Abstract
This review article describes the significant recent advances in analysis of proteins by capillary and microchip electrophoresis during the period mid 2014 to early 2017. The review highlights the progressions, new methodologies, innovative instrumental modifications, and challenges for efficient protein analysis in human specimens, animal tissues, and plant samples. Protein analysis fields covered in this review include analysis of native, reduced, and denatured proteins in addition to Western blotting, protein therapeutics and proteomics.
1. Introduction
Since proteins are the functional units of the cellular machinery they provide significant information regarding the molecular basis of health and disease.1 Hence techniques to separate and isolate various proteins in a given sample are critical to studying and understanding their functional characteristics. It has been known for more than a century that proteins can travel under the influence of an electric field.2–4 However, the use of this phenomenon for routine chemical separation did not occur until the late 1930s when Arne Tiselius demonstrated that electrophoresis could be applied for separation of serum proteins (albumin, α-, β-, and γ-globulin).5, 6 This approach by Tiselius was later recognized by a Nobel Prize in 1948.7 In the last four decades massive advancements in detection modes, operation speed, automation, and miniaturization of electrophoresis have taken place. Consequently, electrophoresis became a considerable analytical technique in several basic, environmental, biomedical, forensic, and clinical laboratories.8–12 The technique in its major platforms, capillary electrophoresis (CE) and microchip electrophoresis (MCE), is characterized by high separation efficiency, fast analysis time, ease of operation, and low sample and electrolyte consumption.13–15 Different modes of electrophoresis including capillary zone electrophoresis (CZE), capillary gel electrophoresis (CGE), isotachophoresis (ITP), micellar electrokinetic chromatography (MEKC), and isoelectric focusing (IEF) interfaced with various detection modes including laser-induced fluorescence (LIF), ultraviolet (UV), and mass spectrometry (MS) have been used for protein separations and analyses in different samples.16
The aim of this review is to present substantial developments within all above fields of protein analysis by CE in the last three years. This review does not aim to be comprehensive but rather to show the major progress in protein analysis by CE and MCE. Recent review articles have been published highlighting protein analyses by CE,16 MCE,17 and in proteomics.18, 19 Our review highlights the most important developments regarding protein analysis using CE and MCE. Specifically, new advances in overcoming protein adsorption for CE and MCE methods, the use of various CE modes for protein analysis, the development of novel materials for MCE, interfacing CE with MS detection for protein analysis, further enhancements in preconcentration technique for improved sensitivity, and developments of CE/MCE in the applications of proteomics and protein therapeutics. It is hoped that by gathering these important developments into one review, it will guide future workers with respect to the current state-of-the-art for protein analysis and what is needed for these techniques to expand beyond their current niche.
2. Separation Modes and Conditions
Electrophoresis is a separation technique that separates analytes based on their charge-to-size ratios.20 While the separation typically takes place in a narrow-bore silica capillary in CE, it is usually performed in a planar microfluidic device in MCE. CE and MCE occupy a distinctive position within the separation science field and are considered in many ways as a hybrid between slab gel electrophoresis and LC and are usually compared with these two.21 Due to the high throughput, in addition to the above mentioned advantages of electrophoresis, CE and MCE are now routinely used for separation of different types of analytes ranging from small ions to large proteins and DNA fragments.
One of the most notorious limitations in protein analysis by CE and MCE is the adsorption of the analytes onto the walls of the separation channel. This adsorption is basically due to the electrostatic or hydrophobic interactions of proteins with the charged fused silica capillaries.22, 23 This adsorption is reported to contaminate the surface and cause a progressive non-uniform ζ potential across the separation compartment which leads to unavoidable band broadening and/or peak asymmetry.22 As a consequence, protein adsorption results in poor repeatability of electroosmotic flow (EOF) and migration time, reduced detector response, skewed peak shapes, and decreased resolution and separation efficiency.24–27 The adsorption to the inner surface of the separation compartment creates a major obstacle in all applications of CE with protein-containing samples whether proteins are the target analytes or not.28 Much effort in the past few years has been devoted to circumventing this problem as will be detailed below. For further detailed explanations of this phenomenon the readers are advised to access the foundational literature in the field of CE for protein analysis.29–32
2.1 CE
2.1.1 Advancements in Coatings for CZE
Capillary coatings are crucial for high-performance separations of proteins in CE and CE-MS. Intuitively, coatings are required to prevent protein adsorption phenomena onto the capillary wall and to increase the repeatability of the separations as discussed above. Capillary coatings should thus ensure high separation efficiency and good repeatability. Different coating approaches have been explored and reviewed,33, 34 including covalently linked polymer coatings, physically adsorbed polymers, and dynamic coatings using either polymeric or small molecule additives in the background electrolyte.35, 36 One of the disadvantages of dynamic coatings is the possible interference with the separation and/or detection systems, especially with MS.37 The migration of the coating material to the MS causes signal suppression and contamination of the ion source.33 For this reason, permanent coatings are more versatile and attractive than dynamic ones. However, the recent advancements in dynamic coating materials are discussed below.
It is worth noting that coating is a powerful approach, however, other strategies could be utilized for alleviation of the adsorption issue. The most feasible of these is employing an extremely high- or low- pH background electrolyte (BGE) for the separation. This approach circumvents the interaction between proteins and the capillary wall through manipulation of the net charge on the wall and the protein molecules with low or high pH BGEs, respectively.37 However, in the last two years there has been no advancements in this approach while several papers described the improvement in capillary coatings.
Among these coating approaches, the successive multiple ionic-polymer layers (SMIL) represents an attractive alternative to covalently attached polymer coatings due to the simple and straightforward preparation and possible regeneration. The application of a multilayer ionic polymer for the improvement of protein separation performances in CZE mode was investigated.38 A systematic study comparing a broad library based on nine different polyanions and eight different polycations was undertaken. Among the 72 possible polycation/polyanion couples, one third (23 couples) were evaluated for the separation of a test mixture of five intact proteins. The best results were obtained with a system composed of poly(diallyldimethyl ammonium) (PDADMA) as a polycation and Poly(L-lysine citramide) (Plc) as a polyanion. The non cross-linked PDADMA/Plc 5-layers coatings provided RSDs on migration time as low as 0.1% and about 10% on N/l (where N is the plate number and l is the capillary length) for 100 successive injections. Capillary-to-capillary reproducibility (n=3) was 1.2% on migration time and 7% on N/l. The separation performance of PDADMA/Plc was compared to that of a cross-linked poly(lysine)/polyacrylamide (Pls/PAA) 5-layers coating as shown in Figure 1. The same group studied the influence of the BGE ionic strength on the electrophoretic behavior of five model proteins.39 The separations were performed in acidic (pH 2.5) environment and counter-EOF mode using a 5 layer PDADMA/poly(styrene sulfonate) (PSS) SMIL coating. It was found that the ionic strength has a strong influence on the mobility of proteins and on the EOF. It was also evidenced that the EOF mobility decreases faster (in absolute value) than that of proteins leading to a longer analysis time. However, the effective electrophoretic mobility window was reduced with increasing ionic strength which provided unexpectedly enhanced resolution at low ionic strength (3–10 mM). Additionally, fully volatile buffers based on formic acid/ammonium formate at various concentrations were successfully used aiming for future investigation of the coating with MS detection.
Figure 1.
Electropherograms obtained for 100 successive separations of the protein test mixture using A. non cross-linked PDADMA/Plc 5-layers coating and B. cross-linked Pls/PAA 5-layers coating. For better clarity, every 10th injection is shown on the graph. Experimental conditions: capillary 40 cm (29.6 cm to the detector) × 50 μm I.D. Electrolyte: 0.5 M acetic acid at pH 2.5. Applied voltage:−30 kV. 3 min rinse with the background electrolyte between runs. Hydrodynamic injection: 30 mbar, 4 s. Temperature: 25°C. (Reproduced from reference38 with permission)
In a comparative study for separation of plant proteins, the performance of three coating types was evaluated.40 The authors systematically compared the analytical performance of hydroxypropylmethylcellulose (HPMC) coated capillaries, polyacrylamide (PAA) coated capillaries (commercially available), and capillaries modified with 1-(4-iodobutyl) 4-aza- 1-azoniabicyclo[2,2,2]octane iodide (M7C4I). The authors reported that the highest separation efficiency and repeatability were obtained with the PAA capillary. M7C4I was destroyed by the high organic solvent content in the BGE which limited its applicability for plant protein separation. HPMC was comparable to PAA, however, the RSD values for migration times were significantly higher than those for PAA.
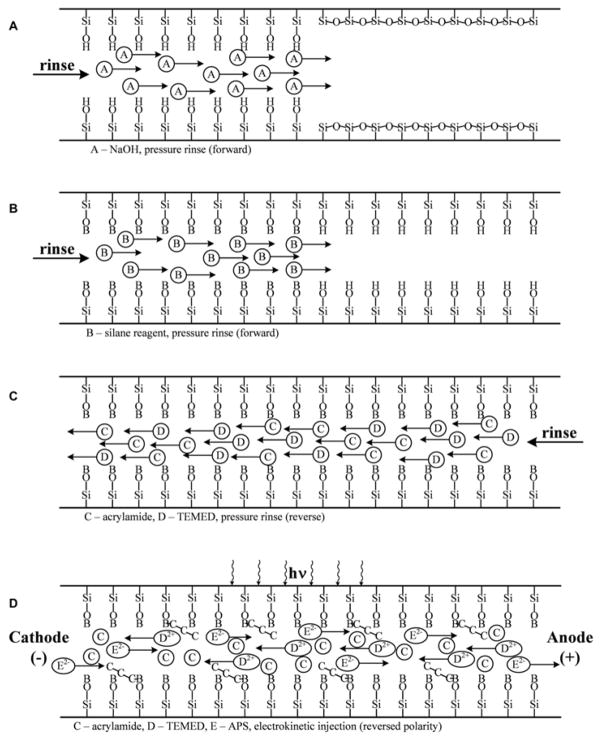
In a short communication, a fully automated covalent linear polyacrylamide (LPA) coating procedure was investigated for efficient EOF suppression and separation of proteins.41 The authors adapted the Hjerten method42 for capillary coating; however, in the current approach, the capillary stays in the commercial CE instrument during the entire coating and regeneration processes. The fully automated process took advantage of the opposite electromigration properties of the negatively charged ammonium persulfate (APS) initiator and the positively charged TEMED catalyst with the uncharged acrylamide monomer filled capillary tubing. Prior to the polymerization reaction, the 3-(trimethoxysilyl)propyl methacrylate bifunctional reagent was covalently attached to the activated (NaOH treated) inner silica capillary wall via its silane functional group, while the free methacrylate group took part in the acrylamide polymerization process. A schematic of the automated process is shown in Figure 2. The resulted capillary coating was stable for up to 100 runs and was successfully applied for the separation of a test mixture of three proteins.
Figure 2.
Schematic representation of the automated capillary coating and regeneration process (A) Rinse with 1 M NaOH solution; (B) rinse with freshly prepared aqueous solution of 4% 3-(trimethoxysilyl)propyl methacrylate; (C) filling the capillary with 4% acrylamide monomer, containing 0.05% TEMED; and (D) applying the electric field in reversed polarity mode to force the electromigration of the negatively charged persulfate ions toward the detection end and the positively charged TEMED molecules toward the injection end. UV irradiation through the detection window was used to start the free-radical polymerization reaction. (Reproduced from reference41 with permission)
In a simple capillary wall coating, the assembly of polydopamine/polyacrylamide (PDA/PAA) was examined for a single step surface modification.43 The rationale behind such combination depends on the fact that PAA contains several amide groups capable of functioning as hydrogen donor and acceptor. Consequently, intermolecular interaction is expected between PAA and a slightly alkaline dopamine solution to form PDA/PAA coating. The codeposition as well as the integrity and thickness of the coat were confirmed by atomic force microscopy (AFM), ellipsometry, water contact angle (WCA) measurements, and X-ray photoelectron spectrometry (XPS). The coat provided effective EOF suppression and was exploited for the analysis of a mixture of acidic, basic and neutral proteins. The major limitation of the work is the significantly long time to prepare the mixed coat in order to reach a relatively constant value of the coating thickness (up to 24 h for PDA/PAA-10 kDa and at least 36 h for PDA/PAA-80 kDa to reach the saturation point of the coat).
In a two layer covalent coating approach, the use of a capillary coating of poly(N-vinyl aminobutyric acid) (PVAA) was demonstrated using diazoresin (DR) as a coupling agent.44 The ionic bonding between PVAA and DR was converted into covalent bonding through UV-initiated cross-linking. The covalent coat showed good stability and repeatability (RSD% of EOF was < 0.9 run to run and < 3.6 capillary to capillary, n=5) and was applied for the separation of lysozyme, cytochrome c, amyloglucosidase, myoglobin, and BSA. The same group investigated a similar UV-initiated cross-linking of DR with cyclodextrin-derived (CD) dendrimer for creation of a covalent coat.45 The authors found that the highly branched (CD) dendrimer improved the separation efficiency when compared to the PVAA/DR system described earlier. Concomitantly, they compared the performance of DR/carboxyl fullerenes (C60-COOH) and DR/graphene oxide (GO).46 The covalently coated DR/C60-COOH capillary coatings presented good chemical stability and repeatability. The RSDs of migration times for the proteins were less than 2.5% after 100 continuous runs. Compared with DR/GO coatings prepared by the same method, the DR/C60-COOH capillary coating provided better stability due to the outstanding self-lubrication and low surface energy of C60 in the C60-COOH coatings. Recently, the same group reported a new method for fabrication of covalently cross-linked poly(ethylene glycol) (PEG) in a capillary.47 The authors used a diazotized PEG (Diazo-PEG) as a new photosensitive coating agent. The Diazo-PEG provided higher and more stable EOF suppression than DR/PEG previously reported by the same group.48
Phospholipid bilayers (PLBs) have proven useful as a capillary coating in CE analysis of proteins due to the inherent biocompatibility of the hydrated phosphorylcholine headgroup, which is highly protein resistant.49, 50 The primary drawback of this class of coatings remains their limited long term chemical and physical stability.24 In order to enhance the coating stability, the utility of hybrid phospholipid bilayers (HPBs) was studied and applied for the separation of cationic proteins.51 HPBs are formed by covalently modifying a support with a hydrophobic monolayer into which a self-assembled lipid monolayer is deposited. The authors employed 3-cyanopropyldimethylchlorosilane (CPDCS) or n-octyldimethylchlorosilane (ODCS) for modification of the capillary surface to provide a nominally hydrophobic surface upon which lipid fusion could occur. For the purpose of fusion, they used either 1,2dilauroyl-sn-glycero-3-phosphocholine (DLPC) or 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) vesicles that self-assemble resulting in a HPB. The HPB exhibited similar separation efficiencies and reduction in nonspecific adsorption compared to PLB, but with higher stability. The highest separation efficiency was obtained with CPDCS/DLPC capillaries and the lowest efficiency was obtained with ODCS/DLPC ones.
