Abstract
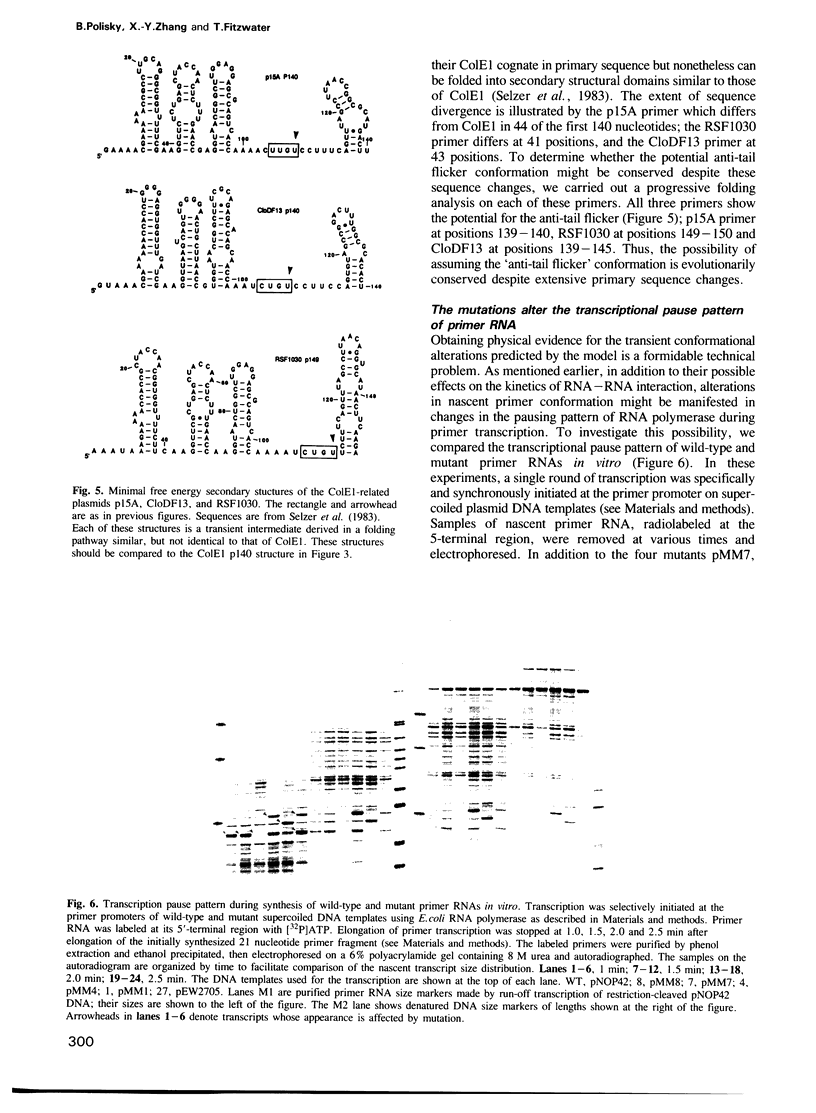
The control of plasmid ColE1 copy number is mediated by the kinetics of interaction of two complementary plasmid-encoded RNAs. One RNA is the primer precursor and the other is a small counter-transcript called RNA I. The interaction of these highly structured RNAs results in inhibition of formation of mature primer RNA necessary for replication initiation. We have studied several plasmid copy number mutants which have single base changes in the primer which render the primer resistant to inhibition by RNA I despite the fact that the mutations are located outside the overlap between primer and RNA I. We propose a model to account for the resistance of the mutant primers which is based on the differential folding of the nascent primer transcripts during transcription. We propose that the mutant primers diverge in structure from their wild-type counterparts during a discrete period during transcription. During this brief divergence, they are proposed to interact kinetically more slowly with RNA I than wild-type primer because a particular domain (the anti-tail) required for efficient interaction with RNA I is buried in a stem-loop structure while this same domain is predicted to be single-stranded in the wild-type. Despite substantial sequence divergence from ColE1, the primer precursors of the related plasmids CloDF13, RSF1030 and p15A also have retained the potential to expose their anti-tail in a similar manner to ColE1, suggesting that the folding pathway has been conserved in evolution.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF

