Abstract
Background
Hantaan and Seoul viruses, in the Hantavirus genus, are known to cause hemorrhagic fever with renal syndrome (HFRS). The plaque reduction neutralization test (PRNT), as conventional neutralization test for hantaviruses, is laborious and time-consuming. Alternatives to PRNT for hantaviruses are required.
Methods
In this study, the methods for Hantaan and Seoul viruses serological typing including microneutralization test (MNT), pseudoparticle neutralization test (PPNT) and immunofluorescence assay based on viral glycoproteins (IFA-GP) were developed and compared with PRNT using a panel of 74 sera including 44 convalescent sera of laboratory confirmed HFRS patients and 30 patients sera of non-hantavirus infection. Antibody titres and serotyping obtained with different methods above were analyzed by paired-t, linear correlation, McNemar χ2 and Kappa agreement tests.
Results
Antibody titres obtained with MNT50, PPNT50 and IFA-GP were significantly correlated with that obtained with PRNT50 (p < 0.001). GMT determined by PPNT50 was statistically higher than that determined by PRNT50 (p < 0.001), while GMT determined by MNT50 and IFA-GP were equal with (p > 0.05) and less than (p < 0.001) that obtained with PRNT50 respectively. Serotyping obtained with MNT50 and PRNT50, PPNT50 and PRNT50 were highly consistent (p < 0.001), whereas that obtained with IFA-GP and PRNT50 were moderately consistent (p < 0.001). There were no significant differences for serotyping between PRNT50 and MNT50, as well as PRNT50 and PPNT50 (p > 0.05). IFA-GP was less sensitive than PRNT50 and MNT50 for serotyping of hantaviruses infection (p < 0.05). However, for 79.5% (35/44) samples, serotyping determined by IFA-GP and PRNT50 were consistent.
Conclusions
MNT50 and PPNT50 both can be used as simple and rapid alternatives to PRNT50, and MNT50 is more specific while PPNT50 is more sensitive than other assays for neutralizing antibody determination. So far, this work has been the most comprehensive comparison of alternatives to PRNT.
Keywords: Hantaan, Seoul, Serotyping, Plaque reduction neutralization test, Microneutralization test, Pseudoparticle neutralization test, Immunofluorescence assay, Glycoproteins
Background
Hantaviruses belong to the Hantavirus genus in the Bunyaviridae family [1]. Hantaviruses are enveloped, negative-stranded RNA viruses containing three single-stranded RNA genome segments designated as small (S), medium (M) and large (L); they encode nucleocapsid protein (N), envelope glycoproteins (Gn and Gc) and RNA-dependent RNA polymerase, respectively [2, 3]. Among the viral proteins, nucleocapsid protein possesses an immunodominant antigen, and the antigenicitiy of N protein is conserved compared with that of envelope glycoproteins [4, 5]. Gn and Gc form oligomers on the surface of the virion and are the targets of neutralizing antibodies [6–8].
Hantavirus causes two human diseases: hemorrhagic fever with renal syndrome (HFRS) in Eurasia and hantavirus pulmonary syndrome (HPS) in the Americas. At least four hantaviruses cause HFRS: Hantaan, Seoul, Puumala, and Dobrava viruses caused most of HFRS cases in Eurasia [9, 10]. Hantaan virus (HTNV) and Seoul virus (SEOV) are major causative agents of HFRS in China [11], During the last decade, about 10,000 cases of HFRS were registered annually in China [12]. In general, hantaviruses are host-restricted that Hantaan virus isolates are carried by Apodemus and Seoul virus isolates by Rattus [1].
The plaque reduction neutralization test (PRNT), is laborious and time-consuming (takes about 2 ~ 3 weeks), and is unsuitable for high-throughput testing [13–15]. Therefore, alternative methods to PRNT are needed.
Microneutralization test (MNT) has been developed for viruses such as influenza virus, Puumala virus, etc. [16–22]. By using 96-well microplates in combination with enzyme immunoassay, MNT is simple, rapid, and adaptable to high-throughput formats.
Pseudotyped reporter viruses containing the envelope glycoprotein of one virus and the core and genome of vector virus such as vesicular stomatitis virus (VSV), murine leukemia virus (MuLV) or lentivirus have been developed for many other viruses [23–27]. Since pseudoparticle is unable to produce infectious progeny viruses unless the envelope proteins are provided in trans, pseudoparticle neutralization test (PPNT) is a safe alternative to neutralization test using live viruses. Pseudoparticles bearing glycoproteins of hantaviruses have also been developed and used in PPNT [28–33] for hantaviruses.
Here, we compared the MNT and PPNT data with those obtained with PRNT using 44 convalescent sera from laboratory confirmed patients of HFRS and 30 sera negative for hantavirus infection. Moreover, the effective expressions of glycoproteins of HTNV and SEOV in 293T cells enable us to develop a method of immunofluorescence assay based on viral glycoproteins (IFA-GP) to detect antibody titres against recombinant glycoproteins of the two viruses. The IFA-GP titres may correlate with the neutralizing antibody titres obtained by PRNT, thus IFA-GP has the possibility to be a simpler alternative to PRNT. Here, results obtained with IFA-GP were also compared with that obtained with PRNT using the same panel of sera mentioned above.
Methods
Cells, viruses, antibody
Vero-E6 cells (ATCC, C1008 CRL1586) were propagated in growth medium (Eagle’s MEM supplemented with 10% heat-inactivated fetal bovine serum [FBS], 2 mM L-glutamine, 100 U/ml Penicillin, 100 μg/ml Streptomycin, and 1.5 g/L Sodium Bicarbonate solution). HEK 293T human embryo kidney cells (ATCC, CRL11268) and Huh-7.5 human hepatocellular carcinoma cells [34] were propagated in Dulbecco’s Modified Eagle Medium [DMEM] containing 10% heat-inactivated FBS, 100 U/ml Penicillin, 100 μg/ml Streptomycin.
HTNV strain 84FLi and SEOV strain L99 maintained in our laboratory were propagated on Vero E6 cells.
The mouse monoclonal antibodies (mAbs) L13F3 directed against N protein of SEOV and HTNV were generated in our laboratory [35]. Mouse mAbs 8B6 directed against hantavirus Gn glycoprotein [6] and human recombinant mAbs Y5 directed against hantavirus Gc glycoprotein [36] were stored in our laboratory.