Physical adsorption of the coating material into the capillary wall is an alternative approach to covalent binding for formation of a permanent coat. Chitin is a natural polysaccharide that is known for its biocompatibility, low toxicity, and biodegradability.52 The use of native chitin as a physically adsorbed coat was reported for the first time 53. Native chitin was dissolved hexafluoroisopropanol (HFIP) to make a concentration of 0.5-mg/mL. The solution was aspirated into the capillary by application of vacuum at the capillary outlet for 40 s, then filtered air was pushed through the capillary inlet for 20 min. The physically adsorbed coat afforded efficiently stable and reversed EOF (RSD was 1.55% for within-batch coated capillaries and was 2.31% for between-batch coated ones), and this coat was applied for separation of four basic proteins. One critique to this work is that the high hydrophobicity of HFIP is neglected as a source of the success of the coating.
In a dynamic modification of capillary wall, a novel cationic coat was used for separation of model basic and neutral proteins (lysozyme, cytochrome c, ribonuclease A, and myoglobin).54 The positively charged quaternary amine type surfactant N-dodecyl-N,N-dimethyl-(1,2-propandiol) ammonium chloride (DDPAC) was synthesized and added to the BGE to function as a dynamic coating for the capillary wall. The comparison of analyses conducted with similar cationic surfactants, cetyltrimethylammonium bromide (CTAB) and dodecyltrimethylammonium bromide (DTAB), showed that DDPAC provides the best efficiency and selectivity for the separation of the studied proteins in 100 mM acetate buffer pH 5.5 with addition of 10 mM of the studied compound.
The use of dendritic glycopolymers as dynamic and covalent coatings for separation and detection of nanogram levels of albumin in biological samples was developed.55 The authors investigated the potential use of maltose-modified hyperbranched poly(ethylene imine) (PEI-Mal) either as a dynamic or a covalent capillary coat. When used as a dynamic coat, the EOF provided a satisfactory repeatability at three pH values (2.2, 8.5 and 10.2) with RSD% (n=50) ≤ 3.3, and the separation of a model mixture of four proteins (albumin, insulin, lysozyme, and myoglobin) was improved. However, the addition of the PEI-Mal to the BGE can cause unfavorable interaction between protein and the glycopolymer. Thus, the covalent approach showed an additional advantage over the dynamic one and was further used for preconcentration of analytes as further described in section 2.3.
In another important application of dynamic coating, the simultaneous separation of acidic and basic proteins has been reported.56 The authors employed Gemini pyrrolidinium (Cn-4-Cn PB) surfactants as a sole dynamic coat and compared its performance to that with hexafluoroisopropanol (HFIP) as a buffer additive. They found that the hydrophobic chain length of the Cn-4-Cn PB can distinctly affect the separation efficiency and selectivity. Moreover, the presence of HFIP can promote the formation of more homogeneous and more compact bilayer coating which was evidenced by the higher efficiency of the surfactant in combination with HFIP than the sole surfactant.
2.1.2 IEF
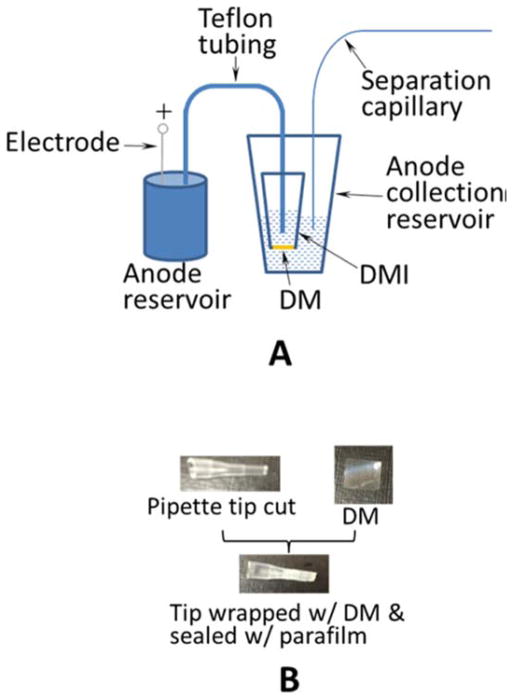
In a novel IEF mode, a simple electrokinetic means for fractionation of protein samples based on their pI values was developed.57 The key element in the apparatus described in the paper is a dialysis membrane interface (DMI) that eliminates electrolysis-caused protein oxidation/reduction and constrains proteins in the desired places. The configuration of the apparatus is presented in Figure 3 where the anode side is shown in Figure 3A and the construction of the DMI is presented in Figure 3B. Most significantly, the fractionated sample is MS-friendly since the system eliminates the use of ampholytes that interfere with MS detection. Despite its simplicity, the method suffers from low throughput which could be avoided by implementation of duplicated structures on the device to perform multiple electrokinetic fractionation in parallel.
Figure 3.
Apparatus for electrokinetic fractionation (A) Apparatus configuration of the anode-side. The apparatus on the cathode-side is image-symmetric to that on the anode-side. DM, dialysis membrane; DMI, dialysis membrane interface. Separation capillary, 45 cm coated capillary with an o.d. of 360 μm and an i.d. of 150 μm; Teflon tubing, 22 cm with an o.d. of 1.05 mm and an i.d. of 0.55 mm; anode/cathode reservoir, 100 mL container; anode/cathode collection reservoir, 600 μL tube. (B) Preparation of DMI. (Reprinted from reference57 with permission)
In a recent IEF report, a modified OFFGEL electrophoresis (OGE) for fractionation of protein extracts from Phragmites leaves was exploited.58 The samples were fractionated using a 24-well setup and the OFFGEL kit 3–10 (Agilent Technologies) and a 3100 OFFGEL Fractionator. Contrary to the manufacturer’s instructions, protein denaturing and reducing agents were not added to the OGE stock solution. The non-denaturing conditions served the purpose of maintaining the integrity of protein samples which was evaluated by quantification of peroxidase activity. Although this platform is not capillary based it is a new application of IEF and could be translated to a CE format.
2.1.3 CGE
Capillary gel electrophoresis (CGE) shows many advantages over conventional sodium dodecyl sulfate polyacrylamide slab gel electrophoresis (SDS-PAGE). These advantages include fast separation times, high capability of recovery and regeneration, on-line detection, increased analysis throughput, and ease of operation.59, 60 CGE methods allow for more automated analysis at higher voltages which leads to faster and more efficient separations. The surface-to-volume ratio of small inner diameter capillaries allows for better heat dissipation compared to SDS-PAGE and therefore higher voltages can be applied. The use of entangled polymers for CGE allows for regeneration and reuse of capillaries. In comparison to SDS-PAGE, less sample volume is required for CGE methods and that combined with on-line detection allows for much improvement on sample analysis throughput. These advantages of CGE have been shown useful for the following applications and research.
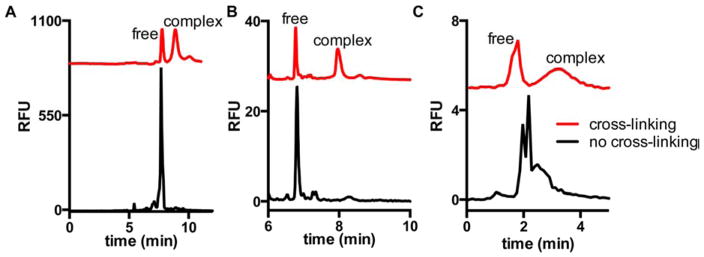
CGE has recently been used as an effective tool for the analysis of protein-protein interactions (PPI) in which binding affinities of PPIs can been more easily obtained by CE methods utilizing a protein cross linking method.61 In order to eliminate the adsorption to the capillary wall and to maintain the protein interaction during the separation, protein cross-linking capillary electrophoresis (PXCE) in a denaturing separation environment was developed. Formaldehyde was chosen as the cross-linking agent because of its short reaction time and reactivity toward many amino acid residues and the cross-linking reaction was performed prior to injection into the CGE. The PXCE technique was investigated for determining the Kd of three protein-protein complexes. The cross-linking was essential for this application, otherwise the denaturing conditions of the SDS-CGE separation will disrupt the PPI as shown in Figure 4.
Figure 4.
Electropherograms with (red trace) or without (black trace) cross-linking of protein complexes of (A) Hsp70–488–Bag3, (B) Hsp90-488 dimer, and (C) FITC-lysozyme-antilysozyme. Separation was performed with (A,B) dextran gel and (C) free solution with pH 10, 10 mM borate buffer. Cross-linking was with 1% formaldehyde for 10 min in HEPES buffer. (Reprinted from reference61 with permission)
High-throughput CGE (HT-CGE) was used for the quantification of PEGylated proteins with varying degrees of conjugation.62 Structurally stable proteins were analyzed by a standard protocol specified by the manufacturer. For proteins subjected to structural degradation, an improved analytical protocol was presented which combines trichloroacetic acid (TCA) precipitation and HT-CGE. The main benefit of integrating TCA precipitation into the analytical protocol is the immediate inactivation of the proteases and the resulting preservation of the sample composition. Moreover, TCA is assumed to precipitate all proteins in solution whereas salts and other buffer components remain in solution. Consequently, TCA precipitation reduces the concentration of interfering components from the sample which otherwise may alter electrophoretic mobility of proteins in HT-CGE.
In a newer application, CGE was investigated for analysis of viral proteins in influenza virus and virosome samples.63 The analyses were performed in uncoated capillaries using a commercial gel kit (AB Sciex). In the optimized CGE described in this paper, the influenza proteins were injected hydrodynamically at 100 mbar for 100 s which provided similar results (based on peak area, S/N, and migration time) compared to electrokinetic injection (EKI) at -18 kV for 100 s. However, hydrodynamic injection (HDI) is less affected by sample matrix and provides better precision.
2.1.4 FFE
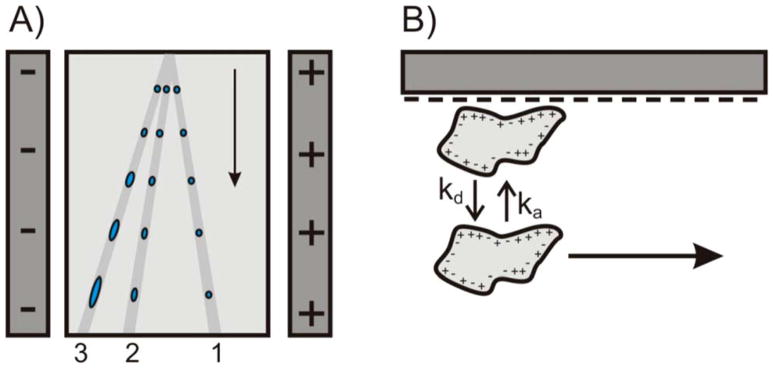
Free-flow electrophoresis (FFE) continuously separates target molecules from a sample that is exposed to an electric field.64 The electric field is directed orthogonally to a pressure-driven hydrodynamic electrolyte flow which carries the sample.65 The technique has been extensively used for protein purification and -to a less extent-protein analysis.66–68 FFE was revisited for the separation and preparation of target protein from complex sample.69 The main purpose of this work was to develop a fundamental strategy for separation of a target basic protein (cytochrome c) from other two proteins (myoglobin and BSA). To achieve this, the charge-to-mass ratio analysis was performed using CLC Protein Workbench Software which guided the prediction of the pH of chamber buffer (pH 8.0) to separate cytochrome c from the crude sample (in swine heart). In a micro FFE (μFFE) system, it was shown that surface adsorption has no effect on peak position or spatial broadening.70 Surface adsorption affected the time it takes an analyte to travel through the μFFE separation channel and therefore contributes to temporal broadening. These results were confirmed using μFFE separations of fluorescein, rhodamine 110, and rhodamine 123 in a low ionic strength buffer to promote surface adsorption. Peak widths and asymmetries were measured in both the temporal and spatial dimensions. Under these conditions rhodamine 123 exhibited significant interactions with the separation channel surface, causing increased peak broadening and asymmetry in the temporal dimension. Broadening or asymmetry in the spatial dimension was not significantly different than that of fluorescein, which did not interact with the capillary surface. The effect of strong surface interactions was assessed using μFFE separations of Chromeo P503 labeled myoglobin and cytochrome c. Myoglobin and cytochrome c were well resolved and gave rise to symmetrical peaks in the spatial dimension even under conditions where permanent adsorption onto the separation channel surface occurred. μFFE may therefore be well-suited for analysis of proteins that exhibit strong surface interactions. A schematic showing the flow paths through a μFFE separation is illustrated in Figure 5.
