Abstract
A prerequisite of temporal lobe epilepsy (TLE) surgery is to lateralize the disease. Recent studies have shown the capability of diffusion weighted MRI (DWMRI) in lateralizing TLE patients. This has been achieved by analyzing diffusion parameters of specific white matter tracts or regions known to be involved in the disease; however, other brain regions and connections have not been investigated for TLE lateralization. Whole brain structural connectivity using DWMRI provides a wealth of information regarding the structural connections in the brain. This information can be explored to find the most effective connections for TLE lateralization. In this work, we investigate the connectivity matrices calculated from DWMRI of 10 left and 10 right TLE patients to find the most effective connections for lateralizing the disease. Linear support vector machine (LSVM) classifier and leave-one-out cross validation scheme are used to estimate classification performance of the connectivity feature subsets. A subset of three connections with 100% classification accuracy is found. The corresponding LSVM classifier may be used to lateralize prospective TLE patients.
I. INTRODUCTION
Seizures in epilepsy are associated with the impairment of the connectivity of the brain regions. 60% of the epilepsy patients are diagnosed as temporal lobe epilepsy (TLE) patients. Many TLE patients do not respond favorably to drugs. Surgical resection of the temporal structures is a treatment option for drug resistant TLE patients. Determining the abnormal temporal lobe, called lateralization, is a prerequisite of the surgery and may require further investigation using MRI, EEG, SPECT, PET, fMRI, WADA, and neuropsychological testing [1].
Diffusion Weighted MRI (DWMRI) can reveal white matter structural changes [2] due to a disease such as epilepsy [3–5]. So far, most of the studies employing DWMRI in TLE have investigated the alternation of diffusion tensor imaging (DTI) scalar parameters such as apparent diffusion coefficient (ADC) or fractional anisotropy (FA) in specific tracts or regions. Some studies have used this information in lateralizing TLE patients [6–8]. However, these studies either combined the information acquired from DTI and other imaging modalities or used the DTI information of specific tracts to literalize the disease.
Investigating the alternation of connectivity between the brain regions due to the disease requires whole brain connectivity or connectome [9], in which the brain is modeled as a matrix, network, or graph. The brain regions constitute the nodes and the structural connections between them build the links. One can compare the whole connectivity matrix or some specific connections between two groups of individuals. Whole brain connectivity analysis can reveal major differences between the two groups. A few studies have employed this method in group analysis of TLE patients [9–11]. The findings of these studies have shown the capability of this approach in discovering and understating the structural connectivity changes due to the disease. Yet, additional information may be explored and interpreted since the connectivity matrix typically provides hundreds of measures of the brain connectivity.
Investigating features of the connectivity matrices may provide a means of discriminating left-TLE from right-TLE patients and identifying the connections that are different between the two groups. This may advance our understanding of the differences of the seizures that occur in these patients. More importantly, the features may be used in a classifier to noninvasively lateralize prospective TLE patients and reduce the clinical diagnosis cost.
In our proposed approach, first, we calculate the structural connectivity matrices for each case from T1-weighted and diffusion-weighted images. Then, using a semi-exhaustive feature selection scheme, three connections are selected that have the best performance in discriminating the two groups. Finally, a linear support vector machine (LSVM) classifier is built to lateralize prospective TLE patients noninvasively.
II. MATERIALS AND METHODS
A. Data
10 left TLE (mean age: 38.8 ± 12.4 years; 5 females) as well as 10 right TLE patients (mean age: 41.4 ± 16.6 years; 5 females) were investigated in this study. coronal T1-weighted images were acquired employing a single 3.0T MRI system (GE Medical Systems, Milwaukee, U.S.A.) using the spoiled gradient echo protocol (SPGR) pulse sequence with TR/TI/TE = 10400/4500/300 ms, flip angle = 15°, voxel size = 0.9375×0.9375 × 1.00 mm^3, imaging matrix 256×256, and field-of-view (FOV) of 240×240 mm^2. In addition, DTI images (b-value of 1000 s/mm^2) were acquired using echo planar imaging (EPI) with TR/TI/TE = 7500/0/76 ms, flip angle = 90°, voxel size = 0.98×0.98×2.6 mm^3, imaging matrix 256×256, FOV of 250 × 250 mm^2, and 25 diffusion gradient directions as well as a baseline non-diffusion-weighted image.
