptibility (7–12). Nonetheless, the majority of estimated heritability for risk to COPD remains unexplained (13). In addition, the effect of several recognized risk alleles on lung function or risk of COPD, particularly in cohorts of severely affected subjects, has not been well studied. Meta-analysis of genetic associations across multiple cohorts has the advantage of improving power to detect additional susceptibility risk variants by combining information across studies, which may add to our understanding of disease mechanisms (14), as well as providing potential new targets for COPD therapy development (15, 16).
This study had two primary goals. First, we wished to investigate a panel of variants in a larger meta-analysis of cross-sectional data to increase our power to detect associations (17) with moderate-to-severe and severe COPD. The marker panel was composed of two groups of single-nucleotide polymorphisms (SNPs). The first group included top associations from previous genome-wide association studies (GWASs), including SNPs that did not reach genome-wide significance (18), and the second group included genetic variants hypothesized to affect COPD (19), including SNPs previously associated with lung function (20–22). We hypothesized some of these loci would reach predefined levels of statistical significance with our additional sample size in this meta-analysis.
Because genetic variation is fixed at birth, genetic risk scores in cross-sectional data may offer a way to consolidate genetic information (23) into a clinically meaningful tool that could help clinicians to predict disease susceptibility, progression, and outcomes (24, 25). Our second goal was to determine the relevance of genetic risk scores to COPD by modeling the effect of COPD- and lung function–associated risk alleles on clinical status, severe COPD-affection status, and forced expiratory volume in 1 second (FEV1) % predicted. We hypothesized that a combined risk score composed of SNPs shown to influence risk to COPD and lung function would explain the genetic contribution to COPD-related outcomes in a clinically useful manner.
Materials and Methods
We performed genetic meta-analysis using eight cohorts, including a total of 16,707 subjects (Table 1). We genotyped 3,346 SNPs (see the online supplement) in 5,358 subjects from 4 cohorts: the Transcontinental COPD Genetics Study (TCGS)–Korea and TCGS-Poland (26), the International COPD Genetics Network (ICGN) (27, 28), and the Boston Early-Onset COPD Study (29). To maximize power for meta-analysis, we combined these results with existing data from five additional cohorts: COPDGene non-Hispanic whites and African Americans (30), Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-Points (ECLIPSE) (31), National Emphysema Treatment Trial (NETT) (32)/Normative Aging Study (NAS) (33), and Genetics of COPD in Norway (GenKOLS) (34). Detailed descriptions of these cohorts have been previously published (35).
Table 1.
Baseline Characteristics of Meta-analysis Cohorts
| Characteristics | COPDGene NHW | COPDGene AA | ICGN (1,103 Pedigrees) | ECLIPSE | GenKOLS | NETT/NAS | EOCOPD (201 Pedigrees) | TCGS-Poland | TCGS-Korea |
|---|---|---|---|---|---|---|---|---|---|
| Control | |||||||||
| n | 2,534 | 1,749 | 696 | 178 | 808 | 435 | 560 | 307 | 219 |
| Sex, % male | 49.3 | 58.1 | 48.3 | 57.9 | 50.1 | 100 | 41.6 | 67.4 | 96.8 |
| Age | 59.5 (8.7) | 52.8 (6.0) | 54.4 (8.9) | 57.5 (9.4) | 55.6 (9.7) | 69.8 (7.5) | 40.8 (17.5) | 58.8 (7.3) | 52.9 (8.41) |
| Pack-years | 37.8 (20.3) | 36.4 (20.1) | 29.4 (19.8) | 32.1 (24.8) | 19.7 (13.6) | 40.7 (27.9) | 10.8 (18.4) | 34 (15.2) | 27.3 (14.9) |
| FEV1 % predicted | 96.8 (11) | 98.4 (12.2) | 99.1 (14.4) | 107.8 (13.6) | 94.9 (9.2) | 100.0 (13.2) | 95.7 (11.5) | 103 (12.7) | 94.4 (9.4) |
| Moderate-to-severe COPD (GOLD II–IV) | 2,812 | 821 | 1,769 | 1,764 | 863 | 373 | 366 | 304 | 149 |
| n | |||||||||
| Sex, % male | 55.7 | 55.2 | 58.6 | 67.0 | 60.1 | 63.8 | 39.9 | 70.1 | 99.3 |
| Age | 64.7 (8.2) | 59.0 (8.2) | 59.2 (6.9) | 63.6 (7.1) | 65.5 (10.0) | 67.5 (5.8) | 53.2 (12) | 62.6 (7.41) | 68.9 (6.21) |
| Pack-years | 56.3 (28.0) | 42.4 (23.0) | 51.3 (28.2) | 50.3 (27.4) | 32.0 (18.5) | 66.4 (30.7) | 41.1 (24.4) | 44.5 (22.4) | 44.9 (24.5) |
| FEV1 % predicted | 49.6 (18.0) | 52.2 (17.8) | 40.5 (16.7) | 47.6 (15.6) | 50.6 (17.4) | 28.1 (7.4) | 35.1 (20) | 29.1 (9.22) | 33.8 (8.28) |
| Severe COPD (GOLD III–IV) | 1390 | 352 | 1099 | 999 | 383 | 373 | 251 | 304 | 149 |
| n | |||||||||
| Sex, % male | 57.8 | 58 | 60.9 | 69.9 | 61.5 | 63.8 | 33.1 | 70.1 | 99.3 |
| Age, yr | 65.2 (7.8) | 60.6 (8.1) | 59.2 (6.27) | 63.5 (7.0) | 66.7 (9.7) | 67.5 (5.8) | 51.3 (10.1) | 62.6 (7.41) | 68.9 (6.21) |
| Pack-years | 58.7 (28.4) | 43.9 (23.4) | 53.6 (28.8) | 50.7 (26.3) | 33.0 (19.9) | 66.4 (30.7) | 41.7 (22.6) | 44.5 (22.4) | 44.9 (24.5) |
| FEV1 % predicted | 34.0 (9.9) | 34.8 (10.4) | 30 (9.96) | 36.5 (8.6) | 34.4 (10.3) | 28.1 (7.4) | 23.3 (9.44) | 29.1 (9.22) | 33.8 (8.28) |
Definition of abbreviations: AA, African American; COPD, chronic obstructive pulmonary disease; ECLIPSE, Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points; EOCOPD, Boston Early-Onset COPD Study; GenKOLS, Genetics of Chronic Obstructive Lung Disease, Norway; GOLD, Global Initiative for Chronic Obstructive Lung Disease; ICGN, International COPD Genetics Network; NAS, Normative Aging Study; NETT, National Emphysema Treatment Trial; NHW, non-Hispanic white; TCGS,Transcontinental COPD Genetics Study.
Number of subjects is presented as n, sex is presented as percent male. Mean values for age, pack-years, and FEV1 % predicted are shown as mean (±SD). Moderate-to-severe COPD represents GOLD II–IV COPD cases, whereas severe COPD represents GOLD III–IV COPD cases. Number of pedigrees is presented below the study name for the pedigree-based studies, ICGN and EOCOPD.
All subjects were current or former cigarette smokers with and without COPD, except for the Early-Onset COPD Study, which included a small number of nonsmokers. We defined “moderate to severe” COPD as GOLD (Global Initiative for Chronic Obstructive Lung Disease) (2) spirometric grade II–IV COPD (post-bronchodilator FEV1/forced vital capacity [FVC] < 0.7, FEV1 < 80% predicted), whereas “severe” COPD was defined as grades III–IV COPD (FEV1/FVC < 0.7, FEV1 < 50% predicted). Controls had normal spirometry (FEV1/FVC ≥ 0.7, FEV1 ≥ 80%). Previously diagnosed alpha-1 antitrypsin deficiency was an exclusion criterion for all cohorts.
Genetic Analysis
We used PLINK v1.9 (36) and GWAF (37) for case–control and family-based data, respectively, to perform multiple logistic regression within each dataset and then performed fixed-effect meta-analysis using METAL (38). Given that many of our SNPs were chosen from top findings from prior GWASs (see Table E1 in the online supplement for full list of SNPs and provenance), we required an overall P value of less than 5 × 10−8 for statistical significance. We also considered a more liberal suggestive threshold based on a Bonferroni correction for the number of tested SNPs (P < 1.49 × 10−5).
Genetic Risk Scores
We used PLINK v1.9 to create genetic scores based on significant associations from prior GWASs of COPD and lung function (20, 21, 39). We oriented risk alleles to be consistent with prior reports, and gave each allele equal weight. We applied these scoring systems to the ICGN cohort, the largest individual cohort not used in the discovery of any of the risk score variants. Risk scores were also applied to the COPDGene and TCGS Poland cohorts using analogous methods.