Figure 5.
(A) Schematic illustrating three hypothetical flow paths through a μFFE separation channel. Analyte 1 does not interact with the separation channel surface. Analytes 2 and 3 interact with the surface with increasing affinity giving rise to temporal broadening. (B) Illustration of the adsorption/desorption equilibrium of an analyte interacting with the glass surface of the separation channel. Note that the analyte is immobilized while adsorbed onto the surface. (Reproduced from reference 70 with permission)
2.1.5 CE-MS
CE coupled with MS has proven to be a powerful analytical tool for the characterization of proteins. It combines the high separation efficiency, short analysis time, small sample and electrolyte consumption, and simple instrumentation with a variety of separation modes available with the mass selectivity and sensitivity offered by MS detection.71,72 Coupling of CE and MS is most commonly carried out by electrospray ionization (ESI) using either sheath liquid, sheathless, or liquid junction interfaces. Although CE-ESI-MS coupling is less straightforward than, for example, LC coupled to ESI-MS, it is has been applied in a significant number of laboratories for intact protein analysis and applications of proteomics and protein therapeutics as discussed in section 4.73 Many challenges exist with interfacing CE to MS, but advances have been made to overcome voltage interferences and current incompatibilities with CE circuit when pairing CE with ESI-MS.74 Different CE-ESI-MS setups were explored for the separation and characterization of the two isoforms of beta2-microglobulin (β2-m) under denaturing and non-denaturing conditions.75 The authors evaluated CE-ESI-MS either coupled with sheath liquid or sheathless interfacing. The sheath liquid is known for its high robustness despite the fact that the high flow rate might result in analyte dilution which leads to inferior sensitivity. Alternatively, the sheathless approach does not lead to analyte dilution, however, it is more difficult to implement compared to the sheath liquid approach because of the circuit closure requirement for a CE separation. The sheath liquid interface was coupled either to single quadrupole (SQ) MS or time-of-flight (TOF) MS and the sheathless one was coupled as nanoESI-QTOF MS. The work demonstrated that the sheathless approach was successful in preserving the protein structure integrity, meanwhile the sheath liquid approach may not preserve the integrity due to exposure of the protein to an organic solvent. ESI-TOF-MS can discriminate conformers and reliably assign protein forms that only differ by 1 Da.
The possibility of sheath liquids to tune and modify the ionization without affecting CE selectivity and efficiency is particularly interesting in CE-MS of proteins. This way, their charge-state distribution (CSD) can be modulated while keeping the separation performance unchanged.76 To evaluate this effect, several supercharging reagents (added to the sheath liquid) with different BGEs were studied for CE-ESI-MS of intact proteins.77 The authors used three model proteins, human insulin (5.8 kDa and pI 5.2), human growth hormone (22.2 kDa and pI 5.3), and hemoglobin A0 (64.4 kDa and pI 8.4), because they present different properties in terms of molecular weight, structure, and flexibility (insulin and growth hormone are rigid and hemoglobin A0 is flexible). Among the tested supercharging reagents, 3-nitrobenzyl alcohol (m-NBA) and sulfolane permitted shift towards a higher charge state in an acidic BGE. Under basic conditions, sulfolane did not succeed in supercharging proteins while m-NBA only allowed a slight shift of the protein CSDs. This finding emphasizes that proteins that are negatively charged in solution cannot experience in-spray supercharging effect.
Affinity CE (ACE) hyphenated with MS was exploited for the first time for the assessment of PPI.78 In this study, the protease trypsinogen (24 kDa) and its inhibitor aprotinin (6.5 kDa) were selected as a model system and the separation was performed in a capillary coated with polybrene-dextran sulfate-polybrene to prevent the adsorption to capillary wall. Both CE and MS data acquired with ACE-MS, i.e. effective electrophoretic mobilities and ratio of signal intensities of free and complexed protein, can be used to determine dissociation constants. By measuring effective electrophoretic mobility shift of trypsinogen as a function of aprotinin concentration, similar Kd’s were found for all the interacting trypsinogen variants indicating comparable affinity. However, the MS-based Kd assessment suffered from a relative large spread in ESI-MS signal intensities and deviation in ESI efficiencies among free and complexed protein.
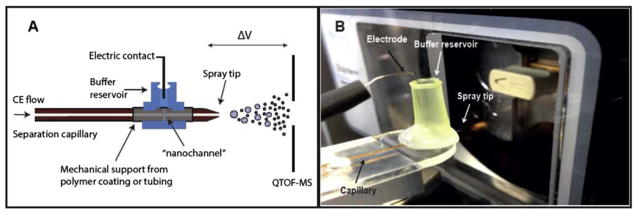
A novel and simple sheathless interface was recently described for the analysis of proteins and peptides.79 The interface represents a considerable improvement over traditional interfaces in the way that the electric contact for both the CE and ESI voltage is applied through a significantly narrow fracture close to the end of the CE capillary. The fracture has a dimension of less than 2 μm width and provides the electric contact needed for both CE and ESI. Since the capillary sections before and after the fracture are perfectly aligned and with no excess dead volume, this interface illustrates a novel approach for achieving stable electrical contact with minimum band broadening and unique field-free pumping properties. Furthermore, the current CE-MS interface can be fabricated in a simple, robust and fast manner thereby significantly reducing costs. Thus, these features of the developed CE-MS set-up should make it useful and versatile approach for high sensitivity analysis of both protein samples. A schematic of the CE-MS interface is shown in Figure 6.
Figure 6.
(A) A schematic of the novel CE-MS interface and (B) a photograph of the CE-MS interface implemented on a hybrid Q-TOF mass spectrometer (Synapt G2). (Reproduced from reference 79 with permission)
Recently, cIEF-MS based on sequential injection method was reported for separation of cytochrome c, myoglobin, β-lactoglubin, and carbonic anhydrase.80 The cIEF system was coupled to ion trap MS with an electrokinetically driven sheath flow nanospray interface (previously developed by the same group).81, 82 The authors used an acidic sheath electrolyte in the nanoelectrospray emitter and injected a short plug (3 cm) of NH4OH before the sample and ampholyte mixture (50 cm). The NH4OH section acted as a catholyte during focusing, and as the focusing progressed the plug was titrated by the acidic sheath flow electrolyte to a lower pH. Upon consumption of NH4OH by the acidic sheath flow electrolyte, chemical mobilization started automatically by the aid of the acidic electrolyte. The automated cIEF-MS/MS was performed in a LPA-coated capillary which showed low precision for migration times. This is likely due to the hydrolysis of the coat by the basic ampholyte near the distal end of the capillary which causes progressive EOF mismatch for the subsequent runs. The same group performed CZE-ESI-MS in a LPA-coated capillary for proteomics and intact protein analysis.83 In this work, the authors compared the performance of a commercial electrokinetically pumped sheath flow nanospray interface (EMASS II) with their homemade instrument.81, 82 The two systems produced nearly identical numbers of protein and peptide identifications: 399 protein identifications/1372 peptide identifications vs 388 protein identifications/1258 peptide identifications. The combination of EMASS-II with a commercial autosampler (PrinCE) was then tested for analysis of five intact standard proteins, and only 5 μL of sample was required for reproducible results.
2.2 MCE
2.2.1 Advancements in Polymers for MCE
The use of microfluidics for electrophoresis provides many advantages including short analysis time, reduced sample/reagent consumption, and high-throughput capability.21, 84–86 Microchip electrophoresis was founded on fabrication in glass microchips,87 but fabrication has been expanded to include the use of various polymers and materials including polydimethylsiloxane (PDMS),88, 89 poly(methyl methacrylate) (PMMA), 90, 91 and cyclic olefin copolymer (COC).92, 93 Advantages to fabricating microchips from polymer materials includes ease of fabrication (some without the use of a clean room), lower costs of materials, and mass-replication technologies that can allow for wider use of microfluidic devices.94–96 Such advances can improve the feasibility of microchip electrophoresis methods for clinical point-of-care applications, and polymeric materials for MCE allow for the selection of the most suitable material for particular applications.97
PDMS microchips have been the predominant material for the expansion of substrates for microchip electrophoresis, but challenges of sample loss, peak broadening, poor resolution, unstable EOF and long migration times accompany with these PDMS microchips due to factors including analyte adsorption to the inner walls of the microchannels and EOF suppression/control.98–100 Coatings have been implemented to overcome these issues;101 however, many of these methods include drawbacks of complicated procedures, expensive materials and processing, time consuming steps, and inadequate stability of the coating.102 Recent microchip electrophoresis research addresses these issues.
Polymer-based MCE, specifically PDMS microchip separation, involves the use of surface modifications typically accomplished by covalently immobilized or physically adsorbed means, including dynamic and permanent coatings.98, 103 As an alternative method, a vacuum-drying process has been implemented for coating of PDMS microchannels in a PDMS-to-glass microchip.102 This method involves a rapid procedure of 10 min vacuum-drying time resulting in suppressed analyte adsorption and stabilized EOF. A reduction in RSDs from 10.2% to 2.5% of repeated separations was achieved compared to separations in the bare microchannel. The vacuum-drying coating method is shown to be comparable to more conventional dynamic coating methods but is simpler, faster, and more durable. Several neutral and charged polymers were utilized including poly(vinylpyrrolidone) (PVP), poly(vinylalcohol) (PVA), hydoxyethyl cellulose (HEC), poly(dimethylacrylamide) (PDMA), dextran sulfate (DxS), and poly(ehylenimine) (PEI) where PVP, PVA, PDMA, and HEC provided EOF-suppressed separation conditions and DxS and PEI provided separation conditions with accelerated and reversed EOF. The electrophoretic analysis of BSA was used to compare a bare PDMS microchannel and a PVA-coated microchannel. An improvement on band-broadening and reduction of analyte adsorption occurs in the PVA-coated microchannel. The authors state that quality coating is achieved in 10 min, however the data presented for this comparison was done with a vacuum-drying coated microchip that used a 60-min coating time.
A USB flash drive style microfluidic device for electrophoretic separations at low voltages (i.e. +5–6 volts) has been described.46 The device integrates a PDMS microchip (soft lithography over capillary producing rounded channels) and printed circuit board using electrochemical detection with microelectrodes. This work allows for microchip electrophoresis on a portable platform and is powered by a laptop computer. The USB PDMS microchip separates three proteins in under 80 s. The separation channel of this device is 375-μm wide by 25 mm long causing concern about separation performance when expanded to other applications due to the short channel length and the voltage limitation. The authors stated that a separation performance improvement for a baseline separation of three proteins in under 80 s was achieved with the when compared to traditional CE. The CE apparatus they compared their MCE method to utilized a 75-μm i.d. by 41 cm capillary at +12 kV which only shows separation of two of the proteins in 12 min. However, the comparisons made by the authors between the USB PDMS-microchip system and conventional CE methods are not equal comparisons as the microchip system uses an EKI and electrochemical detection and the CE system uses a gravity-driven injection and UV absorbance detection. A PDMS microchip has been applied to the electrophoretic separation of pre-term birth biomarker and is further described in section 2.3.104
The analysis of clinically relevant biomarkers by MCE with PMMA microchips provides low cost, portability, and disposability for the potential of clinical setting implementation. The use of PMMA MCE for the separation of immune complexes was investigated where various buffers were evaluated for proper surface modification while determining the effect of these buffers on immune complex formation.105 To reduce microchannel wall hydrophobicity and prevent protein adsorption, various cellulose-based polymer dynamic coatings were assessed. Use of 1% 88 kDa methylcellulose with 0.5X phosphate-buffered saline was identified as an appropriate sieving matrix and buffer presenting an electrophoretic separation that is suitable for detection of thymidine kinase 1 in serum for early diagnosis of cancer.
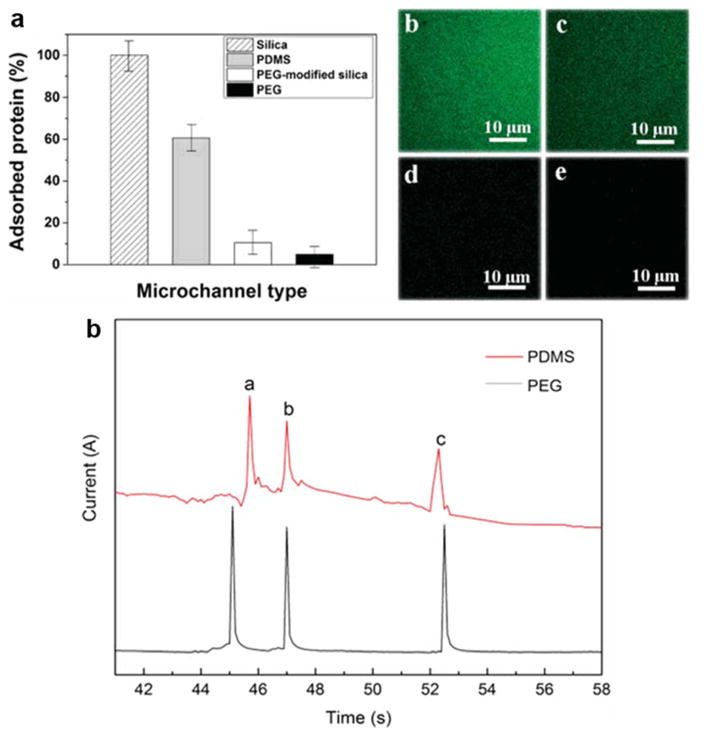
A unique approach to solving issues of protein adsorption on microchip channel inner walls has been shown with fabrication of microchips from polyethylene glycol (PEG) prepolymers.99 PEG is a commonly used polymer for coating PDMS and PMMA microchannels to prevent protein adsorption, and in this work it is employed as a material for the microchannel fabrication. The authors state that a sandwich photolithography approach simplifies the fabrication process in comparison to PDMS microchips, however it is still a multi-step process. The performance of the microchip was evaluated for protein adsorption compared to bare silica, PEG-modified silica, and PDMS. Separation performance was demonstrated with the analysis of three model proteins in under 53 s and showed superior separation performance for the PEG microchip in comparison to an unmodified PDMS microchip (Figure 7).
