Abstract
Bronchiolitis obliterans (BO) is an increasingly important lung disease characterized by fibroproliferative airway lesions and decrements in lung function. Occupational exposure to the artificial food flavoring ingredient diacetyl, commonly used to impart a buttery flavor to microwave popcorn, has been associated with BO development. In the occupational setting, diacetyl vapor is first encountered by the airway epithelium. To better understand the effects of diacetyl vapor on the airway epithelium, we used an unbiased proteomic approach to characterize both the apical and basolateral secretomes of air–liquid interface cultures of primary human airway epithelial cells from four unique donors after exposure to an occupationally relevant concentration (∼1,100 ppm) of diacetyl vapor or phosphate-buffered saline as a control on alternating days. Basolateral and apical supernatants collected 48 h after the third exposure were analyzed using one-dimensional liquid chromatography tandem mass spectrometry. Paired t tests adjusted for multiple comparisons were used to assess differential expression between diacetyl and phosphate-buffered saline exposure. Of the significantly differentially expressed proteins identified, 61 were unique to the apical secretome, 81 were unique to the basolateral secretome, and 11 were present in both. Pathway enrichment analysis using publicly available databases revealed that proteins associated with matrix remodeling, including degradation, assembly, and new matrix organization, were overrepresented in the data sets. Similarly, protein modifiers of epidermal growth factor receptor signaling were significantly altered. The ordered changes in protein expression suggest that the airway epithelial response to diacetyl may contribute to BO pathogenesis.
Keywords: proteomics; diacetyl; 2,3-butanedione; occupational lung disease; bronchiolitis obliterans
Clinical Relevance
How diacetyl causes popcorn lung has not been elucidated. Previous studies have shown profound epithelial injury in rodent models of popcorn lung followed by the development of constrictive and obstructive airway lesions. The current report reveals new insights into the airway epithelial response to diacetyl vapor exposure, which could contribute to the airway fibroproliferative response.
Bronchiolitis obliterans (BO) is an increasingly important human disease that is now recognized in a variety of clinical contexts, including autoimmune disease, as a consequence of lung or bone marrow transplantation, or as a result of occupational exposures. Histologically, BO is characterized by airway-centered fibrosis that can cause partial or total airway occlusion. Clinically, BO results in significant decrements in lung function and can progress to disability or death.
Diacetyl (DA; 2,3-butanedione) is a volatile α-diketone that occurs naturally as a result of fermentation and has most commonly been used to impart a buttery aroma and flavor to microwave popcorn, flavored coffee, and e-cigarettes. Growing evidence now shows that occupational exposure to DA vapor is associated with the development of BO in the microwave popcorn industry (1–3), in the food-flavoring manufacturing industry (4), and in the manufacture of diacetyl itself (5). Despite the increasing recognition of occupational BO, the mechanisms that lead to the development of BO remain poorly understood. BO-inducing toxins such as DA are first encountered by the airway epithelium. As such, the early secretory response of the airway epithelium after DA exposure may play a central role in the pathogenesis of BO.
To better understand the airway epithelial response to DA and the role it may play in the lesion development that is characteristic of BO, we employed an unbiased proteomic approach using primary human airway epithelial cells from multiple independent donors. We previously used a proteomic approach to successfully identify proteins unique to the lung fluid of patients with idiopathic pulmonary fibrosis (6). More recently, a similar proteomic approach has been used to provide new insights into alterations in proteins secreted by the airway epithelium in cystic fibrosis (7), and into the polarized nature of protein secretion by human airway epithelial cells (8). The goal of the current hypothesis-generating studies was to identify soluble factors that could be diagnostic of exposures most closely associated with a high risk of developing BO and thus could be used as biomarkers, or that could play a role in lesion development. Here then, for the first time, we report changes in protein secretion in the apical and basolateral compartments of fully differentiated primary human airway epithelial cells after repeated exposure to occupationally relevant high concentrations of DA vapor, providing new insights into early events that may contribute to flavoring-induced airways disease.
Materials and Methods
A complete description of the materials and methods used in this work is available in the online supplement.
Cell Culture
Air-liquid interface (ALI) cultures of fully differentiated primary tracheobronchial epithelial cells with a mucociliary phenotype from four independent, healthy, nonsmoking donors were purchased from MatTek (Ashland, MA) and cultured in 6-well Transwell plates at 37°C in 5% CO2.
Vapor Exposure
The cells were exposed to an occupationally relevant concentration (∼1100 ppm) of DA vapor for 1 h on days 0, 2, and 4 essentially as previously described (9, 10) to model the repeated high-concentration exposures that would be encountered by workers who prepare and mix flavoring compounds in microwave popcorn factories (1). Day 6 apical washes were centrifuged and the basolateral supernatants were collected, and all were retained at −80°C for proteomic analysis. We performed dose-response studies by testing increasing concentrations of DA vapor using lactate dehydrogenase (LDH) activity as an indicator of injury. The results demonstrate that a single DA vapor exposure caused no increase in LDH activity at any vapor concentration. However, after the second and third exposures, LDH activity increased at the higher concentration, which is consistent with some degree of cellular injury. For a detailed rationale for this exposure system, please refer to Materials and Methods in the online supplement.
Proteomic Analysis
Sample preparation, quantitative mass spectrometry, and measures of analytical versus biological variability are described in detail in the Materials and Methods section in the online supplement.
Quantitative analysis of protein expression in the secretomes of PBS- and DA-exposed cells
The data were imported into Rosetta Elucidator for mass and retention time alignment, database searching of tandem mass spectrometry spectra, and quantitation of the area-under-the-curve of identified features. Peptide scoring and annotation identified 4,273 apical peptides and 1,046 apical proteins. Similarly, we identified 6,067 basolateral peptides and 1,327 basolateral proteins. Filtering the data to remove low-quality peptides and scaling the data to the robust median across all samples resulted in 3,077 apical peptides (Table E1 in the online supplement) and 1,046 proteins (Table E2), and 6,067 basolateral peptides (Table E3) and 1,327 proteins (Table E4).
Statistical analyses
Peptide- and protein-level expression values were log-2 transformed, and features with missing or low-abundance measurements were not subjected to further analysis. Proteins having greater than 30% technical variability, those quantified with a single peptide, and experimental control proteins were not subjected to further analyses. A total of 541 proteins in the apical secretome and 793 in the basolateral secretome met these criteria. Differential expression between PBS and DA exposure was analyzed by paired t test by donor in each secretome. The resulting P values were corrected for multiple testing by controlling the false discovery rate (FDR) of 0.10 with the Benjamini-Hochberg method (11).
For unsupervised agglomerative clustering of the significantly differentially expressed apical and basolateral protein sets, we used the Euclidean distance metric and Ward linkage method. Quantitative pathway enrichment analyses of the protein sets were performed with the REACTOME pathway analysis tool (12). Putative protein–protein interactions within the data sets were visualized using the publicly available STRING suite of analysis tools (13).
RT-PCR
Total mRNA for protein tyrosine phosphatase, receptor type S, fibulin 3 (FBLN3; also known as epidermal growth factor [EGF]-containing fibulin-like extracellular matrix [ECM] protein 1 [EFEMP1]), DNA damage-binding protein 1 (DDB1), ECM protein 1 (ECM1), and growth differentiation factor 15 (GDF15) was analyzed by Taqman using β-actin as an endogenous control. Changes in expression were calculated using the 2-Ct method.
Results
Proteomic Analysis of DA-Exposed, Fully Differentiated Primary Human Airway Epithelial Cells in ALI Culture Reveals that Variability in Protein Expression Is Driven by the Exposure
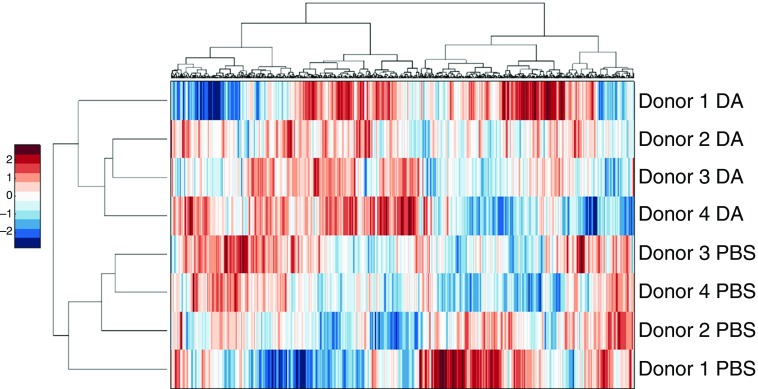
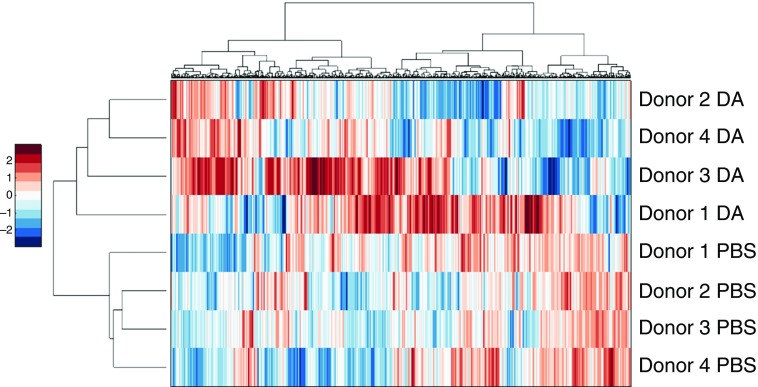
Our proteomic analysis identified 541 apical and 793 basolateral proteins by more than two individual peptides and with less than 30% technical variability, as described in Materials and Methods. Unsupervised hierarchical clustering analysis of these proteins resulted in the apical secretome segregating by exposure (Figure 1). The basolateral secretome also segregated by exposure, with the exception of donor 9831 (Figure 2). These results show that DA exposure largely underlies the variability in the protein expression data. Similarly, in a principal component analysis, both the apical (Figure E2) and basolateral (Figure E3) secretomes segregated by exposure. Although the proteomic response of donor 9831 varied somewhat from the responses of the other donors, showing the presence of variation between donors in the airway epithelial response to DA, the principal component analysis supports the conclusion that the greatest variability in the data was due to DA exposure.