The resultant risk scores were used as predictors in a linear mixed model of FEV1 % predicted, as well as logistic regression models of both moderate-to-severe and severe COPD incorporating generalized estimating equations. Models were controlled for age, pack-years of smoking, principal components of genetic ancestry, and for familial correlation. In addition, we used the pROC (40) and GenABEL (41) packages in R to compare the accuracy of two models (i.e., model with genetic risk factors and clinical predictors versus the clinical predictors alone) through receiver operator characteristic curves and net reclassification index (NRI). Subjects were divided into three tiers of COPD risk (low, 0–33.3%; intermediate, 33.4–66.7%; and high, 66.8–100%) for NRI analysis to assess the discriminatory benefit of adding genetic information to the clinical risk model of age and pack-years of smoking alone.
Additional details regarding the SNPs and cohorts used in this study—genotype-, marker-, and subject-level quality control, and risk score modeling and NRI analysis—are available in the Materials and Methods section and the online supplement.
Results
The baseline characteristics of all cohorts are shown in Table 1. Notably, the TCGS-Korea, TCGS-Poland, and NETT/NAS studies were designed to contain only severe COPD cases, which is reflected in the low average FEV1 % predicted seen among cases for these studies.
Genetic Association Analysis
The moderate-to-severe COPD analysis included 9,221 cases and 7,486 control subjects, and confirmed signals in the previously described TGFB2, FAM13A, HHIP, CHRNA3/CHRNA5/IREB2, and RIN3 regions; in addition, an SNP in 16p11.2, recently described in an exome chip analysis of these same cohorts (42), was associated with moderate-to-severe COPD (rs40834, P = 1.90 × 10−8, estimated odds ratio [OR] = 1.17; Table E2). The analysis of severe COPD (Table 2) included 5,300 cases and 7,486 control subjects. We confirmed significance of SNPs in the TGFB2, FAM13A, HHIP, MMP3/MMP12, and CHRNA3/CHRNA5/IREB2 regions. We also identified two SNPs at loci not previously described as genome-wide significant: 5q23.2 between the PRDM6 and PPIC genes (rs6860095, P = 1.01 × 10−8, estimated OR = 1.24), and an intronic SNP within the PPP4R4 gene (rs112458284, P = 1.28 × 10−8, estimated OR = 1.69) in 14q32.13.
Table 2.
Genome-Wide Significant Severe Chronic Obstructive Pulmonary Disease Associations
| rs ID | Chromosome | Base Position | Effect Allele | P Value | OR (95% CI) | Effect Allele Frequency | Nearest Gene(s) |
|---|---|---|---|---|---|---|---|
| rs1890995 | 1 | 218604678 | A | 3.79 × 10−11 | 1.27 (1.37–1.19) | 0.73 | TGFB2 |
| rs4416442 | 4 | 89866713 | T | 5.38 × 10−17 | 1.32 (1.39–1.23) | 0.43 | FAM13A |
| rs13141641 | 4 | 145506456 | T | 1.69 × 10−21 | 1.38 (1.29–1.48) | 0.61 | HHIP |
| rs6860095 | 5 | 122405957 | A | 1.01 × 10−8 | 1.24 (1.15–1.33) | 0.74 | PRDM6/PPIC |
| rs679620 | 11 | 102713620 | T | 1.87 × 10−8 | 1.19 (1.12–1.27) | 0.54 | MMP3/MMP12 |
| rs112458284 | 14 | 94672731 | T | 1.28 × 10−8 | 1.69 (2.04–1.41) | 0.04 | PPP4R4 |
| rs17486278 | 15 | 78867482 | A | 1.70 × 10−27 | 1.43 (1.54–1.35) | 0.37 | CHRNA5 |
Definition of abbreviations: CI, confidence interval; OR, odds ratio.
Significant associations for Global Initiative for Chronic Obstructive Lung Disease spirometric stages III–IV chronic obstructive pulmonary disease (COPD), organized by chromosome. In each case, the lead single-nucleotide polymorphism for the locus is presented. Effect alleles represent the allele that is associated with the stated odds ratio for COPD-risk. Base position was calculated using human genome assembly 19 (hg19) coordinates.
We examined these loci using the GTEx expression quantitative trait loci database (43) and Haploreg v4.1 (44). SNP rs6860095 affected gene expression levels of PPIC, snoU13, SNX2, and RN7SL689P in multiple tissues, although not in lung tissue. No significant expression quantitative trait loci were found for SNP rs112458284; however, it lies approximately 200 kb away from SERPINA1, which encodes the protein responsible for alpha-1 antitrypsin deficiency (45, 46).
We investigated whether rs112458284 could be tagging alleles of SERPINA1 known to contribute to risk of COPD (e.g., the Z allele, rs28929474, or S-allele, rs17580). rs112458284 showed linkage disequilibrium (LD) with the Z allele in directly genotyped (i.e., not imputed) samples from COPDGene non-Hispanic white subjects (r2 = 0.41, normalized coefficient of linkage disequilibrium [D′] = 0.78) and, to a lesser extent, the S allele (r2 = 8.63 × 10−5, D′ = 0.25). Consistent with this hypothesis, the Z allele was associated with COPD at near–genome-wide significance in our primary analysis using imputed data in COPDGene (P = 1.53 × 10−7, OR = 1.78, confidence interval [CI] = 1.44–2.21); this signal improved using genotyping data (P = 2.05 × 10−8, OR = 1.84, CI = 1.49–2.27), although rs112458284 was still the strongest association signal in the region. To further investigate whether there was any association signal at the rs112458284 that was independent from the Z allele, we also conditioned on the Z allele in a meta-analysis model, and found that the association signal for rs112458284 was attenuated (P = 0.0087).
Known alpha-1 antitrypsin deficiency was an exclusion criterion in our study; however, our genotyping (and imputed data) identified three previously unrecognized Z allele homozygotes in the Poland cohort (35) and six additional Z allele homozygotes in the ECLIPSE cohort (47). After removing these subjects, the rs112458284 association was only mildly attenuated (P = 7.22 × 10−8), as was the association with the Z allele (P = 9.29 × 10−8, OR = 1.80, CI = 1.45–2.23). Thus, heterozygous carriers of the Z allele appear to be driving a large proportion of the association, consistent with prior studies showing an increased risk of COPD for MZ heterozygotes (48). In addition, these results suggest that if we had not specifically excluded subjects with known alpha-1 antitrypsin deficiency in our other populations, the association with SNP rs112458284 would likely be even more extreme (49). Allele frequencies for the Z allele in each cohort are provided in Table E3. The poor imputation quality of the S allele in our cohorts prevented us from further assessing its impact on the rs112458284 association.
We next examined our other SNPs at a more liberal P value threshold. Using a Bonferroni significance threshold for 3,346 SNPs (P < 1.49 × 10−5), we identified suggestive associations at three loci (THSD4, AGER/PPT2, and ADAM19). All of these were in regions previously associated in GWASs of lung function. We then examined linkage disequilibrium within each candidate locus to further explore whether these associations represented the same variants as previous associations. We defined “lead SNP” as the association yielding the lowest P value in a given region, and the “candidate SNP” as the previously described variant. In 14 of these lead SNPs, LD with the candidate SNP measured by D′ was greater than 0.8, whereas eight also had an r2 greater than 0.3 (Table 3). Notably, SNPs associated with lower lung function showed a directionally consistent increased risk for COPD in 23 of 25 previously reported SNPs directly genotyped in our meta-analysis (binomial for enrichment, P = 9.7 × 10−6). These 23 lung function risk alleles included 12 showing a nominally statistically significant (P < 0.05) effect on COPD risk (see Table 3). Only lung function risk alleles in the ZKSCAN3 and NCR3-AIF1 genes showed a directionally discordant effect on COPD susceptibility (lower risk of COPD), although these discordant association results were not statistically significant. Additional results for other variants are reported in Table E4.
Table 3.