B. Calculating Connectivity Matrices
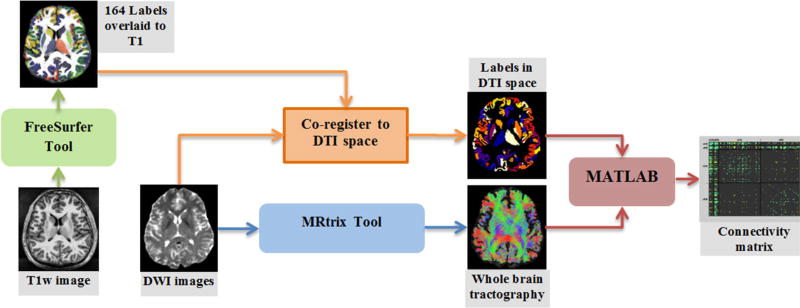
The following steps, as shown in Fig. 1 schematically, were employed to calculate the structural connectivity matrix for each case: automatic brain parcelation, tractography, co-registration, and connectivity measurement.
Figure 1.
Diagram of the method used to estimate the brain structural connectivity matrix using T1-weighted and DTI images. The steps shown in this diagram are usually employed in most of the connectivity analysis studies although the employed tools or technical parameters may vary.
Automatic brain parcelation was done on T1-weighted images employing the FreeSurfer package [12]. Using this tool, for each case, T1-weighted images were processed and labeled into several subcortical and cortical regions. In this work, we select 82 regions per hemisphere including 74 cortical and 8 subcortical regions in each hemisphere. The subcortical regions include thalamus, caudate nucleus, putamen, globus pallidus, hippocampus, amygdala, nucleus accumbens, and ventral diencephalon.
DWMRI images were processed using MRtrix package [13] to perform whole brain tractography. First, a mask was generated from DWI images to extract the brain. Then, diffusion tensor was constructed followed by calculating the fractional anisotropy (FA) map. Voxels with higher FA value than 0.7 were considered to belong to single fibers and used to build the diffusion weighted signal response profile of a single fiber. By de-convolving this response from DWI images using spherical harmonics (with the order of 6), a DWI orientation map was calculated. Using the FA and orientation maps and starting from a dilated white matter mask, the program performed a whole brain probabilistic tractography to generate 150,000 fibers longer than 10 mm and avoided from entering voxels with FA value lower than 0.1. This threshold will let the tractography algorithm to diffuse into the gray matter; where most of our interested regions are.
Skull stripped T1-weighted image (intensity inverted) was co-registered to B0 image using coarse registration in FSL [14]. The resulting transform is applied to the parcellation label map to transform the labels to the DTI space.
Having the whole fibers and the co-registered label image, the connectivity matrix was calculated for each subject by measuring the connectivity strength between each pair of regions (i,j). The FA based connectivity measure was calculated as the averaged FA value of all the fibers passing or connecting the two regions:
| (1) |
C. Feature Selection and Classification
In this work, we chose the linear support vector machines (LSVM) [15] for classification of the L-TLE patients from the R-TLE patients. LSVM has the advantage of a fast and efficient training phase as well as a tangible interpretation of the results if the number of features is low (e.g., two or three features). To reduce the number of features, we used the following semi-exhaustive searching approach to find three features from the connectivity matrix (which has 164×163/2=13,366 different entries/features) that can be used in LSVM to well classify the two groups. Note that each feature represents the connectivity between two regions of the brain.
To reduce the computational complexity, we removed the connections that are equal to zero in more than 50% of the cases in each of the groups. Moreover, we eliminated the cortical-to-cortical connections knowing that in TLE the subcortical regions are the most affected structures. The initial feature reduction step reduced the features to 1,600 for the subset selection.
In the feature subset selection procedure, first, the classification error for all subsets with one feature was calculated and the best 400 features were selected. After that, among these 400 features, all possible two-member subsets were tested and the classification error for each subset was computed. Here, only the best 200 two-subsets were selected and passed to the next step. Finally, all combinations of the selected 200 two-member subsets and the selected 400 one-member subsets were tested to find the best three-member subset. At each trial, to assess the prediction performance of the classifier corresponding to each subset, a leave-one-out cross validation test was employed.
All these steps were implemented in MATLAB (MATLAB and Statistics and Machine Learning Toolbox Release 2012b, The MathWorks, Inc., Natick, Massachusetts, United States).
III. RESULTS AND DISCUSSION
A. Selected Features
The following three features (connections) were selected from the connectivity matrix as the most effective features to discriminate L-TLE from R-TLE patients with 100% accuracy.
