Abstract
Background aims
Mesenchymal stromal cells (MSCs) are being investigated for use in cell therapy. The extensive in vitro expansion necessary to obtain sufficient cells for clinical use increases the risk that genetically abnormal cells will arise and be propagated during cell culture. Genetic abnormalities may lead to transformation and poor performance in clinical use, and are a critical safety concern for cell therapies using MSCs.
Methods
We used spectral karyotyping (SKY) to investigate the genetic stability of human MSCs from ten donors during passaging.
Results
Our data indicate that chromosomal abnormalities exist in MSCs at early passages and can be clonally propagated. The karyotypic abnormalities observed during our study diminished during passage.
Conclusions
Karyotyping of MSCs reveals characteristics which may be valuable in deciding the suitability of cells for further use. Karyotypic analysis is useful for monitoring the genetic stability of MSCs during expansion.
Keywords: Spectral Karyotyping, SKY, Mesenchymal Stromal Cells, Mesenchymal Stem Cells, genomic stability, MSC
Introduction
Human mesenchymal stromal or stem cells (hMSCs) are derived from several tissues including bone marrow. They are plastic-adherent in tissue culture, can differentiate into osteoblasts, chondrocytes, and adipocytes, and express cell surface markers CD73, CD90, CD105, but not CD11b, CD14, CD19, CD34, CD45, and CD79α [1]. MSCs are being investigated in clinical trials for a variety of indications.
MSCs are often significantly expanded to obtain sufficient cells for therapy, which increases the probability that genetic changes may arise. Genetic stability and potential transformation leading to tumor growth are safety concerns for stem cell therapies [2]. Chromosomal alterations are associated with increased tumorigenicity and the inability to reach desired differentiation states [3, 4]. Additionally, there is donor to donor variability in the genetic stability of cells used for therapy [5]. To explore effects of extended passage and donor source on genetic stability, we performed karyotypic analysis of hMSCs as part of a larger analysis of attributes of these cells that might predict safety and performance in clinical use [6–12].
High rates of aneuploidy, escape from replicative senescence, and transformation in rodent MSCs prompted concern for use of hMSCs [13, 14]. However, studies for hMSCs report low levels of chromosomal aberrations among donor samples [15, 16] and during culture expansion [5, 17, 18]. Low levels of non-clonal chromosomal aberrations in MSCs used in clinical trials have been observed, but there are no reports of malignant transformation [19]. The evidence of malignant transformation during in vitro expansion is mixed [5, 20], as initial reports for hMSCs were a result of contaminated cell lines [21, 22]. However, a recent paper reported that verified hMSCs exhibited spontaneous tumorigenic transformation associated with genomic alterations during culture [23], thus MSCs tumorigenic potential remains unresolved. Additionally, the significance of chromosomal alterations or the effect of genetic instability on therapeutic performance is poorly understood.
Multiple methods are available to assess chromosomal stability of cells including Giemsa (G) banding, fluorescence in situ hybridization (FISH), spectral karyotyping (SKY), and comparative genomic hybridization (CGH). SKY is a rapid FISH based method in which chromosome-specific fluorescent labels are used to visualize all chromosomes in a single hybridization [24, 25]. Using SKY, we identified genomic aberrations in bone marrow-derived hMSCs from multiple donors, at multiple passages, to analyze the chromosomal stability of each cell line during culture. We found that chromosomal aberrations both exist and arise in culture expanded MSCs, and could be clonally propagated. However, in all cases, the aberrations diminished with extended culture, suggesting that they did not provide a replicative advantage, providing further evidence of the general genetic stability of hMSCs.
Materials and Methods
Cell Culture
Bone marrow-derived hMSCs from ten donors were purchased from All Cells (Alameda, CA) or Lonza (Walkersville, MD) as passage 1 vials and designated with identifiers indicating donor. Both AllCells and Lonza use standard protocols for isolation of MSCs from bone marrow and culture cells in Mesencult (Stem Cell Technologies), or MSCGM (Lonza), respectively. MSCs were expanded in culture medium composed of α-minimum essential medium (Life Technologies), 1% L-glutamine (Life Technologies), 1% penicillin and streptomycin (Life Technologies), and 16.5% fetal bovine serum (JM Bioscience) to passages 3, 5, and 7 (P3, P5, and P7), as previously described [7]. Passage number equals times cells were trypsinized until cryopreservation. Expansion lots PCBM1641, PCBM1632, 167696, 110877, 8F3560, PCBM1662, and 127756 were tested for expression of cell surface markers by flow cytometry [7], and growth kinetics using an IncuCyte Live Cell Imager [8]. Additionally, CFUs, quantitative adipogenic differentiation, proteomic profiling, transcriptome analysis, T-cell immunomodulation, and changes in chromatin modifications have been described for these lots [6–12].