Figure 7.
Efficacy of PEG microchannel in preventing protein adsorption (a) and fluorescent images of FITC-BSA adsorption in silica (b), PDMS (c), PEG-modified silica (d), and PEG (e) microchannels. Comparison of the anti-protein-fouling PEG MCE with traditional PDMS MCE in protein separation performance (f). Separation conditions: buffer, 40mM phosphate (pH = 6.0); applied voltage, +200 V; sample, 0.5 mg/ml for each protein; microchannel, 14 cm × 50 μm × 50 μm (13.3 cm effective); temperature, 25 °C. Peak identification: (a) Cyt-c; (b) Lys; (c) BSA. (Modified from reference99 with permission)
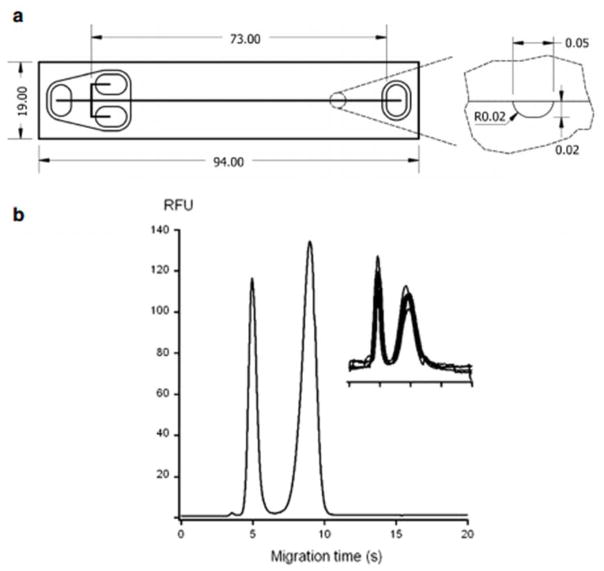
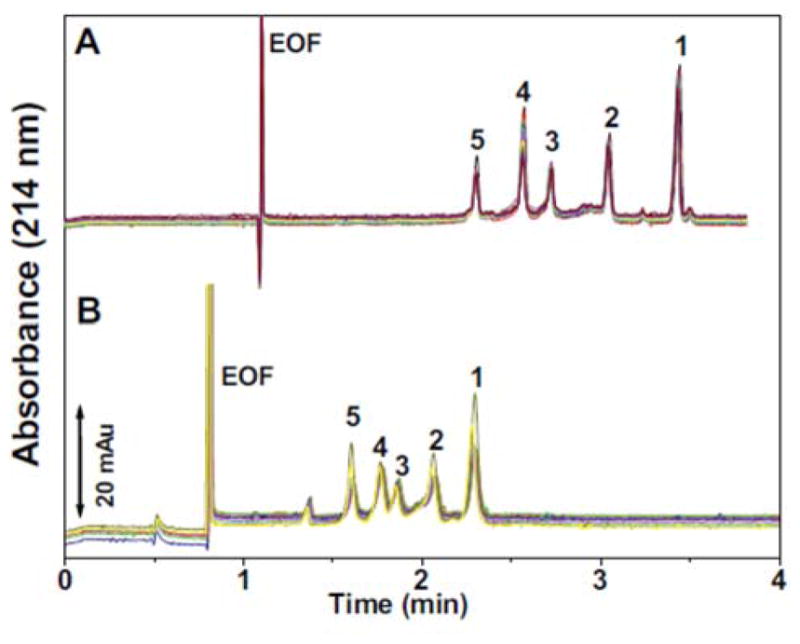
Although current polymer materials for MCE meet criteria of good separation performance and transparency for optical detection, new microchip materials can overcome issues of high fluorescence background, swelling in solvents, and provide opportunities for easier fabrication and more efficient microchannel coating. Thiol-ene substrates have been used in microfluidic techniques recently due to properties including good solvent resistance, tunable rigidity, rapid polymerization kinetics, good optical transparency, excellent chemical resistance to solvents, good adhesion to glass and metal, and temperature resistance (from -150°C to 125°C).106, 107 The material was introduced for microchip electrophoresis separations recently,107 and thiol-ene microchip electrophoresis involving microchannel surface modification via neutral copolymers for protein separations was performed.106 In this work, the initial SU-8 mold fabrication step is performed in a cleanroom. The casting of a PDMS mold, fabrication of thiol-ene microchannel and cover layers, and bonding of the thiol-ene layers is performed outside of the cleanroom.107 To obtain thiol-ene microchannels with a neutral, permanent coating, copolymerization of N,N-dimethylacrylamide (DMA), (3-(methacryloyl-oxy)propyl) -trimethoxysilane (PMA) and 3-trimethylsilanyl-prop-2-yne methacrylate (MAPS) was investigated. Several thiol-ene formulations were compared and thiol-ene with 20% excess thiols and a permanent coating of p-(DMA-PMA-MAPS) was found to have the best surface modification for increasing the hydrophilicity and minimizing protein interactions providing good EOF stability. Separations of proteins on these microchips can be done in a pH range of 3.5–9.5 and fast (5–10 s) separations were demonstrated with BSA, TI, and histone proteins. Figure 8 displays the thiol-ene microchip dimensions and channel cross-section and the separation of two proteins in under 10 s. Good repeatability is also shown with an electropherogram of five overlapping separations of two proteins.
Figure 8.
Dimensions (mm) and shape of the single-cross thiol-ene microchip (a) and electropherogram obtained from the separation of two acidic fluorescently labeled proteins (b). Analytes (in the migration time order): (1) TI-Alexa 488 at 12.5 μM and (2) BSA-FITC at 20 μM prepared in 5 mM PBS (pH 7.4). BGE: 50 mM PBS (pH 7.4). The insert in the figure displays electropherograms from five repetitive separations of TI-Alexa 488 (1.5 μM) and BSA-FITC (3 μM). (Modified from reference106 with permission)
Dyneon THV has been implemented for MCE in modes of CZE for protein analysis and for non-aqueous capillary electrophoresis (NACE).108 Dyneon THV is a transparent fluorinated thermoplastic composed of tetrafluoroethylene (TFE), hexafluoropropylene (HFP), and vinylidenefluoride (VDF), and it is a good electrical insulator with a simple, non-cleanroom fabrication processes utilizing hot embossing.109 Dyenon THV has lower EOF than fused-silica capillary and an inert surface that is easy to modify with a semi-permanent, noncovalent coating that was physically adsorbed to prevent protein adsorption. Pluronic F127 is a triblock copolymer with two hydrophilic PEO segments and one central hydrophobic PPO that was previously developed for COC microchip coating.110 This coating was compared with a PEG stearate 40 polymer, a new coating material for plastic microchips, for Dyenon THV MCE. The PEO stearate 40 coating provided an 86% reduction of protein adsorption in comparison to a 55% reduction for Pluronic F127 with the Dyneon THV microchip. The coating must be regenerated after each MCE run. With use of a pinched injection a separation of TI and BSA proteins was achieved in less than 3 min with a resolution of 1.55; RSDs of migration times were 2.9% and 1.5%, respectively.
A concentration gradient detection method based on Schlieren optics has been employed with a microfluidic device coupling electrophoresis and a multi-channel detection system for the determination of proteins diffusion coefficients.111 The diffusion coefficient is calculated in the frequency domain which improves upon conventional peak height measurements. This work uses a COC microfluidic device with multi-channels for increased sample throughput and has advantages of improved sensitivity and enhanced resolution.
Droplet microfluidics was interfaced to capillary 112 and microchip 113 electrophoresis as a method of separation and detection for the contents of individual droplets. The use of droplets with CE allows for reduction of sample consumption due to the ability to only utilize the volume of sample that needs to be injected (i.e. 10 nL) without wasting larger volumes of samples that would otherwise not be analyzed in traditional CE/MCE injection modes.114 The recent application of droplets interfaced to microchip electrophoresis for the analysis of SIRT5 enzymatic screening assay demonstrates subsecond separations using only nanoliter volumes with increased analysis throughout, and this work shows great robustness and reliability with over 11,000 MCE injections from droplets made. 115 Over 1400 samples were screened improving the throughput 3-fold relative to previous MCE-based screens and 25-fold compared to previous MCE/CE screens for sirtuin.
To improve separation efficiency compared to slab-gel electrophoresis, protein sizing on-chip via electrophoretic separation has been demonstrated using a silica particle array-based sieving mechanism for proteins sized from 6.5 to 66 kDa.116 This method of fabrication is simple for achieving pore-size ranges compared to nanolithography-derived structures. This work introduces an evaluation of pore size range from 22–103 nm for an improved separation for SDS-protein complexes compared to standard SDS-PAGE methods. The colloidal self-assembly bed was theoretically described and evaluated showing a uniform pore size and improved separation and resolution with smaller pore sizes.
2.2.2 Microfluidic Western Blotting
Western blotting is a gold standard technique widely used in biochemical and life science research for the analysis of proteins in complex biological samples. The technique in its conventional form involves the separation of proteins by SDS-PAGE, transfer of separated proteins from the gel to a membrane (typically polyvinylidene fluoride (PVDF) or nitrocellulose membranes), and an immunoassay for detection of target proteins.117 Although Western blotting has been employed for more than 30 years, constraints still exist for its throughput due to minimal automation, long analysis times, limited ability to multiplex, and large sample and reagent requirements.118 Miniaturizing and automating the Western blotting technique would increase the throughput and decrease the cost of analyzing biological samples for target proteins content, and advances have been made with MCE for this purpose.
2.2.2.1 Microchip Western Blotting
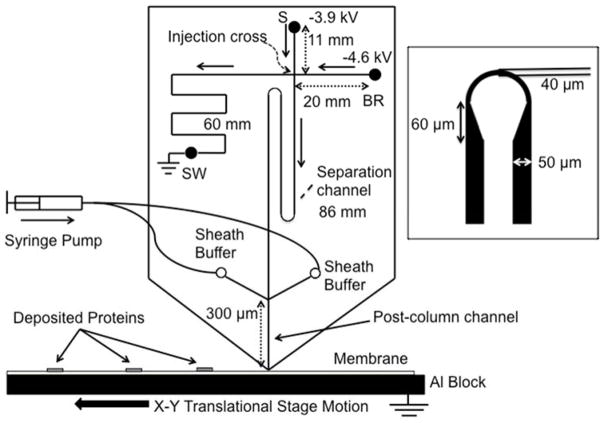
Microfluidic Western blotting can be approached by performing a gel electrophoresis separation on a microchip with deposition of proteins directly onto a PVDF or nitrocellulose blotting membrane. Sieving gel electrophoresis is performed with denatured proteins, and the microchip is diced at the end of the separation channel to be interfaced with a moving membrane on an XY translational stage that captures separated proteins.119, 120 Not only does this microfluidic device speed up the separation and reduce the sample consumption compared to conventional Western blotting, the gel-to-membrane transfer step is greatly simplified. Recent advancements in the technique have included improved separation parameters for increased resolution and the ability to easily multiplex for several targets with greatly reduced sample consumption. Detection of eleven proteins was achieved with 400 ng protein giving 100-fold decrease in sample consumption compared to three triplicate conventional Western blots.120 Resolution was improved by optimizing channel dimensions, and a channel length of 8.6 cm resulted in the ability to separate ERK1/2 which have a 5% difference in molecular weight. Injection conditions were also optimized with the addition of a preconcentration buffer using a 10 s long injection. The microchip design is displayed in Figure 9 where channel lengths are labeled. The 86 mm long separation channel was fabricated by the use of asymmetrically tapered turns to reduce dispersion. A separation field of 400 V/cm are used for microchip Western blotting.
Figure 9.
Microchip overview. Sample was injected using gated injection method. SDS-protein complexes separated by size were captured in discrete zones on the PVDF membrane moving beneath the chip outlet to preserve separation information. Sieving media was pumped through the sheath channels to ensure a stable current. Channel lengths are indicated by double arrow lines and direction of flow during separation is indicated by solid, single arrows. Separation channel was between the injection cross and the end of the channel, and length was 86 mm. Drawing is not to scale. 300 μm post channel was drawn long for clarity. The inset figure shows the asymmetric turn used to reduce geometric dispersion. (Reproduced from reference120 with permission)
A method of MCE involving on-chip immunoprobing has been developed as an alternative to conventional Western blotting.121 This microfluidic platform utilizes a photoactive gel to capture sieved proteins by photoinduced cross-linking of the proteins to the sieving gel followed by in situ immunoprobing for target proteins where multiple μWesterns can be done in parallel. Recent expansion of this work has involved optimizing the sieving gel composition to achieve separation of low molecular weight proteins.122 Previously, μWesterns of low molecular weight proteins was challenging due to the need for a sieving gel to effectively separate small proteins and also allow larger molecular weight antibodies (150 kDa compared to 6.5–28 kDa proteins) to migrate through the gel for in-situ immunoprobing. In this work, a Tris tricine discontinuous buffer is implemented for improved destacking and subsequent separation of low molecular weight proteins. The gel fabrication was altered to introduce a swept antibody plug that prevents antibody accumulation throughout the analysis, and separation of a molecular mass range of 6.5–116 kDa was achieved. The Tris tricine system also resulted in 40% higher separation resolution with a 100-fold improvement on signal-to-noise ratio for small pore gels.