The connection between right accumbens area and right middle temporal gyrus
The connection between right caudate and right anterior cingulate gyrus and sulcus
The connection between left thalamus and right cuneus gyrus
First, it can be seen that two-third of the connections are in the right hemisphere, which means that the difference of the right side of the brains of the two groups is higher than the difference of the left side. Secondly, the role of some of the structures building these connections has been identified in epilepsy and discussed previously such as thalamus in [16], middle temporal gyrus in [17], caudate and cingulate gyrus in [18], and cuneus gyrus in [19]. Therefore, the selected features are consistent with the previous findings. However, to fully investigate the impact of each connection on the disease, a group comparison between each TLE group and normal subjects is needed.
B. Proposed Classifier for TLE lateralization
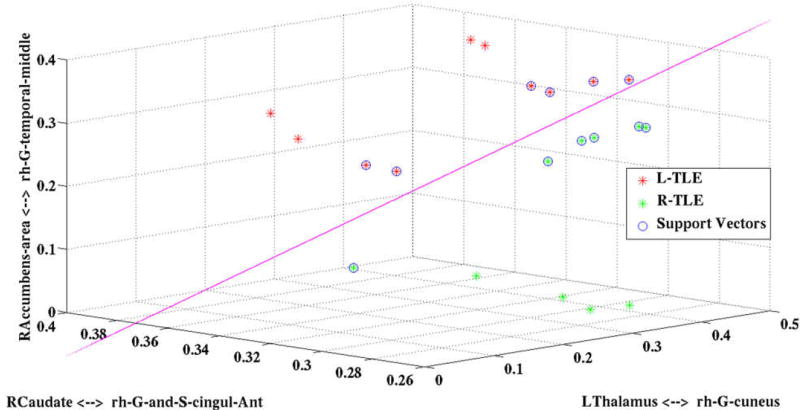
The LSVM classifier constructed based on the selected features is shown in Fig. 2 along with the support vectors. Note that the decision plane is parallel to the angle of view; therefore, it is seen as a 2D line. It can be seen that the resulting decision plane can discriminate the two groups, properly. Now, any prospective TLE patient can be mapped into this diagram to decide if the patient is L-TLE or R-TLE. Moreover, the decision can be qualified visually, the closer the patient to the decision plane, the lower confidence in the decision.
Figure 2.
The LSVM classifier developed for lateralizing TLE patients. Note that the decision plane is parallel to the angle of view; therefore, it is seen as a 2D line. The decision plane can classify L-TLE (red stars) from R-TLE (green stars) using the support vectors (blue circles). DTI connectivity features of the prospective TLE patients can be input to this classifier for lateralization of the disease.
IV. CONCLUSION
In this work, we performed whole brain structural connectivity analysis in temporal lobe epilepsy disease in order to explore effective connections that can be used in lateralization of the disease. To this end, the T1-weighted image of each case was segmented to 164 regions and the structural connectivity between each two region pairs was calculated using probabilistic tractography. From the resulting structural connectivity matrices, we extracted three highly discriminant connections and developed a linear SVM classifier to discriminate between the two classes. The classifier could classify L-TLE from R-TLE patients with 100% accuracy. The developed classifier can be used to visualize the feature values and predict the diseased side of prospective TLE patients. The decision can also be qualified visually by mapping the TLE data to the feature space. Although most of the structures in the selected features (connections) have been reported previously to be affected in TLE, more investigation and experiments are needed so one can fully interpret the role of extracted features in the lateralization. An advantage of the proposed approach is that it considered a wide population of connections throughout the whole brain. As a final note, by increasing the patient population, the proposed classifier may be modified to include other types of epilepsy like extra-temporal lobe epilepsy.
Acknowledgments
Research supported in part by NIH grant R01-EB013227.
Contributor Information
Esmaeil Davoodi-Bojd, Departments of Research Administration and Radiology, Henry Ford Health System, Detroit, MI, USA (phone: 313-874-4503.
Kost V. Elisevich, Department of Clinical Neurosciences, Spectrum Health Medical Group, Grand Rapids, MI, USA
Jason Schwalb, Departments of Neurosurgery and Research Administration, Henry Ford Health System, Detroit, MI, USA.
Ellen Air, Departments of Neurosurgery and Research Administration, Henry Ford Health System, Detroit, MI, USA.
Hamid Soltanian-Zadeh, Departments of Neurosurgery and Research Administration, Henry Ford Health System, Detroit, MI, USA; School of Cognitive Sciences, Institute for Research in Fundamental Sciences (IPM) and Control and Intelligent Processing Center of Excellence (CIPCE), School of Electrical and Computer, University of Tehran, Tehran, Iran.