Metaphase Spreads
Cryopreserved cells were cultured to 70–80% confluence, then 10 μl/ml of 10% Demecolcine (Sigma Aldrich) was added. After 4–5 hours, cells were trypsinized, centrifuged, resuspended in a few drops of media, and a hypotonic solution (0.2 % Potassium Chloride, 0.2% Sodium Citrate, and 0.01% Fetal Bovine Serum) was added dropwise. Cells were incubated at 37°C for 20 minutes, centrifuged, and resuspended in hypotonic solution. Fixative (3:1 solution of Methanol:Acetic Acid) was added dropwise. Cells were incubated 15 minutes at RT, centrifuged and resuspended in fixative twice. Resuspended cells were dropped onto clean slides over a 60°C water bath and air dried. Slides were microscopically examined for metaphase spreading and stored 5 to 15 days at room temperature before Spectral Karyotyping.
Karyotyping
Chromosome hybridization for spectral karyotyping was performed using the human SkyPaint and CAD detection kits (Applied Spectral Imaging) according to the company protocol. Metaphase chromosomes were analyzed on a Nikon Eclipse E800 microscope equipped with the HISKY system and software (Applied Spectral Imaging). We observed variable random loss of chromosomes, likely due to the technical preparation of slides. Unless multiple karyotypes exhibited the same chromosomal loss, these data were excluded from the analysis. Karyotyping followed the International System for Chromosome Nomenclature 2009 [26].
Giemsa stained mitotic chromosome preparation (G-banding) was performed by the WiCell cytogenetics laboratory. Twenty metaphase spreads were analyzed for each sample.
Statistical Analysis
The percent abnormal karyotypes were calculated for all samples and technical replicates averaged. This resulted in heteroscedastic data requiring nonparametric analysis. Friedman’s test, a repeated measures ANOVA on ranks, was performed. To test sampling bias within donors, contingency tables were constructed from counts of normal and abnormal spreads and two-tailed p-values calculated using Fisher’s exact test for 2×2 tables or the Freeman-Halton test for larger tables. Data was analyzed using SAS system for Windows, Version 9.3 (Copyright 2012, SAS Institute Inc., Cary, NC).
Results
Primary characterization of MSCs
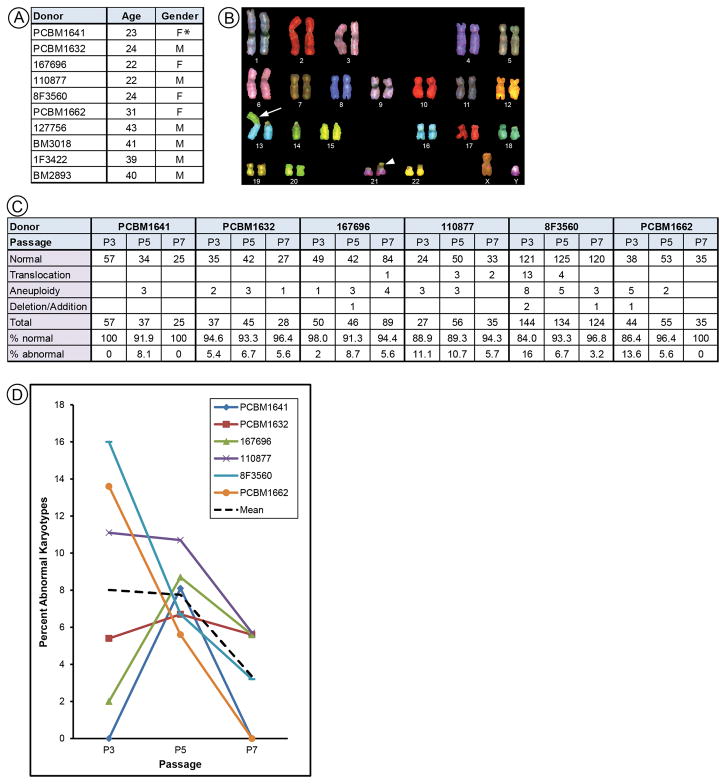
We used SKY to analyze chromosomal stability of MSCs derived from different donors expanded through 7 passages. The International System for Human Cytology Nomenclature 2009 report [26] recommends scoring a minimum of 20 mitotic spreads, and defines a clonal population as two of the same abnormal karyotype. To increase the robustness of our data, we attempted to collect a minimum of forty mitotic figures per sample; however, this was not always feasible due to diminishing proliferative potential of some MSC lines. MSCs were derived from young or middle-aged donors (Fig. 1A).
Figure 1.