Figure 1.
Two-dimensional (2D) agglomerative clustering of apical proteins. Protein expression values were converted to Z-scores, followed by 2D agglomerative clustering using the Ward method. DA, diacetyl.
Figure 2.
2D agglomerative clustering of basolateral proteins. Protein expression values were converted to Z-scores, followed by 2D agglomerative clustering using the Ward method.
Proteomic Analysis of DA-Exposed Primary Human Airway Epithelial Cells in ALI Culture Reveals a Highly Polarized Secretory Response
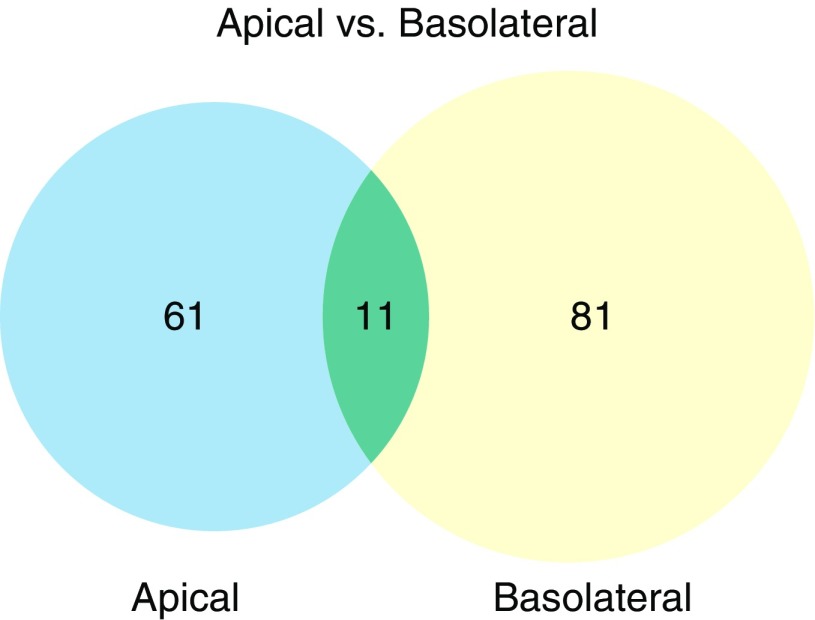
As shown in Figure 3, by using a paired t test to evaluate changes in protein expression across all four donors, and an FDR-adjusted P value of less than 0.1 as described in Materials and Methods, we found that after DA exposure there were 61 significantly differentially expressed proteins unique to the apical secretome (Table 1) and 81 significantly differentially expressed proteins unique to the basolateral secretome (Table 2). We identified an additional 11 proteins that were present in both the apical and basolateral secretomes (Table 3).
Figure 3.
The apical and basolateral secretomes of DA-exposed primary human airway epithelial cells are highly polarized. The Venn diagram shows the overlap of differentially expressed protein sets (significant at the level of false discovery rate [FDR] < 0.1).
Table 1.
Apical Proteins with Significantly Altered Expression in DA versus PBS
| Primary Protein Name | Protein Description | Peptide Count | %CV QC | Fold Change DA versus PBS | t test P value | t test P value w/FDR Correction |
|---|---|---|---|---|---|---|
| KLK6_HUMAN | Kallikrein-related peptidase 6 | 4 | 6.7 | 7.0 | 0.004 | 0.068 |
| ECM1_HUMAN | Extracellular matrix protein 1 | 7 | 19.9 | 6.8 | 0.010 | 0.089 |
| H4_HUMAN | Histone cluster 4, H4 | 6 | 15.3 | 6.3 | 0.004 | 0.065 |
| TRFE_HUMAN | Transferrin | 11 | 17.2 | 4.7 | 0.002 | 0.054 |
| H2B1M_HUMAN | Histone H2B type F-S | 3 | 10.2 | 4.3 | 0.005 | 0.078 |
| LAMA3_HUMAN | Laminin subunit α-3 | 13 | 18.4 | 4.0 | 0.007 | 0.080 |
| DSC2_HUMAN | Desmocollin-2 | 2 | 15.3 | 3.9 | 0.003 | 0.065 |
| DKK1_HUMAN | Dickkopf-related protein 1 | 2 | 9.5 | 3.6 | 0.002 | 0.054 |
| H2AY_HUMAN | Core histone macro-H2A.1 | 4 | 6.2 | 3.3 | 0.011 | 0.091 |
| CCD80_HUMAN | Coiled-coil domain-containing protein 80 | 2 | 2.2 | 3.0 | 0.001 | 0.054 |
| FLNA_HUMAN | Filamin-A | 12 | 14.3 | 2.8 | 0.008 | 0.085 |
| LAMB3_HUMAN | Laminin subunit β-3 | 9 | 11.7 | 2.7 | 0.004 | 0.065 |
| MMP9_HUMAN | Matrix metalloproteinase-9 | 8 | 14.6 | 2.7 | 0.000 | 0.054 |
| PGBM_HUMAN | Basement membrane-specific heparan sulfate proteoglycan core protein | 15 | 5.6 | 2.6 | 0.006 | 0.080 |
| LEG1_HUMAN | Galectin-1 | 3 | 27.6 | 2.6 | 0.011 | 0.095 |
| CEAM6_HUMAN | Carcinoembryonic antigen-related cell adhesion molecule 6 | 5 | 17.0 | 2.5 | 0.008 | 0.083 |
| LAMC2_HUMAN | Laminin subunit γ-2 | 16 | 14.3 | 2.5 | 0.007 | 0.080 |
| ST14_HUMAN | Suppressor of tumorigenicity 14 protein | 2 | 7.6 | 2.3 | 0.010 | 0.086 |
| GDF15_HUMAN | Growth/differentiation factor 15 | 4 | 19.6 | 2.2 | 0.002 | 0.054 |
| PSB5_HUMAN | Proteasome subunit-β type 5 | 3 | 9.0 | 2.2 | 0.013 | 0.098 |
| MMP10_HUMAN | Stromelysin-2 | 3 | 18.5 | 2.1 | 0.009 | 0.086 |
| LYPD3_HUMAN | Ly6/PLAUR domain-containing protein 3 | 5 | 4.8 | 2.1 | 0.000 | 0.016 |
| APLP2_HUMAN | Amyloid-like protein 2 | 2 | 1.6 | 2.0 | 0.001 | 0.054 |
| RSMN_HUMAN | Small nuclear ribonucleoprotein-associated protein N | 2 | 17.9 | 2.0 | 0.006 | 0.078 |
| EPS8_HUMAN | Epidermal growth factor receptor kinase substrate 8 | 7 | 29.5 | 2.0 | 0.012 | 0.097 |
| TENA_HUMAN | Tenascin C | 11 | 7.4 | 1.9 | 0.002 | 0.054 |
| DSG2_HUMAN | Desmoglein-2 | 5 | 10.3 | 1.9 | 0.003 | 0.065 |
| PEPD_HUMAN | Xaa-Pro dipeptidase | 3 | 27.8 | 1.8 | 0.002 | 0.054 |
| SSBP_HUMAN | Single-stranded DNA-binding protein mitochondrial | 4 | 6.6 | 1.8 | 0.009 | 0.086 |
| TIMP1_HUMAN | Metalloproteinase inhibitor 1 | 5 | 8.5 | 1.8 | 0.009 | 0.086 |
| CSTN1_HUMAN | Calsyntenin-1 | 5 | 5.0 | 1.7 | 0.002 | 0.054 |
| PSA3_HUMAN | Proteasome subunit-α type 3 | 3 | 7.4 | 1.6 | 0.010 | 0.086 |
| ANXA8_HUMAN | Annexin A8 | 5 | 3.6 | 1.6 | 0.012 | 0.097 |
| PSB6_HUMAN | Proteasome subunit-β type 6 | 3 | 10.9 | 1.6 | 0.012 | 0.097 |
| PSA7_HUMAN | Proteasome subunit-α type 7 | 7 | 1.4 | 1.5 | 0.005 | 0.078 |
| TKT_HUMAN | Transketolase | 13 | 8.3 | 1.5 | 0.007 | 0.080 |
| CD14_HUMAN | Monocyte differentiation antigen CD14 | 3 | 4.7 | 1.5 | 0.005 | 0.078 |
| CADH1_HUMAN | Cadherin-1 | 8 | 9.4 | 1.4 | 0.007 | 0.080 |
| SPTN1_HUMAN | Spectrin α chain non-erythrocytic 1 | 25 | 3.1 | 1.4 | 0.013 | 0.098 |
| PRDX1_HUMAN | Peroxiredoxin-1 | 14 | 3.3 | 1.3 | 0.009 | 0.086 |
| SPTB2_HUMAN | Spectrin β chain brain 1 | 10 | 12.5 | 1.3 | 0.007 | 0.080 |
| GNAI2_HUMAN | Guanine nucleotide-binding protein G(i) α-2 subunit | 3 | 1.2 | 1.1 | 0.009 | 0.086 |
| PSA_HUMAN | Puromycin-sensitive aminopeptidase | 11 | 13.7 | −1.1 | 0.003 | 0.065 |
| DDB1_HUMAN | DNA damage-binding protein 1 | 8 | 4.2 | −1.2 | 0.002 | 0.058 |
| SLPI_HUMAN | Antileukoproteinase | 7 | 3.3 | −1.4 | 0.009 | 0.086 |
| CBX3_HUMAN | Chromobox protein homolog 3 | 2 | 3.5 | −1.5 | 0.001 | 0.054 |
| GELS_HUMAN | Gelsolin | 31 | 6.7 | −1.5 | 0.006 | 0.078 |
| HSP71_HUMAN | Heat shock 70 kDa protein 1 | 6 | 5.4 | −1.6 | 0.004 | 0.065 |
| FUBP1_HUMAN | Far upstream element-binding protein 1 | 3 | 10.4 | −1.7 | 0.012 | 0.097 |