Lung Function Variants
| Chromosome | Previously Reported Variant |
Lead Variant in Meta-analysis Window |
Linkage Disequilibrium between Previously Reported and Lead Variants |
|||||
|---|---|---|---|---|---|---|---|---|
| rs ID | Base Position | Nearest Gene | Meta-analysis P Value | rs ID | Meta-analysis P Value | r2 | D′ | |
| 1 | rs2284746 | 17306675 | MFAP2 | 0.12 | rs3170740 | 0.10 | 0.91 | 0.98 |
| 1 | rs993925 | 218860068 | TGFB2-LYPLAL1 | 0.56 | rs72738847 | 4.56 × 10−6 | 0.00 | 0.34 |
| 2 | rs2571445 (rs918949) | 218683153 | TNS1 | 0.07 | rs3791953 | 1.75 × 10−2 | 0.00 | 0.12 |
| 2 | rs7594321 | 230224031 | DNER | 0.09 | rs12995479 | 0.02 | 0.00 | 0.02 |
| 2 | rs12477314 | 239877148 | HDAC4-FLJ43879 | 2.37 × 10−3 | rs35877146 | 1.26 × 10−3 | 0.72 | 0.90 |
| 3 | rs1529672 | 25520582 | RARB | 3.08 × 10−4 | rs1529672 | 3.08 × 10−4 | N/A | N/A |
| 3 | rs1344555 | 169300219 | MECOM/EVI1 | 0.68 | rs933607 | 2.29 × 10−4 | 0.03 | 0.24 |
| 4 | rs7671167 | 89883979 | FAM13A | 2.45 × 10−15 | rs4416442 | 1.84 × 10−17 | 0.65 | 0.99 |
| 4 | rs10516526 | 106688904 | GSTCD/INTS12/NPNT | 7.39 × 10−4 | rs11735213 | 5.12 × 10−5 | 0.67 | 0.91 |
| 4 | rs1032296 | 145434688 | HHIP | 4.13 × 10−10 | rs13141641 | 1.26 × 10−18 | 0.41 | 0.89 |
| 5 | rs153916 | 95036700 | SPATA9-RHOBTB3 | 2.90 × 10−3 | rs153916 | 2.90 × 10−3 | N/A | N/A |
| 5 | rs11168048 | 147842353 | HTR4 | 0.01 | rs17720155 | 4.41 × 10−4 | 0.33 | 0.78 |
| 5 | rs11134779 (rs1422795) | 156936766 | ADAM19 | 7.98 × 10−3 | rs62390771 | 4.16 × 10−7 | 0.02 | 0.38 |
| 6 | rs6903823* | 28322296 | ZKSCAN3 | 0.75 | rs3800326 | 0.10 | 0.09 | 1.00 |
| 6 | rs2857595* | 31568469 | NCR3-AIF1 | 0.71 | rs2844479 | 0.03 | 0.03 | 0.51 |
| 6 | rs2070600 | 32151443 | AGER/PPT2 | 7.05 × 10−6 | rs2070600 | 7.05 × 10−6 | N/A | N/A |
| 6 | rs7765379 | 32680928 | HLA-DQB1 | 0.12 | rs9275141 | 5.67 × 10−3 | 0.14 | 1.00 |
| 6 | rs2798641 | 109268050 | ARMC2 | 1.15 × 10−4 | rs2848598 | 2.06 × 10−5 | 0.31 | 0.89 |
| 6 | rs3817928 | 142750516 | GPR126 | 5.71 × 10−3 | rs9399401 | 1.91 × 10−4 | 0.63 | 0.96 |
| 9 | rs16909898 | 98231008 | PTCH1 | 0.12 | rs357523 | 7.77 × 10−3 | 0.47 | 0.73 |
| 10 | rs7068966 | 12277992 | CDC123 | 0.05 | rs10906083 | 0.03 | 0.01 | 0.13 |
| 10 | rs11001819 | 78315224 | C10orf11 | 0.39 | rs7904646 | 2.08 × 10−3 | 0.00 | 0.33 |
| 12 | rs11172113 | 57527283 | LRP1 | 2.28 × 10−4 | rs2122692 | 9.12 × 10−5 | 0.44 | 0.80 |
| 12 | rs1036429 (rs7307510) | 96271427 | CCDC38 | 6.35 × 10−3 | rs7306887 | 7.35 × 10−4 | 0.13 | 0.87 |
| 15 | rs12899618 | 71645120 | THSD4 | 0.01 | rs10459646 | 4.37 × 10−7 | 0.09 | 1.00 |
| 16 | rs12447804 | 58075282 | MMP15 | 0.16 | rs2550370 | 9.55 × 10−3 | 0.03 | 0.63 |
| 16 | rs2865531 (rs4888380) | 75390315 | CFDP1 | 3.09 × 10−3 | rs37586 | 4.88 × 10−4 | 0.13 | 1.00 |
| 17 | rs11654749 | 69125606 | KCNJ2 | 0.39 | rs35883109 | 0.01 | 0.00 | 0.08 |
| 21 | rs9978142 | 35652239 | KCNE2-LINC00310/C21orf82 | 0.98 | rs73205216 | 8.96 × 10−5 | 0.02 | 1.00 |
Definition of abbreviations: D′, normalized coefficient of linkage disequilibrium; N/A, not applicable.
For each previously reported variant and lead variant, the P value refers to the association with moderate-to-severe chronic obstructive pulmonary disease in our analysis. Nominally significant associations (P < 0.05) among previously reported variants are shown in bold. Linkage disequilibrium values (r2 [between-locus correlation coefficient]) between the previously reported variant and the lead variant in meta-analysis window were obtained using data from 1,000 Genomes Project Phase 1 v3. Proxies for variants not available in our dataset are in parentheses, and P values displayed are for the proxy variant.
Risk alleles showing a discordant association direction of effect for chronic obstructive pulmonary disease risk and decreased lung function risk.
Genetic Risk Scores
We next examined the ability of genetic risk scores to explain both FEV1 % predicted and COPD affection status. Based on our results presented previously here, we constructed risk scores using genome-wide significant SNPs associated with COPD (COPD7 score, comprised of seven COPD risk SNPs) and also including genome-wide significant SNPs associated with lung function in population-based studies (LUNG30 score, composed of thirty lung function associated risk SNPs); Tables 4 and E5 describe the loci involved in each score. We evaluated the risk scores using the ICGN cohort, the largest available cohort not used in the discovery of these risk loci. Results from the unadjusted model are shown in Figure 1. In a linear mixed model, adjusting for age, pack-years of smoking, principal components of ancestry, and a within-family component, we found the COPD7 risk score (ranging from 0 to 14 scoring alleles) was associated with a 1.86% reduction in FEV1 % predicted for each additional risk allele carried (Table 5). Using generalized estimating equations for models of moderate-to-severe and severe COPD (Table 6), each additional risk allele in the COPD7 risk score was associated with an OR of 1.18 for moderate-to-severe COPD and 1.19 for severe COPD (P = 4.1 × 10−8 and P = 4.4 × 10−8, respectively). We found nearly identical results for a standard logistic regression (OR = 1.17 and OR = 1.19, respectively) without family adjustment, and therefore used these simpler models to generate receiver operator characteristic curves for affection status using genetic variants alone, age and pack-years, and the combination of age, pack-years, and genetic information. The area under the curve (AUC) for the genetic model was 0.58 for moderate-to-severe COPD and 0.59 for severe COPD. In addition, adding genetic risk scores (COPD7) only modestly increased the AUC (Figure 2) over the AUC of the clinical model. Three-tiered categorical analysis of reclassification (50) after addition of the COPD7 risk score and adjustment for genetic components of ancestry into the clinical model (containing only age and pack-years of smoking) resulted in an NRI of 0.053 (P = 2.32 × 10−3) for the combined model risk stratification of moderate-to-severe COPD, and an NRI of 0.047 for risk stratification of severe COPD (P = 0.01). For the expanded LUNG30 score, we found a lower per-allele effect, but larger overall effect, because more factors went into the score (Tables 5 and 6). We also tested risk scores in the TCGS-Poland and COPDGene cohorts and found comparable results (see Materials and Methods and the online supplement).
Table 4.
Genetic Risk Score Loci
| Gene | SNP Identifier |
|---|---|
| CHRNA3 | rs12914385 |
| RAB4B/EGLN2/MIA/CYP2A6 | rs7937 |
| RIN3 | rs754388 |
| TGFB2 | rs4846480 |
| MMP12 | rs626750 |
| HHIP | rs13141641 |
| FAM13A | rs4416442 |
| PTCH1 | rs16909898 |
| C10orf11 | rs11001819 |
| HTR4 | rs11168048 |
| DNER | rs7594321 |
| HDAC4-FLJ43879 | rs12477314 |
| MFAP2 | rs2284746 |
| ARMC2 | rs2798641 |
| LRP1 | rs11172113 |
| RARB | rs1529672 |
| GSTCD/INTS12/NPNT | rs10516526 |
| NCR3-AIF1 | rs2857595 |
| MECOM/EVI1 | rs1344555 |
| AGER/PPT2 | rs2070600 |
| ZKSCAN3 | rs6903823 |
| SPATA9-RHOBTB3 | rs153916 |
| MMP15 | rs12447804 |
| HLA-DQB1 | rs7765379 |
| KCNJ2/CASC17 | rs11654749 |
| GPR126 | rs3817928 |
| TGFB2-LYPLAL1 | rs993925 |
| CDC123 | rs7068966 |
| KCNE2-LINC00310/C21orf82 | rs9978142 |
| THSD4 | rs12899618 |
Definition of abbreviations: COPD7, genetic risk score composed of seven COPD risk SNPs (ranging from 0 to 14 scoring alleles); LUNG30, genetic risk score compsed of thirty lung function associated risk SNPs (ranging from 0 to 60 scoring alleles); SNP, single-nucleotide polymorphism.