SKY analysis for MSCs from donors aged 22 to 31 years. (A) All MSCs used in this study are listed by donor identifier, age, and reported gender. Asterisk indicates vendor information not in agreement with karyotype. Data for donors aged 39 to 43 years shown in Figure 3. (B) Spectral karyotype of PCBM1632 shows a Robertsonian translocation (arrow) in all cells, for subsequent analysis this karyotype is classified as normal for this cell line. (C) Raw data for each donor displaying the number for total, normal, or abnormal karyotypes. Percentages for normal/abnormal karyotypes were calculated. (D) Scatter plot showing percent abnormal karyotypes for MSCs from each of six donors at P3, P5, and P7. The dashed black line indicates the mean.
The primary karyotype of PBMC1632 displayed a Robertsonian Translocation of chromosomes t(13;14)(Fig. 1B). We also identified two MSC lines with karyotypically abnormal subpopulations at passage 3 (P3); 127756 displayed a translocation with karyotype 46,XY,t(12;13), present in 10% of cells examined and 8F3560 displayed two translocations with karyotypes 46,XX,t(1;7) and 46,XX,t(4;17) at approximately 4% and 2% of the total population, respectively.
Karyotypic changes during culture
Six MSC lines were analyzed by SKY at P3, P5, and P7. Karyotypes were categorized as: normal, translocation (exchange of material between chromosomes), aneuploid (gain or loss of an entire or multiple chromosomes), or deletion/addition (gain or loss of material from a single chromosome)(Fig. 1C). Aneuploidy was the most frequent abnormality observed, and was random, with different karyotypes observed in each instance. Percent abnormal karyotypes were calculated for each donor (Fig. 1D). MSCs from 8F3560, PCBM1662, and 110877 showed a decrease in abnormal karyotypes from P3 to P7. MSCs from 167696 and PCBM1641 showed an increase in abnormal karyotypes at P5 then a decrease at P7. Cells from PCBM1632 showed no change in the percentage of abnormal karyotypes at any passage. A greater range in the number of abnormal karyotypes was observed among all samples in P3 as compared to P5 and P7. This difference in variance (range of 16% within P3 as compared to 5–6% for P5 and P7) suggests an underlying biological difference at P3. Due to the large difference in variance we ran Friedman’s test to check for consistent changes across the three passages. No statistically significant difference (p = 0.154) between passages was found. The mean percent abnormal karyotypes at each passage showed a trend toward decreasing abnormal karyotypes from P3 (mean = 8.0) through P5 (mean = 7.8), to P7 (mean = 3.4)(Fig. 1D). Although this small data set did not reach statistical significance, we believe they show a trend towards a decreasing number of abnormal karyotypes with passage.
Extended karyotypic analysis of 8F3560
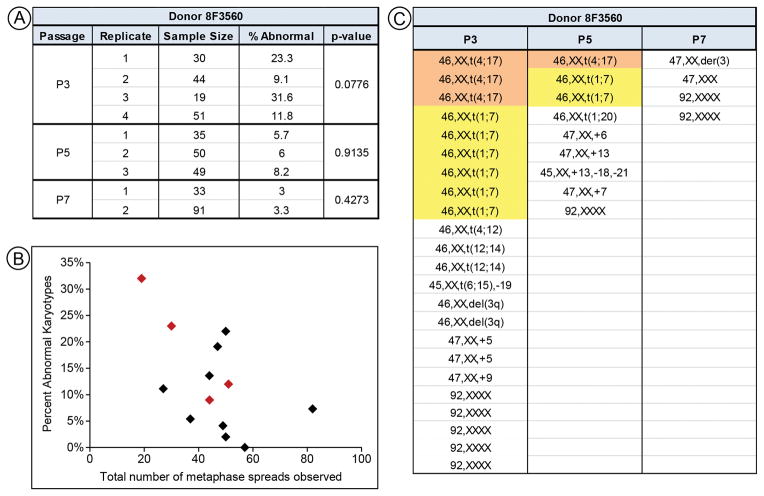
Cell line 8F3560 had two clonal subpopulations of abnormal karyotypes at P3, in contrast to other donor samples, which had random primarily aneuploid karyotypes. We further examined the karyotypic profile of 8F3560 by preparing and analyzing 144, 134, and 124 metaphase spreads for P3, P5, and P7, respectively. As our study was part of a larger analysis of MSCs using various methods [6–12], we were able to test the robustness of SKY by comparing technical replicates. Expansion involved multiple pools of cells aliquoted into individual cryovials and raised a concern that different pools might exhibit different karyotypes. We performed six repeat experiments to augment the original data. These showed a similar trend to that shown in Figure 1, including a large variance in the percentages of karyotypic abnormalities for P3 and less among those for P5 or P7 (Fig. 2A).