| AL1A1_HUMAN | Retinal dehydrogenase 1 | 15 | 7.9 | −1.7 | 0.001 | 0.054 |
| THIL_HUMAN | Acetyl-CoA acetyltransferase mitochondrial | 2 | 24.5 | −1.8 | 0.006 | 0.078 |
| CFAB_HUMAN | Complement factor B | 26 | 21.6 | −1.8 | 0.003 | 0.065 |
| S10A4_HUMAN | Protein S100-A4 | 2 | 12.6 | −1.8 | 0.003 | 0.065 |
| PEDF_HUMAN | Pigment epithelium-derived factor | 15 | 8.4 | −2.1 | 0.012 | 0.097 |
| CD59_HUMAN | CD59 glycoprotein | 4 | 6.4 | −2.2 | 0.001 | 0.054 |
| PIGR_HUMAN | Polymeric immunoglobulin receptor | 36 | 7.4 | −2.3 | 0.007 | 0.080 |
| BPIA1_HUMAN | BPI fold-containing family A member 1 | 5 | 12.7 | −2.4 | 0.007 | 0.080 |
| PLTP_HUMAN | Phospholipid transfer protein | 4 | 10.1 | −2.6 | 0.002 | 0.054 |
| FBLN3_HUMAN | EGF-containing fibulin-like extracellular matrix protein 1 | 14 | 12.1 | −2.7 | 0.002 | 0.054 |
| AACT_HUMAN | α-1-antichymotrypsin | 7 | 20.0 | −3.4 | 0.004 | 0.065 |
| MANBA_HUMAN | β-mannosidase | 4 | 17.0 | −3.4 | 0.006 | 0.078 |
| B2MG_HUMAN | β-2-microglobulin | 6 | 25.5 | −3.5 | 0.001 | 0.054 |
| NUCB2_HUMAN | Nucleobindin-2 | 5 | 19.5 | −3.9 | 0.006 | 0.078 |
| IBP7_HUMAN | Insulin-like growth factor-binding protein 7 | 10 | 10.6 | −3.9 | 0.013 | 0.098 |
| A1AT_HUMAN | α-1-antitrypsin | 7 | 6.9 | −5.6 | 0.001 | 0.054 |
| CEL_HUMAN | Bile salt-activated lipase | 5 | 15.5 | −5.6 | 0.002 | 0.056 |
| CYTM_HUMAN | Cystatin-M | 3 | 15.5 | −5.9 | 0.001 | 0.054 |
| IBP2_HUMAN | Insulin-like growth factor-binding protein 2 | 11 | 6.3 | −6.5 | 0.005 | 0.078 |
| CLUS_HUMAN | Clusterin | 9 | 4.6 | −11.0 | 0.001 | 0.054 |
| ISK5_HUMAN | Serine protease inhibitor Kazal-type 5 | 2 | 8.6 | −11.9 | 0.008 | 0.086 |
| CO4A_HUMAN | Complement C4-A | 5 | 7.4 | −14.8 | 0.002 | 0.057 |
| CDHR3_HUMAN | Cadherin-related family member 3 | 3 | 5.3 | −20.7 | 0.002 | 0.056 |
Definition of abbreviations: BPI, bactericidal permeability-increasing protein; CV, coefficient of variation; DA, diacetyl; EGF, epidermal growth factor; FDR, false discovery rate; PLAUR, urokinase plasminogen activator surface receptor; QC, quality control.
Table 2.
Basolateral Proteins with Significantly Altered Expression in DA versus PBS
| Primary Protein Name | Protein Description | Peptide Count | %CV QC | Fold Change DA versus PBS | t test P value | t test P value w/FDR Correction |
|---|---|---|---|---|---|---|
| A2ML1_HUMAN | α-2-macroglobulin-like protein 1 | 7 | 16.6 | 11.9 | 0.011 | 0.099 |
| UPAR_HUMAN | Urokinase plasminogen activator surface receptor | 3 | 26.2 | 10.5 | 0.009 | 0.095 |
| ECM1_HUMAN | Extracellular matrix protein 1 | 24 | 10.5 | 9.9 | 0.001 | 0.061 |
| PRS27_HUMAN | Serine protease 27 | 3 | 11.1 | 9.2 | 0.011 | 0.099 |
| DAF_HUMAN | Complement decay-accelerating factor | 4 | 3.1 | 8.8 | 0.008 | 0.094 |
| EPHA2_HUMAN | Ephrin type-A receptor 2 | 6 | 15.1 | 6.4 | 0.001 | 0.061 |
| TIMP1_HUMAN | Metalloproteinase inhibitor 1 | 5 | 7.0 | 5.7 | 0.001 | 0.067 |
| ZG16B_HUMAN | Zymogen granule protein 16 homolog B | 3 | 12.7 | 5.2 | 0.003 | 0.083 |
| PCDGK_HUMAN | Protocadherin γ-C3 | 2 | 9.8 | 4.6 | 0.010 | 0.099 |
| GOLM1_HUMAN | Golgi membrane protein 1 | 8 | 4.2 | 4.5 | 0.005 | 0.090 |
| IL36G_HUMAN | Interleukin-36 γ | 3 | 10.3 | 4.5 | 0.008 | 0.094 |
| BSSP4_HUMAN | Brain-specific serine protease 4 | 4 | 12.4 | 4.5 | 0.001 | 0.067 |
| DNJA4_HUMAN | DnaJ homolog subfamily A member 4 | 2 | 13.0 | 4.4 | 0.003 | 0.083 |
| MARCS_HUMAN | Myristoylated alanine-rich C-kinase substrate | 8 | 10.5 | 4.2 | 0.003 | 0.083 |
| CAP1_HUMAN | Adenylyl cyclase-associated protein 1 | 7 | 2.3 | 4.1 | 0.008 | 0.094 |
| VASN_HUMAN | Vasorin | 7 | 2.8 | 4.0 | 0.010 | 0.099 |
| PSCA_HUMAN | Prostate stem cell antigen | 3 | 10.5 | 4.0 | 0.002 | 0.075 |
| NRP1_HUMAN | Neuropilin-1 | 3 | 24.1 | 4.0 | 0.006 | 0.094 |
| DDAH1_HUMAN | N(G) N(G)-dimethylarginine dimethylaminohydrolase 1 | 4 | 13.8 | 3.8 | 0.003 | 0.083 |
| KLK10_HUMAN | Kallikrein-10 | 6 | 15.8 | 3.7 | 0.005 | 0.093 |
| BASP1_HUMAN | Brain acid soluble protein 1 | 10 | 11.3 | 3.7 | 0.003 | 0.083 |
| CXL16_HUMAN | C-X-C motif chemokine 16 | 2 | 10.8 | 3.6 | 0.005 | 0.090 |
| EFNB1_HUMAN | Ephrin-B1 | 4 | 16.5 | 3.6 | 0.004 | 0.090 |
| PCDH1_HUMAN | Protocadherin-1 | 13 | 9.3 | 3.6 | 0.000 | 0.061 |
| GPX3_HUMAN | Glutathione peroxidase 3 | 5 | 9.0 | 3.4 | 0.009 | 0.099 |
| VSIG2_HUMAN | V-set and immunoglobulin domain-containing protein 2 | 2 | 5.6 | 3.4 | 0.008 | 0.094 |
| S100P_HUMAN | Protein S100-P | 4 | 8.0 | 3.4 | 0.007 | 0.094 |
| EPCR_HUMAN | Endothelial protein C receptor | 4 | 10.4 | 3.3 | 0.011 | 0.099 |
| GRN_HUMAN | Granulins | 12 | 6.9 | 3.3 | 0.000 | 0.008 |
| LAYN_HUMAN | Layilin | 6 | 2.0 | 3.2 | 0.001 | 0.068 |
| CATD_HUMAN | Cathepsin D | 14 | 6.7 | 3.2 | 0.003 | 0.083 |
| GDF15_HUMAN | Growth/differentiation factor 15 | 11 | 11.0 | 3.2 | 0.001 | 0.067 |
| TIMP2_HUMAN | Metalloproteinase inhibitor 2 | 8 | 10.4 | 3.1 | 0.004 | 0.090 |
| LRRF1_HUMAN | Leucine-rich repeat flightless-interacting protein 1 | 3 | 6.8 | 3.1 | 0.007 | 0.094 |
| VIME_HUMAN | Vimentin | 8 | 10.4 | 3.0 | 0.001 | 0.061 |
| SPIT1_HUMAN | Kunitz-type protease inhibitor 1 | 27 | 7.0 | 3.0 | 0.003 | 0.083 |
| SEM7A_HUMAN | Semaphorin-7A | 6 | 15.9 | 3.0 | 0.000 | 0.004 |
| PIP_HUMAN | Prolactin-inducible protein | 9 | 15.7 | 2.9 | 0.006 | 0.094 |
| CATB_HUMAN | Cathepsin B | 17 | 11.7 | 2.7 | 0.001 | 0.070 |
| SPIT2_HUMAN | Kunitz-type protease inhibitor 2 | 2 | 15.3 | 2.6 | 0.001 | 0.067 |