Serum samples
A panel of 74 human sera was used in this study, including 44 convalescent sera from laboratory confirmed patients of HFRS in China, 15 sera from healthy individuals and 15 sera from patients of dengue fever or severe fever with thrombocytopenia syndrome which were negative for hantavirus infection. Sera from patients of HFRS were tested by recombinant nucleocapsid proteins based IgM capture ELISA [37] or IgG ELISA [38] for HTNV and SEOV. Hantavirus infection was confirmed by hantavirus IgM positive or IgG titre ≥4-fold increase between acute-phase and convalescent-phase sera. Real-time RT-PCR developed in our laboratory by Pang et al. [39] was used to differentiate infection of HTNV or SEOV. As shown in Table 1, all 44 HFRS patients were IgM antibodies against hantavirus positive in acute phase sera. 33 of the 44 acute phase sera were tested using real-time RT-PCR, only 5 samples were confirmed to be HTNV or SEOV infection.
Table 1.
PRNT, MNT, PPNT, IFA-virus and IFA-GP titres of convalescent sera from patients of HFRS
| Sero-typea | Patient ID | Diagnosis of HFRSb | Reciprocal PRNT titrec | Reciprocal MNT titrec | Reciprocal PPNT titrec | Reciprocal IFA titrec | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acute serum | Convalescent serum | IC50 | IC80 | IC50 | IC80 | IC50 | IC80 | IFA-virus | IFA-GP | ||||||||||||||
| IgM | IgG | RNA | IgG | SEOV | HTNV | SEOV | HTNV | SEOV | HTNV | SEOV | HTNV | SEOV | HTNV | VSV | SEOV | HTNV | VSV | SEOV | HTNV | SEOV | HTNV | ||
| HTN | SD11 | + | + | NT | + | 20 | 80 | <20 | 20 | 20 | 80 | <20 | 20 | 80 | 320 | <20 | 20 | 80 | <20 | 80 | 320 | 20 | 80 |
| JX46 | + | + | − | + | <20 | 80 | <20 | 20 | <20 | 80 | <20 | 80 | 80 | 320 | <20 | 20 | 80 | <20 | 80 | 80 | 80 | 80 | |
| SD15 | + | + | NT | + | <20 | 80 | <20 | 20 | <20 | 80 | <20 | 80 | 80 | 320 | <20 | 20 | 80 | <20 | 80 | 80 | 20 | 80 | |
| LN264 | + | − | NT | + | <20 | 80 | <20 | 20 | <20 | 80 | <20 | 20 | 20 | 320 | <20 | 20 | 80 | <20 | 80 | 80 | <20 | 20 | |
| JX11 | + | + | − | + | 20 | 80 | <20 | 80 | <20 | 80 | <20 | 80 | 20 | 320 | 20 | <20 | 80 | <20 | 80 | 80 | 20 | 80 | |
| JX16 | + | − | − | + | 20 | 320 | <20 | 80 | 20 | 320 | <20 | 80 | 80 | 1280 | <20 | 20 | 320 | <20 | 80 | 80 | 20 | 80 | |
| JX53 | + | + | − | + | 20 | 320 | <20 | 80 | 20 | 320 | 20 | 80 | 80 | 320 | <20 | 20 | 80 | <20 | 20 | 20 | 20 | 80 | |
| JX39 | + | + | − | + | 20 | 320 | <20 | 80 | <20 | 320 | <20 | 80 | 20 | 1280 | <20 | 20 | 320 | <20 | 80 | 80 | 20 | 80 | |
| JX36 | + | + | − | + | <20 | 320 | <20 | 80 | <20 | 320 | <20 | 80 | 20 | 320 | 20 | 20 | 80 | <20 | 80 | 320 | 20 | 80 | |
| SD7 | + | + | − | + | <20 | 320 | <20 | 80 | <20 | 320 | <20 | 80 | 320 | 320 | <20 | 80 | 80 | <20 | <20 | 80 | 80 | 80 | |
| SD14 | + | + | − | + | <20 | 320 | <20 | 80 | <20 | 320 | <20 | 80 | 80 | 1280 | <20 | 20 | 320 | <20 | <20 | 80 | 20 | 80 | |
| SD39 | + | + | − | + | <20 | 320 | <20 | 80 | <20 | 80 | <20 | 80 | 80 | 320 | <20 | 20 | 80 | <20 | 80 | 80 | 20 | 80 | |
| SD21 | + | + | H+ | + | 20 | 320 | 20 | 80 | <20 | 320 | <20 | 80 | 320 | 1280 | <20 | 80 | 320 | <20 | 320 | 320 | 80 | 80 | |
| SD31 | + | + | − | + | 20 | 320 | <20 | 80 | 20 | 320 | <20 | 80 | 320 | 5120 | <20 | 80 | 320 | <20 | 1280 | 1280 | 80 | 320 | |
| SD38 | + | + | − | + | <20 | 320 | <20 | 80 | <20 | 320 | <20 | 80 | 80 | 1280 | <20 | 20 | 320 | <20 | 80 | 80 | 20 | 80 | |
| JX12 | + | + | − | + | <20 | 320 | <20 | 80 | <20 | 320 | <20 | 80 | 80 | 1280 | <20 | 20 | 320 | <20 | 20 | 20 | 20 | 80 | |
| JX60 | + | + | NT | + | 20 | 1280 | <20 | 320 | 20 | 1280 | <20 | 320 | 80 | 5120 | <20 | 20 | 1280 | <20 | 320 | 320 | 80 | 320 | |
| JX35 | + | + | − | + | <20 | 1280 | <20 | 320 | 20 | 1280 | <20 | 320 | 320 | 1280 | <20 | 80 | 320 | <20 | 80 | 80 | 20 | 80 | |
| SD27 | + | + | − | + | 20 | 1280 | 20 | 320 | 20 | 5120 | 20 | 1280 | 80 | 5120 | <20 | 20 | 1280 | <20 | 80 | 320 | 80 | 320 | |
| SD25 | + | + | − | + | 80 | 1280 | 20 | 320 | 80 | 1280 | 20 | 320 | 320 | 1280 | <20 | 80 | 320 | <20 | 320 | 320 | 80 | 320 | |
| SD33 | + | − | − | + | 80 | 1280 | 20 | 320 | 20 | 1280 | <20 | 320 | 320 | 1280 | <20 | 80 | 320 | <20 | 320 | 320 | 80 | 320 | |
| SD36 | + | + | − | + | <20 | 1280 | <20 | 320 | 20 | 1280 | <20 | 320 | 320 | 1280 | <20 | 80 | 320 | <20 | 80 | 80 | 80 | 80 | |
| SD26 | + | + | − | + | <20 | 1280 | <20 | 320 | <20 | 1280 | <20 | 320 | 80 | 1280 | <20 | 20 | 320 | <20 | 20 | 80 | 20 | 80 | |
| JX3 | + | + | − | + | 20 | 1280 | <20 | 320 | <20 | 1280 | <20 | 320 | 80 | 1280 | <20 | 20 | 320 | <20 | 320 | 320 | 80 | 320 | |
| SD29 | + | + | H+ | + | 80 | 1280 | 20 | 320 | 20 | 1280 | 20 | 320 | 320 | 5120 | 20 | 80 | 1280 | <20 | 80 | 320 | 20 | 80 | |
| SD20 | + | + | − | + | 320 | 1280 | 80 | 320 | 320 | 1280 | 80 | 320 | 1280 | 5120 | <20 | 320 | 1280 | <20 | 80 | 80 | 320 | 320 | |