Genetic risk scores were composed using previous chronic obstructive pulmonary disease (COPD) and lung function–associated loci. The LUNG30 score included all of the loci listed in the above table; the COPD7 score included only those in the bold. Loci names are based on previously reported SNP associations annotated to the nearest gene or region.
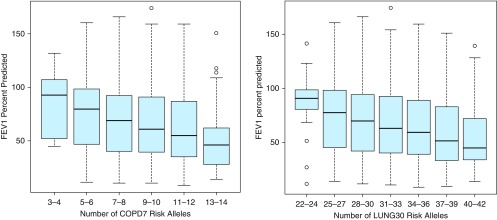
Figure 1.
Unadjusted FEV1% predicted by number of COPD7 and LUNG30 risk alleles. Boxplots showing FEV1 % predicted stratified by number of risk alleles in the International COPD Genetics Network pedigree-based cohort. For each boxplot, the black line represents the median data point, the upper and lower edges of the light blue box represent data within the 25th to 75th percentile of the distribution, the upper and lower “whiskers” represent the upper and lower limits of the data, and open circles represent outliers. The figure on the left shows the COPD7 risk score, whereas the figure on the right shows the LUNG30 risk score. COPD7, genetic risk score composed of seven COPD risk SNPs (ranging from 0 to 14 scoring alleles); LUNG30, genetic risk score compsed of thirty lung function associated risk SNPs (ranging from 0 to 60 scoring alleles); SNP, single-nucleotide polymorphism.
Table 5.
Genetic Risk Scores: Lung Function in International Chronic Obstructive Pulmonary Disease Genetics Network
| Risk Score | Unadjusted FEV1 % per Risk Allele (95% CI) | P Value | Adjusted FEV1 % per Risk Allele (95% CI) | P Value |
|---|---|---|---|---|
| COPD7 | −2.02 (−1.34 to −2.70) | 6.74 × 10−9 | −1.86 (−1.24 to −2.50) | 7.90 × 10−9 |
| LUNG30 | −1.18 (−0.83 to −1.53) | 4.70 × 10−11 | −1.10 (−0.78 to −1.43) | 3.78 × 10−11 |
Definition of abbreviations: CI, confidence interval; COPD7, genetic risk score composed of seven COPD risk SNPs (ranging from 0 to 14 scoring alleles); LUNG30, genetic risk score composed of thirty lung function associated risk SNPs (ranging from 0 to 60 scoring alleles); SNP, single-nucleotide polymorphism.
For each risk score (chronic obstructive pulmonary disease [COPD] 7 and LUNG30), the linear mixed-model coefficient is presented with 95% CI and P value. Final model included adjustment for age, pack-years, familial correlation, and principal components for genetic ancestry, whereas the unadjusted model was not adjusted for age and pack-years.
Table 6.
Genetic Risk Scores: Affection Status in International Chronic Obstructive Pulmonary Disease Genetics Network
| Moderate COPD | P Value | Severe COPD | P Value | |
|---|---|---|---|---|
| COPD7 | ||||
| OR per risk allele (95% CI) | 1.18 (1.11–1.25) | 4.10 × 10−8 | 1.19 (1.12–1.27) | 4.43 × 10−8 |
| AUC (95% CI) | 0.58 (0.56–0.61) | 0.59 (0.56–0.61) | ||
| Total NRI (95% CI) | 0.053 (0.019–0.086) | 2.32 × 10−3 | 0.047 (0.01–0.084) | 1.32 × 10−2 |
| Event NRI, % | 0.23 | 0.83 | ||
| Nonevent NRI, % | 5.03 | 3.88 | ||
| LUNG30 | ||||
| OR per risk allele (95% CI) | 1.12 (1.09–1.15) | 1.25 × 10−13 | 1.12 (1.09–1.15) | 1.25 × 10−13 |
| AUC (95% CI) | 0.60 (0.57–0.62) | 0.60 (0.57–0.63) | ||
| NRI (95% CI) | 0.090 (0.053–0.126) | 1.72 × 10−6 | 0.047 (0.007–0.087) | 2.22 × 10−2 |
| Event NRI, % | 2.35 | 0.65 | ||
| Nonevent NRI, % | 6.61 | 4.67 |
Definition of abbreviations: AUC, area under the curve; CI, confidence interval; COPD, chronic obstructive pulmonary disease; COPD7, genetic risk score composed of seven COPD risk SNPs (ranging from 0 to 14 scoring alleles); LUNG30, genetic risk score composed of thirty lung function associated risk SNPs (ranging from 0 to 60 scoring alleles); NRI, net reclassification index; OR, odds ratio; SNP, single-nucleotide polymorphism.
For each risk score (COPD7 and LUNG30), OR is for each additional risk allele on the outcome of either moderate COPD (GOLD [Global Initiative for Chronic Obstructive Lung Disease] II–IV) or severe COPD (GOLD III–IV). AUC is for a model including only the genetic data of risk score alleles adjusted for principal components of genetic ancestry. NRI represents the three-tiered value of the model combining genetic risk score, age, pack-years of smoking, and principal components of genetic ancestry compared to the model containing age and pack-years alone. Event NRI represents the percentage of subjects with the outcome of COPD correctly reclassified to a higher risk group after adding genetic data. Nonevent NRI represents the percentage of subjects without the outcome of COPD correctly reclassified to a lower risk group after adding genetic data.
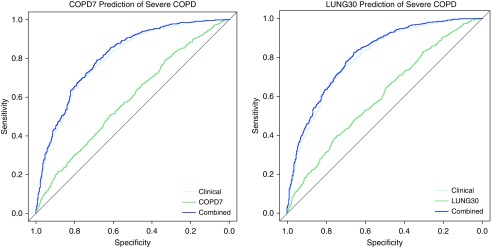
Figure 2.
Severe COPD diagnosis using COPD7 and LUNG30. Receiver operator characteristic curves showing diagnostic accuracy of models based on clinical variables (age and pack-years of smoking alone, shown in light blue), COPD7 or LUNG30 risk allele data alone (green), and the combination of clinical and genetic risk score data (dark blue) for predicting Global Initiative for Chronic Obstructive Lung Disease spirometric stages III–IV COPD affection status in the International COPD Genetics Network cohort. The differences between the clinical and combined curves were statistically significant in both the COPD7 (difference 0.010; P = 4.4 × 10−3) and the LUNG30 scores (difference 0.012; P = 4.7 × 10−3).
Discussion
Genetic association studies in COPD have identified well replicated genome-wide associations with COPD, but the majority of genetic susceptibility remains unexplained. In a large-scale genetic association meta-analysis of nine cohorts, analyzing both moderate-to-severe and severe COPD, we identified two new associations at genome-wide significance with severe COPD, including one in strong LD with SERPINA1, and associations at a more liberal significance threshold in regions previously associated with population-based lung function. We found consistent directions of effect on risk to COPD in 23 previously identified markers associated with lung function, consistent with recent reports (7). We also constructed genetic risk scores that showed compelling relationships for quantitative measures of lung function and modest discrimination for COPD affection status. Our results further inform the discussion of how genetic variants could influence COPD susceptibility.
The discovery that variants in LD with SERPINA1 are associated with severe COPD demonstrates how genetic association studies can confirm known disease mechanisms. This rs112458284 variant is also in strong LD with rs45505795 near SERPINA10 (r2 = 0.96 and D′ = 1.0) in 1,000 Genomes EUR Phase I v3 data (www.internationalgenome.org), which we recently described in a GWAS of quantitative measures of emphysema (47). The 5q23.2 region containing SNP rs6860095 is strongly associated with severe COPD risk. Both PPIC and PRDM6 lie in this region. PPIC (also known as cyclophilin C) has functions related to mitochondrial metabolism, inflammation, and immune response through its interactions with cyclosporine A. Although the related protein, cyclophilin A, has been associated with both COPD (51) and lung cancer (52), to our knowledge, no prior study has shown significant association between PPIC and risk of COPD. The PRDI-BF1 and RIZ homology domain containing 6 (PRDM6) protein is involved in chromatin remodeling and transcriptional control of smooth muscle gene expression (53). Expression of PRDM6 has been implicated in the pseudoglandular and canalicular stages of lung morphogenesis in murine models, and expression has been documented in smooth muscle of the developing murine trachea, bronchi, and pulmonary trunk (53). Additional studies are needed to confirm association between markers in 5q23.2 and severe COPD.
We examined genomic loci previously associated with FEV1, FEV1/FVC, and additional variants previously hypothesized to be associated with COPD (19). SNPs that met criteria for suggestive association were found in regions previously associated with lung function, including the AGER/PPT2 and the THSD4 regions. In addition, the majority of variants associated with quantitative measures of lung function showed consistent directions of effect for both COPD and low lung function.