| APLP2_HUMAN | Amyloid-like protein 2 | 8 | 10.5 | 2.5 | 0.007 | 0.094 |
| MMP14_HUMAN | Matrix metalloproteinase-14 | 5 | 9.5 | 2.3 | 0.011 | 0.099 |
| DIAC_HUMAN | Di-N-acetylchitobiase | 5 | 17.9 | 2.3 | 0.008 | 0.094 |
| PNPH_HUMAN | Purine nucleoside phosphorylase | 5 | 26.0 | 2.3 | 0.011 | 0.099 |
| QSOX1_HUMAN | Sulfhydryl oxidase 1 | 18 | 3.9 | 2.2 | 0.002 | 0.075 |
| RBSK_HUMAN | Ribokinase | 2 | 16.7 | 2.2 | 0.001 | 0.061 |
| TXND5_HUMAN | Thioredoxin domain-containing protein 5 | 7 | 6.4 | 2.1 | 0.011 | 0.099 |
| MMP9_HUMAN | Matrix metalloproteinase-9 | 16 | 10.6 | 2.1 | 0.010 | 0.099 |
| A4_HUMAN | Amyloid β A4 protein | 13 | 19.6 | 2.0 | 0.003 | 0.083 |
| HEBP2_HUMAN | Heme-binding protein 2 | 9 | 9.4 | 2.0 | 0.003 | 0.083 |
| CAH13_HUMAN | Carbonic anhydrase 13 | 2 | 19.9 | 2.0 | 0.002 | 0.072 |
| CYTB_HUMAN | Cystatin-B | 10 | 11.6 | 1.9 | 0.005 | 0.092 |
| TRXR1_HUMAN | Thioredoxin reductase 1 cytoplasmic | 5 | 19.6 | 1.9 | 0.010 | 0.099 |
| TRFE_HUMAN | Serotransferrin | 36 | 5.8 | 1.8 | 0.002 | 0.082 |
| CPPED_HUMAN | Calcineurin-like phosphoesterase domain-containing protein 1 | 4 | 23.6 | 1.8 | 0.007 | 0.094 |
| DDR1_HUMAN | Epithelial discoidin domain-containing receptor 1 | 9 | 9.2 | 1.8 | 0.008 | 0.094 |
| NEO1_HUMAN | Neogenin | 8 | 5.6 | 1.8 | 0.010 | 0.099 |
| GDIR2_HUMAN | Rho GDP-dissociation inhibitor 2 | 2 | 24.7 | 1.7 | 0.007 | 0.094 |
| AATM_HUMAN | Aspartate aminotransferase mitochondrial | 15 | 11.6 | 1.7 | 0.011 | 0.099 |
| GLRX1_HUMAN | Glutaredoxin-1 | 3 | 12.0 | 1.7 | 0.008 | 0.094 |
| K22E_HUMAN | Keratin type II cytoskeletal 2 epidermal | 10 | 3.7 | 1.7 | 0.006 | 0.094 |
| SPTB2_HUMAN | Spectrin β chain brain 1 | 7 | 12.9 | 1.6 | 0.011 | 0.099 |
| PSB6_HUMAN | Proteasome subunit-β type 6 | 4 | 6.0 | 1.4 | 0.011 | 0.099 |
| PSB1_HUMAN | Proteasome subunit-β type 1 | 4 | 25.7 | 1.2 | 0.001 | 0.068 |
| THOP1_HUMAN | Thimet oligopeptidase | 4 | 5.7 | −1.3 | 0.009 | 0.094 |
| ACPH_HUMAN | Acylamino-acid-releasing enzyme | 7 | 8.7 | −1.4 | 0.006 | 0.094 |
| PTPRF_HUMAN | Receptor-type tyrosine-protein phosphatase F | 14 | 9.9 | −1.7 | 0.005 | 0.090 |
| ADH7_HUMAN | Alcohol dehydrogenase class 4 μ/σ chain | 18 | 8.6 | −1.9 | 0.007 | 0.094 |
| DDB1_HUMAN | DNA damage-binding protein 1 | 13 | 9.8 | −1.9 | 0.003 | 0.083 |
| HIBCH_HUMAN | 3-hydroxyisobutyryl-CoA hydrolase mitochondrial | 3 | 5.6 | −2.1 | 0.005 | 0.090 |
| INO1_HUMAN | Inositol-3-phosphate synthase 1 | 3 | 4.6 | −2.2 | 0.007 | 0.094 |
| BCAM_HUMAN | Basal cell adhesion molecule | 14 | 8.7 | −2.2 | 0.007 | 0.094 |
| H2A1B_HUMAN | Histone H2A type 1-B/E | 6 | 9.0 | −2.9 | 0.007 | 0.094 |
| RCC2_HUMAN | Regulator of chromosome condensation 2 | 10 | 9.2 | −3.1 | 0.006 | 0.094 |
| RNAS4_HUMAN | RNase 4 | 3 | 2.6 | −3.3 | 0.006 | 0.094 |
| FBLN3_HUMAN | EGF-containing fibulin-like extracellular matrix protein 1 | 12 | 8.9 | −3.4 | 0.008 | 0.094 |
| GLNA_HUMAN | Glutamine synthetase | 6 | 15.9 | −3.6 | 0.007 | 0.094 |
| EGFR_HUMAN | Epidermal growth factor receptor | 3 | 10.0 | −3.7 | 0.004 | 0.090 |
| GPNMB_HUMAN | Transmembrane glycoprotein NMB | 3 | 9.5 | −3.7 | 0.004 | 0.090 |
| CO7A1_HUMAN | Collagen α-1(VII) chain | 8 | 14.2 | −3.8 | 0.009 | 0.094 |
| CO5A2_HUMAN | Collagen α-2(V) chain | 2 | 23.6 | −3.9 | 0.005 | 0.090 |
| LEG7_HUMAN | Galectin-7 | 13 | 10.3 | −3.9 | 0.011 | 0.099 |
| CO1A1_HUMAN | Collagen α-1(I) chain | 3 | 9.8 | −4.1 | 0.009 | 0.094 |
| PHYD1_HUMAN | Phytanoyl-CoA dioxygenase domain-containing protein 1 | 2 | 17.0 | −4.1 | 0.006 | 0.094 |
| C1S_HUMAN | Complement C1s subcomponent | 2 | 2.5 | −4.1 | 0.003 | 0.083 |
| C1R_HUMAN | Complement C1r subcomponent | 7 | 6.1 | −4.2 | 0.002 | 0.082 |
| EPHB2_HUMAN | Ephrin type-B receptor 2 | 5 | 2.5 | −4.2 | 0.001 | 0.067 |
| LMNB2_HUMAN | Lamin-B2 | 7 | 24.0 | −4.7 | 0.008 | 0.094 |
| IBP7_HUMAN | Insulin-like growth factor-binding protein 7 | 22 | 13.8 | −5.0 | 0.000 | 0.008 |
| XRCC6_HUMAN | X-ray repair cross-complementing protein 6 | 2 | 11.6 | −6.2 | 0.004 | 0.089 |
| CELR1_HUMAN | Cadherin EGF LAG seven-pass G-type receptor 1 | 2 | 9.8 | −17.4 | 0.012 | 0.099 |
| IMPA2_HUMAN | Inositol monophosphatase 2 | 2 | 11.8 | −31.9 | 0.001 | 0.067 |
Definition of abbreviations: CoA, co-enzyme A; CV, coefficient of variation; DA, diacetyl; EGF, epidermal growth factor; FDR, false discovery rate; GDP, guanosine 5’-diphosphate; LAG, laminin-G; NMB, neuromedin-B; QC, quality control.
Table 3.
Proteins Identified in Both Apical and Basolateral Secretome with Significantly Altered Expression in DA versus PBS
| Primary Protein Name | Protein Description | Apical Fold Change DA versus PBS | Basolateral Fold Change DA versus PBS |
|---|---|---|---|
| ECM1_HUMAN | Extracellular matrix protein 1 | 6.756 | 10.988 |
| TRFE_HUMAN | Serotransferrin | 4.687 | 1.950 |
| MMP9_HUMAN | Matrix metalloproteinase-9 | 2.690 | 2.415 |
| GDF15_HUMAN | Growth/differentiation factor 15 | 2.230 | 2.623 |
| APLP2_HUMAN | Amyloid-like protein 2 | 2.029 | 1.970 |
| TIMP1_HUMAN | Metalloproteinase inhibitor 1 | 1.801 | 4.957 |
| PSB6_HUMAN | Proteasome subunit-β type 6 | 1.560 | 1.185 |
| SPTB2_HUMAN | Spectrin β chain brain 1 | 1.277 | 1.421 |
| DDB1_HUMAN | DNA damage-binding protein 1 | −1.182 | −2.027 |
| FBLN3_HUMAN | EGF-containing fibulin-like extracellular matrix protein 1 | −2.738 | −2.818 |
| IBP7_HUMAN | Insulin-like growth factor-binding protein 7 | −3.910 | −5.367 |
Definition of abbreviations: DA, diacetyl; EGF, epidermal growth factor.
