Abstract
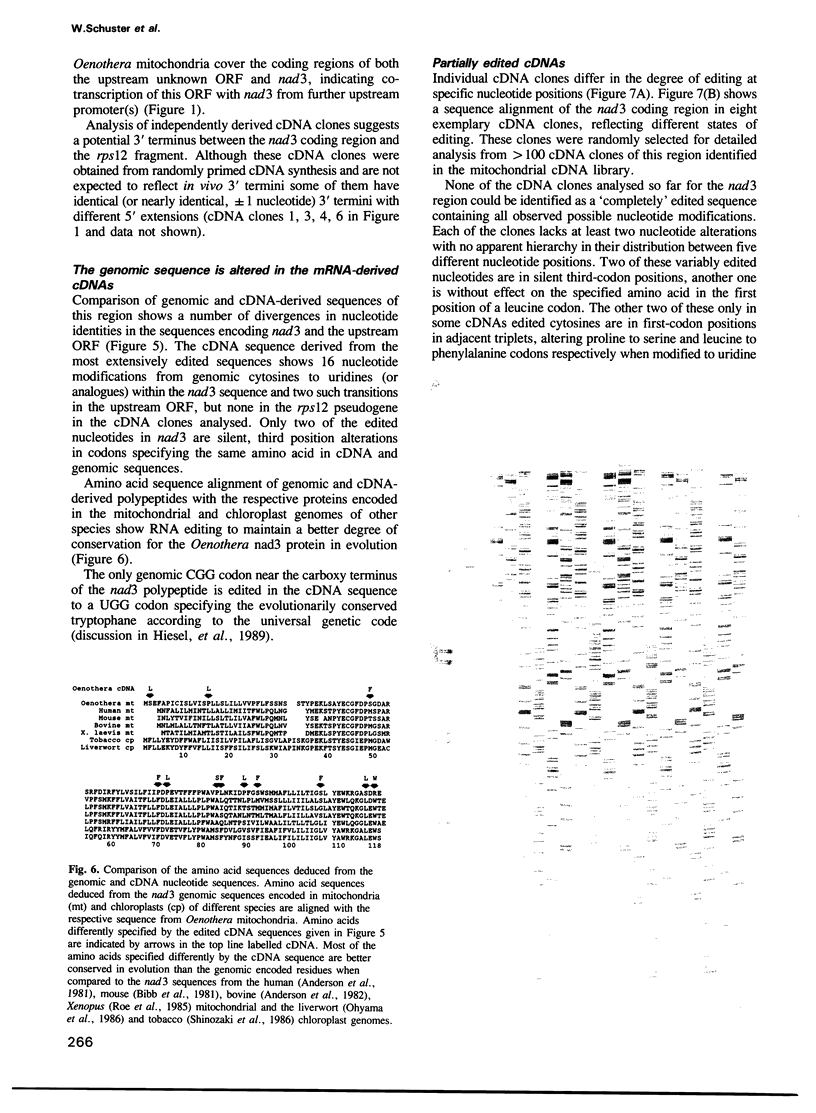
A number of cytosines are altered to be recognized as uridines in transcripts of the nad3 locus in mitochondria of the higher plant Oenothera. Such nucleotide modifications can be found at 16 different sites within the nad3 coding region. Most of these alterations in the mRNA sequence change codon identities to specify amino acids better conserved in evolution. Individual cDNA clones differ in their degree of editing at five nucleotide positions, three of which are silent, while two lead to codon alterations specifying different amino acids. None of the cDNA clones analysed is maximally edited at all possible sites, suggesting slow processing or lowered stringency of editing at these nucleotides. Differentially edited transcripts could be editing intermediates or could code for differing polypeptides. Two edited nucleotides in an open reading frame located upstream of nad3 change two amino acids in the deduced polypeptide. Part of the well-conserved ribosomal protein gene rps12 also encoded downstream of nad3 in other plants, is lost in Oenothera mitochondria by recombination events. The functional rps12 protein must be imported from the cytoplasm since the deleted sequences of this gene are not found in the Oenothera mitochondrial genome. The pseudogene sequence is not edited at any nucleotide position.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Bass B. L., Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988 Dec 23;55(6):1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Benne R. RNA-editing in trypanosome mitochondria. Biochim Biophys Acta. 1989 Mar 1;1007(2):131–139. doi: 10.1016/0167-4781(89)90031-6. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Borst P., Benne R., Tabak H. F. A fused chimeric protein made in human cells. Cell. 1989 Aug 11;58(3):421–422. doi: 10.1016/0092-8674(89)90419-4. [DOI] [PubMed] [Google Scholar]
- Brennicke A. Mitochondrial DNA from Oenothera berteriana: PURIFICATION AND PROPERTIES. Plant Physiol. 1980 Jun;65(6):1207–1210. doi: 10.1104/pp.65.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. H., Habib G., Yang C. Y., Gu Z. W., Lee B. R., Weng S. A., Silberman S. R., Cai S. J., Deslypere J. P., Rosseneu M. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987 Oct 16;238(4825):363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- Driscoll D. M., Wynne J. K., Wallis S. C., Scott J. An in vitro system for the editing of apolipoprotein B mRNA. Cell. 1989 Aug 11;58(3):519–525. doi: 10.1016/0092-8674(89)90432-7. [DOI] [PubMed] [Google Scholar]