Figure 2.
Extended analysis on MSCs from donor 8F3560. (A) Results for individual replicates done on cells from donor 8F3560. P-values > 0.05 indicate a lack of significant differences between technical replicates. (B) Plot of the total number of metaphase spreads vs. percent abnormal karyoypes for all donors at P3. No correlation is observed. 8F3560 is indicated by red diamonds. (E) Table identifying all abnormal karyotypes from all replicates for each passage of cell line 8F3560. Clonal populations are highlighted.
The P3 data in Figure 2A suggested a possible relationship between cell populations with higher numbers of abnormal karyotypes and the yield of overall metaphase spreads. These data were plotted for all P3 lines and no linear correlation was seen (Fig. 2B).
To test the hypothesis that there are differences among 8F3560 technical replicates, we constructed contingency tables for each passage to compare normal and abnormal metaphase spreads. We failed to find evidence of sample bias in these data, as the p-values were non-significant, rejecting the hypothesis that differences existed among samples (p>0.05; Fig. 2A). This conclusion is strengthened by the observation that the same clonal populations of abnormal karyotype cells were identified in multiple pools, suggesting that they were present in the original sample.
We also obtained G-banding data for cell line 8F3560. Only P3 and P5 cells could be propagated by the karyotyping vendor. At P3, a single non-clonal abnormality, 47,XX,+5, was observed. No abnormal karyotypes were observed at P5.
Based on cumulative 8F3560 data, the total number of abnormal karyotypes decreased with passage and clonal subpopulations observed in early passages disappeared during culture. All clonal subpopulations identified in the larger analysis decrease in number from P3 to P5; none were present at P7 (Fig. 2C). Tetraploid karyotypes (92,XXXX) were observed in all passages, but they could not be identified as clonal as these can arise independently. Fewer random abnormal karyotypes were observed at P7 than at either P3 or P5. In summary, while MSCs from this donor exhibited more abnormal karyotypes, the trend towards decreasing both clonal and random abnormal karyotypes with increasing passage was consistent with other cell lines analyzed in this study.
Karyotype analysis of older donors
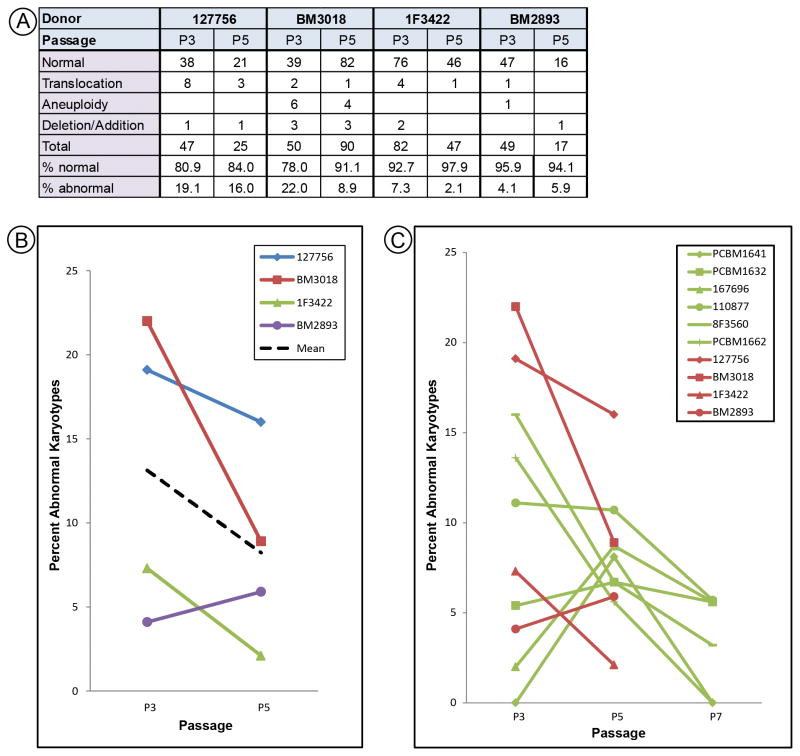
Cells from 127756 (43 y/o) were karyotyped at P3 and P5; after which they senesced. More abnormal karyotypes were seen compared to cells from six younger donors (Fig. 1C). We hypothesized that older donors may have more cells harboring abnormal karyotypes, or that cells from older donors might be less genetically stable. To test this, cells were obtained from three additional “older” donors age 39, 40, and 41, as compared to our original “younger” donors aged 22 to 31. Karyotype analysis was only performed at P3 and P5 due to senescence (Fig. 3A). Abnormal karyotypes seen in MSCs from older donors were primarily translocations, whereas cells from younger donors were predominantly aneuploid. Similar to the trends observed with younger donors, greater variance was observed among the P3 data compared to P5, although the difference was not as pronounced (Fig. 3B). In addition, the number of abnormal karyotypes dropped from P3 to P5 in three of the four lines examined. Comparison of karyotypic abnormalities from cells of all donors (Fig. 3C) did not support the hypothesis that older donors have a higher number of abnormal karyotypes.