| LN34 | + | + | NT | + | 1280 | 20,480 | 320 | 5120 | 1280 | 20,480 | 320 | 5120 | 5120 | 20,480 | 20 | 1280 | 5120 | <20 | 320 | 320 | 5120 | 5120 | |
| SEO | LN216 | + | + | NT | + | 80 | 20 | 20 | <20 | 320 | 20 | 80 | <20 | 320 | 80 | <20 | 80 | 20 | <20 | 80 | 80 | 80 | 20 |
| LN28 | + | − | NT | + | 80 | 20 | 20 | <20 | 80 | 20 | 20 | 20 | 80 | <20 | <20 | 20 | <20 | <20 | 320 | 80 | 80 | <20 | |
| LN266 | + | + | NT | + | 80 | 20 | 20 | <20 | 320 | <20 | 80 | <20 | 320 | 80 | <20 | 80 | 20 | <20 | 80 | 80 | 80 | 20 | |
| LN265 | + | + | − | + | 80 | 20 | 20 | 20 | 80 | 20 | 80 | 20 | 320 | 20 | <20 | 80 | <20 | <20 | 80 | 80 | 80 | <20 | |
| LN263 | + | + | − | + | 80 | 20 | 20 | 20 | 320 | <20 | 80 | <20 | 320 | 80 | <20 | 80 | 20 | <20 | 80 | 80 | 80 | <20 | |
| LN262 | + | + | S+ | + | 80 | 20 | 20 | 20 | 320 | <20 | 80 | <20 | 1280 | <20 | <20 | 320 | <20 | <20 | 320 | 80 | 320 | 20 | |
| JX1 | + | − | − | + | 320 | 20 | 80 | 20 | 320 | 20 | 80 | <20 | 1280 | 80 | 20 | 80 | 20 | <20 | 320 | 320 | 80 | 20 | |
| SD4 | + | + | − | + | 320 | 80 | 80 | 20 | 320 | 20 | 320 | <20 | 1280 | 20 | 20 | 320 | <20 | <20 | 80 | 80 | 80 | <20 | |
| SD2 | + | + | − | + | 320 | 20 | 80 | <20 | 320 | <20 | 80 | <20 | 320 | <20 | 20 | 80 | <20 | <20 | 320 | 320 | 80 | <20 | |
| JX41 | + | + | NT | + | 320 | <20 | 80 | <20 | 1280 | <20 | 320 | <20 | 320 | 80 | <20 | 80 | 20 | <20 | 80 | 20 | 80 | 20 | |
| SD30 | + | + | − | + | 320 | <20 | 80 | <20 | 320 | <20 | 80 | <20 | 1280 | 20 | 20 | 320 | <20 | <20 | 80 | 20 | 80 | 20 | |
| SD1 | + | + | S+ | + | 320 | <20 | 80 | <20 | 320 | 20 | 320 | 20 | 1280 | 20 | <20 | 320 | <20 | <20 | 320 | 320 | 320 | 80 | |
| LN27 | + | + | NT | + | 1280 | 20 | 320 | <20 | 1280 | <20 | 320 | <20 | 1280 | <20 | <20 | 320 | <20 | <20 | 1280 | 80 | 320 | 20 | |
| LN249 | + | + | S+ | + | 1280 | 320 | 320 | 80 | 1280 | 80 | 1280 | 20 | 5120 | 80 | <20 | 1280 | 20 | <20 | 320 | 320 | 1280 | 320 | |
| SD37 | + | + | − | + | 5120 | 320 | 5120 | 80 | 20,480 | 1280 | 5120 | 320 | 20,480 | 320 | 20 | 5120 | 80 | <20 | 80 | 80 | 1280 | 320 | |
| Unknown | JX22 | + | + | NT | + | 20 | 20 | <20 | <20 | <20 | 20 | <20 | 20 | 80 | 80 | <20 | 20 | 20 | <20 | 80 | 80 | 20 | 20 |
| SD40 | + | + | − | + | 320 | 320 | 80 | 80 | 320 | 320 | 80 | 80 | 1280 | 1280 | 20 | 320 | 320 | <20 | 80 | 80 | 80 | 80 | |
aSerotype of infection was determined to be HTN (Hantaan), SEO (Seoul) or Unknown by whether antibody level against the homotypic virus was ≥4-fold higher than that of the heterotypic virus
bHFRS patients were confirmed by using recombinant nucleocapside proteins based IgM capture ELISA [37] or IgG ELISA [38] for Hantaan virus (HTNV) and Seoul virus (SEOV), and viral RNA was detected by real-time RT-PCR [39] to confirm genotype of infection. +, Positive result. −, Negative result. NT, Not tested. H+, HTNV RNA positive. S+, SEOV RNA positive
cConvalescent sera from laboratory confirmed patients of hemorrhagic fever with renal syndrome (HFRS) were titrated against HTNV strain 84FLi and SEOV strain L99 by plaque reduction neutralization test (PRNT), microneutralization test (MNT), pseudoparticle neutralization test (PPNT), immunofluorescence assay based on virus-infected cells(IFA-virus) and immunofluorescence assay based on viral glycoproteins (IFA-GP). PRNT, MNT, PPNT and IFA titres were expressed as the reciprocal of the highest dilution of serum resulting in specific reduction of the number of virus plaques or absorbance value or luciferase activity or fluorescence by 50% (IC50) and 80% (IC80), respectively. IC50, 50% inhibition concentration. IC80, 80% inhibition concentration
Plaque reduction neutralization test
Methods used for PRNT were essentially the same as previously described [40]. The viral stocks of 84FLi and L99 were titred three times by semi-log10 dilutions by plaque forming assay. The same virus stocks were used in MNT. Then serial 4-fold dilutions of human sera (starting at 1:20) were incubated with 100 PFU of 84FLi or L99 at the final volume of 100 μl at 4 °C overnight. The mixtures were added to confluent monolayers of Vero-E6 cells grown in 6-well plates in duplicates and incubated at 37 °C in a humidified 5% CO2 atmosphere for 2 h. Then 4 ml of growth medium containing 10% FBS, 0.6% agarose and 1% DMSO (Sigma) was added to each well. After an incubation at 37 °C in a humidified 5% CO2 atmosphere for 7 days (SEOV) and 9 days (HTNV), the plaques were visualized by adding a second overlay identical to the first but containing 2% FBS, 3% neutral red and without DMSO (4 ml per well). The plates were incubated at 37 °C, 5% CO2 atmosphere and plaques were observed for 2 ~ 4 days. The 50% inhibition concentration (IC50) and 80% inhibition concentration (IC80) were determined as the reciprocal of the highest dilution of serum resulting in 50% (PRNT50) and 80% (PRNT80) reduction of plaques as compared to the virus control, respectively.