2.2.2.2 Single Cell Western Blotting
The μWestern technique has been further expanded to single-cell Westerns providing the ability to measure cell-to-cell variation with high specificity.123 Single-cell Westerns are achieved with in situ lysis in microwells where a 30-μm-thick photoactive polyacrylamide gel layer allows for μWesterns of proteins from the lysed cells. The single-cell Western achieved multiplexed detection of eleven protein targets/single cell where simultaneous assays of about 2000 cells were achieved in 4 h where separation of molecular mass differences of about 50% was achieved. This work also has the ability to interface fluorescence activated cell sorting (FACS) for analyzing subpopulations of about 200 cells with single-cell resolution.
Signaling proteins that are often of interest in single-cell analysis contain a broad molecular mass range. To further enhance single-cell μWesterns, improvement upon resolving issues for large molecular weight ranges was made. 124 This was achieved by development of a sieving matrix with a spatially modulated pore-size gradient. A fabrication approach combining grayscale photolithography and glass-SU8 mold was used for patterning high spatial resolution gel densities. In a 1 mm long separation lane proteins spanning 25–289 kDa were efficiently separated due to the pore-size gradient. The use of a nonuniform pore-size gel alone would cause nonuniform, spatially biased immunoassay performance depending on the molecular mass of target protein. To overcome this, a temporally modulate cross-linker density is used to expand the hydrogel pores to prevent biased in-gel immunoprobing of immobilized proteins.
The same group recently reported a two-layer, lab-on-a-disk device has been created to integrate cell handling for cytology of sparingly available cells with single-cell Western blotting.125 Use of centrifugation for cell preparation allows for a more robust integration with single-cell Westerns for cell samples with as a few as 200 cells/low density cell suspensions with minimal cell loss. An array of microwells is molded to a polyacrylamide gel electrophoresis layer. U-shaped “cell trapping” dams are located above and around microwells. A centrifugal force deposits cells in the microwells, and in-situ lysis and electrophoretic separation occurs with in-situ antibody probing of targets to follow.
2.3 Improving Sensitivity with Preconcentration Techniques
One of the most cited limitations in CE and MCE is the poor detection sensitivity; particularly when compared to traditional separation techniques such as HPLC.126–129 Due to this inferior concentration sensitivity, various preconcentration methods are employed before or during the electrophoretic separation for enrichment of proteins. The development of efficient preconcentration strategies is of absolute necessity especially when dealing with biological samples where target proteins are usually present in low concentration.20
Large volume sample stacking (LVSS) and field-amplified sample injection (FASS) have been investigated for the preconcentration of a mixture of four proteins.55 LVSS allows for the injection of multifold larger sample volume than traditional HDI.130 Thus, a large volume of a sample plug dissolved in a low conductivity matrix (water solution) was hydrodynamically injected into a PEI-Mal-coated capillary which was filled prior with the acidic BGE solution (described in section 2.1.1). Since the investigated proteins were positively charged at acidic pH values, they migrated to the end of the capillary under the positive voltage. Meanwhile, the reversed EOF slowly pushed the aqueous matrix to the injection side. This counter movement led to a concentration of the positively charged analytes on the boundary between the sample and buffer solutions. LVSS provided enhancement in detection sensitivity of up to 82-fold and the LODs were down to 1 μg/mL. In FASI, the sample (prepared in water) was injected electrokinetically at 15 kV for 90 s. This provided an enhancement in sensitivity up to 1300-fold and the LODs were down to 0.1 μg/mL.
A powerful dual approach for the enrichment of dilute BSA tryptic digest and E. Coli digest has been reported.131 The authors employed a strong cation exchange (SCX) solid phase extraction (SPE) wherein samples were loaded in acidic buffer and eluted in using a basic buffer. Samples were then analyzed using an acidic BGE in CZE mode. This buffer combination resulted in the formation of a dynamic pH junction which further enhanced the detection sensitivity. This combination resulted in 3000-fold in sensitivity enhancement when compared to standard EKI. Recently, they almost repeated the extraction and preconcentration system in LPA-coated capillary.132 They were successfully able to obtain about 1000 protein identification from Xenopus laevis zygote homogenate as detailed in section 3.1.
In a two-dimensional gel electrophoresis (2DE) approach, a simple and efficient extraction protocol was optimized for enrichment of lipophilic proteins of Mycobacterium tuberculosis.133 The authors added Triton X-100 in the protein extraction buffer followed by Triton X-114 (for phase separation) directly which eliminated the need for membranes preisolation. The addition of the surfactant to the extraction buffer improved the solubilization of the lipophilic proteins which resulted in 2–3-fold enrichment in MALDI-TOF MS detection of target proteins.
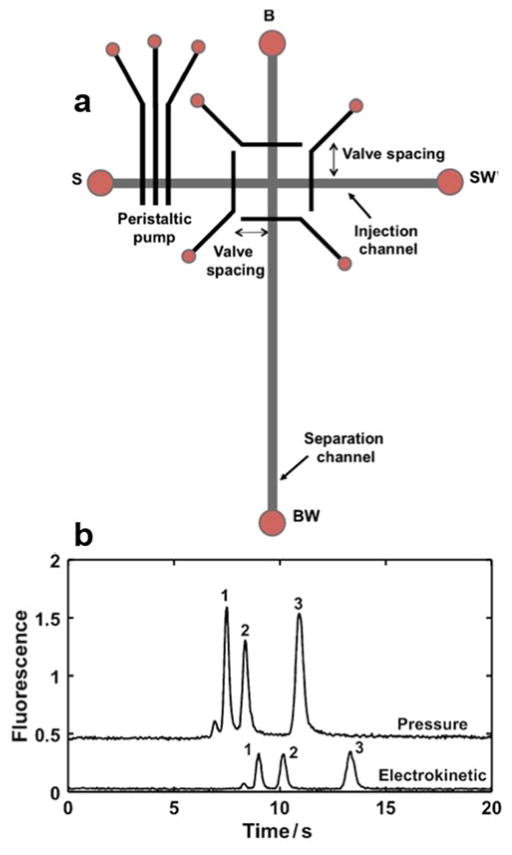
Despite the fact that EKI is the major sample introduction mode in MCE, a novel system for pressure injection and stacking of pre-term birth biomarkers (PTB) in a microfluidic device was reported.104 As illustrated in Figure 10, the device features a multilayer PDMS microchip. The fluidic layer contains injection and separation channels, and the control layer contains a peristaltic pump and four pneumatic valves around the T-intersection to carry out sample injection and plug capture. An unpatterned PDMS membrane was sandwiched between the fluidic and control layers as the actuated component in the pump and valves. The authors studied the effects of the peristaltic pump actuation rate and injection time and chose a 50-ms actuation rate and a 30-s injection time that offered a good combination of speed, peak height, and number of theoretical plates (as much as 500,000 plates/m or more). They evaluated four different valve spacings (100, 200, 300, and 400 μm) and chose 200 μm for providing the best peak height. Interestingly, when compared to EKI, the HDI scheme enhanced the detection sensitivity of the PTBs by a factor-of-4 with efficient elimination of the injection bias.
Figure 10.
(A) Top-down view of the microchip design showing peristaltic pump and pneumatic valves around the T-intersection. Four different valve spacings were used. S sample, SW sample waste, B buffer, BW buffer waste. (B) Comparison of pressure driven Vs electrokinetic injection with 30 s injection times. Electropherograms are offset vertically. For pressure injection, a 50-ms actuation rate and 200-μm valve spacing were used. (Reproduced from reference104 with permission)
The integration of SPE and MCE on a single device was exploited for the enrichment of a model protein (ferritin).134 The system features a multilayer microfluidic device consisting of a pneumatic peristaltic pump, fluid control valves, a porous polymer monolith for SPE and a microchannel for electrophoresis. The porous polymer monolith column (acrylate-based monolith with C8 functional groups) was synthesized using UV photopolymerization. Fluidic and control channel dimensions were optimized to actuate valves with 30 psi and produce reasonable flow rates. The preconcentration capability of the SPE unit showed a 4-fold enhancement and the loading capacity of the polymer monolith was 56 fM (25 ng) of ferritin.
Nanoporous membranes have been used as a preconcentration device in microfluidics for several years.135–137 The membranes can provide a powerful tool to interface vertically separated microfluidic channels to create a 3-D fluid architecture.138 Recently, a microfluidic device featuring two nanoporous membranes with different sizes was employed for preconcentration of albumin in urine samples.139 The upper membrane containing larger (100 nm) nanopores acted as a size-selective filter passing out particles, cells and larger proteins, ensuring high robustness and reliability for analyzing untreated samples. The lower, smaller pore sized membrane (10 nm) permitted the transport of inorganic ions and small organic ones but not proteins allowing them to be concentrated. By using this dual membrane device, the analysis of HSA in urine samples was feasible and enabled quantitative determination for the diagnosis of albuminuria. Later on, a fiber-based microfluidic methodology was developed for the isolation, separation and preconcentration of biomolecules.140 R-phycoerythrin (R-PE), a 240 kDa protein, was used as a test target solute. The main advantage of fibers over classical and closed microfluidic channels is the simplicity of being sectioned (cut) into parts to transport or store the desired sample zone as the solutes and fluids remain spatially separated upon the surface.
A rapid and efficient assessment of therapeutic protein (TP) activity based on electrokinetic concentration and molecular charge modulation was described.141 In a previous report,142 the authors stated that electrokinetic concentration could simultaneously preconcentrate and separate bound and unbound species in aptamer-protein binding assay. However, the usefulness of sole electrokinetic concentration for TP and TP receptor (TPR) binding assays is quite limited. This is mostly due to the minimal mobility differences in case of TP-TPR assays. Molecular charge modulation overcomes the inferior separation resolution of electrokinetic concentration by artificially enhancing the mobility difference with mobility modulators, disregarding the intrinsic mobility of TP, TPR, and the TP-TPR complex.
3. Applications of CE and MCE
3.1 Proteomics
The use of CE for proteomics represents a large body of research. Recent reviews highlight the use of CZE and CE-ESI-MS for bottom-up proteomics,18, 19, 143, 144 and CE and MCE methods for top-down proteomics including investigation of post-translational modifications (PTMs).18 These reviews include advances that have been made for CE interfacing to ESI-MS, electrospray ionization-tandem mass spectrometry (ESI-MS/MS), and matrix-assisted laser desorption/ionization mass spectrometry (MADLI-MS). A review in early 2017 covered the combination of electrokinetic and chromatographic techniques for proteomics to highlight the use of these complimentary techniques for advancing protein identification. Literature is cited to show coupling of “top-down” and “bottom-up” proteomics for efficient large-scale profiling with a study of specific sub-proteomics applications.145 The most recent advances made in this application are described below.
CZE-MS/MS of protein digest has been utilized for a bottom-up approach for proteomic analysis.132 In this work the CZE was combined with an online sample reduction, alkylation and digestion to identify 975 protein groups and 3749 peptide groups. This work has low sample consumption, which combined with the reduction of sample preparation time which greatly improves the throughput of proteomic analysis.
Analysis of 580 yeast proteoforms for intact protein characterization was accomplished with CZE-ESI-MS/MS using a top-down approach.146 The proteoform was prefractionated into 23 fractions and with use of CZE and intact detection and fragmentation 580 proteoforms and 180 protein groups were identified. This CZE method employed dynamic pH junction and LPA-coated capillary to improve loading volume and peak capacity. An advantage of CZE for proteomics work is that minor sequence variations/PTMs have charge differences but with similar hydrophobicities that can be separated by CZE that complementary reverse phase LC techniques would be unable to separate. Identification of PTMs including N-terminal acetylation, signal peptide removal, and oxidation was accomplished with this method. CZE coupled with high resolution MS was also demonstrated for the characterization of intact proteoforms with charge and glycan heterogeneity using a top-down approach.147 CE was performed using a cross-linked polyethylenimine coating for separation based on charge and hydrodynamic volume differences where column efficiencies were between 350,000 – 450,000 plates. This work shows advancements towards an in-depth characterization and quantitative analysis of biopharmaceutical proteoforms. This is demonstrated by the separation and resolution of deamination and siltation charge differences and for isobaric positional isomers of a single sialic acid on biantennary glycan antennae.
3.2 Protein Therapeutics
The field of monoclonal antibody (mAb) based therapeutics largely contributes to successful drugs for health issues including inflammation, infectious diseases, autoimmune disorders, and some cancers. These therapeutics are suitable due to high specificity, high efficacy, and great pharmacokinetic properties making efficient analysis tools vital to therapeutic and drug discovery research.148, 149 With the implementation of recombinant protein therapeutics due to their high specificity and potency come the need for purity and quality control analysis.150 mAb’s contain tetrameric glycoproteins, and the Fc regions of heavy chains contain N-glycosylation sites that contribute to heterogeneity of these molecules.151 Complex bio-synthesis pathways and PTMs cause changes in structure and function that impact mAb stability and efficiency. Thus, improvement upon analysis techniques to enable fast and accurate characterization of biotherapeutics is necessary.71, 150
A recent review on CE characterization of mAbs highlights many recent publications for the advancements of CE for protein therapeutics, and the publication includes a table of separation conditions for the analysis of IgG, biosimilars, Fc-fusion proteins, and antibody-drug conjugates (ADCs) in the years 2010–2015.152 In the highlighted work from 2015 and early 2016 CE-ESI-MS analysis was developed for the analysis of reduced mAb with heavy and light chains153 and analysis of intact IgG1.149 A CZE method with cationic capillary coating for the identification of mAb in routine mAb for cancer therapy in hospitals was reported.154 Characterization of ADC was performed by sheathless CE-MS/MS with nanoESI infusion for detection of species with different number of conjugated drugs has also been described.155 A characterization of therapeutic antibodies by CE-ESI-MS allows for rapid sequencing and degradation hotspot analysis, and this work achieved enhanced sensitivity for poorly ionizing molecular species like glycoproteins.156
Glycan analysis is an important direction for clinical and biomedical research because of the deep involvement of glycans in biological processes. Recently, a multiplexing CE mapping method for glycan analysis was developed.157 The authors investigated the potential of combined parallel application of CZE, MEKC, and CGE for rapid and accurate identification of glycan. The covariance and correlation coefficient study indicated that CZE and MEKC mechanisms provided low covariance and high orthogonalities, and therefore, a larger space for the accurate identification of glycan using multiplexing platform. The CZE and MEKC cross-validation method was applied to the identification of N-glycans released from two human IgG samples.