Figure 3.
SKY analysis for MSCs from donors aged 39 to 43 years. (A) Data for total, normal, or abnormal karyotypes. (B) Percent abnormal karyotypes were graphed for each donor at P3 and P5. The dashed black line indicates the mean. (C) Combined data from cells from all donors and all passages. Old donors (red) and young donors (green) are displayed on the same plot for comparison.
Discussion and Conclusions
We used SKY to monitor chromosomal stability of hMSCs from ten donors during in vitro expansion. Karyotypes observed at P3 revealed characteristics which might be useful in deciding the suitability of cells for further use. Cells from two of ten donors exhibited clonal subpopulations of abnormal karyotypes from 2% to 10% of the total population at P3. The International Society for Cellular Therapy has suggested the presence of two identical abnormal metaphases out of 20 should be an exclusion limit for clinical use of MSCs [27].
MSCs at P3 displayed more variability in the quantity of abnormal karyotypes found than was observed for P5 or P7. We speculate this reflects the adjustment of MSCs to culture conditions as compared to the bone marrow. Others have shown that genetic and epigenetic changes occur as cells adapt to cell culture [28–30].
We examined the karyotypic profile of MSCs during extended passage. Data from P3, P5, and P7 suggest a trend towards loss of abnormal karyotypes with increasing passage. Clonal populations of abnormal karyotypes observed at P3 disappeared in later passages and random abnormal karyotypes were also less prevalent. Previous studies also report loss of aneuploidy [19, 31, 32] or abnormalities detected by CGH [33] with increased passage number. Disappearance of karyotypically abnormal cells could be due to apoptosis, cell cycle arrest or replicative senescence caused by deleterious genetic mutation. Analysis of MSCs using FISH or CGH indicates a high level of aneuploidy is present in senesced cells at late passage [5, 19, 34]. However, their lack of replicative ability suggests that they do not present a risk of tumorigenesis. Our results for ten MSCs donors indicated there is variability among donors in both the quantity and type of karyotypic abnormalities found, suggesting donor to donor variability in genetic stability. A recent paper reports nonrandom aneuploidy in 5 of 20 MSC cultures, and reoccurring abnormalities present in multiple samples from the same donor [19], further supporting that chromosomal stability may be donor-dependent. Studies of spontaneous background frequencies of DNA damage in peripheral lymphocytes find differences between gender, age groups, and smokers vs. non-smokers [35, 36]. The impact of donor variability on MSC genetic stability and function is unknown. We note that cells from donor 8F3560, which exhibited clonal populations of abnormal karyotypes, also show weak differentiation potential in a quantitative assay for adipogenesis [7].
Our data suggest that MSCs from donors of age 39 to 43 years did not uniformly have more karyotypic abnormalities than cells from donors aged 22 to 31 years, although more translocations were observed in the older age group. These data support that donor variability may be greater than any trend toward decreased genetic stability in MSCs with increasing donor age. MSCs are being studied for autologous use in regenerative medicine, so the genetic stability of cells from older donors is an important consideration.
Karyotyping is a practical way to assess genome stability and can be useful as part of an initial characterization of an MSC population. We found SKY to be a feasible method for karyotype analysis. It identifies small (1–2 Mb) or complex chromosomal rearrangements better than standard karyotyping [37, 38]. It readily identifies aberrant chromosomes even if their structural abnormalities or small size render them hard to recognize by G-banding [39]. SKY readily identifies translocations and complex chromosomal rearrangements between chromosomes [24, 25, 38]. However, it cannot evaluate certain structural abnormalities, such as inversions, deletions, and duplications within the same chromosome. Complex structural rearrangements which connect multiple non-homologous chromosomes, as are seen in tumor cells, can also give misleading interpretations with SKY [40]. Also, all karyotypic methods, which are dependent on mitosis, under represent genetic abnormality in a population that includes non-dividing cells. We and others [41] have addressed the problem of obtaining sufficient mitotic spreads as a result of the relatively low proliferative index of MSCs by protocol optimization, suggesting the method has general applicability for MSCs. Our repeat analysis of MSCs from donor 8F3560 indicates that SKY is robust and shows no signs of significant sample bias. However, the choice of the karyotyping method should be based on the type of information needed, and SKY in some circumstances should be used in combination with other methods, such as G-banding.