DA Exposure of Primary Human Airway Epithelial Cells in ALI Culture Induces a Matrix-Remodeling Proteomic Signature
To determine whether broad categories of proteins were overrepresented in our data set, we performed a pathway enrichment analysis of the proteins identified in Tables 1 and 2 using the publicly available REACTOME database. In the apical secretome, we observed that proteins with increased expression were overrepresented in pathways that are important for ECM degradation, matrix organization, and cell–cell interactions (Table 4). These included laminins (LAMC2, LAMA3, and LAMB3), perlecan (PGBM), E-cadherin (CADH1), matrix metalloproteinase 9 (MMP9), MMP10, and metalloproteinase inhibitor 1 (TIMP1). Conversely, we observed that proteins with decreased expression in the apical secretome were overrepresented in pathways that are important for complement activation (including complement factor B [CFAB], CD59, and complement component 4A [CO4A]) and lipid metabolism (including phospholipid transfer protein [PLTP] and bile salt–activated lipase [CEL]; Table 5). In the basolateral secretome, proteins with increased expression were overrepresented in pathways that are important for ECM degradation and organization (including cathepsin B [CATB], CATD, TIMP1, TIMP2, MMP9, and MMP14) and platelet function (including adenylyl cyclase-associated protein 1 [CAP1], transferrin [TRFE], amyloid β A4 [A4], and TIMP1; Table 6). Finally, in the basolateral secretome, proteins with decreased expression were overrepresented in pathways that are important for ECM synthesis and assembly (including the fibrillar collagens CO1A1, CO5A2, and CO7A1, and FBLN3; Table 7).
Table 4.
Top 10 REACTOME Pathways Enriched in Proteins Up-Regulated in Apical Secretome by DA Exposure
| Pathway Name | No. of Proteins | No. of Proteins in Pathway | P value | FDR | Proteins Identified |
|---|---|---|---|---|---|
| Degradation of the extracellular matrix | 8 | 135 | 1.28E-07 | 5.43E-05 | LAMC2; CADH1; LAMA3; MMP10; LAMB3; PGBM; MMP9; TIMP1 |
| Extracellular matrix organization | 10 | 291 | 4.37E-07 | 7.68E-05 | LAMC2; CADH1; CEAM6; LAMA3; MMP10; LAMB3; PGBM; MMP9; TIMP1; TENA |
| Apoptosis | 8 | 165 | 5.80E-07 | 7.68E-05 | CADH1; PSA3; DSG2; CD14; PSB6; PSA7; PSB5; SPTN1 |
| Programmed cell death | 8 | 170 | 7.24E-07 | 7.68E-05 | CADH1; PSA3; DSG2; CD14; PSB6; PSA7; PSB5; SPTN1 |
| Cell-cell communication | 7 | 139 | 2.48E-06 | 2.08E-04 | LAMC2; CADH1; LAMA3; LAMB3; SPTB2; SPTN1; FLNA |
| Non-integrin membrane–ECM interactions | 5 | 59 | 6.51E-06 | 4.56E-04 | LAMC2; LAMA3; LAMB3; PGBM; TENA |
| Laminin interactions | 4 | 30 | 1.01E-05 | 6.08E-04 | LAMC2; LAMA3; LAMB3; PGBM |
| Type I hemidesmosome assembly | 3 | 11 | 1.73E-05 | 9.15E-04 | LAMC2; LAMA3; LAMB3 |
| TCF-dependent signaling in response to WNT | 7 | 198 | 2.44E-05 | 1.00E-03 | PSA3; H2B1M; H4; PSB6; PSA7; DKK1; PSB5 |
| Anchoring fibril formation | 3 | 15 | 4.32E-05 | 1.00E-03 | LAMC2; LAMA3; LAMB3 |
| Cell junction organization | 5 | 90 | 4.86E-05 | 1.00E-03 | LAMC2; CADH1; LAMA3; LAMB3; FLNA |
Definition of abbreviations: DA, diacetyl; ECM, extracellular matrix; FDR, false discovery rate; TCF, T-cell factor.
Table 5.
Top 10 REACTOME Pathways Enriched in Proteins Down-Regulated in Apical by DA Exposure
| Pathway name | No. of Proteins | No. of Proteins in Pathway | P value | FDR | Proteins Identified |
|---|---|---|---|---|---|
| Regulation of complement cascade | 3 | 27 | 9.74E-05 | 9.74E-03 | CFAB; CD59; CO4A |
| Activation of C3 and C5 | 2 | 7 | 2.49E-04 | 1.25E-02 | CFAB; CO4A |
| Lipid digestion, mobilization, and transport | 3* | 70 | 1.54E-03 | 4.95E-02 | PLTP; CEL |
| HDL-mediated lipid transport | 2* | 20 | 1.98E-03 | 4.95E-02 | PLTP |
| Complement cascade | 4 | 201 | 4.00E-03 | 5.49E-02 | CLUS; CFAB; CD59; CO4A |
| Platelet degranulation | 3 | 105 | 4.82E-03 | 5.49E-02 | A1AT; CLUS; AACT |
| Lipoprotein metabolism | 2* | 32 | 4.95E-03 | 5.49E-02 | PLTP |
| Cargo concentration in the ER | 2 | 33 | 5.25E-03 | 5.49E-02 | A1AT; CD59 |
| Response to elevated platelet cytosolic Ca2+ | 3 | 110 | 5.49E-03 | 5.49E-02 | A1AT; CLUS; AACT |
| Utilization of ketone bodies | 1 | 3 | 9.75E-03 | 8.77E-02 | THIL |
| Lysosomal oligosaccharide catabolism | 1 | 4 | 1.30E-02 | 1.03E-01 | MANBA |
Definition of abbreviations: DA, diacetyl; ER, endoplasmic reticulum; FDR, false discovery rate; HDL, high-density lipoprotein; NB, nota bene.
NB: PLTP has two natural variants.
Table 6.
Top 10 REACTOME Pathways Enriched in Proteins Up-Regulated in Basolateral by DA Exposure
| Pathway Name | No. of Proteins | No. of Proteins in Pathway | P value | FDR | Proteins Identified |
|---|---|---|---|---|---|
| Platelet degranulation | 7 | 105 | 7.24E-06 | 1.90E-03 | ECM1; QSOX1; CAP1; TIMP1; APLP2; A4; TRFE |
| Response to elevated platelet cytosolic Ca2+ | 7 | 110 | 9.77E-06 | 1.90E-03 | ECM1; QSOX1; CAP1; TIMP1; APLP2; A4; TRFE |
| Activation of matrix metalloproteinases | 4 | 32 | 6.98E-05 | 9.07E-03 | TIMP2; MMP14; MMP9; TIMP1 |
| Degradation of the extracellular matrix | 6 | 135 | 3.06E-04 | 2.97E-02 | CATB; TIMP2; CATD; MMP14; MMP9; TIMP1 |
| Signaling by MST1 | 2 | 5 | 5.47E-04 | 4.27E-02 | SPIT2; SPIT1 |
| Extracellular matrix organization | 8 | 291 | 7.46E-04 | 4.85E-02 | CATB; TIMP2; CATD; MMP14; DDR1; MMP9; TIMP1; A4 |
| Collagen degradation | 4 | 63 | 8.97E-04 | 4.93E-02 | CATB; CATD; MMP14; MMP9 |
| Platelet activation, signaling, and aggregation | 7 | 256 | 1.68E-03 | 8.05E-02 | ECM1; QSOX1; CAP1; TIMP1; APLP2; A4; TRFE |
| Axon guidance | 10 | 551 | 3.67E-03 | 1.58E-01 | EPHA2; NEO1; PSB1; PSB6; CAP1; SPTB2; EFNB1; NRP1; MMP9; SEM7A |
| EPH-ephrin–mediated repulsion of cells | 3 | 50 | 4.79E-03 | 1.87E-01 | EPHA2; EFNB1; MMP9 |
Definition of abbreviations: DA, diacetyl; EPH, ephrin; FDR, false discovery rate; MST1, macrophage stimulating 1.
Table 7.