Microneutralization test
Microneutralization tests for HTNV and SEOV were developed according to MNT for influenza virus with minor modifications [21]. Direct ELISA and indirect ELISA using mAbs L13F3 were compared by evaluating their P/N ratios.
The viral stocks of 84FLi and L99 were titred at semi-log10 dilutions and the TCID50 were calculated by the method of Reed-Munch. Serial 4-fold diluted human sera were mixed with 100 TCID50 of 84FLi or L99 in duplicate in microplates and incubated at 4 °C overnight. The confluent monolayers of Vero-E6 cells were trypsinized and 4 × 104 cells in 0.1 ml volume complete EMEM containing 10% FBS, were added to each well containing the virus-serum mixtures in microplates, and incubated at 37 °C in a humidified 5% CO2 atmosphere for 7 days. Then the supernatant was removed and monolayers were fixed with precooled 80% (v/v) solution of acetone-PBS for 20 min at 4 °C. Subsequently, the acetone-PBS was removed. Then HRP-conjugated mAbs L13F3 diluted with PBS containing 10% FBS and 0.05% Tween 20 were added to the acetone fixed monolayers. After an incubation of 45 min at 37 °C, the plates were washed and TMB was added to develop color. The absorbance values at 450 nm (OD450) were observed by Varioskan LUX multimode microplate reader (Thermo, USA). The IC50 and IC80 were determined as the reciprocal of the highest dilution of serum resulting in 50% (MNT50) and 80% (MNT80) reduction of OD450 values as compared to the virus control. Positive, negative controls and virus titrations were included in each assay.
Expression of viral glycoproteins and IFA-GP
RNA was extracted from culture supernatants of 84FLi and L99 infected Vero-E6 cells using QIAamp Viral RNA kit (Qiagen, Germany), and cDNA was prepared by reverse transcription using random primers. To generate glycoprotein expression plasmids, full-length M genes were amplified and cloned into the pCAGGS-MCS vector to generate pCHTNM and pCSEOM.
The HTNV and SEOV glycoproteins were expressed in 293T cells and identified by IFA. Briefly, 293T cells grown on 100-mm dish were transfected with plasmids of pCHTNM, pCSEOM and pCAGGS separately. After 48 h, the 293T cells were trypsinized, fixed with acetone for 20 min, and treated with mouse mAbs 8B6 against hantavirus Gn glycoprotein and human recombinant mAbs Y5 against hantavirus Gc glycoprotein, serum from laboratory confirmed patient of HFRS, and serum from healthy human for 45 min at 37 °C. After washing with PBS, fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG or goat anti-human IgG diluted in PBS containing 5% skim milk, were added. After 45 min incubation at 37 °C, cells were washed with PBS and examined with a fluorescence microscope.
The immunofluorescence assay using recombinant glycoproteins of HTNV and SEOV as antigen to detect anti-glycoprotein antibodies described in this part was designated as IFA-GP.
Production of lentiviral pseudotyped particles bearing glycoproteins of HTNV and SEOV and pseudoparticle neutralization test
293T cells were seeded in 100-mm diameter dishes 8 ~ 18 h prior to transfection. 12 μg of the glycoprotein expression plasmids of HTNV or SEOV were co-transfected with 12 μg of a lentivirus-based luciferase reporter construct pNL4–3.Luc.R-E- [41] with X-treme GENE HP DNA Transfection Reagent (Roche, Switzerland). Vesicular stomatitis virus glycoprotein envelope (VSV-G)-pseudotyped lentiviral particles were made and used as control to evaluate non-specific neutralization caused by inhibition materials to transduction of lentivirals. Transfection reagent was added to plasmids diluted in opti-MEM (Gibco, USA), and incubated at room temperature for 15 to 20 min, then added dropwise to 293T cells and incubated at 37 °C for 8 h. Then the medium was replaced with fresh medium containing 2% FBS. Culture medium containing pseudoparticles of HTNV (HTNVpp) and SEOV (SEOVpp) were collected at 24 and 48 h after transfection and clarified by centrifugation at 900 x g for 10 min. To make virus stocks used in PPNT, 1 volume of lenti-X concentrator (Clontech, USA) was combined with 3 volumes of the clarified supernatant, the mixture were incubated at 4 °C overnight, and then centrifuged at 1500 x g for 45 min at 4 °C. After centrifugation, the supernatant was removed carefully, the pellet was suspended in DMEM containing 10% FBS. Single use aliquots were frozen with liquid nitrogen and stored at −80 °C.
Pseudoparticle neutralization tests for HTNV and SEOV were carried out as previously described for SFTSV with some modifications [27]. HTNVpp and SEOVpp stocks were titrated. Then, serial 4-fold dilutions of heat inactivated human sera were mixed with 3000 relative luciferase unit (RLU) of HTNVpp or SEOVpp in duplicate in 96-well plate for 1 h at 37 °C in a total volume of 100 μl. Then the virus-sera mixtures were added to Huh-7.5 cells plated in black 96-well plates 24 h before. After 48 h, the luciferase activities in cell lysates were measured with One-Glo Luciferase Assay System and Varioskan LUX multimode microplate reader (Thermo, USA). The IC50 and IC80 were determined as the reciprocal of the highest dilution resulting in 50% (PPNT50) and 80% (PPNT80) reduction of luminescence values as compared to positive control, respectively.
Immunofluorescence assay using virus-infected cells (IFA-virus)
Vero-E6 cells infected with 84FLi or L99 were maintained in complete EMEM containing 2% heat-inactivated FBS and incubated for 7 days at 37 °C. Then cells were trypsinized and spotted onto 8-well slides, air dried and fixed with acetone. These slides were incubated with 4-fold serial dilutions of human sera (starting at 1:20 dilution), and probed with FITC-conjugated goat anti-human IgG. IFA titres were expressed as the reciprocal of the highest dilution of serum resulting in specific fluorescence of hantaviruses.