References
- 1.Engel J. Biomarkers in epilepsy: introduction. Biomarkers in medicine. 2011 Oct;5:537–544. doi: 10.2217/bmm.11.62. [DOI] [PubMed] [Google Scholar]
- 2.Assaf Y, Pasternak O. Diffusion tensor imaging (DTI)-based white matter mapping in brain research: a review. Journal of Molecular Neuroscience. 2008;34:51–61. doi: 10.1007/s12031-007-0029-0. [DOI] [PubMed] [Google Scholar]
- 3.Eriksson S, Rugg-Gunn F, Symms M, Barker G, Duncan J. Diffusion tensor imaging in patients with epilepsy and malformations of cortical development. Brain. 2001 Mar;124:617–626. doi: 10.1093/brain/124.3.617. [DOI] [PubMed] [Google Scholar]
- 4.Thivard L, Lehéricy S, Krainik A, Adam C, Dormont D, Chiras J, et al. Diffusion tensor imaging in medial temporal lobe epilepsy with hippocampal sclerosis. Neuroimage. 2005 Nov 15;28:682–690. doi: 10.1016/j.neuroimage.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 5.Lemkaddem A, Daducci A, Kunz N, Lazeyras F, Seeck M, Thiran J-P, et al. Connectivity and tissue microstructural alterations in right and left temporal lobe epilepsy revealed by diffusion spectrum imaging. NeuroImage: Clinical. 2014;5:349–358. doi: 10.1016/j.nicl.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nazem-Zadeh MR, Schwalb JM, Elisevich KV, Bagher-Ebadian H, Soltanian-Zadeh H. Lateralization of Temporal Lobe Epilepsy by Analysis of Asymmetric Fractional Anisotropy in the Cingulum. presented at the 67th Annual Meeting of the American Epilepsy Society (AES); Washington, DC, USA: 2013. [Google Scholar]
- 7.Nazem-Zadeh MR, Schwalb JM, Elisevich KV, Bagher-Ebadian H, Soltanian-Zadeh H. Lateralization of Temporal Lobe Epilepsy using a Novel Uncertainty Analysis of MR Diffusion in Hippocampus, Cingulum, and Fornix, and Hippocampal Volume and FLAIR Intensity. presented at the Gordon Research Conference on Mechanisms of Epilepsy & Neural Synchronization; West Dover, VT, USA: 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pustina D, Avants B, Sperling M, Gorniak R, He X, Doucet G, et al. Predicting the laterality of temporal lobe epilepsy from PET, MRI, and DTI: A multimodal study. NeuroImage: clinical. 2015;9:20–31. doi: 10.1016/j.nicl.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sporns O. The human connectome: a complex network. Ann N Y Acad Sci. 2011 Apr;1224:109–25. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- 10.Besson P, Dinkelacker V, Valabregue R, Thivard L, Leclerc X, Baulac M, et al. Structural connectivity differences in left and right temporal lobe epilepsy. Neuroimage. 2014 Oct 15;100:135–44. doi: 10.1016/j.neuroimage.2014.04.071. [DOI] [PubMed] [Google Scholar]
- 11.DeSalvo MN, Douw L, Tanaka N, Reinsberger C, Stufflebeam SM. Altered Structural Connectome in Temporal Lobe Epilepsy. Radiology. 2014 Mar;270:842–848. doi: 10.1148/radiol.13131044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002 Jan 31;33:341–55. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 13.Tournier JD, Calamante F, Connelly A. MRtrix: Diffusion tractography in crossing fiber regions. International Journal of Imaging Systems and Technology. 2012;22:53–66. [Google Scholar]
- 14.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 15.Cortes C, Vapnik V. Support-Vector Networks. Machine Learning. 20:273–297. [Google Scholar]
- 16.Barron DS, Tandon N, Lancaster JL, Fox PT. Thalamic structural connectivity in medial temporal lobe epilepsy. Epilepsia. 2014 Jun;55:e50–5. doi: 10.1111/epi.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bandt SK, Werner N, Dines J, Rashid S, Eisenman LN, Hogan RE, et al. Trans-middle temporal gyrus selective amygdalohippocampectomy for medically intractable mesial temporal lobe epilepsy in adults: seizure response rates, complications, and neuropsychological outcomes. Epilepsy Behav. 2013 Jul;28:17–21. doi: 10.1016/j.yebeh.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riley JD, Moore S, Cramer SC, Lin JJ. Caudate atrophy and impaired frontostriatal connections are linked to executive dysfunction in temporal lobe epilepsy. Epilepsy Behav. 2011 May;21:80–7. doi: 10.1016/j.yebeh.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu M, Concha L, Lebel C, Beaulieu C, Gross DW. Mesial temporal sclerosis is linked with more widespread white matter changes in temporal lobe epilepsy. NeuroImage: clinical. 2012;1:99–105. doi: 10.1016/j.nicl.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]