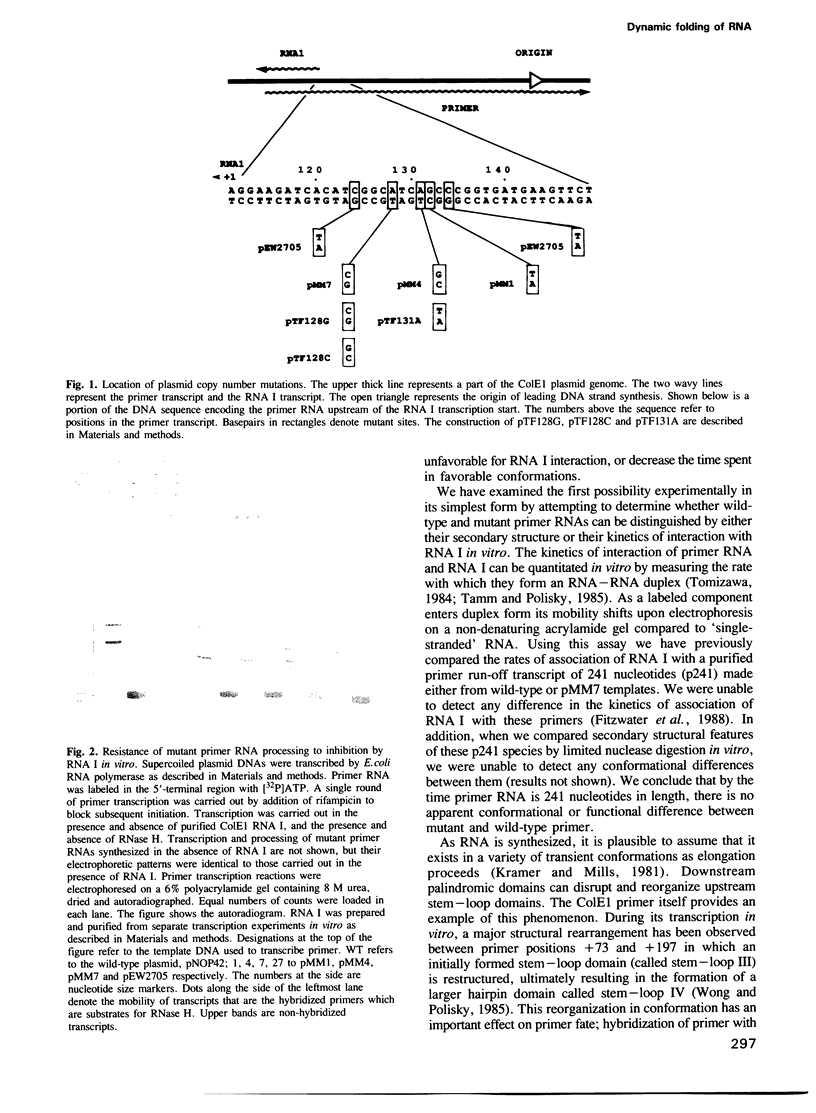
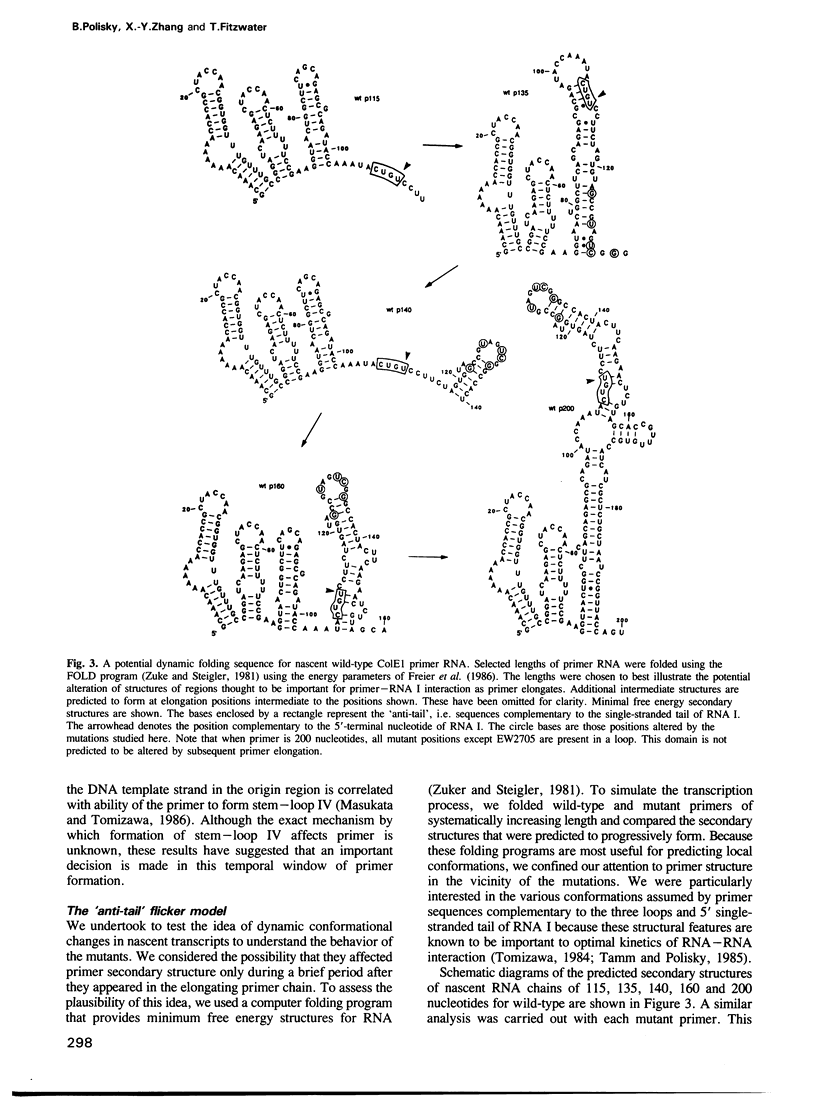
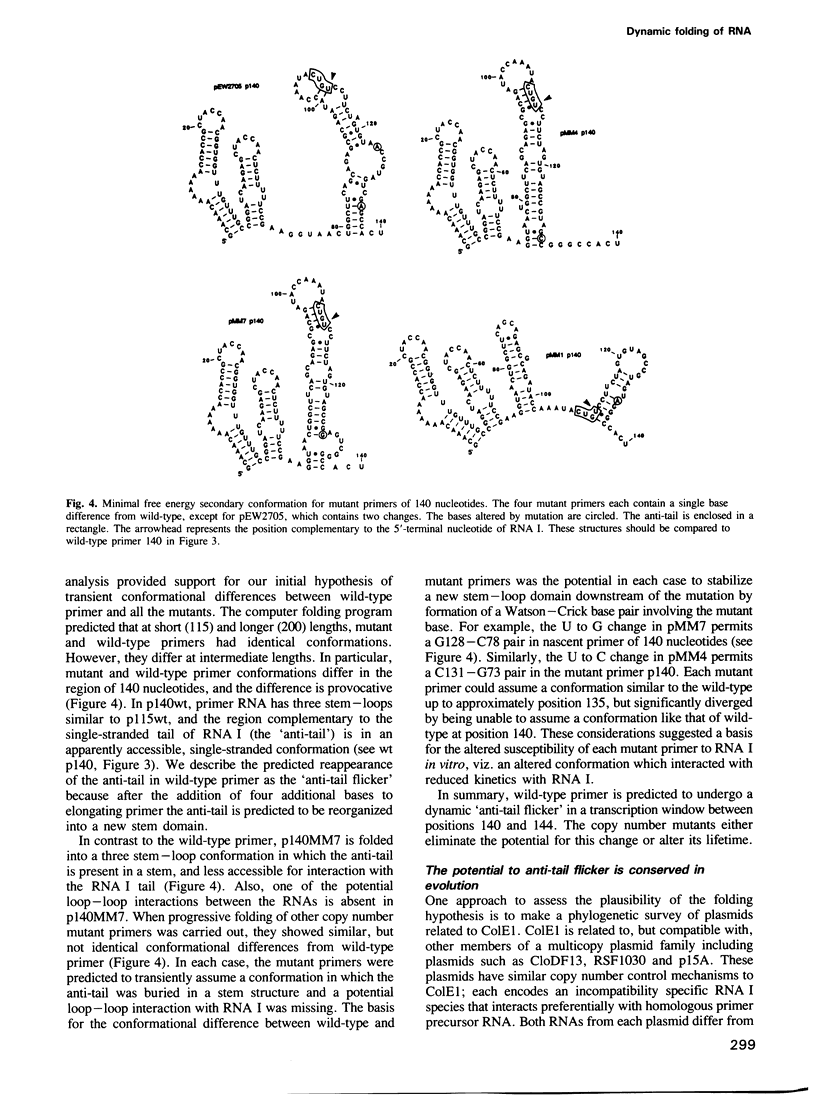




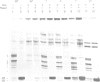
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cesareni G., Cornelissen M., Lacatena R. M., Castagnoli L. Control of pMB1 replication: inhibition of primer formation by Rop requires RNA1. EMBO J. 1984 Jun;3(6):1365–1369. doi: 10.1002/j.1460-2075.1984.tb01978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley T. P., Polisky B. Suppression of ColE1 RNA-RNA mismatch mutations in vivo by the ColE1 Rop protein. Plasmid. 1987 Jul;18(1):24–34. doi: 10.1016/0147-619x(87)90075-8. [DOI] [PubMed] [Google Scholar]
- Fitzwater T., Zhang X. Y., Elble R., Polisky B. Conditional high copy number ColE1 mutants: resistance to RNA1 inhibition in vivo and in vitro. EMBO J. 1988 Oct;7(10):3289–3297. doi: 10.1002/j.1460-2075.1988.tb03196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980 May;77(5):2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Chamberlin M. J. Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J Biol Chem. 1981 Mar 25;256(6):2777–2786. [PubMed] [Google Scholar]