Genetic risk scores using selected risk variants from COPD-based cohorts could provide a clinically relevant context to individual-level genetic data, and could have applications in assessing risk to COPD and its severity (25). We investigated the ability of genetic risk scores to explain COPD risk and FEV1 % predicted. Genetic data alone only achieved an AUC of approximately 0.6 in our modeling of moderate-to-severe COPD risk. This finding is comparable to the AUC of genetic risk scores in other complex diseases, such as coronary artery disease (54) and type II diabetes (55). This low AUC of our risk score may be due to the fact that genetic data do not account for the contributions to COPD of other significant risk factors, such as age and environmental exposures, such as pack-years of smoking. The addition of genetic data to the clinical model including age and pack-years of smoking resulted in statistically significant, but small, increases in AUCs and in statistically significant NRI values when classifying risk for severe COPD. Interpretation of the NRI is more straightforward for clinically actionable consensus endpoints, such as primary prevention statin therapy for coronary artery disease outcomes, which are less well defined for COPD. Despite these concerns, the clinical relevance of this model is most apparent in the risk score coefficient itself. The LUNG30 model implies that a subject with 35 risk alleles would show a threefold increase in risk of COPD compared with a subject with a score of 25, all other variables being equal.
Similarly, in our modeling of FEV1 % predicted, we found a small, but detectable, effect of each individual risk allele, although the cumulative effect of this score may be clinically relevant. For example, within the ICGN dataset, we had subjects with as few as 16 and as many as 45 alleles in their LUNG30 score. Based on our model, this difference in alleles would account for an approximately 30% difference in FEV1 % predicted, holding all other variables equal. Such a 30% FEV1 % predicted difference implies that two people (with similar age and pack-years of smoking) may fall into different GOLD severity classes due to the effect of these risk alleles alone. Although the COPD-based ascertainment of the ICGN pedigrees may have led to enrichment of these risk alleles in this cohort, the significance of the risk scores was robust when tested in two additional case–control cohorts.
Despite having analyzed over 16,000 subjects, our study and the experience in other GWAS suggest that power is still a major limitation in detecting additional COPD associations. The definition of COPD phenotypes and its severity by spirometric criteria alone (2) was consistent in our meta-analysis; however, this does not address other aspects of heterogeneity in COPD that may be under genetic control (such as emphysema or exacerbations). The study was cross-sectional in design, with lung function assessment at only one point of time, so we cannot assess the impact of lung function trajectories (56) in our models. This study was not a comprehensive survey of genome-wide data, and its ability to detect new associations was limited to previously identified loci and their surrounding regions. Four of the datasets in our meta-analysis were previously investigated for genetic associations for COPD status (18), so our results are enriched for previously discovered associations. In addition, genotyping was performed before the results of recent COPD and lung function GWAS studies by the UK BiLEVE group (7) and Soler Artigas and colleagues (57, 58) were published, and the additional risk loci for COPD and lung function found in these studies were not included in our analysis. We chose to use a simple model for our genetic (and clinical) risk scores. More sophisticated models using these SNPs, based on genome-wide results, and incorporating additional clinical factors, may result in improved prediction. In pathway analysis in Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, and the Reactome (see the supplemental Materials and Methods and Results sections), the closest gene(s) to the LUNG30 risk score variants showed enrichment in gene sets related to structural components of lung development, control of lung development, inflammatory response, and response to steroid hormone, among others. These enriched terms and pathways may help to provide insight into the pathophysiologic mechanisms of COPD pathogenesis that are represented by the LUNG30 association signal. However, our risk scores focused on SNPs previously associated with lung function and COPD; most of the causal genes at these loci are not known, and these signals capture only a minority of relevant genetic mechanisms contributing to COPD pathogenesis. The performance of our genetic risk scores in other racial groups and in never-smokers has not been tested, although this is an area of interest for follow-up investigations. Although associations with SNPs rs112458284 and rs6860095 did achieve genome-wide levels of significance in our analysis of severe COPD, these results still need to be replicated in independent populations.
In summary, we performed a meta-analysis of markers in selected genes, and discovered two new SNPs associated with severe COPD that reached significance levels equivalent to accepted thresholds of genome-wide significance, one of which tags recognized risk alleles in SERPINA1. Our study is one of the largest genetic association studies of severe COPD, and the first to identify SERPINA1 at genome-wide significance for COPD. Our study supports the idea that loci associated with lung function play some role in susceptibility to COPD. We also showed the clinical applicability of simple genetic risk scores for explaining COPD spirometric severity in an independent cohort. This study adds to the growing body of genetic knowledge about COPD, including efforts at subtyping, prediction, and mechanistic investigation, which may ultimately inform patient counseling, clinical decision-making, and lead to new therapies for this disease.
Acknowledgments
Acknowledgments
The authors acknowledge and thank Augustine M. K. Choi, M.D. (Weill Cornell Medicine, New York, NY), for his support of this project. In addition, they thank the following members of the individual study cohorts, without whom this work would not have been possible.
The COPDGene Investigators Are:
Administrative Core: James Crapo, M.D. (Principal Investigator), Edwin Silverman, M.D., Ph.D. (Principal Investigator), Barry Make, M.D., Elizabeth Regan, M.D., Ph.D.
Genetic Analysis Core: Terri Beaty, Ph.D., Nan Laird, Ph.D., Christoph Lange, Ph.D., Michael Cho, M.D., Stephanie Santorico, Ph.D., John Hokanson, M.P.H., Ph.D., Dawn DeMeo, M.D., M.P.H., Nadia Hansel, M.D., M.P.H., Craig Hersh, M.D., M.P.H., Peter Castaldi, M.D., M.Sc., Merry-Lynn McDonald, Ph.D., Emily Wan, M.D., Megan Hardin, M.D., Jacqueline Hetmanski, M.S., Margaret Parker, M.S., Marilyn Foreman, M.D., Brian Hobbs, M.D., Robert Busch, M.D., Adel El-Bouiez, M.D., Dandi Qiao, Ph.D., Elizabeth Regan, M.D., Eitan Halper-Stromberg, Ph.D., Ferdouse Begum, Ph.D., Sungho Won, Ph.D., Sharon Lutz, Ph.D.
Imaging Core: David A. Lynch, M.B., Harvey O. Coxson, Ph.D., MeiLan K. Han, M.D., M.S., Eric A. Hoffman, Ph.D., Stephen Humphries, M.S., Francine L. Jacobson, M.D., Philip F. Judy, Ph.D., Ella A. Kazerooni, M.D., John D. Newell, Jr., M.D., Elizabeth Regan, M.D., James C. Ross, Ph.D., Raul San Jose Estepar, Ph.D., Berend C. Stoel, Ph.D., Juerg Tschirren, Ph.D., Eva van Rikxoort, Ph.D., Bram van Ginneken, Ph.D., George Washko, M.D., Carla G. Wilson, M.S., Mustafa Al Qaisi, M.D., Teresa Gray, Alex Kluiber, Tanya Mann, Jered Sieren, Douglas Stinson, Joyce Schroeder, M.D., Edwin Van Beek, M.D., Ph.D.
Pulmonary Function Test Quality Assurance Core, Salt Lake City, UT: Robert Jensen, Ph.D.
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, Ph.D., Anna Faino, M.S., Matt Strand, Ph.D., Carla Wilson, M.S.
Epidemiology Core, University of Colorado Anschutz Medical Campus, Aurora, CO: John E. Hokanson, M.P.H., Ph.D., Gregory Kinney, M.P.H., Ph.D., Sharon Lutz, Ph.D., Kendra Young Ph.D., Katherine Pratte, M.S.P.H., Lindsey Duca, M.S.
Ann Arbor Veterans Administration (VA), Ann Arbor, MI: Jeffrey L. Curtis, M.D., Carlos H. Martinez, M.D., M.P.H., Perry G. Pernicano, M.D.
Baylor College of Medicine, Houston, TX: Nicola Hanania, M.D., M.S., Philip Alapat, M.D., Venkata Bandi, M.D., Mustafa Atik, M.D., Aladin Boriek, Ph.D., Kalpatha Guntupalli, M.D., Elizabeth Guy, M.D., Amit Parulekar, M.D., Arun Nachiappan, M.D.
Brigham and Women’s Hospital, Boston, MA: Dawn DeMeo, M.D., M.P.H., Craig Hersh, M.D., M.P.H., George Washko, M.D., Francine Jacobson, M.D., M.P.H.
Columbia University, New York, NY: R. Graham Barr, M.D., Dr.P.H., Byron Thomashow, M.D., John Austin, M.D., Belinda D’Souza, M.D., Gregory D. N. Pearson, M.D., Anna Rozenshtein, M.D., M.P.H.
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., M.D., Lacey Washington, M.D., H. Page McAdams, M.D.
Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, M.D., M.P.H., Joseph Tashjian, M.D.
Johns Hopkins University, Baltimore, MD: Robert Wise, M.D., Nadia Hansel, M.D., M.P.H., Robert Brown, M.D., Karen Horton, M.D., Nirupama Putcha, M.D., M.H.S.