The majority of current literature suggests MSCs have stable karyotypes [15, 42, 43]; several publications showing transformation of MSC lines have been retracted [21, 22]. However, a recent report of genetic abnormalities and tumorigenesis in hMSCs, verified by STR analysis, indicates that this issue is not yet settled [23]. Furthermore, questions remain about the contribution of individual donors to MSC genetic stability, what role the manufacturing process has on cytogenetic abnormalities, and the effect these abnormalities may have on the ability of these cells to perform their intended function while remaining safe [27]. Our analysis indicates that SKY provides a robust method to characterization MSC lines as part of an assessment of their suitability for further use. Our data also support the general contention that MSCs expanded in culture have a relatively stable karyotype. However, karyotypic abnormalities were found and can persist clonally, suggesting that monitoring genetic stability for this cell type should be part of the characterization of lines intended for clinical use.
Acknowledgments
The authors thank Dr. Lei Huang for help in developing the metaphase spread protocol, and Dr. Thomas Ried and his laboratory for advice in the initial stages of this project. We thank Drs. Jiang Hu and Estelle Russek-Cohen for providing early statistical advice. Elaine Thompson and Kathleen McGinnis were supported by appointments to the Research Participation Program at the Center for Biologics Evaluation and Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration. We thank Drs. Cheng-Hong Wei, Brent McCright, Haritha Vallabhaneni, Patrick Lynch and Mark Mortin for comments that improved the manuscript. This work was supported by grants from the FDA Modernizing Science Initiative, the Medical Countermeasures Initiative, and research funds from the Center of Biologics Evaluation and Research, FDA.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Dominici M, Le BK, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–17. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 2.Casiraghi F, Remuzzi G, Abbate M, Perico N. Multipotent mesenchymal stromal cell therapy and risk of malignancies. Stem Cell Rev. 2013;9:65–79. doi: 10.1007/s12015-011-9345-4. [DOI] [PubMed] [Google Scholar]
- 3.Moon SH, Kim JS, Park SJ, Lim JJ, Lee HJ, Lee SM, et al. Effect of chromosome instability on the maintenance and differentiation of human embryonic stem cells in vitro and in vivo. Stem Cell Res. 2011;6:50–59. doi: 10.1016/j.scr.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Sundaram S, One J, Siewert J, Teodosescu S, Zhao L, Dimitrievska S, et al. Tissue-engineered vascular grafts created from human induced pluripotent stem cells. Stem Cells Transl Med. 2014;3:1535–43. doi: 10.5966/sctm.2014-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Zhang Z, Chi Y, Zhang Q, Xu F, Yang Z, et al. Long-term cultured mesenchymal stem cells frequently develop genomic mutations but do not undergo malignant transformation. Cell Death Dis. 2013;4:e950. doi: 10.1038/cddis.2013.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellayr IH, Catalano JG, Lababidi S, Yang AX, Lo Surdo JL, Bauer SR, et al. Gene markers of cellular aging in human multipotent stromal cells in culture. Stem Cell Res Ther. 2014;5:59. doi: 10.1186/scrt448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo Surdo JL, Millis BA, Bauer SR. Automated microscopy as a quantitative method to measure differences in adipogenic differentiation in preparations of human mesenchymal stromal cells. Cytotherapy. 2013;15:1527–40. doi: 10.1016/j.jcyt.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo Surdo JL, Bauer SR. Quantitative approaches to detect donor and passage differences in adipogenic potential and clonogenicity in human bone marrow-derived mesenchymal stem cells. Tissue Eng Part C Methods. 2012;18:877–89. doi: 10.1089/ten.tec.2011.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch PJ, Thompson EE, McGinnis K, Rovira Gonzalez YI, Lo Surdo JL, Bauer SR, et al. Chromatin Changes at the PPAR-gamma2 Promoter During Bone Marrow-Derived Multipotent Stromal Cell Culture Correlate with Loss of Gene Activation Potential. Stem Cells. 2015:10. doi: 10.1002/stem.1967. [DOI] [PubMed] [Google Scholar]