A CZE method utilizing a neutral coated capillary and a simple buffer system was developed for the separation of glycoforms of glycoprotein.158 The method has been shown to have excellent specificity, precision, and accuracy. The peaks separated by CZE were characterized using orthogonal methods, and peak identification was determined by a combination of MALDI-MS and high performance anion exchange chromatography. The peaks demonstrated to correlate with isoforms containing different numbers of sialic acids within the protein. The CZE method has been widely applied as an in-process purity method during the development of this glycoprotein; providing much needed purity information in a high throughput manner. Finally, the method has also been used for stability testing of the purified glycoprotein.
Recent publications described the analysis of host-cell protein impurities in recombinant mAb therapeutics.159, 160 CZE-ESI-MS/MS analysis is used and variations on techniques were applied including coupling a strong cation exchange-solid phase extractor. A comparison to LC is made highlighting that CZE generates more protein identifications. Method development for CZE of therapeutic mAb was done utilizing an ε-aminocaproic acid (EACA) buffer system with a top-down and middle-up analysis approach.161 The phrase “two-phase-four-step” was introduced for this method that is capable of resolving isoforms of mAbs. The two phases include a screening phase with pH and ionic strength adjustments and a fine tuning phase with adjustments on organic additives and viscosity enhancers for optimal peak resolving and low migration times. Separation of a commercially available mAb isoforms was done and the repeatability was represented by 0.8% RSD of peak area and 5% RSD of migration time.
The Fc region of mAbs has consequences for Fc-mediated effect or functions that might be desirable for therapeutic applications. CZE-MS with native MS infusion was performed for precise characterization of variants in cetuximab.150 The role of N-glycosylation and C-terminal lysine truncations were evaluated with CZE-UV/MALDI-MS and CZE-UV/ESI-MS and Fc/2 dimers were also characterized. Figure 11 displays separation of Fc/2 region and the impact on resolution from glycosylation and lysine truncation.
Figure 11.
Analysis of the Fc region of the mAb and with off line CZE-UV/ESI-MS separation of middle-up cetuximab charge variants (a). Experimental conditions: Inlet BGE: EACA 200 mM ammonium acetate 25 mM pH 5.70 and outlet BGE: ammonium acetate 25 mM pH 5.70; HPC-coated capillary, total/effective length 82/75.5 cm × 75 μm i.d.; voltage, 20 kV; UV absorbance at 200 nm. Impact of glycosylation and lysine truncation on electrophoretic resolution. Electrophoretic separation of (1) cetuximab with IdeS treatment, (2) cetuximab with IgGZERO + IdeS treatment and (3) cetuximab with CP-B + IdeS treatment (b). Inlet BGE composed by a mixture of EACA 200 mM and ammonium acetate 25 mM pH 5.70 and outlet BGE by ammonium acetate 25 mM at pH 5.70. (Modified from reference150 with permission)
cIEF-CZE-MS for fast characterization of therapeutic antibodies has been done; overcoming challenges of cIEF and MS compatibility with a separation step to remove the nonvolatile species.162, 163 For compatibility with small transfer volumes and cIEF high voltages a multi-port valve and sample loop system was implemented,163 and an evaluation of sample transfer loop volumes for efficiency and peak width was done.162 IEF separation for pI range of 5–10 is applied to the analysis of charge variants of mAbs. In 3 h up to 6 analytes were transferred from cIEF to CZE-MS with the ability to detect a mass differences of 2 Da between mAb variants.
Advances in electrophoretic methods for analysis of protein therapeutics have been shown using MCE with online ESI-MS detection. This technique has been applied to assess structural variations of therapeutic mAbs that can impact their immunogenic properties.164 In order to get proper analysis of mAb modifications that vary in net charge, Zwitterionic BGE additives typically used in the separation were replaced with microchannel modification by PEG through a chemical vapor deposition process for a compatible interface of MCE with ESI-MS.165 In this work, variants of Infliximab with a 128 Da difference were analyzed. Analysis of ADC variants166 and glycated blood proteins for a diagnostic tool with diabetes have been accomplished with similar approaches.167
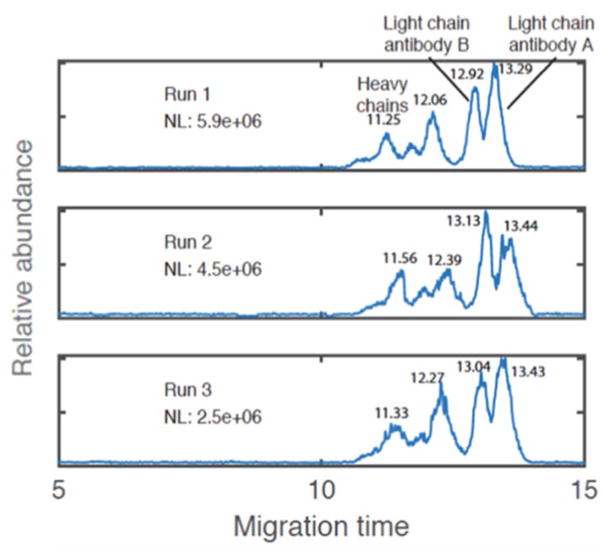
Recent advances have been made with CZE-ESI-MS for separation of reduced mAbs into their heavy and light chain components utilizing a commercially available LPA coated capillary.168 In less than 10 min, heavy and light chains were baseline resolved and the addition of 5% acetic acid to the BGE further improved resolution for antibody mixtures. Figure 12 displays the separation of two antibodies with identification of the heavy and light chains with the improved separation conditions and resolution.
Figure 12.
Base peak electropherograms for a two-antibody mixture (mouse anti-human IgG4 Fc-UNLB and human/mouse/rat activin A beta A subunit antibody), including migration times for each heavy chain and light chain. Triplicate CZE–ESI–MS analysis of a two-antibody mixture. 50 cm long, 30 μm I.D. capillary was used for separation. About 30 nL liquid aliquot was injected for each analysis. Sheath liquid: 0.5% FA, 10% methanol. Voltages: 23.7 kV/1.2 kV. LTQ-XL mass spectrometer was used for detection. NL is the normalization level. (Modified from reference168 with permission)
Commercially available microchip electrophoresis systems have been utilized for protein therapeutic analysis. The 2100 Bioanalyzer MCGE system has been expanded for glycoprotein analysis to analyze PTM-proteins.169 The MCGE system was applied to immunoprecipitation for detection of C-reactive protein (CRP) as a sepsis marker.170 Immunoprecipitation allows for purification of the CRP specifically, and in combination with the MCGE system can serve as an early detection method for sepsis. Clinical serum samples were analyzed in 3 h with reduced sample (50 μL) and consumable consumption. Another application of the MCGE system is for the analysis of PEG modified proteins, which has been reported to increase the therapeutic efficacy of protein drugs. The MCGE system was utilized to study electrophoretic behavior of linear and branched PEG-conjugated proteins where differentiating between the two proteins is a valuable tool for the biopharmaceutical development field.171
The Caliper LabChip® GXII instrument has been utilized by for mAb product quality analysis to expand the use of a commercialized system in the biopharmaceutical development field with fast separation times of 45 sec/analysis.172 In this work, it was found that applying this separation tool to mAbs required different minimal levels of SDS for complete denaturation depending on the subclass of mAb.
4. Conclusions
This review shows that CE and MCE methods are powerful tools for analysis of proteins in various samples and technological adaptations to these techniques are reported regularly. They provide high separation efficiency, considerable detection sensitivity, fast analysis time, and require low amounts of samples and reagents. Furthermore, electrophoretic methods are used in a wide range of applications. They are successfully employed for analysis of protein therapeutics, for determination of proteins in complex biomatrices (e.g. biological fluids, tissue extracts, and plant samples), and for physicochemical characterization of proteins enabling determination of parameters including relative molecular mass, pI, effective charge and binding constants of various protein complexes. Nevertheless, electrophoretic techniques also have some limitations.
One critical issue in analysis of proteins by CE/MCE is the adsorption of analytes on the wall of the separation compartment. Therefore, a great deal of research is devoted to the development and application of novel, effective capillary coating materials. Currently, the field needs more effort to be directed to the automation and high throughput implementation of surface modification for efficient suppression of protein adsorption.
Another limitation of CE is the low concentration sensitivity which can sometimes be problematic especially when dealing with biological samples. However, the sensitivity of CE methods can be substantially improved by the application of various preconcentration techniques and/or by employing more sensitive detection methods (LIF, MS). Despite the fact that more than three orders of magnitude have been achieved for sensitivity enhancement, there is still a need for more sensitive strategies that allow the detection of target proteins with minimal sample treatment.
Presently, gels are more widely used for protein analysis compared to CE/MCE, however there is much value in expanding the use of CE/MCE for these analyses including increased automation, better reproducibility, lower sample consumption, and faster analysis times. Particularly, the widely-used technique of Western blotting has seen minimal advances since its development over 30 years ago, and the field of MCE can further benefit from more commercialization.
Many of the research articles discussed here covered protein analysis related to biomedical studies and applications. CE/MCE technology is being developed to improve the throughput of analysis for biomarker proteins and diagnostic tools that can be implemented in hospitals and other point-of-care applications. Additionally, the work that has been accomplished with CE/MCE in proteomics and protein therapeutics shows the power of using these techniques for a variety of protein analysis applications. Continuing to improve the efficiency of biotherapeutic separations and analyses is of much importance, and CE/MCE can fill these needs.
Supplementary Material
Acknowledgments
The authors are supported by the National Institute of Health grants R37 DK046960, RO1 GM102236, and 1R43GM112289-01. M.D. thanks the support of the Postdoctoral Translational Scholars Program (PTSP) of MICHR (UL1TR000433).
Footnotes
Conflict of Interest
The authors have declared no conflict of interest.