- Feagin J. E., Jasmer D. P., Stuart K. Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei. Cell. 1987 May 8;49(3):337–345. doi: 10.1016/0092-8674(87)90286-8. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Wintz H., Weil J. H., Grienenberger J. M. The genes coding for subunit 3 of NADH dehydrogenase and for ribosomal protein S12 are present in the wheat and maize mitochondrial genomes and are co-transcribed. Mol Gen Genet. 1988 Dec;215(1):118–127. doi: 10.1007/BF00331312. [DOI] [PubMed] [Google Scholar]
- Hiesel R., Brennicke A. Cytochrome oxidase subunit II gene in mitochondria of Oenothera has no intron. EMBO J. 1983;2(12):2173–2178. doi: 10.1002/j.1460-2075.1983.tb01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan G. A., Neuhard J. Pyrimidine metabolism in microorganisms. Bacteriol Rev. 1970 Sep;34(3):278–343. doi: 10.1128/br.34.3.278-343.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L. M., Wallis S. C., Pease R. J., Edwards Y. H., Knott T. J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987 Sep 11;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Rasmussen J., Hanson M. R. A NADH dehydrogenase subunit gene is co-transcribed with the abnormal Petunia mitochondrial gene associated with cytoplasmic male sterility. Mol Gen Genet. 1989 Jan;215(2):332–336. doi: 10.1007/BF00339738. [DOI] [PubMed] [Google Scholar]
- Roe B. A., Ma D. P., Wilson R. K., Wong J. F. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985 Aug 15;260(17):9759–9774. [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L., Shaw J. RNA editing and the mitochondrial cryptogenes of kinetoplastid protozoa. Cell. 1989 May 5;57(3):355–366. doi: 10.1016/0092-8674(89)90911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K. RNA editing: new insights into the storage and expression of genetic information. Parasitol Today. 1989 Jan;5(1):5–8. doi: 10.1016/0169-4758(89)90211-1. [DOI] [PubMed] [Google Scholar]
- Wisdom G. B., Orsi B. A. The purification and properties of cytidine aminohydrolase from sheep liver. Eur J Biochem. 1969 Jan;7(2):223–230. doi: 10.1111/j.1432-1033.1969.tb19595.x. [DOI] [PubMed] [Google Scholar]
- Zalkin H. CTP synthetase. Methods Enzymol. 1985;113:282–287. doi: 10.1016/s0076-6879(85)13039-9. [DOI] [PubMed] [Google Scholar]