Top 10 REACTOME Pathways Enriched in Proteins Down-Regulated in Basolateral by DA Exposure
| Pathway Name | No. of Proteins | No. of Proteins in Pathway | P value | FDR | Proteins Identified |
|---|---|---|---|---|---|
| PTK6 promotes HIF1A stabilization | 2 | 6 | 1.49E-04 | 4.54E-02 | GPNMB; EGFR |
| Synthesis of IP2, IP, and Ins in the cytosol | 2 | 11 | 4.97E-04 | 4.83E-02 | IMPA2; INO1 |
| Assembly of collagen fibrils and other multimeric structures | 3 | 55 | 5.71E-04 | 4.83E-02 | CO5A2; CO7A1; CO1A1 |
| Collagen degradation | 3 | 63 | 8.45E-04 | 4.83E-02 | CO5A2; CO7A1; CO1A1 |
| Anchoring fibril formation | 2 | 15 | 9.18E-04 | 4.83E-02 | CO7A1; CO1A1 |
| Collagen biosynthesis and modifying enzymes | 3 | 66 | 9.66E-04 | 4.83E-02 | CO5A2; CO7A1; CO1A1 |
| Integrin cell–surface interactions | 3 | 86 | 2.06E-03 | 8.34E-02 | CO5A2; CO7A1; CO1A1 |
| Collagen formation | 3 | 88 | 2.19E-03 | 8.34E-02 | CO5A2; CO7A1; CO1A1 |
| Syndecan interactions | 2 | 27 | 2.91E-03 | 9.60E-02 | CO5A2; CO1A1 |
| Signaling by overexpressed wild-type EGFR in cancer | 1 | 2 | 5.88E-03 | 1.59E-01 | EGFR |
Definition of abbreviations: DA, diacetyl; EGFR, epidermal growth factor receptor; FDR, false discovery rate; HIF1A, hypoxia inducible factor 1A; Ins, inositol; IP, inositol phosphate; PTK6, protein tyrosine kinase 6.
DA Exposure of Primary Human Airway Epithelial Cells in ALI Culture Results in Dysregulation of ECM Organization and EGF Receptor Activation and Signaling
To further identify relationships between the differentially expressed proteins in our data set, we used the STRING suite of online analysis tools to identify known protein–protein interactions (13). Several nodal interactions were revealed in the apical secretome, including those between MMP9, MMP10, TIMP1 and tenascin C (TENA); between proteasome subunit-α types 3 and 7 (PSMA3 and PSMA7) and puromycin-sensitive aminopeptidase (NPEPPS); between the laminins A3, B3 and C2; and between the histone proteins H2B1, H2AY and histone cluster 4 (Figure E4).
In our STRING analysis of protein associations within the basolateral secretome, we observed a nodal interaction centered on the EGF receptor (EGFR) and proteins that have been shown or hypothesized to regulate the EGFR (Figure E5). This is consistent with our previous report that DA exposure causes shedding of amphiregulin, a canonical ligand for the EGFR (9). In our proteomic analysis, FBLN3, DDB1, GDF15, and ECM1 were significantly differentially expressed. FBLN3 has been shown to activate the EGFR in pancreatic carcinoma cells (14). DDB1 has been shown to contribute to EGFR signaling attenuation in Caenorhabditis elegans (15). GDF15 is a member of the transforming growth factor β (TGF-β) superfamily of proteins that has been shown to potentiate EGFR signaling in the hippocampus in mice (16). ECM1 is a member of the IL-1 family of proteins that associates with perlecan in the basement membrane and can potentiate EGFR signaling (17).
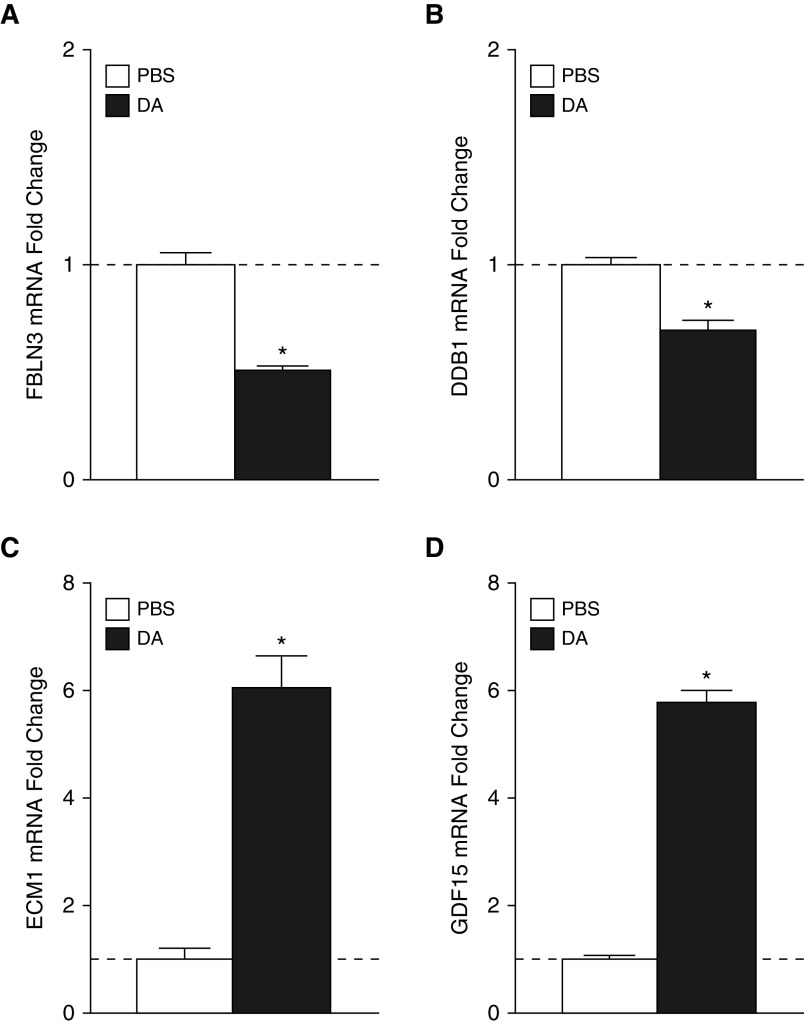
To further investigate the importance of this pathway, we measured individual intracellular transcript levels by RT-PCR. We show in Figure 4 that DA exposure induced significant decreases in mRNA for FBLN3 and DDB1 (Figures 4A–4C), which are known or hypothesized negative regulators of EGFR signaling. Similarly, mRNA for ECM1 and GDF15 (Figures 4D and 4E), which have been shown to potentiate EGFR signaling, was increased. These changes in protein secretion and intracellular transcript levels suggest that DA exposure of primary human airway epithelial cells in ALI culture results in altered EGFR signaling.
Figure 4.
Known or hypothesized modifiers of EGFR signaling show mRNA expression consistent with protein changes identified by proteomic analysis. FBLN3 (A) and DDB1 (B) mRNA expression is significantly attenuated by DA exposure, whereas ECM1 (C) and GDF15 (D) mRNA expression is significantly increased by DA exposure. *P < 0.05 versus PBS. DDB1, DNA damage-binding protein 1; ECM1, extracellular matrix protein 1; EGFR, epidermal growth factor receptor; FBLN3, EGF-containing fibulin-like extracellular matrix protein 1; GDF15: growth/differentiation factor 15.
Discussion
Clinical and experimental evidence links the development of BO to DA exposure. However, little is known about the underlying biological mechanisms that contribute to disease pathogenesis. In a recently published study (10), we added to our understanding of early events after DA vapor exposure by showing that fully differentiated human airway epithelial cells grown in ALI culture lose cilia and appear to dedifferentiate to a squamous-like phenotype. As in that study, here we focused on airway epithelial cells because they are the first cell type to be encountered by inhaled toxins such as DA. To begin to understand the processes that might occur after DA exposure, we exposed fully differentiated primary airway epithelial cells from four unique donors to DA vapor concentrations or PBS as a control to model repeated occupational exposure. We then used discovery-based proteomics to elucidate DA-induced airway epithelial secretory responses in the apical and basolateral compartments. This study shows that the apical and basolateral secretomes of DA-exposed airway epithelial cells in ALI culture are highly polarized and distinct, with minimal overlap. Furthermore, we demonstrated that the epithelial secretory response to DA in four independent donors was remarkably consistent, particularly in the apical secretome. Finally, we identified novel proteins in both the apical and basolateral secretomes that provide new insights into mechanisms that may drive the development of BO.
In the present analysis, we identified many proteins that have not been previously associated with BO but could play a biologically plausible role in the pathogenesis of this disease. For example, CXCL16 showed increased expression in the basolateral secretome. Elevated CXCL16 has previously been associated with epithelial-derived lung cancers and has been shown to modulate the expression of MMP and TIMP (18), including MMP9 and TIMP2, both of which showed altered expression in our data set. Similarly, IL-36γ showed increased expression in the basolateral secretome (Table 2). IL-36γ is a member of the IL-1 superfamily of cytokines and has been shown to promote inflammation or fibrosis in chronic fibrotic skin (19) and bowel disease (20), but not in lung disease. However, exogenous IL-36 instillation in the rodent lung has been associated with neutrophil recruitment (19, 21), a common feature of BO in preclinical rodent models and in human BO.
A particularly novel aspect of the present study is that proteins and transcripts known or thought to negatively regulate EGFR signaling, including FBLN3 and DDB1, were decreased, whereas those known or thought to increase EGFR signaling, including ECM1 and GDF15, were increased. These results strongly suggest that EGFR signaling may be highly active in DA-exposed epithelial cells. We previously showed that after DA instillation in rats, significant epithelial injury and a rapid, potentially aberrant repair process preceded lesion development. Similarly, we showed that DA exposure of fully differentiated primary human airway epithelial cells in culture shed robust quantities of the EGFR ligand amphiregulin into the culture media. It is well established that EGFR signaling is important for epithelial regeneration and repair after injury. Therefore, this is consistent with the observation from our data set that negative regulators of EGFR signaling are decreased while positive regulators of EGFR signaling are increased during the repair process that begins after DA exposure. In two separate reports, FBLN3 was hypothesized to bind to the EGF binding site in the EGFR (14, 22). DDB1 is another protein that has been identified in both apical and basolateral secretomes. DDB1 has been shown in C. elegans to be part of a ubiquitin ligase complex that negatively regulates EGFR signaling (15). Conversely, ECM1 was recently hypothesized to facilitate signaling through the EGFR (17). GDF15 is a member of the TGF-β superfamily of growth factors and has been hypothesized to be a positive regulator of EGFR signaling (16). Although it does not meet analytical requirements for low technical variability, PTPRS was the most robustly down-regulated protein in the basolateral secretome and is a negative regulator of EGFR signaling (48.29% coefficient of variation and −46.32 average fold change). PTPRS interacts directly with and dephosphorylates the EGFR (23, 24), and genetic ablation of PTPRS has been associated with robust EGFR-PI3K pathway activation (25). The observation that both negative and positive regulators of EGFR signaling were dysregulated strongly supports the notion that EGFR signaling plays an important role in the airway epithelial response to DA exposure.