References
- 1.Srinivas PR. Journal. 2012;869:23–28. doi: 10.1007/978-1-61779-821-4_2. [DOI] [PubMed] [Google Scholar]
- 2.Michaelis L. Biochemische Zeitschrift. 1909;16:81–86. [Google Scholar]
- 3.Michaelis L. Biochemische Zeitschrift. 1909;16:486–488. [Google Scholar]
- 4.Reiner L. Biochemische Zeitschrift. 1927;191:158–174. [Google Scholar]
- 5.Tiselius A. Biochemical Journal. 1937;31:313–317. doi: 10.1042/bj0310313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiselius A. Biochemical Journal. 1937;31:1464–1477. doi: 10.1042/bj0311464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smithies O. In: Protein Electrophoresis: Methods and Protocols. Kurien BT, Scofield RH, editors. Humana Press; Totowa, NJ: 2012. pp. 1–21. [DOI] [Google Scholar]
- 8.Ouimet CM, D’amico CI, Kennedy RT. Expert Opinion on Drug Discovery. 2016:1–12. doi: 10.1080/17460441.2017.1268121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Deeb S, Waetzig H, El-Hady DA, Sanger-van de Griend C, Scriba GKE. Electrophoresis. 2016;37:1591–1608. doi: 10.1002/elps.201600058. [DOI] [PubMed] [Google Scholar]
- 10.Harstad RK, Johnson AC, Weisenberger MM, Bowser MT. Analytical Chemistry. 2016;88:299–319. doi: 10.1021/acs.analchem.5b04125. [DOI] [PubMed] [Google Scholar]
- 11.Lacna J, Foret F, Kuban P. Electrophoresis. 2017;38:203–222. doi: 10.1002/elps.201600354. [DOI] [PubMed] [Google Scholar]
- 12.Neaga IO, Bodoki E, Hambye S, Blankert B, Oprean R. Talanta. 2016;148:247–256. doi: 10.1016/j.talanta.2015.10.077. [DOI] [PubMed] [Google Scholar]
- 13.Dawod M, Breadmore MC, Guijt RM, Haddad PR. J Chromatogr A. 2008;1189:278–284. doi: 10.1016/j.chroma.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 14.Dawod M, Breadmore MC, Guijt RM, Haddad PR. J Chromatogr A. 2009;1216:3380–3386. doi: 10.1016/j.chroma.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Dawod M, Breadmore MC, Guijt RM, Haddad PR. Electrophoresis. 2010;31:1184–1193. doi: 10.1002/elps.200900420. [DOI] [PubMed] [Google Scholar]
- 16.Stepanova S, Kasicka V. Analytica Chimica Acta. 2016;933:23–42. doi: 10.1016/j.aca.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Štěpánová S, Kašička V. Journal of Separation Science. doi: 10.1002/jssc.201600962. n/a–n/a. [DOI] [Google Scholar]
- 18.Stepanova S, Kasicka V. Journal of Separation Science. 2016;39:198–211. doi: 10.1002/jssc.201500973. [DOI] [PubMed] [Google Scholar]
- 19.Sun LL, Zhu GJ, Yan XJ, Zhang ZB, Wojcik R, Champion MM, Dovichi NJ. Proteomics. 2016;16:188–196. doi: 10.1002/pmic.201500339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawod M, Chung DS. J Sep Sci. 2011;34:2790–2799. doi: 10.1002/jssc.201100384. [DOI] [PubMed] [Google Scholar]
- 21.Breadmore MC. Journal of Chromatography A. 2012;1221:42–55. doi: 10.1016/j.chroma.2011.09.062. [DOI] [PubMed] [Google Scholar]
- 22.Gilges M, Kleemiss MH, Schomburg G. Analytical Chemistry. 1994;66:2038–2046. [Google Scholar]
- 23.Novotny MV, Cobb KA, Liu JP. Electrophoresis. 1990;11:735–749. doi: 10.1002/elps.1150110911. [DOI] [PubMed] [Google Scholar]
- 24.Adem SM, Mansfield E, Keogh JP, Hall HK, Aspinwall CA. Analytica Chimica Acta. 2013;772:93–98. doi: 10.1016/j.aca.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez I, Li SFY. Analytica Chimica Acta. 1999;383:1–26. [Google Scholar]
- 26.MacDonald AM, Lucy CA. Journal of Chromatography A. 2006;1130:265–271. doi: 10.1016/j.chroma.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 27.Lucy CA, MacDonald AM, Gulcev MD. Journal of Chromatography A. 2008;1184:81–105. doi: 10.1016/j.chroma.2007.10.114. [DOI] [PubMed] [Google Scholar]
- 28.de Jong S, Krylov SN. Analytical Chemistry. 2012;84:453–458. doi: 10.1021/ac2030333. [DOI] [PubMed] [Google Scholar]
- 29.Dolnik V. Electrophoresis. 1997;18:2353–2361. doi: 10.1002/elps.1150181226. [DOI] [PubMed] [Google Scholar]
- 30.Dolnik V. Electrophoresis. 1999;20:3106–3115. doi: 10.1002/(SICI)1522-2683(19991001)20:15/16<3106::AID-ELPS3106>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Dolnik V. Electrophoresis. 2006;27:126–141. doi: 10.1002/elps.200500567. [DOI] [PubMed] [Google Scholar]
- 32.Dolnik V. Electrophoresis. 2008;29:143–156. doi: 10.1002/elps.200700584. [DOI] [PubMed] [Google Scholar]
- 33.Huhn C, Ramautar R, Wuhrer M, Somsen GW. Analytical and Bioanalytical Chemistry. 2010;396:297–314. doi: 10.1007/s00216-009-3193-y. [DOI] [PubMed] [Google Scholar]
- 34.Horvath J, Dolnik V. Electrophoresis. 2001;22:644–655. doi: 10.1002/1522-2683(200102)22:4<644::AID-ELPS644>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Katayama H, Ishihama Y, Asakawa N. Analytical Chemistry. 1998;70:5272–5277. doi: 10.1021/ac980522l. [DOI] [PubMed] [Google Scholar]
- 36.Katayama H, Ishihama Y, Asakawa N. Analytical Chemistry. 1998;70:2254–2260. doi: 10.1021/ac9708755. [DOI] [PubMed] [Google Scholar]
- 37.Hajba L, Guttman A. TrAC Trends in Analytical Chemistry. 2017;90:38–44. [Google Scholar]
- 38.Bekri S, Leclercq L, Cottet H. Journal of Chromatography A. 2015;1399:80–87. doi: 10.1016/j.chroma.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 39.Bekri S, Leclercq L, Cottet H. Journal of Chromatography A. 2016;1432:145–151. doi: 10.1016/j.chroma.2015.12.060. [DOI] [PubMed] [Google Scholar]
- 40.Pobozy E, Sentkowska A, Piskor A. Journal of Separation Science. 2014;37:2388–2394. doi: 10.1002/jssc.201301236. [DOI] [PubMed] [Google Scholar]
- 41.Bodnar J, Hajba L, Guttman A. ELECTROPHORESIS. 2016;37:3154–3159. doi: 10.1002/elps.201600405. [DOI] [PubMed] [Google Scholar]
- 42.Hjerten S. Journal of Chromatography. 1985;347:191–198. [Google Scholar]
- 43.Chen LJ, Zhang YL, Tan L, Liu ST, Wang YM. Journal of Separation Science. 2015;38:2915–2923. doi: 10.1002/jssc.201500346. [DOI] [PubMed] [Google Scholar]
- 44.Yu B, Jiao MM, Cong HL, Shu X, Yang SJ. Journal of Separation Science. 2014;37:725–730. doi: 10.1002/jssc.201301117. [DOI] [PubMed] [Google Scholar]
- 45.Yu B, Chi M, Han YX, Cong HL, Tang JB, Peng QH. Talanta. 2016;152:76–81. doi: 10.1016/j.talanta.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 46.Yu B, Shu X, Cong HL, Chen X, Liu HW, Yuan H, Chi M. Journal of Chromatography A. 2016;1437:226–233. doi: 10.1016/j.chroma.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Yu B, Chen X, Cong HL, Shu X, Peng QH. Analytical and Bioanalytical Chemistry. 2016;408:6781–6788. doi: 10.1007/s00216-016-9804-5. [DOI] [PubMed] [Google Scholar]
- 48.Yu B, Cui W, Cong HL, Jiao MM, Liu P, Yang SJ. Rsc Advances. 2013;3:20010–20015. [Google Scholar]
- 49.Cunliffe JM, Baryla NE, Lucy CA. Analytical Chemistry. 2002;74:776–783. doi: 10.1021/ac015627u. [DOI] [PubMed] [Google Scholar]
- 50.Gulcev MD, Lucy CA. Analytical Chemistry. 2008;80:1806–1812. doi: 10.1021/ac702408u. [DOI] [PubMed] [Google Scholar]
- 51.Gallagher ES, Adem SM, Bright LK, Calderon IAC, Mansfield E, Aspinwall CA. Electrophoresis. 2014;35:1099–1105. doi: 10.1002/elps.201300537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sueyoshi K, Hori Y, Otsuka K. Microfluidics and Nanofluidics. 2013;14:933–941. [Google Scholar]
- 53.Guo XF, Guo XM, Wang H, Zhang HS. Talanta. 2015;144:110–114. doi: 10.1016/j.talanta.2015.05.080. [DOI] [PubMed] [Google Scholar]
- 54.Znaleziona J, Drahonovsky D, Drahos B, Sevcik J, Maier V. Journal of Separation Science. 2016;39:2406–2412. doi: 10.1002/jssc.201501349. [DOI] [PubMed] [Google Scholar]
- 55.Polikarpov N, Potolytsyna V, Bessonova E, Tripp S, Appelhans D, Voit B, Kartsova L. Journal of Chromatography A. 2015;1378:65–73. doi: 10.1016/j.chroma.2014.11.051. [DOI] [PubMed] [Google Scholar]
- 56.Tian Y, Li YF, Mei J, Cai B, Dong JF, Shi ZG, Xiao YX. Journal of Chromatography A. 2015;1412:151–158. doi: 10.1016/j.chroma.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 57.Chen H, Zhu ZF, Yu HQ, Lu JJ, Liu SR. Analytical Chemistry. 2016;88:9293–9299. doi: 10.1021/acs.analchem.6b02856. [DOI] [PubMed] [Google Scholar]
- 58.Ferreira RA, Martins-Dias S. Analytical Biochemistry. 2016;509:100–103. doi: 10.1016/j.ab.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 59.Zhu Z, Lu JJ, Liu S. Anal Chim Acta. 2012;709:21–31. doi: 10.1016/j.aca.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu D, Regnier FE. Journal of Chromatography. 1992;608:349–356. doi: 10.1016/0021-9673(92)87142-u. [DOI] [PubMed] [Google Scholar]
- 61.Ouimet CM, Shao H, Rauch JN, Dawod M, Nordhues B, Dickey CA, Gestwicki JE, Kennedy RT. Analytical Chemistry. 2016;88:8272–8278. doi: 10.1021/acs.analchem.6b02126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morgenstern J, Busch M, Baumann P, Hubbuch J. Journal of Chromatography A. 2016;1462:153–164. doi: 10.1016/j.chroma.2016.07.078. [DOI] [PubMed] [Google Scholar]
- 63.van Tricht E, Geurink L, Pajic B, Nijenhuis J, Backus H, Germano M, Somsen GW, Sanger-van de Griend CE. Talanta. 2015;144:1030–1035. doi: 10.1016/j.talanta.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 64.Agostino FJ, Krylov SN. Trac-Trends in Analytical Chemistry. 2015;72:68–79. [Google Scholar]
- 65.Krivankova L, Bocek P. Electrophoresis. 1998;19:1064–1074. doi: 10.1002/elps.1150190704. [DOI] [PubMed] [Google Scholar]
- 66.Hannig K. Journal of Chromatography. 1978;159:183–191. doi: 10.1016/s0021-9673(00)98555-8. [DOI] [PubMed] [Google Scholar]
- 67.Hannig K. Electrophoresis. 1982;3:235–243. [Google Scholar]
- 68.Wildgruber R, Weber G, Wise P, Grimm D, Bauer J. Proteomics. 2014;14:629–636. doi: 10.1002/pmic.201300253. [DOI] [PubMed] [Google Scholar]
- 69.Shen QY, Guo CG, Yan J, Zhang Q, Xie HY, Jahan S, Fan LY, Xiao H, Cao CX. Journal of Chromatography A. 2015;1397:73–80. doi: 10.1016/j.chroma.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 70.Geiger M, Harstad RK, Bowser MT. Analytical Chemistry. 2015;87:11682–11690. doi: 10.1021/acs.analchem.5b02262. [DOI] [PubMed] [Google Scholar]
- 71.Zhao SS, Chen DDY. Electrophoresis. 2014;35:96–108. doi: 10.1002/elps.201300372. [DOI] [PubMed] [Google Scholar]
- 72.Domínguez-Vega E, Haselberg R, Somsen GW. In: Capillary Electrophoresis of Proteins and Peptides: Methods and Protocols. Tran NT, Taverna M, editors. Springer; New York, New York, NY: 2016. pp. 25–41. [DOI] [Google Scholar]
- 73.Haselberg R, de Jong GJ, Somsen GW. Electrophoresis. 2013;34:99–112. doi: 10.1002/elps.201200439. [DOI] [PubMed] [Google Scholar]
- 74.Xia JQ. In: Capillary Electrophoresis-Mass Spectrometry: Therapeutic Protein Characterization. Xia JQ, Zhang L, editors. Springer International Publishing; Cham: 2016. pp. 7–11. [DOI] [Google Scholar]
- 75.Bertoletti L, Schappler J, Colombo R, Rudaz S, Haselberg R, Domínguez-Vega E, Raimondi S, Somsen GW, De Lorenzi E. Analytica Chimica Acta. 2016;945:102–109. doi: 10.1016/j.aca.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 76.Klepárník K. ELECTROPHORESIS. 2015;36:159–178. doi: 10.1002/elps.201400392. [DOI] [PubMed] [Google Scholar]
- 77.Bonvin G, Rudaz S, Schappler J. Analytica Chimica Acta. 2014;813:97–105. doi: 10.1016/j.aca.2013.12.043. [DOI] [PubMed] [Google Scholar]
- 78.Dominguez-Vega E, Haselberg R, Somsen GW, de Jong GJ. Analytical Chemistry. 2015;87:8781–8788. doi: 10.1021/acs.analchem.5b01701. [DOI] [PubMed] [Google Scholar]
- 79.Nguyen T, Petersen NJ, Rand KD. Analytica Chimica Acta. 2016;936:157–167. doi: 10.1016/j.aca.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 80.Zhu G, Sun L, Dovichi NJ. Journal of Separation Science. 2016 doi: 10.1002/jssc.201601051. [DOI] [PubMed] [Google Scholar]
- 81.Sun LL, Zhu GJ, Zhang ZB, Mou S, Dovichi NJ. Journal of Proteome Research. 2015;14:2312–2321. doi: 10.1021/acs.jproteome.5b00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wojcik R, Dada OO, Sadilek M, Dovichi NJ. Rapid Communications in Mass Spectrometry. 2010;24:2554–2560. doi: 10.1002/rcm.4672. [DOI] [PubMed] [Google Scholar]