Comparison of antibody titres and serotyping obtained by PRNT, MNT, PPNT, IFA-GP
Serotype of infection was determined by whether antibody level against the homotypic virus was ≥4-fold higher than that of the heterotypic virus. Antibody titres and serotyping results obtained by methods above were compared using statistical analysis.
Statistics
The difference of geometric mean titres (GMTs) were evaluated by paired-t test and the correlations between methods were analyzed by linear correlation test using GraphPad Prism 5 and IBM SPSS statistics 19.
Results of serotyping between methods were compared by McNemar χ2 tests on cross-table (for categorical variable) and Kappa agreement tests using IBM SPSS statistics 19.
A P value less than 0.05 was considered statistically significant.
Results
Virus titration by plaque forming assay
Figure 1 shows the results of plaque forming assay. The titre (PFU/ml) was 1.6 × 106.0 for 84FLi and 5.1 × 106.0 for L99.
Fig. 1.
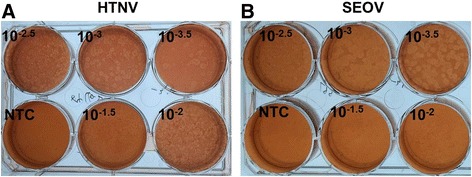
Virus titration by plaque forming assay. a Hantaan virus strain 84FLi; b, Seoul virus strain L99
Microneutralization test
To choose a better detection method in MNT, direct ELISA and indirect ELISA in MNT were compared.
1:2000 diluted mAb L13F3 achieved the best P/N ratio for both direct ELISA and indirect ELISA (Fig. 2), and direct ELISA produced higher P/N ratio than indirect ELISA (Fig. 2). Since direct ELISA is simpler and more time-saving than indirect ELISA, it was used in MNT.
Fig. 2.
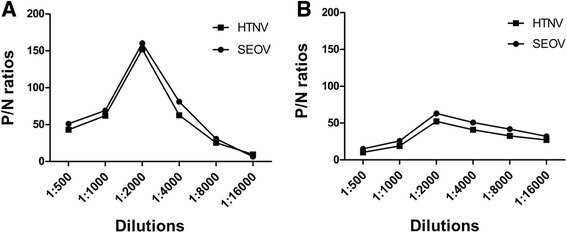
Comparison of detection methods in MNT. a direct ELISA; b indirect ELISA. P/N ratios, positive/negative ratios
Viral stocks were titrated using direct ELISA. The titres of both virus stocks were 2.0 × 106.0 TCID50/ml for 84FLi and 6.3 × 106.0 TCID50/ml for L99 (Fig. 3).
Fig. 3.
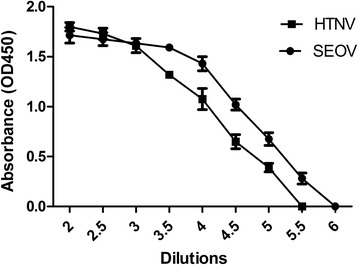
Microtitration of Hantaan virus strain 84FLi and Seoul virus strain L99 used in MNT. Data are means of three experiments. The error bars indicate standard deviations (SD)
IFA detection of HTNV and SEOV glycoproteins expressed in 293T cells
IFA assays showed that both Gn and Gc glycoproteins of HTNV and SEOV were effectively expressed in transiently transfected 293T cells. Thus, the recombinant viral glycoproteins can be used as antigen to detect hantavirus-specific antibodies in human sera by IFA (Fig. 4).
Fig. 4.
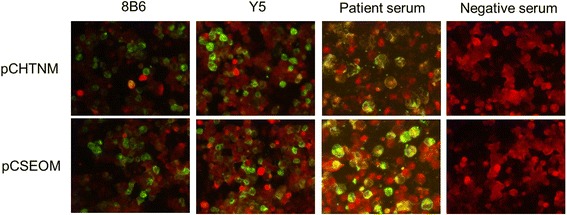
Immunofluorescence detections of glycoproteins of Hantaan and Seoul viruses expressed in 293T cells. pCHTNM, Recombinant expressing plasmid for glycoprotein of Hantaan virus strain 84FLi. pCSEOM, Recombinant expressing plasmid for glycoprotein of Seoul virus strain L99
Titration of pseudoparticles of HTNV and SEOV
HTNVpp and SEOVpp were titrated on Huh-7.5 cells. The infectious titre (RLU/ml) was 2.0 × 105 for HTNVpp and 8.0 × 105 for SEOVpp (Fig. 5). A comparable amount of HTNVpp or SEOVpp giving an RLU of 3000 was incubated with various dilutions of sera was used in PPNT.
Fig. 5.
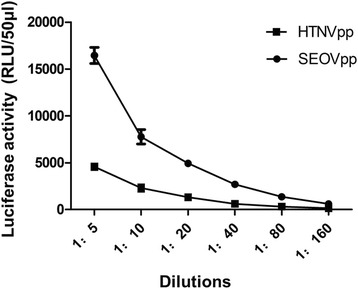
Titration of HTNVpp and SEOVpp on Huh-7.5 cells. Data are means of three experiments. The error bars indicate standard deviations (SD)
Comparisons of antibody titres obtained by PRNT, MNT, PPNT, IFA-GP and IFA-virus
All 74 human sera were assayed by PRNT, MNT, PPNT, IFA-GP and IFA-virus. There were no non-specific neutralization for all negative sera at 1:20 dilution for PRNT, MNT, IFA-GP and IFA-virus. For PPNT50, 46.7% (14/30) of negative sera yielded inhibition of HTNVpp, SEOVpp and VSVpp, no non-specific neutralization found at dilutions ≥1:80 of the sera. The cutoff level for a positive test was set at 1:80 for PPNT50.
For 44 convalescent sera of HFRS patients, antibody titres determined by PRNT, MNT, PPNT, IFA-GP and IFA-virus are listed in Table 1.
As shown in Table 1, antibody titres determined by 50% inhibition concentration were relatively higher than that determined by 80% inhibition concentration. PRNT50, MNT50 and PPNT50 were used for the evaluation of these methods.
To evaluate the correlations of titres obtained with mentioned methods, linear correlation analyses were performed. As shown in Fig. 6a and b, titres obtained with MNT50 and PRNT50, PPNT50 and PRNT50, were both highly correlated (r > 0.8, p < 0.001), reflecting the reliability of these methods. As shown in Fig. 6c, titres determined by IFA-GP and PRNT50 were moderately correlated (r = 0.736, p < 0.001). There were no significant differences among all correlation coefficients as judged by their 95% confidence intervals (Fig. 6).
Fig. 6.