Another novel observation in the current data set was revealed by the REACTOME analyses, which showed that proteins involved in matrix organization, degradation, and turnover were abundantly present in both the apical and basolateral secretomes. As an example, we identified PGBM (perlecan) to be robustly increased in the apical secretome. Perlecan is a basement membrane protein that has been hypothesized to be fundamental for organ and tissue integrity (26), and has been shown to have increased expression in BO after lung transplantation (27). Similar to our observation that perlecan was differentially expressed in the apical secretome, three other components of the basement membrane—LAMA3, LAMB3, and LAMC2—were differentially expressed in the apical secretome (Table 1). Interestingly, the fibrillar collagens that make up the basement membrane, including CO5A2, CO7A1, and CO1A1 (28), showed decreased expression in the basolateral secretome (Table 7), whereas proteins that are known to degrade ECM and facilitate matrix turnover, including MMP9, TIMP1, and TIMP2, were increased. DDR1 is another novel protein identified in our data set. DDR1 is a cell-surface receptor for fibrillar collagen, with tyrosine kinase activity that regulates cell attachment to and remodeling of the ECM, as well as cell migration, differentiation, survival, and proliferation (29–34). Taken together, these observations support the notion that soluble factors derived from the airway epithelium could drive BO lesion development by fundamentally altering the matrix in the subepithelial space. Further studies exploring the effects of secreted soluble proteins directly on fibroblasts would be of interest to elucidate the mechanism by which DA-induced epithelial injury contributes to airway fibrosis in BO.
Although the current data support the notion that the airway epithelium could play an active role in driving remodeling of the matrix in developing BO lesions, a number of matrix-remodeling proteins identified in our analysis have already been implicated in BO, thus providing additional validation for the biological relevance of our results. For example, MMP-9 and TIMP1 were elevated in both the apical and basolateral secretomes after DA exposure, and have previously been observed to be increased in BO, particularly in the clinical context of lung transplantation and in preclinical experimental models (35). Similarly, we previously showed experimentally in our rodent model of DA-induced BO that TENA, which was up-regulated apically in the current study, had increased protein expression in the subepithelial space of developing BO lesions (36). These observations that independently replicate our results in other preclinical and clinical studies of BO further support the notion that the airway epithelium may actively participate in directing matrix remodeling after DA exposure.
Similarly to other in vitro investigations, this study has clear strengths and limitations. A unique strength of the current study is that we used brief, repeated exposures to DA vapor to replicate exposure to occupationally relevant concentrations of inhaled DA. Another strength of the present study is that we examined the response of airway epithelial cells from four independent human donors. Because human subjects are genetically diverse, this approach had the potential to yield highly variable results. However, we observed that the responses of the individual donors were strikingly consistent, making our results much more generalizable than those obtained in many previous studies that relied on cells from a single human donor. Despite these strengths, we recognize that our work also has several limitations. First, although growing primary airway epithelial cells at the ALI accurately reproduces their in vivo environment, the basement membrane they synthesize is likely incomplete when compared with the basement membrane on which they reside in vivo. Similarly, the mesenchymal cells that would normally lie beneath this basement membrane are absent. This was also an advantage because we were able to evaluate the responses attributed only to the epithelium. In vitro proteomic studies on ALI epithelial cultures, in which basement-membrane–like and underlying mesenchymal structures are present, are now possible and will be pursued in the future. Second, we did not perform a direct validation of specific differentially expressed proteins. However, we did directly validate intracellular transcript changes that correlated with changes in proteins known or hypothesized to modulate EGFR signaling. In addition, several proteins we identified as elevated (MMP9 and TIMP1) have also been reported to be elevated in preclinical, providing additional confidence in the current results. Third, some of the observed differentially expressed proteins could be an indicator of cell injury rather than active secretion, which could be consistent with epithelial changes observed in our previous in vitro (10) and in vivo (36) studies. The presence of histone cluster proteins, which are associated with ubiquitination and DDB1, is consistent with this idea and with previous reports showing that DA causes changes in epithelial barrier function (37) or ion transport (38), and that it can form adducts with 2-deoxyguanosine (39) that lead to cell death or epithelial apoptosis (40). Finally, we recognize the limitations of resolution inherent to proteomic technology, and we acknowledge that low-abundance proteins may be below the limits of detection.
In summary, we have observed polarized and highly regulated changes in protein expression in response to DA vapor exposure of human airway epithelial cells. Importantly, the significant changes in protein expression observed in the apical and basolateral secretomes were reproduced in all four independent human donors. In response to DA, the secretome was enriched for proteins that are associated with matrix remodeling and regulate signaling through the EGFR. Taken as a whole, these results support the notion that the epithelium may actively direct the fibroproliferative response of the underlying mesenchyme and may serve as a regulator of BO pathobiology. The present results further suggest several potential proteins and/or pathways that could be targeted in future studies to further understand disease pathobiology or to intervene in early disease development. In addition, our work provides several novel protein targets that could be pursued as potential biomarkers of DA exposure to identify workers at higher risk for BO.
Footnotes
This work was supported by National Institutes of Health grant R21-OH010490 and the Lung Transplant Foundation.
Author Contributions: D.M.B. contributed to the experimental design and data analysis, and wrote the manuscript. W.M.G. and F.L.K. performed the cell culture. A.M.V. performed statistical analyses. C.D.B. and A.E.N. performed laboratory analyses of samples. M.A.M. and D.L.M. provided oversight and contributed to the experimental design. S.M.P. and M.W.F. performed proteomic analyses and contributed to oversight, experimental design, and data analysis and interpretation. All co-authors substantially revised the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2016-0372OC on March 1, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Kreiss K, Gomaa A, Kullman G, Fedan K, Simoes EJ, Enright PL. Clinical bronchiolitis obliterans in workers at a microwave-popcorn plant. N Engl J Med. 2002;347:330–338. doi: 10.1056/NEJMoa020300. [DOI] [PubMed] [Google Scholar]
- 2.Lockey JE, Hilbert TJ, Levin LP, Ryan PH, White KL, Borton EK, Rice CH, McKay RT, LeMasters GK. Airway obstruction related to diacetyl exposure at microwave popcorn production facilities. Eur Respir J. 2009;34:63–71. doi: 10.1183/09031936.00050808. [DOI] [PubMed] [Google Scholar]
- 3.Akpinar-Elci M, Travis WD, Lynch DA, Kreiss K. Bronchiolitis obliterans syndrome in popcorn production plant workers. Eur Respir J. 2004;24:298–302. doi: 10.1183/09031936.04.00013903. [DOI] [PubMed] [Google Scholar]
- 4.Kim TJ, Materna BL, Prudhomme JC, Fedan KB, Enright PL, Sahakian NM, Windham GC, Kreiss K. Industry-wide medical surveillance of California flavor manufacturing workers: cross-sectional results. Am J Ind Med. 2010;53:857–865. doi: 10.1002/ajim.20858. [DOI] [PubMed] [Google Scholar]
- 5.van Rooy FG, Rooyackers JM, Prokop M, Houba R, Smit LA, Heederik DJ. Bronchiolitis obliterans syndrome in chemical workers producing diacetyl for food flavorings. Am J Respir Crit Care Med. 2007;176:498–504. doi: 10.1164/rccm.200611-1620OC. [DOI] [PubMed] [Google Scholar]
- 6.Foster MW, Morrison LD, Todd JL, Snyder LD, Thompson JW, Soderblom EJ, Plonk K, Weinhold KJ, Townsend R, Minnich A, et al. Quantitative proteomics of bronchoalveolar lavage fluid in idiopathic pulmonary fibrosis. J Proteome Res. 2015;14:1238–1249. doi: 10.1021/pr501149m. [DOI] [PubMed] [Google Scholar]