- 10.Mindaye ST, Ra M, Lo Surdo JL, Bauer SR, Alterman MA. Global proteomic signature of undifferentiated human bone marrow stromal cells: evidence for donor-to-donor proteome heterogeneity. Stem Cell Res. 2013;11:793–805. doi: 10.1016/j.scr.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Mindaye ST, Ra M, Lo Surdo JL, Bauer SR, Alterman MA. Improved proteomic profiling of the cell surface of culture-expanded human bone marrow multipotent stromal cells. J Proteomics. 2013;78:1–14. doi: 10.1016/j.jprot.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 12.Nazarov C, Lo Surdo J, Bauer SR, Wei CH. Assessment of immunosuppressive activity of human mesenchymal stem cells using murine antigen specific CD4 and CD8 T cells in vitro. Stem Cell Res Ther. 2013;4:128. doi: 10.1186/scrt339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foudah D, Redaelli S, Donzelli E, Bentivegna A, Miloso M, Dalpra L, et al. Monitoring the genomic stability of in vitro cultured rat bone-marrow-derived mesenchymal stem cells. Chromosome Res. 2009;17:1025–39. doi: 10.1007/s10577-009-9090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miura M, Miura Y, Padilla-Nash HM, Molinolo AA, Fu B, Patel V, et al. Accumulated chromosomal instability in murine bone marrow mesenchymal stem cells leads to malignant transformation. Stem Cells. 2006;24:1095–103. doi: 10.1634/stemcells.2005-0403. [DOI] [PubMed] [Google Scholar]
- 15.Ben-David U, Mayshar Y, Benvenisty N. Large-scale analysis reveals acquisition of lineage-specific chromosomal aberrations in human adult stem cells. Cell Stem Cell. 2011;9:97–102. doi: 10.1016/j.stem.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Capelli C, Pedrini O, Cassina G, Spinelli O, Salmoiraghi S, Golay J, et al. Frequent occurrence of non-malignant genetic alterations in clinical grade mesenchymal stromal cells expanded for cell therapy protocols. Haematologica. 2014;99:e94–e97. doi: 10.3324/haematol.2014.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Froelich K, Mickler J, Steusloff G, Technau A, Ramos TM, Scherzed A, et al. Chromosomal aberrations and deoxyribonucleic acid single-strand breaks in adipose-derived stem cells during long-term expansion in vitro. Cytotherapy. 2013;15:767–81. doi: 10.1016/j.jcyt.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Nikitina VA, Chausheva AI, Zhanataev AK, Osipova EY, Durnev AD, Bochkov NP. Assessment of DNA damage in human bone marrow cells and multipotent mesenchymal stromal cells. Bull Exp Biol Med. 2011;151:550–52. doi: 10.1007/s10517-011-1379-8. [DOI] [PubMed] [Google Scholar]
- 19.Tarte K, Gaillard J, Lataillade JJ, Fouillard L, Becker M, Mossafa H, et al. Clinical-grade production of human mesenchymal stromal cells: occurrence of aneuploidy without transformation. Blood. 2010;115:1549–53. doi: 10.1182/blood-2009-05-219907. [DOI] [PubMed] [Google Scholar]
- 20.Bernardo ME, Zaffaroni N, Novara F, Cometa AM, Avanzini MA, Moretta A, et al. Human bone marrow derived mesenchymal stem cells do not undergo transformation after long-term in vitro culture and do not exhibit telomere maintenance mechanisms. Cancer Res. 2007;67:9142–49. doi: 10.1158/0008-5472.CAN-06-4690. [DOI] [PubMed] [Google Scholar]
- 21.Garcia S, Bernad A, Martin MC, Cigudosa JC, Garcia-Castro J, de la Fuente R. Pitfalls in spontaneous in vitro transformation of human mesenchymal stem cells. Exp Cell Res. 2010;316:1648–50. doi: 10.1016/j.yexcr.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 22.Torsvik A, Rosland GV, Svendsen A, Molven A, Immervoll H, McCormack E, et al. Spontaneous malignant transformation of human mesenchymal stem cells reflects cross-contamination: putting the research field on track - letter. Cancer Res. 2010;70:6393–96. doi: 10.1158/0008-5472.CAN-10-1305. [DOI] [PubMed] [Google Scholar]
- 23.Pan Q, Fouraschen SM, de Ruiter PE, Dinjens WN, Kwekkeboom J, Tilanus HW, et al. Detection of spontaneous tumorigenic transformation during culture expansion of human mesenchymal stromal cells. Exp Biol Med (Maywood ) 2014;239:105–15. doi: 10.1177/1535370213506802. [DOI] [PubMed] [Google Scholar]
- 24.Schrock E, du MS, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–97. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 25.McNeil N, Ried T. Novel molecular cytogenetic techniques for identifying complex chromosomal rearrangements: technology and applications in molecular medicine. Expert Rev Mol Med. 2000;2000:1–14. doi: 10.1017/S1462399400001940. [DOI] [PubMed] [Google Scholar]
- 26.Willatt L, Morgan S, Shaffer LG, Slovak ML, Campbell LJ. Human Genetics. Springer-Verlag; 2009. ISCN 2009 an international system for human cytogenetic nomenclature; pp. 603–04. [Google Scholar]