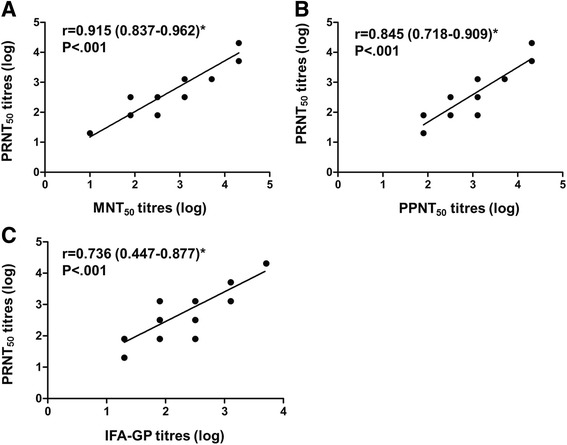
Comparisons of antibody titres. Each dot represents the log10 antibody titre of a serum against Hantaan virus strain 84FLi or Seoul virus strain L99. Total number of sera assayed is 44. The best-fit lines are shown on each graph. On the graphs, r and P indicate the correlation coefficient with 95% confidence interval and the P value of significance, respectively. *95% confidence interval of correlation coefficient
When comparing the GMTs obtained with these methods by paired-t test, as shown in Fig. 7, GMT determined by PPNT50 was significantly higher than that obtained with PRNT50 (p < 0.001), while GMT determined by MNT50 was statistically equal with that obtained by PRNT50 (p > 0.05), and GMT determined by IFA-GP was significantly less than that of PRNT50 (p < 0.001). These results indicate that PPNT50 was the most sensitive method for titrating antibody.
Fig. 7.
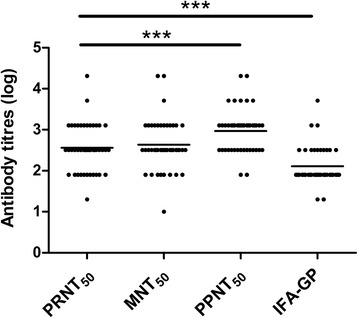
Comparisons of MNT50, PPNT50, and IFA-GP titres with PRNT50 titres. Each dot represents the antibody titres (log) of a serum against Hantaan virus strain 84FLi or Seoul virus strain L99. Total number of sera assayed is 44. Horizontal bars indicate the respective group mean. ***P < .001
To determine the consistency of serotyping obtained with these methods, Kappa agreement tests were performed. Results of serotyping determined by MNT50 and PRNT50, PPNT50 and PRNT50 were both highly consistent (kappa = 1.00 or 0.96; p < 0.001); whereas that determined by IFA-GP and PRNT50 were only moderately consistent (kappa = 0.73, p < 0.001).
Finally, no statistically significant differences were observed for serotyping between PRNT50 and MNT50, as well as PRNT50 and PPNT50 (p > 0.05). It was found that IFA-GP was statistically less sensitive (p < 0.05) than PRNT50 and MNT50 for serotyping of hantaviruses infections. Seven sera, which could be serotyped by PRNT50 and MNT50, could not be serotyped by IFA-GP.
Overall, MNT50 was more specific while PPNT50 was more sensitive for titrating neutralizing antibody than other assays. IFA-GP was less sensitive than other neutralization tests for serotyping.
Discussion
HFRS was caused by HTNV and SEOV in China, no other hantavirus infection of human has been comfirmed. HTNV and SEOV have discriminative antigenicities, and are carried by different rodents. For vaccine development, diagnosis and rodents control, it is necessary to identify the type of hantavirus that cause human disease. Molecular genotyping including conventional PCR and real-time RT-PCR have been developed for Hantavirus to detect viral RNA and genotyping in clinical samples [42, 43]. Because of very low virus load and shortness of viremia of HFRS patients, the nucleic acid test is not sensitive enough to be implemented in clinical diagnosis of HFRS routinely [42, 43]. In this study, 33 acute phase sera of HFRS patients were tested by real-time RT-PCR, only 5 sera have been genotyped. Instead, serological assays to detect IgM and/or IgG antibodies by ELISA or IFA have been broadly used. Chu et al. explored the antigenic relationships among 32 hantavirus isolates, they found most of the SEO-like and HTN-like viruses displayed strong two-way cross-reactivity tested by ELISA [40]. The analysis of mAbs against N protein of HTNV and SEOV showed the most mAbs react with both HTNV and SEOV [5]. As the “gold standard” for Hantavirus serotyping, PRNT has been broadly used. Determination of neutralizing antibody is necessary for doing population based seroepidemiological survey assessing the exposure of population to these viruses or evaluating immune effect of vaccine. PRNT is time-consuming, laborious and need to handle with living virus. Some easy accessing and safe methods for neutralizing antibody detection were established and preliminary evaluated.
As compared with PRNT, MNT is more objective, simpler and time-saving for high throughput detection of virus neutralizing antibodies. The same virus stocks were used in both MNT and PRNT, so neutralizing antibody titres determined by MNT50 and PRNT50 were highly correlated.
However, the MNT for Hantavirus still need to handle with living viruses. PPNTs for HTNV and SEOV have been developed, which can be completed in 3 ~ 4 days and don't need high biosafety level facility since the pseudotyped virus is unable to produce infectious viruses. In this study, neutralizing antibody titres obtained with PPNT50 were significantly higher than that obtained with PRNT50. Although one serum (SD7) was determined as infection of HTNV by PRNT50 and MNT50, it cannot be serotyped by PPNT50. For most sera from HFRS patients, the neutralizing antibody determined by PPNT were consistent with that determined by neutralization tests using live viruses.
While IFA-virus can’t differentiate infection of HTNV and SEOV is a common opinion [44], IFA-GP has not been reported for hantaviruses serotyping. Here, IFA-GP was less sensitive than PRNT50 and MNT50 for serotyping infections of hantaviruses. Seven sera, which could be serotyped by PRNT50 and MNT50, could not be serotyped by IFA-GP. Some of the envelope glycoprotein antigenic determinants are not involved in virus neutralization [6], so anti-glycoprotein antibody titres may not be fully correlated with neutralizing antibody titres. However, for 79.5% (35/44) samples, serotyping obtained with IFA-GP and PRNT50 were consistent. In view of the simplicity of IFA-GP, it is meaningful for serotyping.
Conclusion
While PRNT is the standard neutralization test for hantaviruses, MNT50 and PPNT50 both can be used as simple and rapid alternatives to PRNT50 for Hantaan and Seoul viruses. MNT50 is more specific while PPNT50 is more sensitive than other assays for neutralizing antibody determination. IFA-GP is meaningful for serotyping in view of its simplicity. So far, this work has been the most comprehensive comparison of alternatives to PRNT.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Science and Technology Major Project of China (2017ZX10101001–001).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Declarations
The views and opinions expressed herein are those of the authors only and do not necessarily represent the views and opinions of the China CDC or Beijing CDC.