Los Angeles Biomedical Research Institute at Harbor University of California Los Angeles Medical Center, Torrance, CA: Richard Casaburi, Ph.D., M.D., Alessandra Adami, Ph.D., Janos Porszasz, M.D., Ph.D., Hans Fischer, M.D., Ph.D., Matthew Budoff, M.D., Harry Rossiter, Ph.D.
Michael E. DeBakey VAMC, Houston, TX: Amir Sharafkhaneh, M.D., Ph.D., Charlie Lan, D.O.
Minneapolis VA, Minneapolis, MN: Christine Wendt, M.D., Brian Bell, M.D.
Morehouse School of Medicine, Atlanta, GA: Marilyn Foreman, M.D., M.S., Gloria Westney, M.D., M.S., Eugene Berkowitz, M.D., Ph.D.
National Jewish Health, Denver, CO: Russell Bowler, M.D., Ph.D., David Lynch, M.D.
Reliant Medical Group, Worcester, MA: Richard Rosiello, M.D., David Pace, M.D.
Temple University, Philadelphia, PA: Gerard Criner, M.D., David Ciccolella, M.D., Francis Cordova, M.D., Chandra Dass, M.D., Gilbert D’Alonzo, D.O., Parag Desai, M.D., Michael Jacobs, Pharm.D., Steven Kelsen, M.D., Ph.D., Victor Kim, M.D., A. James Mamary, M.D., Nathaniel Marchetti, D.O., Aditi Satti, M.D., Kartik Shenoy, M.D., Robert M. Steiner, M.D., Alex Swift, M.D., Irene Swift, M.D., Maria Elena Vega-Sanchez, M.D.
University of Alabama, Birmingham, AL: Mark Dransfield, M.D., William Bailey, M.D., J. Michael Wells, M.D., Surya Bhatt, M.D., Hrudaya Nath, M.D.
University of California, San Diego, CA: Joe Ramsdell, M.D., Paul Friedman, M.D., Xavier Soler, M.D., Ph.D., Andrew Yen, M.D.
University of Iowa, Iowa City, IA: Alejandro Comellas, M.D., John Newell, Jr., M.D., Brad Thompson, M.D.
University of Michigan, Ann Arbor, MI: MeiLan Han, M.D., Ella Kazerooni, M.D., Carlos Martinez, M.D.
University of Minnesota, Minneapolis, MN: Joanne Billings, M.D., Tadashi Allen, M.D.
University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, M.D., Divay Chandra, M.D., M.Sc., Joel Weissfeld, M.D., M.P.H., Carl Fuhrman, M.D., Jessica Bon, M.D.
University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, M.D., Sandra Adams, M.D., Diego Maselli-Caceres, M.D., Mario E. Ruiz, M.D.
The International COPD Genetics Network Investigators Are:
Edwin K. Silverman, Brigham & Women’s Hospital, Boston, MA; David A. Lomas, Cambridge Institute for Medical Research, University of Cambridge, Cambridge, UK; Barry J. Make, National Jewish Medical and Research Center, Denver, CO; Alvar Agusti and Jaume Sauleda, Hospital Universitari Son Dureta, Fundación Caubet-Cimera and Ciber Enfermedades Respiratorias, Islas Baleares and Palma de Mallorca, Spain; Peter M. A. Calverley, University of Liverpool, Liverpool, UK; Claudio F. Donner, Division of Pulmonary Disease, S. Maugeri Foundation, Veruno (NO), Italy; Robert D. Levy, University of British Columbia, Vancouver, BC, Canada; Peter D. Paré, University of British Columbia, Vancouver, BC, Canada; Stephen Rennard, Section of Pulmonary & Critical Care, University of Nebraska Medical Center, Omaha, NE; Jørgen Vestbo, Department of Cardiology and Respiratory Medicine, Hvidovre Hospital, Copenhagen, Denmark.
Principal Investigators and Centers Participating in Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-Points (ECLIPSE; NCT00292552, SC0104960) Include:
Bulgaria: Y. Ivanov, Pleven; K. Kostov, Sofia. Canada: J. Bourbeau, Montreal, PQ; M. Fitzgerald, Vancouver, BC; P. Hernandez, Halifax, NS; K. Killian, Hamilton, ON; R. Levy, Vancouver, BC; F. Maltais, Montreal, PQ; D. O’Donnell, Kingston, ON. Czech Republic: J. Krepelka, Prague. Denmark: J. Vestbo, Hvidovre. the Netherlands: E. Wouters, Horn-Maastricht. New Zealand: D. Quinn, Wellington. Norway: P. Bakke, Bergen. Slovenia: M. Kosnik, Golnik. Spain: A. Agusti, J. Sauleda, P. de Mallorca. Ukraine: Y. Feschenko, V. Gavrisyuk, L. Yashina, Kiev; N. Monogarova, Donetsk. United Kingdom: P. Calverley, Liverpool; D. Lomas, Cambridge; W. MacNee, Edinburgh; D. Singh, Manchester; J. Wedzicha, London. United States: A. Anzueto, San Antonio, TX; S. Braman, Providence, RI; R. Casaburi, Torrance, CA; B. Celli, Boston, MA; G. Giessel, Richmond, VA; M. Gotfried, Phoenix, AZ; G. Greenwald, Rancho Mirage, CA; N. Hanania, Houston, TX; D. Mahler, Lebanon, NH; B. Make, Denver, CO; S. Rennard, Omaha, NE; C. Rochester, New Haven, CT; P. Scanlon, Rochester, MN; D. Schuller, Omaha, NE; F. Sciurba, Pittsburgh, PA; A. Sharafkhaneh, Houston, TX; T. Siler, St. Charles, MO; E. Silverman, Boston, MA; A. Wanner, Miami, FL; R. Wise, Baltimore, MD; R. ZuWallack, Hartford, CT.
ECLIPSE Steering Committee:
H. Coxson (Canada), C. Crim (GlaxoSmithKline, USA), L. Edwards (GlaxoSmithKline, USA), D. Lomas (UK), W. MacNee (UK), E. Silverman (USA), R. Tal Singer (Co-chair, GlaxoSmithKline, USA), J. Vestbo (Co-chair, Denmark), J. Yates (GlaxoSmithKline, USA).
ECLIPSE Scientific Committee:
A. Agusti (Spain), P. Calverley (UK), B. Celli (USA), C. Crim (GlaxoSmithKline, USA), B. Miller (GlaxoSmithKline, USA), W. MacNee (Chair, UK), S. Rennard (USA), R. Tal-Singer (GlaxoSmithKline, USA), E. Wouters (the Netherlands), J. Yates (GlaxoSmithKline, USA).
Coinvestigators in the National Emphysema Treatment Trial Genetics Ancillary Study also Include:
J. Benditt, G. Criner, M. DeCamp, P. Diaz, M. Ginsburg, L. Kaiser, M. Katz, M. Krasna, N. MacIntyre, R. McKenna, F. Martinez, Z. Mosenifar, J. Reilly, A. Ries, P. Scanlon, F. Sciurba, and J. Utz.
Footnotes
This work was supported by National Institutes of Health grants T32 HL007427, R01 HL089856 (E.K.S.), R01 HL089897 (J.D.C.), R01 HL113264 (M.H.C. and E.K.S.), P01 HL105339 (E.K.S.), and P01 HL114501 (Augustine M. K. Choi) and the Medical Research Council (UK) and University College London Hospitals National Institute for Health Research Biomedical Research Centre (D.A.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The COPDGene project (NCT00608764) is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Siemens, and Sunovion. The National Emphysema Treatment Trial was supported by NHLBI grants N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, and N01HR76119, the Centers for Medicare and Medicaid Services, and the Agency for Healthcare Research and Quality. The Normative Aging Study is supported by the Cooperative Studies Program/Epidemiologic Research and Information Center of the U.S. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center. The Norway GenKOLS study (Genetics of Chronic Obstructive Lung Disease; GlaxoSmithKline [GSK] code, RES11080), the Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-Points study (clinicaltrials.gov identifier, NCT00292552; GSK code, SCO104960), and the International COPD Genetics Network study are funded by GlaxoSmithKline.