- Kramer F. R., Mills D. R. Secondary structure formation during RNA synthesis. Nucleic Acids Res. 1981 Oct 10;9(19):5109–5124. doi: 10.1093/nar/9.19.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981 Dec 17;294(5842):623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- Levin J. R., Chamberlin M. J. Mapping and characterization of transcriptional pause sites in the early genetic region of bacteriophage T7. J Mol Biol. 1987 Jul 5;196(1):61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell. 1986 Jan 17;44(1):125–136. doi: 10.1016/0092-8674(86)90491-5. [DOI] [PubMed] [Google Scholar]
- Masukata H., Tomizawa J. Effects of point mutations on formation and structure of the RNA primer for ColE1 DNA replication. Cell. 1984 Feb;36(2):513–522. doi: 10.1016/0092-8674(84)90244-7. [DOI] [PubMed] [Google Scholar]
- Morita M., Oka A. The structure of a transcriptional unit on colicin E1 plasmid. Eur J Biochem. 1979 Jul;97(2):435–443. doi: 10.1111/j.1432-1033.1979.tb13131.x. [DOI] [PubMed] [Google Scholar]
- Polisky B. ColE1 replication control circuitry: sense from antisense. Cell. 1988 Dec 23;55(6):929–932. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- Selzer G., Som T., Itoh T., Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983 Jan;32(1):119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Biological regulation by antisense RNA in prokaryotes. Annu Rev Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- Tamm J., Polisky B. Characterization of the ColE1 primer-RNA1 complex: analysis of a domain of ColE1 RNA1 necessary for its interaction with primer RNA. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2257–2261. doi: 10.1073/pnas.82.8.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm J., Polisky B. Structural analysis of RNA molecules involved in plasmid copy number control. Nucleic Acids Res. 1983 Sep 24;11(18):6381–6397. doi: 10.1093/nar/11.18.6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: binding of RNA I to RNA II and inhibition of primer formation. Cell. 1986 Oct 10;47(1):89–97. doi: 10.1016/0092-8674(86)90369-7. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: initial interaction of RNA I and the primer transcript is reversible. Cell. 1985 Mar;40(3):527–535. doi: 10.1016/0092-8674(85)90201-6. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984 Oct;38(3):861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Som T. Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell. 1984 Oct;38(3):871–878. doi: 10.1016/0092-8674(84)90282-4. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Wong E. M., Muesing M. A., Polisky B. Temperature-sensitive copy number mutants of CoIE1 are located in an untranslated region of the plasmid genome. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3570–3574. doi: 10.1073/pnas.79.11.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. M., Polisky B. Alternative conformations of the ColE1 replication primer modulate its interaction with RNA I. Cell. 1985 Oct;42(3):959–966. doi: 10.1016/0092-8674(85)90292-2. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]