Abbreviations
- DMEM
Dulbecco’s Modified Eagle Medium
- ELISA
Enzyme linked immunosorbent assay
- FBS
Fetal bovine serum
- FITC
Fluorescein isothiocyanate
- GMT
Geometric mean titre
- HFRS
Hemorrhagic fever with renal syndrome
- HRP
Horseradish peroxidase.
- HTNV
Hantaan virus
- HTNVpp
Pseudoparticles of HTNV
- IC50
50% inhibition concentration
- IC80
80% inhibition concentration
- IFA
Immunofluorescence assay
- IFA-GP
Immunofluorescence assay based on viral glycoproteins
- IFA-virus
Immunofluorescence assay using virus-infected cells
- mAbs
Monoclonal antibodies
- MNT
Microneutralization test
- MuLV
Murine leukemia virus
- N
Nucleocapsid protein
- OD450
Absorbance value at 450 nm
- P/N
Positive/ Negative
- PFU
Plaque-forming units
- PPNT
Pseudoparticle neutralization test
- PRNT
Plaque reduction neutralization test
- RLU
Relative luciferase unit
- SD
Standard deviation
- SEOV
Seoul virus
- SEOVpp
Pseudoparticles of SEOV
- TCID50
Tissue culture infection dose50
- VSV
Vesicular stomatitis virus
- VSV-G
Vesicular stomatitis virus envelope glycoprotein
- VSVpp
Pseudoparticles of vesicular stomatitis virus
Authors’ contributions
WL, SC designed and performed the experiments and drafted the manuscript. JL, QZ, SZ provided suggestions on the experimental design and helped edit the manuscript. WW, JQ provided advice in this study. ML helped edited this manuscript. DL designed this study, analyzed experimental data and edited this manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This work was approved by the ethics committee of China CDC. Informed consent was obtained from all study participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional.
Contributor Information
Weihong Li, Email: weihonggirl@163.com.
Shouchun Cao, Email: caosc@nifdc.org.cn.
Quanfu Zhang, Email: zhqf@163.com.
Jiandong Li, Email: ldong121@126.com.
Shuo Zhang, Email: zhang_shuo@live.com.
Wei Wu, Email: beilei0702@vip.sina.com.
Jing Qu, Email: ida_dulce@msn.com.
Chuan Li, Email: lcehfcdc@163.com.
Mifang Liang, Email: mifangl@vip.sina.com.
Dexin Li, Email: lidx@chinacdc.cn.
References
- 1.Schmaljohn CS, Hasty SE, Dalrymple JM, et al. Antigenic and genetic properties of viruses linked to hemorrhagic fever with renal syndrome. Science. 1985;227(4690):1041–1044. doi: 10.1126/science.2858126. [DOI] [PubMed] [Google Scholar]
- 2.Elliott RM. Molecular biology of the Bunyaviridae. J Gen Virol. 1990;71:501–522. doi: 10.1099/0022-1317-71-3-501. [DOI] [PubMed] [Google Scholar]
- 3.Walter CT, Barr JN. Recent advances in the molecular and cellular biology of bunyaviruses. J Gen Virol. 2011;92:2467–2484. doi: 10.1099/vir.0.035105-0. [DOI] [PubMed] [Google Scholar]
- 4.Muyangwa M, Martynova EV, Khaiboullina SF, et al. Hantaviral proteins: structure, functions, and role in hantavirus infection. Front Microbiol. 2015;6:1326. doi: 10.3389/fmicb.2015.01326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshimatsu K, Arikawa J. Antigenic properties of N protein of hantavirus. Viruses. 2014;6(8):3097–3109. doi: 10.3390/v6083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arikawa J, Schmaljohn AL, Dalrymple JM, et al. Characterization of Hantaan virus envelope glycoprotein antigenic determinants defined by monoclonal antibodies. J Gen Virol. 1989;70:615–624. doi: 10.1099/0022-1317-70-3-615. [DOI] [PubMed] [Google Scholar]
- 7.Dantas JR, Jr, Okuno Y, Asada H, et al. Characterization of glycoproteins of viruses causing hemorrhagic fever with renal syndrome (HFRS) using monoclonal antibodies. Virology. 1986;151(2):379–384. doi: 10.1016/0042-6822(86)90058-9. [DOI] [PubMed] [Google Scholar]
- 8.Hooper JW, Custer DM, Thompson E, et al. DNA vaccination with the Hantaan virus M gene protects hamsters against three of four HFRS hantaviruses and elicits a high-titre neutralizing antibody response in rhesus monkeys. J Virol. 2001;75:8469–8477. doi: 10.1128/JVI.75.18.8469-8477.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmaljohn C, Hjelle B. Hantaviruses: a global disease problem. Emerg Infect Dis. 1997;3(2):95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart CA, Bennett M. Hantavirus infections: epidemiology and pathogenesis. Microbes Infect. 1999;1(14):1229–1237. doi: 10.1016/S1286-4579(99)00238-5. [DOI] [PubMed] [Google Scholar]
- 11.Song G. Epidemiological progresses of hemorrhagic fever with renal syndrome in China. Chin Med J. 1999;112(5):472–477. [PubMed] [Google Scholar]
- 12.Jonsson CB, Figueiredo LT, Vapalahti O. A global perspective on hantavirus ecology,epidemiology,and disease. Clin Microbiol Rev. 2010;23(2):412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu YK, Jennings G, Schmaljohn A, et al. Cross-neutralization of hantaviruses with immune sera from experimentally infected animals and from hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome patients. J Infect Dis. 1995;172(6):1581–1584. doi: 10.1093/infdis/172.6.1581. [DOI] [PubMed] [Google Scholar]
- 14.Lee PW, Gibbs CJ, Jr, Gajdusek DC, et al. Serotypic classification of hantaviruses by indirect immunofluorescent antibody and plaque reduction neutralization tests. J Clin Microbiol. 1985;22(6):940–944. doi: 10.1128/jcm.22.6.940-944.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu YX, Yao ZH, An Q, et al. Studies on serotypic classification of viruses of hemorrhagic fever with renal syndrome by plaque reduction neutralization test. Bing Du Xue Bao. 1991;7:18–22. [Google Scholar]
- 16.Anderson LJ, Hierholzer JC, Bingham PG, et al. Microneutralization test for respiratory syncytial virus based on an enzyme immunoassay. J Clin Microbiol. 1985;22(6):1050–1052. doi: 10.1128/jcm.22.6.1050-1052.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenny MT, Albright KL, Sanderson RP. Microneutralization test for the determination of mumps antibody in vero cells. Appl Microbiol. 1970;20(3):371–373. doi: 10.1128/am.20.3.371-373.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mannen K, Mifune K, Reid-Sanden FL, et al. Microneutralization test for rabies virus based on an enzyme immunoassay. J Clin Microbiol. 1987;25(12):2440–2442. doi: 10.1128/jcm.25.12.2440-2442.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vorndam V, Beltran M. Enzyme-linked immunosorbent assay-format microneutralization test for dengue viruses. Am J Trop Med Hyg. 2002;66(2):208–212. doi: 10.4269/ajtmh.2002.66.208. [DOI] [PubMed] [Google Scholar]
- 20.Benne CA, Harmsen M, De Jong JC, et al. Neutralization enzyme immunoassay for influenza Virus. J Clin Microbiol. 1994;32(4):987–990. doi: 10.1128/jcm.32.4.987-990.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization. 2010. Serological diagnosis of influenza by microneutralization assay. http://www.who.int/influenza/gisrs_laboratory/2010_12_06_serological_diagnosis_of_influenza_by_microneutralization_assay.pdf?ua=1.