Author Contributions: All of the authors listed have contributed sufficiently to the project to be included as authors by the International Committee of Medical Journal Editors guidelines, including involvement in the conception and design, analysis and interpretation, and drafting the manuscript for important intellectual content; all authors have agreed to be accountable for the work, and all those who are qualified to be authors are listed in the author byline.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2016-0331OC on February 7, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
Contributor Information
Collaborators: James Crapo, Edwin Silverman, Barry Make, Elizabeth Regan, Terri Beaty, Nan Laird, Christoph Lange, Michael Cho, Stephanie Santorico, John Hokanson, Dawn DeMeo, Nadia Hansel, Craig Hersh, Peter Castaldi, Merry-Lynn McDonald, Emily Wan, Megan Hardin, Jacqueline Hetmanski, Margaret Parker, Marilyn Foreman, Brian Hobbs, Robert Busch, Adel El-Bouiez, Dandi Qiao, Elizabeth Regan, Eitan Halper-Stromberg, Ferdouse Begum, Sungho Won, Sharon Lutz, David A. Lynch, Harvey O. Coxson, MeiLan K. Han, Eric A. Hoffman, Stephen Humphries, Francine L. Jacobson, Philip F. Judy, Ella A. Kazerooni, John D. Newell, Jr., Elizabeth Regan, James C. Ross, Raul San Jose Estepar, Berend C. Stoel, Juerg Tschirren, Eva van Rikxoort, Bram van Ginneken, George Washko, Carla G. Wilson, Mustafa Al Qaisi, Teresa Gray, Alex Kluiber, Tanya Mann, Jered Sieren, Douglas Stinson, Joyce Schroeder, Edwin Van Beek, Robert Jensen, Douglas Everett, Anna Faino, Matt Strand, Carla Wilson, John E. Hokanson, Gregory Kinney, Sharon Lutz, Kendra Young, Katherine Pratte, Lindsey Duca, Jeffrey L. Curtis, Carlos H. Martinez, Perry G. Pernicano, Nicola Hanania, Philip Alapat, Venkata Bandi, Mustafa Atik, Aladin Boriek, Kalpatha Guntupalli, Elizabeth Guy, Amit Parulekar, Arun Nachiappan, Dawn DeMeo, Craig Hersh, George Washko, Francine Jacobson, R. Graham Barr, Byron Thomashow, John Austin, Belinda D'Souza, Gregory D. N. Pearson, Anna Rozenshtein, Neil MacIntyre, Jr., Lacey Washington, H. Page McAdams, Charlene McEvoy, Joseph Tashjian, Robert Wise, Nadia Hansel, Robert Brown, Karen Horton, Nirupama Putcha, Richard Casaburi, Alessandra Adami, Janos Porszasz, Hans Fischer, Matthew Budoff, Harry Rossiter, Amir Sharafkhaneh, Charlie Lan, Christine Wendt, Brian Bell, Marilyn Foreman, Gloria Westney, Eugene Berkowitz, Russell Bowler, David Lynch, Richard Rosiello, David Pace, Gerard Criner, David Ciccolella, Francis Cordova, Chandra Dass, Gilbert D'Alonzo, Parag Desai, Michael Jacobs, Steven Kelsen, Victor Kim, A. James Mamary, Nathaniel Marchetti, Aditi Satti, Kartik Shenoy, Robert M. Steiner, Alex Swift, Irene Swift, Maria Elena Vega-Sanchez, Mark Dransfield, William Bailey, J. Michael Wells, Surya Bhatt, Hrudaya Nath, Joe Ramsdell, Paul Friedman, Xavier Soler, Andrew Yen, Alejandro Comellas, John Newell, Jr., Brad Thompson, MeiLan Han, Ella Kazerooni, Carlos Martinez, Joanne Billings, Tadashi Allen, Frank Sciurba, Divay Chandra, Joel Weissfeld, Carl Fuhrman, Jessica Bon, Antonio Anzueto, Sandra Adams, Diego Maselli-Caceres, Mario E. Ruiz, Edwin K. Silverman, David A. Lomas, Barry J. Make, Alvar Agusti, Jaume Sauleda, Peter M. A. Calverley, Claudio F. Donner, Peter D. Paré, Stephen Rennard, Jørgen Vestbo, Y. Ivanov, K. Kostov, J. Bourbeau, M. Fitzgerald, P. Hernandez, K. Killian, R. Levy, F. Maltais, D. O'Donnell, J. Krepelka, J. Vestbo, E. Wouters, D. Quinn, P. Bakke, M. Kosnik, A. Agusti, J. Sauleda, Y. Feschenko, V. Gavrisyuk, L. Yashina, N. Monogarova, P. Calverley, D. Lomas, W. MacNee, D. Singh, J. Wedzicha, A. Anzueto, S. Braman, R. Casaburi, B. Celli, G. Giessel, M. Gotfried, G. Greenwald, N. Hanania, D. Mahler, B. Make, S. Rennard, C. Rochester, P. Scanlon, D. Schuller, F. Sciurba, A. Sharafkhaneh, T. Siler, E. Silverman, A. Wanner, R. Wise, R. ZuWallack, H. Coxson, C. Crim, L. Edwards, D. Lomas, W. MacNee, E. Silverman, R. Tal Singer, J. Vestbo, J. Yates, A. Agusti, P. Calverley, B. Celli, C. Crim, B. Miller, W. MacNee, S. Rennard, R. Tal-Singer, E. Wouters, J. Yates, J. Benditt, G. Criner, M. DeCamp, P. Diaz, M. Ginsburg, L. Kaiser, M. Katz, M. Krasna, N. MacIntyre, R. McKenna, F. Martinez, Z. Mosenifar, J. Reilly, A. Ries, P. Scanlon, F. Sciurba, and J. Utz
References
- 1.Kochanek KD, Murphy SL, Xu J. Deaths: final data for 2011. Natl Vital Stat Rep. 2015;63:1–120. [PubMed] [Google Scholar]
- 2.Vestbo J, Hurd SS, Agustí AG, Jones PW, Vogelmeier C, Anzueto A, Barnes PJ, Fabbri LM, Martinez FJ, Nishimura M, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187:347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- 3.Ingebrigtsen T, Thomsen SF, Vestbo J, van der Sluis S, Kyvik KO, Silverman EK, Svartengren M, Backer V. Genetic influences on chronic obstructive pulmonary disease—a twin study. Respir Med. 2010;104:1890–1895. doi: 10.1016/j.rmed.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 4.McCloskey SC, Patel BD, Hinchliffe SJ, Reid ED, Wareham NJ, Lomas DA. Siblings of patients with severe chronic obstructive pulmonary disease have a significant risk of airflow obstruction. Am J Respir Crit Care Med. 2001;164:1419–1424. doi: 10.1164/ajrccm.164.8.2105002. [DOI] [PubMed] [Google Scholar]
- 5.Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, O’Donnell WJ, Reilly JJ, Ginns L, Mentzer S, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease: risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med. 1998;157:1770–1778. doi: 10.1164/ajrccm.157.6.9706014. [DOI] [PubMed] [Google Scholar]
- 6.Burrows B, Knudson RJ, Cline MG, Lebowitz MD. Quantitative relationships between cigarette smoking and ventilatory function. Am Rev Respir Dis. 1977;115:195–205. doi: 10.1164/arrd.1977.115.2.195. [DOI] [PubMed] [Google Scholar]
- 7.Wain LV, Shrine N, Miller S, Jackson VE, Ntalla I, Soler Artigas M, Billington CK, Kheirallah AK, Allen R, Cook JP, et al. UK Brain Expression Consortium (UKBEC); OxGSK Consortium. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho MH, Castaldi PJ, Wan ES, Siedlinski M, Hersh CP, Demeo DL, Himes BE, Sylvia JS, Klanderman BJ, Ziniti JP, et al. ICGN Investigators; ECLIPSE Investigators; COPDGene Investigators. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21:947–957. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, Feng S, Hersh CP, Bakke P, Gulsvik A, et al. ICGN Investigators. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeMeo DL, Mariani T, Bhattacharya S, Srisuma S, Lange C, Litonjua A, Bueno R, Pillai SG, Lomas DA, Sparrow D, et al. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am J Hum Genet. 2009;85:493–502. doi: 10.1016/j.ajhg.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, DeMeo DL, Hunninghake GM, Litonjua AA, Sparrow D, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou JJ, Cho MH, Castaldi PJ, Hersh CP, Silverman EK, Laird NM. Heritability of chronic obstructive pulmonary disease and related phenotypes in smokers. Am J Respir Crit Care Med. 2013;188:941–947. doi: 10.1164/rccm.201302-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakravarti A, Clark AG, Mootha VK. Distilling pathophysiology from complex disease genetics. Cell. 2013;155:21–26. doi: 10.1016/j.cell.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corvol H, Hodges CA, Drumm ML, Guillot L. Moving beyond genetics: is FAM13A a major biological contributor in lung physiology and chronic lung diseases? J Med Genet. 2014;51:646–649. doi: 10.1136/jmedgenet-2014-102525. [DOI] [PubMed] [Google Scholar]