- 7.Peters-Hall JR, Brown KJ, Pillai DK, Tomney A, Garvin LM, Wu X, Rose MC. Quantitative proteomics reveals an altered cystic fibrosis in vitro bronchial epithelial secretome. Am J Respir Cell Mol Biol. 2015;53:22–32. doi: 10.1165/rcmb.2014-0256RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillai DK, Sankoorikal BJ, Johnson E, Seneviratne AN, Zurko J, Brown KJ, Hathout Y, Rose MC. Directional secretomes reflect polarity-specific functions in an in vitro model of human bronchial epithelium. Am J Respir Cell Mol Biol. 2014;50:292–300. doi: 10.1165/rcmb.2013-0188OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly FL, Sun J, Fischer BM, Voynow JA, Kummarapurugu AB, Zhang HL, Nugent JL, Beasley RF, Martinu T, Gwinn WM, et al. Diacetyl induces amphiregulin shedding in pulmonary epithelial cells and in experimental bronchiolitis obliterans. Am J Respir Cell Mol Biol. 2014;51:568–574. doi: 10.1165/rcmb.2013-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster MW, Gwinn WM, Kelly FL, Brass DM, Valente AM, Moseley MA, Thompson JW, Morgan DL, Palmer SM. Proteomic analysis of primary human airway epithelial cells exposed to the respiratory toxicant diacetyl. J Proteome Res. 2017;16:538–549. doi: 10.1021/acs.jproteome.6b00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benjamini Y. Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 12.Haw R, Hermjakob H, D’Eustachio P, Stein L. Reactome pathway analysis to enrich biological discovery in proteomics data sets. Proteomics. 2011;11:3598–3613. doi: 10.1002/pmic.201100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camaj P, Seeliger H, Ischenko I, Krebs S, Blum H, De Toni EN, Faktorova D, Jauch KW, Bruns CJ. EFEMP1 binds the EGF receptor and activates MAPK and Akt pathways in pancreatic carcinoma cells. Biol Chem. 2009;390:1293–1302. doi: 10.1515/BC.2009.140. [DOI] [PubMed] [Google Scholar]
- 15.Poulin GB, Ahringer J. The Caenorhabditis elegans CDT-2 ubiquitin ligase is required for attenuation of EGFR signalling in vulva precursor cells. BMC Dev Biol. 2010;10:109. doi: 10.1186/1471-213X-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrillo-García C, Prochnow S, Simeonova IK, Strelau J, Hölzl-Wenig G, Mandl C, Unsicker K, von Bohlen Und Halbach O, Ciccolini F. Growth/differentiation factor 15 promotes EGFR signalling, and regulates proliferation and migration in the hippocampus of neonatal and young adult mice. Development. 2014;141:773–783. doi: 10.1242/dev.096131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee KM, Nam K, Oh S, Lim J, Kim YP, Lee JW, Yu JH, Ahn SH, Kim SB, Noh DY, et al. Extracellular matrix protein 1 regulates cell proliferation and trastuzumab resistance through activation of epidermal growth factor signaling. Breast Cancer Res. 2014;16:479. doi: 10.1186/s13058-014-0479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mir H, Singh R, Kloecker GH, Lillard JW, Jr, Singh S. CXCR6 expression in non-small cell lung carcinoma supports metastatic process via modulating metalloproteinases. Oncotarget. 2015;6:9985–9998. doi: 10.18632/oncotarget.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabay C, Towne JE. Regulation and function of interleukin-36 cytokines in homeostasis and pathological conditions. J Leukoc Biol. 2015;97:645–652. doi: 10.1189/jlb.3RI1014-495R. [DOI] [PubMed] [Google Scholar]
- 20.Medina-Contreras O, Harusato A, Nishio H, Flannigan KL, Ngo V, Leoni G, Neumann PA, Geem D, Lili LN, Ramadas RA, et al. Cutting edge: IL-36 receptor promotes resolution of intestinal damage. J Immunol. 2016;196:34–38. doi: 10.4049/jimmunol.1501312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramadas RA, Ewart SL, Iwakura Y, Medoff BD, LeVine AM. IL-36α exerts pro-inflammatory effects in the lungs of mice. PLoS One. 2012;7:e45784. doi: 10.1371/journal.pone.0045784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Gao H, Vo C, Ke C, Pan F, Yu L, Siegel E, Hess KR, Linskey ME, Zhou YH. Anti-EGFR function of EFEMP1 in glioma cells and patient prognosis. Oncoscience. 2014;1:205–215. doi: 10.18632/oncoscience.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suárez Pestana E, Tenev T, Gross S, Stoyanov B, Ogata M, Böhmer FD. The transmembrane protein tyrosine phosphatase RPTPσ modulates signaling of the epidermal growth factor receptor in A431 cells. Oncogene. 1999;18:4069–4079. doi: 10.1038/sj.onc.1202794. [DOI] [PubMed] [Google Scholar]
- 24.Vijayvargia R, Kaur S, Krishnasastry MV. α-Hemolysin-induced dephosphorylation of EGF receptor of A431 cells is carried out by rPTPσ. Biochem Biophys Res Commun. 2004;325:344–352. doi: 10.1016/j.bbrc.2004.10.038. [DOI] [PubMed] [Google Scholar]
- 25.Morris LG, Taylor BS, Bivona TG, Gong Y, Eng S, Brennan CW, Kaufman A, Kastenhuber ER, Banuchi VE, Singh B, et al. Genomic dissection of the epidermal growth factor receptor (EGFR)/PI3K pathway reveals frequent deletion of the EGFR phosphatase PTPRS in head and neck cancers. Proc Natl Acad Sci USA. 2011;108:19024–19029. doi: 10.1073/pnas.1111963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farach-Carson MC, Warren CR, Harrington DA, Carson DD. Border patrol: insights into the unique role of perlecan/heparan sulfate proteoglycan 2 at cell and tissue borders. Matrix Biol. 2014;34:64–79. doi: 10.1016/j.matbio.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersson-Sjöland A, Thiman L, Nihlberg K, Hallgren O, Rolandsson S, Skog I, Mared L, Hansson L, Eriksson L, Bjermer L, et al. Fibroblast phenotypes and their activity are changed in the wound healing process after lung transplantation. J Heart Lung Transplant. 2011;30:945–954. doi: 10.1016/j.healun.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Sakai LY, Keene DR, Morris NP, Burgeson RE. Type VII collagen is a major structural component of anchoring fibrils. J Cell Biol. 1986;103:1577–1586. doi: 10.1083/jcb.103.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leitinger B. Molecular analysis of collagen binding by the human discoidin domain receptors, DDR1 and DDR2. Identification of collagen binding sites in DDR2. J Biol Chem. 2003;278:16761–16769. doi: 10.1074/jbc.M301370200. [DOI] [PubMed] [Google Scholar]
- 30.Vogel W, Gish GD, Alves F, Pawson T. The discoidin domain receptor tyrosine kinases are activated by collagen. Mol Cell. 1997;1:13–23. doi: 10.1016/s1097-2765(00)80003-9. [DOI] [PubMed] [Google Scholar]
- 31.Shrivastava A, Radziejewski C, Campbell E, Kovac L, McGlynn M, Ryan TE, Davis S, Goldfarb MP, Glass DJ, Lemke G, et al. An orphan receptor tyrosine kinase family whose members serve as nonintegrin collagen receptors. Mol Cell. 1997;1:25–34. doi: 10.1016/s1097-2765(00)80004-0. [DOI] [PubMed] [Google Scholar]
- 32.Leitinger B. Transmembrane collagen receptors. Annu Rev Cell Dev Biol. 2011;27:265–290. doi: 10.1146/annurev-cellbio-092910-154013. [DOI] [PubMed] [Google Scholar]
- 33.Fu H-L, Valiathan RR, Arkwright R, Sohail A, Mihai C, Kumarasiri M, Mahasenan KV, Mobashery S, Huang P, Agarwal G, et al. Discoidin domain receptors: unique receptor tyrosine kinases in collagen-mediated signaling. J Biol Chem. 2013;288:7430–7437. doi: 10.1074/jbc.R112.444158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leitinger B. Discoidin domain receptor functions in physiological and pathological conditions. Int Rev Cell Mol Biol. 2014;310:39–87. doi: 10.1016/B978-0-12-800180-6.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy VE, Todd JL, Palmer SM. Bronchoalveolar lavage as a tool to predict, diagnose and understand bronchiolitis obliterans syndrome. Am J Transplant. 2013;13:552–561. doi: 10.1111/ajt.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer SM, Flake GP, Kelly FL, Zhang HL, Nugent JL, Kirby PJ, Foley JF, Gwinn WM, Morgan DL. Severe airway epithelial injury, aberrant repair and bronchiolitis obliterans develops after diacetyl instillation in rats. PLoS One. 2011;6:e17644. doi: 10.1371/journal.pone.0017644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fedan JS, Dowdy JA, Fedan KB, Hubbs AF. Popcorn worker’s lung: in vitro exposure to diacetyl, an ingredient in microwave popcorn butter flavoring, increases reactivity to methacholine. Toxicol Appl Pharmacol. 2006;215:17–22. doi: 10.1016/j.taap.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Zaccone EJ, Goldsmith WT, Shimko MJ, Wells JR, Schwegler-Berry D, Willard PA, Case SL, Thompson JA, Fedan JS. Diacetyl and 2,3-pentanedione exposure of human cultured airway epithelial cells: ion transport effects and metabolism of butter flavoring agents. Toxicol Appl Pharmacol. 2015;289:542–549. doi: 10.1016/j.taap.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.More SS, Raza A, Vince R. The butter flavorant, diacetyl, forms a covalent adduct with 2-deoxyguanosine, uncoils DNA, and leads to cell death. J Agric Food Chem. 2012;60:3311–3317. doi: 10.1021/jf300180e. [DOI] [PubMed] [Google Scholar]
- 40.Hubbs AF, Fluharty KL, Edwards RJ, Barnabei JL, Grantham JT, Palmer SM, Kelly F, Sargent LM, Reynolds SH, Mercer RR, et al. Accumulation of ubiquitin and sequestosome-1 implicate protein damage in diacetyl-induced cytotoxicity. Am J Pathol. 2016;186:2887–2908. doi: 10.1016/j.ajpath.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]