- 22.Hörling J, Lundkvist A, Huggins JW, et al. Antibodies to Puumala virus in humans determined by neutralization test. J Virol Methods. 1992;39(1–2):139–147. doi: 10.1016/0166-0934(92)90132-W. [DOI] [PubMed] [Google Scholar]
- 23.Takada A, Robison C, Goto H, et al. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci U S A. 1997;94(26):14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tani H, Iha K, Shimojima M, et al. Analysis of Lujo virus cell entry using pseudotype vesicular stomatitis virus. J Virol. 2014;88(13):7317–7330. doi: 10.1128/JVI.00512-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia JM, Lai JC. Production of influenza pseudotyped lentiviral particles and their use in influenza research and diagnosis: an update. Expert Rev Anti-Infect Ther. 2016;9(4):443–455. doi: 10.1586/eri.11.25. [DOI] [PubMed] [Google Scholar]
- 26.Garcia JM, Lagarde N, Ma ES, et al. Optimization and evaluation of an influenza a (H5) pseudotyped lentiviral particle-based serological assay. J Clin Virol. 2010;47:29–33. doi: 10.1016/j.jcv.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann H, Li X, Zhang X, et al. Severe fever with thrombocytopenia virus glycoproteins are targeted by neutralizing antibodies and can use DC-SIGN as a receptor for pH-dependent entry into human and animal cell lines. J Virol. 2013;87(8):4384–4394. doi: 10.1128/JVI.02628-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cifuentes-Muñoz N, Darlix JL, Tischler ND. Development of a lentiviral vector system to study the role of the Andes virus glycoproteins. Virus Res. 2010;153(1):29–35. doi: 10.1016/j.virusres.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Qian Z, Haessler M, Lemos JA, et al. Targeting vascular injury using hantavirus-pseudotyped lentiviral vectors. Mol Ther. 2006;13(4):694–704. doi: 10.1016/j.ymthe.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 30.Ma M, Kersten DB, Kamrud KI, et al. Murine leukemia virus pseudotypes of La Crosse and Hantaan Bunyaviruses: a system for analysis of cell tropism. Virus Res. 1999;64(1):23–32. doi: 10.1016/S0168-1702(99)00070-2. [DOI] [PubMed] [Google Scholar]
- 31.Ogino M, Ebihara H, Lee BH, et al. Use of vesicular stomatitis virus pseudotypes bearing hantaan or seoul virus envelope proteins in a rapid and safe neutralization test. Clin Diagn Lab Immunol. 2003;10(1):154–160. doi: 10.1128/CDLI.10.1.154-160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray N, Whidby J, Stewart S, et al. Study of Andes virus entry and neutralization using a pseudovirion system. J Virol Methods. 2010;163(2):416–423. doi: 10.1016/j.jviromet.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Higa MM, Petersen J, Hooper J, et al. Efficient production of Hantaan and Puumala pseudovirions for viral tropism and neutralization studies. Virology. 2012;423(2):134–142. doi: 10.1016/j.virol.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76(24):13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang MF, Song G, Hang CS, et al. Preliminary study on structure protein of epidemic hemorrhagic fever virus using monoclonal antibodies. Bing Du Xue Bao. 1989;5(3):217–223. [Google Scholar]
- 36.Koch J, Liang M, Queitsch I, et al. Human recombinant neutralizing antibodies against hantaan virus G2 protein. Virology. 2003;308:64–73. doi: 10.1016/S0042-6822(02)00094-6. [DOI] [PubMed] [Google Scholar]
- 37.Zhang QF, Li JD, Li WH, et al. Development and application of a two-step MacELISA for the early diagnosis of hemorrhagic fever with renal syndrome. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2008;22(1):6–8. [PubMed] [Google Scholar]
- 38.Zöller L, Yang S, Gött P, et al. Use of recombinant nucleocapsid proteins of the Hantaan and nephropathia epidemica serotypes of hantaviruses as immunodiagnostic antigens. J Med Virol. 1993;39(3):200–207. doi: 10.1002/jmv.1890390305. [DOI] [PubMed] [Google Scholar]
- 39.Pang Z, Li A, Li J, et al. Comprehensive multiplex one-step real-time TaqMan qRT-PCR assays for detection and quantification of hemorrhagic fever viruses. PLoS One. 2014;9(4):e95635. doi: 10.1371/journal.pone.0095635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu YK, Rossi C, Leduc JW, et al. Serological relationships among viruses in the hantavirus genus, family Bunyaviridae. Virology. 1994;198(1):196–204. doi: 10.1006/viro.1994.1022. [DOI] [PubMed] [Google Scholar]
- 41.Connor RI, Chen BK, Choe S, et al. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206(2):935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 42.Vaheri A, Vapalahti O, Plyusnin A. How to diagnose hantavirus infections and detect them in rodents and insectivores. Rev Med Virol. 2008;18(4):277–288. doi: 10.1002/rmv.581. [DOI] [PubMed] [Google Scholar]
- 43.Mattar S, Guzmán C, Figueiredo LT. Diagnosis of hantavirus infection in humans. Expert Rev Anti-Infect Ther. 2015;13(8):939–946. doi: 10.1586/14787210.2015.1047825. [DOI] [PubMed] [Google Scholar]
- 44.Song G, Hang CS, Liao HX, et al. Antigenic difference between viral strains causing classical and mild types of epidemic hemorrhagic fever with renal syndrome in China. J Infect Dis. 1984;150(6):889–894. doi: 10.1093/infdis/150.6.889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.