- 16.Young RP, Hopkins RJ, Hay BA, Whittington CF, Epton MJ, Gamble GD. FAM13A locus in COPD is independently associated with lung cancer—evidence of a molecular genetic link between COPD and lung cancer. Appl Clin Genet. 2010;4:1–10. doi: 10.2147/TACG.S15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 18.Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, Demeo DL, Sylvia JS, Ziniti J, Laird NM, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castaldi PJ, Cho MH, Litonjua AA, Bakke P, Gulsvik A, Lomas DA, Anderson W, Beaty TH, Hokanson JE, Crapo JD, et al. COPDGene and Eclipse Investigators. The association of genome-wide significant spirometric loci with chronic obstructive pulmonary disease susceptibility. Am J Respir Cell Mol Biol. 2011;45:1147–1153. doi: 10.1165/rcmb.2011-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, Zhao JH, Ramasamy A, Zhai G, Vitart V, et al. Wellcome Trust Case Control Consortium; NSHD Respiratory Study Team. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, Zhai G, Zhao JH, Smith AV, Huffman JE, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock DB, Soler Artigas M, Gharib SA, Henry A, Manichaikul A, Ramasamy A, Loth DW, Imboden M, Koch B, McArdle WL, et al. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 2012;8:e1003098. doi: 10.1371/journal.pgen.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S, Song Y. Building genetic scores to predict risk of complex diseases in humans: is it possible? Diabetes. 2010;59:2729–2731. doi: 10.2337/db10-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young RP, Hopkins RJ, Hay BA, Gamble GD. Joint effect of single-nucleotide polymorphisms and smoking exposure in chronic obstructive pulmonary disease risk. Am J Respir Crit Care Med. 2012;185:683. doi: 10.1164/ajrccm.185.6.683. author reply 683–684. [DOI] [PubMed] [Google Scholar]
- 25.Soler Artigas M, Wain LV, Repapi E, Obeidat M, Sayers I, Burton PR, Johnson T, Zhao JH, Albrecht E, Dominiczak AF, et al. Medical Research Council National Survey of Health and Development (NSHD) Respiratory Study Team; SpiroMeta Consortium. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am J Respir Crit Care Med. 2011;184:786–795. doi: 10.1164/rccm.201102-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X, Baron RM, Hardin M, Cho MH, Zielinski J, Hawrylkiewicz I, Sliwinski P, Hersh CP, Mancini JD, Lu K, et al. Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Hum Mol Genet. 2012;21:1325–1335. doi: 10.1093/hmg/ddr569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu G, Warren L, Aponte J, Gulsvik A, Bakke P, Anderson WH, Lomas DA, Silverman EK, Pillai SG International COPD Genetics Network (ICGN) Investigators. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med. 2007;176:167–173. doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 28.Patel BD, Coxson HO, Pillai SG, Agustí AG, Calverley PM, Donner CF, Make BJ, Müller NL, Rennard SI, Vestbo J, et al. International COPD Genetics Network. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;178:500–505. doi: 10.1164/rccm.200801-059OC. [DOI] [PubMed] [Google Scholar]
- 29.Silverman EK, Weiss ST, Drazen JM, Chapman HA, Carey V, Campbell EJ, Denish P, Silverman RA, Celedon JC, Reilly JJ, et al. Gender-related differences in severe, early-onset chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2000;162:2152–2158. doi: 10.1164/ajrccm.162.6.2003112. [DOI] [PubMed] [Google Scholar]
- 30.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vestbo J, Anderson W, Coxson HO, Crim C, Dawber F, Edwards L, Hagan G, Knobil K, Lomas DA, MacNee W, et al. ECLIPSE investigators. Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) Eur Respir J. 2008;31:869–873. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 32.Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, Ries A, Weinmann G, Wood DE National Emphysema Treatment Trial Research Group. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med. 2003;348:2059–2073. doi: 10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 33.Bell B, Rose CL, Damon A. The Normative Aging Study: an interdisciplinary and longitudinal study of health and aging. Int J Aging Hum Dev. 1972;3:5–17. [Google Scholar]
- 34.Sørheim IC, Johannessen A, Grydeland TB, Omenaas ER, Gulsvik A, Bakke PS. Case–control studies on risk factors for chronic obstructive pulmonary disease: how does the sampling of the cases and controls affect the results? Clin Respir J. 2010;4:89–96. doi: 10.1111/j.1752-699X.2009.00154.x. [DOI] [PubMed] [Google Scholar]
- 35.Hobbs BD, Parker MM, Chen H, Lao T, Hardin M, Qiao D, Hawrylkiewicz I, Sliwinski P, Yim JJ, Kim WJ, et al. NETT Genetics Investigators; ECLIPSE Investigators; COPDGene Investigators; International COPD Genetics Network Investigators. Exome array analysis identifies a common variant in IL27 associated with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2016;194:48–57. doi: 10.1164/rccm.201510-2053OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, Franceschini N, van Durme YM, Chen TH, Barr RG, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 42.Hobbs BD, Hardin M, Hawrylkiewicz I, Sliwinski P, Yim JJ, Kim WJ, Kim DK, Agusti A, Make BJ, Calverley PM, et al. Coding variant associations with lung function and COPD using an exome array. Presented at the American Thoracic Society 2015 International Conference, Denver, Colorado; 2015. [Google Scholar]
- 43.GTEx Consortium. Human genomics: the Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrell RW, Jeppsson JO, Laurell CB, Brennan SO, Owen MC, Vaughan L, Boswell DR. Structure and variation of human α1-antitrypsin. Nature. 1982;298:329–334. doi: 10.1038/298329a0. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y, Foreman RC. The molecular genetics of α1 antitrypsin deficiency. BioEssays. 1991;13:163–169. doi: 10.1002/bies.950130404. [DOI] [PubMed] [Google Scholar]
- 47.Cho MH, Castaldi PJ, Hersh CP, Hobbs BD, Barr RG, Tal-Singer R, Bakke P, Gulsvik A, San José Estépar R, Van Beek EJ, et al. NETT Genetics, ECLIPSE, and COPDGene Investigators. A genome-wide association study of emphysema and airway quantitative imaging phenotypes. Am J Respir Crit Care Med. 2015;192:559–569. doi: 10.1164/rccm.201501-0148OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sørheim IC, Bakke P, Gulsvik A, Pillai SG, Johannessen A, Gaarder PI, Campbell EJ, Agustí A, Calverley PM, Donner CF, et al. α1-Antitrypsin protease inhibitor MZ heterozygosity is associated with airflow obstruction in two large cohorts. Chest. 2010;138:1125–1132. doi: 10.1378/chest.10-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thun GA, Imboden M, Ferrarotti I, Kumar A, Obeidat M, Zorzetto M, Haun M, Curjuric I, Couto Alves A, Jackson VE, et al. Causal and synthetic associations of variants in the SERPINA gene cluster with α1-antitrypsin serum levels. PLoS Genet. 2013;9:e1003585. doi: 10.1371/journal.pgen.1003585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929. discussion 207–112. [DOI] [PubMed] [Google Scholar]
- 51.Hu R, Ouyang Q, Dai A, Tan S, Xiao Z, Tang C. Heat shock protein 27 and cyclophilin A associate with the pathogenesis of COPD. Respirology. 2011;16:983–993. doi: 10.1111/j.1440-1843.2011.01993.x. [DOI] [PubMed] [Google Scholar]
- 52.Nigro P, Pompilio G, Capogrossi MC. Cyclophilin A: a key player for human disease. Cell Death Dis. 2013;4:e888. doi: 10.1038/cddis.2013.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis CA, Haberland M, Arnold MA, Sutherland LB, McDonald OG, Richardson JA, Childs G, Harris S, Owens GK, Olson EN. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol Cell Biol. 2006;26:2626–2636. doi: 10.1128/MCB.26.7.2626-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jostins L, Barrett JC. Genetic risk prediction in complex disease. Hum Mol Genet. 2011;20:R182–R188. doi: 10.1093/hmg/ddr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talmud PJ, Hingorani AD, Cooper JA, Marmot MG, Brunner EJ, Kumari M, Kivimäki M, Humphries SE. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ. 2010;340:b4838. doi: 10.1136/bmj.b4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lange P, Celli B, Agustí A, Boje Jensen G, Divo M, Faner R, Guerra S, Marott JL, Martinez FD, Martinez-Camblor P, et al. Lung-function trajectories leading to chronic obstructive pulmonary disease. N Engl J Med. 2015;373:111–122. doi: 10.1056/NEJMoa1411532. [DOI] [PubMed] [Google Scholar]
- 57.Jackson VE, Ntalla I, Sayers I, Morris R, Whincup P, Casas JP, Amuzu A, Choi M, Dale C, Kumari M, et al. Exome-wide analysis of rare coding variation identifies novel associations with COPD and airflow limitation in MOCS3, IFIT3 and SERPINA12. Thorax. 2016;71:501–509. doi: 10.1136/thoraxjnl-2015-207876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soler Artigas M, Wain LV, Miller S, Kheirallah AK, Huffman JE, Ntalla I, Shrine N, Obeidat M, Trochet H, McArdle WL, et al. UK BiLEVE. Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat Commun. 2015;6:8658. doi: 10.1038/ncomms9658. [DOI] [PMC free article] [PubMed] [Google Scholar]