- 27.Barkholt L, Flory E, Jekerle V, Lucas-Samuel S, Ahnert P, Bisset L, et al. Risk of tumorigenicity in mesenchymal stromal cell-based therapies--bridging scientific observations and regulatory viewpoints. Cytotherapy. 2013;15:753–59. doi: 10.1016/j.jcyt.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Nestor CE, Ottaviano R, Reinhardt D, Cruickshanks HA, Mjoseng HK, McPherson RC, et al. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015;16:11. doi: 10.1186/s13059-014-0576-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–15. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 30.Enver T, Soneji S, Joshi C, Brown J, Iborra F, Orntoft T, et al. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet. 2005;14:3129–40. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- 31.Binato R, de Souza FT, Lazzarotto-Silva C, Du RB, Mencalha A, Pizzatti L, et al. Stability of human mesenchymal stem cells during in vitro culture: considerations for cell therapy. Cell Prolif. 2013;46:10–22. doi: 10.1111/cpr.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Redaelli S, Bentivegna A, Foudah D, Miloso M, Redondo J, Riva G, et al. From cytogenomic to epigenomic profiles: monitoring the biologic behavior of in vitro cultured human bone marrow mesenchymal stem cells. Stem Cell Res Ther. 2012;3:47. doi: 10.1186/scrt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahl JA, Duggal S, Coulston N, Millar D, Melki J, Shahdadfar A, et al. Genetic and epigenetic instability of human bone marrow mesenchymal stem cells expanded in autologous serum or fetal bovine serum. Int J Dev Biol. 2008;52:1033–42. doi: 10.1387/ijdb.082663jd. [DOI] [PubMed] [Google Scholar]
- 34.Estrada JC, Torres Y, Benguria A, Dopazo A, Roche E, Carrera-Quintanar L, et al. Human mesenchymal stem cell-replicative senescence and oxidative stress are closely linked to aneuploidy. Cell Death Dis. 2013;4:e691. doi: 10.1038/cddis.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganguly BB. Cell division, chromosomal damage and micronucleus formation in peripheral lymphocytes of healthy donors: related to donor’s age. Mutat Res. 1993;295:135–48. doi: 10.1016/0921-8734(93)90015-u. [DOI] [PubMed] [Google Scholar]
- 36.Wojda A, Zietkiewicz E, Mossakowska M, Pawlowski W, Skrzypczak K, Witt M. Correlation between the level of cytogenetic aberrations in cultured human lymphocytes and the age and gender of donors. J Gerontol A Biol Sci Med Sci. 2006;61:763–72. doi: 10.1093/gerona/61.8.763. [DOI] [PubMed] [Google Scholar]
- 37.Fan YS, Siu VM, Jung JH, Xu J. Sensitivity of multiple color spectral karyotyping in detecting small interchromosomal rearrangements. Genet Test. 2000;4:9–14. doi: 10.1089/109065700316417. [DOI] [PubMed] [Google Scholar]
- 38.Nordgren A, Heyman M, Sahlen S, Schoumans J, Soderhall S, Nordenskjold M, et al. Spectral karyotyping and interphase FISH reveal abnormalities not detected by conventional G-banding. Implications for treatment stratification of childhood acute lymphoblastic leukaemia: detailed analysis of 70 cases. Eur J Haematol. 2002;68:31–41. doi: 10.1034/j.1600-0609.2002.00547.x. [DOI] [PubMed] [Google Scholar]
- 39.Imataka G, Arisaka O. Chromosome analysis using spectral karyotyping (SKY) Cell Biochem Biophys. 2012;62:13–7. doi: 10.1007/s12013-011-9285-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee C, Gisselsson D, Jin C, Nordgren A, Ferguson DO, Blennow E, et al. Limitations of chromosome classification by multicolor karyotyping. Am J Hum Genet. 2001;68:1043–7. doi: 10.1086/319503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muntion S, Sanchez-Guijo FM, Carrancio S, Villaron E, Lopez O, Diez-Campelo M, et al. Optimisation of mesenchymal stromal cells karyotyping analysis: implications for clinical use. Transfus Med. 2012;22:122–27. doi: 10.1111/j.1365-3148.2012.01134.x. [DOI] [PubMed] [Google Scholar]
- 42.Meza-Zepeda LA, Noer A, Dahl JA, Micci F, Myklebost O, Collas P. High-resolution analysis of genetic stability of human adipose tissue stem cells cultured to senescence. J Cell Mol Med. 2008;12:553–63. doi: 10.1111/j.1582-4934.2007.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sensebe L, Tarte K, Galipeau J, Krampera M, Martin I, Phinney DG, et al. Limited acquisition of chromosomal aberrations in human adult mesenchymal stromal cells. Cell Stem Cell. 2012;10:9–10. doi: 10.1016/j.stem.2011.12.005. [DOI] [PubMed] [Google Scholar]