- 83.Peuchen EH, Zhu G, Sun L, Dovichi NJ. Analytical and Bioanalytical Chemistry. 2016:1–7. doi: 10.1007/s00216-016-0122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Castro ER, Manz A. Journal of Chromatography A. 2015;1382:66–85. doi: 10.1016/j.chroma.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 85.Creamer JS, Oborny NJ, Lunte SM. Analytical Methods. 2014;6:5427–5449. doi: 10.1039/C4AY00447G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu DP, Qin JH, Lin BC. Journal of Chromatography A. 2008;1184:542–559. doi: 10.1016/j.chroma.2007.11.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harrison DJ, Fluri K, Seiler K, Fan ZH, Effenhauser CS, Manz A. Science. 1993;261:895–897. doi: 10.1126/science.261.5123.895. [DOI] [PubMed] [Google Scholar]
- 88.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Analytical Chemistry. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 89.Ocvirk G, Munroe M, Tang T, Oleschuk R, Westra K, Harrison DJ. Electrophoresis. 2000;21:107–115. doi: 10.1002/(SICI)1522-2683(20000101)21:1<107::AID-ELPS107>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 90.Lee GB, Chen SH, Huang GR, Sung WC, Lin YH. Sensors and Actuators B: Chemical. 2001;75:142–148. [Google Scholar]
- 91.Bi H, Meng S, Li Y, Guo K, Chen Y, Kong J, Yang P, Zhong W, Liu B. Lab on a Chip. 2006;6:769–775. doi: 10.1039/b600326e. [DOI] [PubMed] [Google Scholar]
- 92.Wei X, Gao X, Zhao L, Peng X, Zhou L, Wang J, Pu Q. Journal of Chromatography A. 2013;1281:148–154. doi: 10.1016/j.chroma.2013.01.039. [DOI] [PubMed] [Google Scholar]
- 93.Osiri JK, Shadpour H, Soper SA. Analytical and Bioanalytical Chemistry. 2010;398:489–498. doi: 10.1007/s00216-010-3914-2. [DOI] [PubMed] [Google Scholar]
- 94.Becker H, Gartner C. Electrophoresis. 2000;21:12–26. doi: 10.1002/(SICI)1522-2683(20000101)21:1<12::AID-ELPS12>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 95.Dolnik V, Liu SR, Jovanovich S. Electrophoresis. 2000;21:41–54. doi: 10.1002/(SICI)1522-2683(20000101)21:1<41::AID-ELPS41>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 96.McDonald JC, Whitesides GM. Accounts of Chemical Research. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 97.Barrios-Romero MD, Crevillen AG, Diez-Masa JC. Journal of Separation Science. 2013;36:2530–2537. doi: 10.1002/jssc.201300275. [DOI] [PubMed] [Google Scholar]
- 98.Belder D, Ludwig M. Electrophoresis. 2003;24:3595–3606. doi: 10.1002/elps.200305648. [DOI] [PubMed] [Google Scholar]
- 99.Cong HL, Xu XD, Yu B, Liu HW, Yuan H. Biomicrofluidics. 2016:10. [Google Scholar]
- 100.Mohamadi MR, Svobodova Z, Verpillot R, Esselmann H, Wiltfang J, Otto M, Taverna M, Bilkova Z, Viovy JL. Analytical Chemistry. 2010;82:7611–7617. doi: 10.1021/ac101337n. [DOI] [PubMed] [Google Scholar]
- 101.Doherty EAS, Meagher RJ, Albarghouthi MN, Barron AE. Electrophoresis. 2003;24:34–54. doi: 10.1002/elps.200390029. [DOI] [PubMed] [Google Scholar]
- 102.Kitagawa F, Nakagawara S, Nukatsuka I, Hori Y, Sueyoshi K, Otsuka K. Analytical Sciences. 2015;31:1171–1175. doi: 10.2116/analsci.31.1171. [DOI] [PubMed] [Google Scholar]
- 103.Zhou JW, Ellis AV, Voelcker NH. Electrophoresis. 2010;31:2–16. doi: 10.1002/elps.200900475. [DOI] [PubMed] [Google Scholar]
- 104.Sahore V, Kumar S, Rogers CI, Jensen JK, Sonker M, Woolley AT. Analytical and Bioanalytical Chemistry. 2016;408:599–607. doi: 10.1007/s00216-015-9141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pagaduan JV, Ramsden M, O’Neill K, Woolley AT. Electrophoresis. 2015;36:813–817. doi: 10.1002/elps.201400436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mesbah K, Mai TD, Jensen TG, Sola L, Chiari M, Kutter JP, Taverna M. Microchimica Acta. 2016;183:2111–2121. [Google Scholar]
- 107.Tahka SM, Bonabi A, Nordberg ME, Kanerva M, Jokinen VP, Sikanen TM. Journal of Chromatography A. 2015;1426:233–240. doi: 10.1016/j.chroma.2015.11.072. [DOI] [PubMed] [Google Scholar]
- 108.Aboud N, Ferraro D, Taverna M, Descroix S, Smadja C, Tran NT. Analyst. 2016;141:5776–5783. doi: 10.1039/c6an00821f. [DOI] [PubMed] [Google Scholar]
- 109.Begolo S, Colas G, Viovy JL, Malaquin L. Lab on a Chip. 2011;11:508–512. doi: 10.1039/c0lc00356e. [DOI] [PubMed] [Google Scholar]
- 110.Perez-Toralla K, Champ J, Mohamadi MR, Braun O, Malaquin L, Viovy JL, Descroix S. Lab on a Chip. 2013;13:4409–4418. doi: 10.1039/c3lc50888a. [DOI] [PubMed] [Google Scholar]
- 111.Zarabadi AS, Pawliszyn J, Hajialamdari M. Journal of Chromatography A. 2017;1484:93–97. doi: 10.1016/j.chroma.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 112.Edgar JS, Pabbati CP, Lorenz RM, He MY, Fiorini GS, Chiu DT. Analytical Chemistry. 2006;78:6948–6954. doi: 10.1021/ac0613131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roman GT, Wang M, Shultz KN, Jennings C, Kennedy RT. Analytical Chemistry. 2008;80:8231–8238. doi: 10.1021/ac801317t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Niu XZ, Pereira F, Edel JB, de Mello AJ. Analytical Chemistry. 2013;85:8654–8660. doi: 10.1021/ac401383y. [DOI] [PubMed] [Google Scholar]
- 115.Guetschow ED, Kumar S, Lombard DB, Kennedy RT. Analytical and Bioanalytical Chemistry. 2016;408:721–731. doi: 10.1007/s00216-015-9206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Azim M, Malekpourkoupaei A, Ye W, Jemere AB, Harrison DJ. ELECTROPHORESIS. 2016 doi: 10.1002/elps.201600339. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 117.Towbin H, Staehelin T, Gordon J. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Anderson GJ, Cipolla CM, Kennedy RT. Analytical Chemistry. 2011;83:1350–1355. doi: 10.1021/ac102671n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jin S, Anderson GJ, Kennedy RT. Analytical Chemistry. 2013;85:6073–6079. doi: 10.1021/ac400940x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jin S, Furtaw MD, Chen HX, Lamb DT, Ferguson SA, Arvin NE, Dawod M, Kennedy RT. Analytical Chemistry. 2016;88:6703–6710. doi: 10.1021/acs.analchem.6b00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hughes AJ, Herr AE. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21450–21455. doi: 10.1073/pnas.1207754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gerver RE, Herr AE. Analytical Chemistry. 2014;86:10625–10632. doi: 10.1021/ac5024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hughes AJ, Spelke DP, Xu ZC, Kang CC, Schaffer DV, Herr AE. Nature Methods. 2014;11:749–U794. doi: 10.1038/nmeth.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Duncombe TA, Kang CC, Maity S, Ward TM, Pegram MD, Murthy N, Herr AE. Advanced Materials. 2016;28:327–334. doi: 10.1002/adma.201503939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim JJ, Sinkala E, Herr AE. Lab on a Chip. 2017 doi: 10.1039/C6LC01333C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Breadmore MC, Shallan AI, Rabanes HR, Gstoettenmayr D, Abdul Keyon AS, Gaspar A, Dawod M, Quirino JP. Electrophoresis. 2013;34:29–54. doi: 10.1002/elps.201200396. [DOI] [PubMed] [Google Scholar]
- 127.Breadmore MC, Thabano JRE, Dawod M, Kazarian AA, Quirino JP, Guijt RM. Electrophoresis. 2009;30:230–248. doi: 10.1002/elps.200800435. [DOI] [PubMed] [Google Scholar]
- 128.Breadmore MC, Tubaon RM, Shallan AI, Phung SC, Keyon ASA, Gstoettenmayr D, Prapatpong P, Alhusban AA, Ranjbar L, See HH, Dawod M, Quirino JP. Electrophoresis. 2015;36:36–61. doi: 10.1002/elps.201400420. [DOI] [PubMed] [Google Scholar]
- 129.Breadmore MC, Wuethrich A, Li F, Phung SC, Kalsoom U, Cabot JM, Tehranirokh M, Shallan AI, Abdul Keyon AS, See HH, Dawod M, Quirino JP. Electrophoresis. 2017;38:33–59. doi: 10.1002/elps.201600331. [DOI] [PubMed] [Google Scholar]
- 130.Burgi DS, Chien RL. Anal Biochem. 1992;202:306–309. doi: 10.1016/0003-2697(92)90110-s. [DOI] [PubMed] [Google Scholar]
- 131.Zhang ZB, Sun LL, Zhu GJ, Yan XJ, Dovichi NJ. Talanta. 2015;138:117–122. doi: 10.1016/j.talanta.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang ZB, Sun LL, Zhu GJ, Cox OF, Huber PW, Dovichi NJ. Analytical Chemistry. 2016;88:877–882. doi: 10.1021/acs.analchem.5b03496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sharma D, Bisht D. Electrophoresis. 2016;37:1187–1190. doi: 10.1002/elps.201600025. [DOI] [PubMed] [Google Scholar]
- 134.Kumar S, Sahore V, Rogers CI, Woolley AT. Analyst. 2016;141:1660–1668. doi: 10.1039/c5an02352a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cannon DM, Kuo TC, Bohn PW, Sweedler JV. Analytical Chemistry. 2003;75:2224–2230. doi: 10.1021/ac020629f. [DOI] [PubMed] [Google Scholar]
- 136.Kovarik ML, Jacobson SC. Analytical Chemistry. 2008;80:657–664. doi: 10.1021/ac701759f. [DOI] [PubMed] [Google Scholar]
- 137.Wu DP, Steckl AJ. Lab on a Chip. 2009;9:1890–1896. doi: 10.1039/b823409d. [DOI] [PubMed] [Google Scholar]
- 138.Kuo TC, Cannon DM, Shannon MA, Bohn PW, Sweedler JV. Sensors and Actuators a-Physical. 2003;102:223–233. [Google Scholar]
- 139.Li F, Guijt RM, Breadmore MC. Analytical Chemistry. 2016;88:8257–8263. doi: 10.1021/acs.analchem.6b02096. [DOI] [PubMed] [Google Scholar]
- 140.Cabot JM, Macdonald NP, Phung SC, Breadmore MC, Paull B. The Analyst. 2016 doi: 10.1039/c6an01515h. [DOI] [PubMed] [Google Scholar]
- 141.Ouyang W, Ko SH, Wu D, Wang AY, Barone PW, Hancock WS, Han J. Analytical Chemistry. 2016;88:9669–9677. doi: 10.1021/acs.analchem.6b02517. [DOI] [PubMed] [Google Scholar]
- 142.Cheow LF, Han JY. Analytical Chemistry. 2011;83:7086–7093. doi: 10.1021/ac201307d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Heemskerk AAM, Deelder AM, Mayboroda OA. Mass Spectrometry Reviews. 2016;35:259–271. doi: 10.1002/mas.21432. [DOI] [PubMed] [Google Scholar]
- 144.Lindenburg PW, Haselberg R, Rozing G, Ramautar R. Chromatographia. 2015;78:367–377. [Google Scholar]
- 145.Di Venere M, Viglio S, Sassera D, Fumagalli M, Bardoni A, Salvini R, Cagnone M, Iadarola P. ELECTROPHORESIS. 2017 doi: 10.1002/elps.201600504. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 146.Zhao YM, Sun LL, Zhu GJ, Dovichi NJ. Journal of Proteome Research. 2016;15:3679–3685. doi: 10.1021/acs.jproteome.6b00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bush DR, Zang L, Belov AM, Ivanov AR, Karger BL. Analytical Chemistry. 2016;88:1138–1146. doi: 10.1021/acs.analchem.5b03218. [DOI] [PubMed] [Google Scholar]
- 148.Beck A, Wagner-Rousset E, Ayoub D, Van Dorsselaer A, Sanglier-Cianferani S. Analytical Chemistry. 2013;85:715–736. doi: 10.1021/ac3032355. [DOI] [PubMed] [Google Scholar]
- 149.Fu X, Xiao HT, Liang S, Bao JJ, Li TX, Zhang Y. Analyst. 2016;141:305–310. doi: 10.1039/c5an01680k. [DOI] [PubMed] [Google Scholar]
- 150.Francois YN, Biacchi M, Said N, Renard C, Beck A, Gahoual R, Leize-Wagner E. Analytica Chimica Acta. 2016;908:168–176. doi: 10.1016/j.aca.2015.12.033. [DOI] [PubMed] [Google Scholar]
- 151.Wojcik R, Zhu GJ, Zhang ZB, Yan XJ, Zhao YM, Sun LL, Champion MM, Dovichi NJ. Bioanalysis. 2016;8:89–92. doi: 10.4155/bio.15.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gahoual R, Beck A, Leize-Wagner E, Francois YN. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2016;1032:61–78. doi: 10.1016/j.jchromb.2016.05.028. [DOI] [PubMed] [Google Scholar]
- 153.Zhu GJ, Sun LL, Dovichi NJ. Talanta. 2016;146:839–843. doi: 10.1016/j.talanta.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jaccoulet E, Smadja C, Prognon P, Taverna M. Electrophoresis. 2015;36:2050–2056. doi: 10.1002/elps.201400603. [DOI] [PubMed] [Google Scholar]
- 155.Said N, Gahoual R, Kuhn L, Beck A, Francois YN, Leize-Wagner E. Analytica Chimica Acta. 2016;918:50–59. doi: 10.1016/j.aca.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 156.Lew C, Gallegos-Perez JL, Fonslow B, Lies M, Guttman A. Journal of Chromatographic Science. 2015;53:443–449. doi: 10.1093/chromsci/bmu229. [DOI] [PubMed] [Google Scholar]
- 157.Feng H-t, Su M, Rifai FN, Li P, Li SFY. Analytica Chimica Acta. doi: 10.1016/j.aca.2016.11.043. http://dx.doi.org/10.1016/j.aca.2016.11.043. [DOI] [PubMed]
- 158.Zhang L, Lawson K, Yeung B, Wypych J. Analytical Chemistry. 2015;87:470–476. doi: 10.1021/ac504187v. [DOI] [PubMed] [Google Scholar]
- 159.Zhang Z, Albanetti T, Linkous T, Larkin CJ, Schoner R, McGivney JB, Dovichi NJ. ELECTROPHORESIS. 2016 doi: 10.1002/elps.201600390. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 160.Zhu GJ, Sun LL, Heidbrink-Thompson J, Kuntumalla S, Lin HY, Larkin CJ, McGivneyiv JB, Dovichi NJ. Electrophoresis. 2016;37:616–622. doi: 10.1002/elps.201500301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Suba D, Urbanyi Z, Salgo A. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2016;1032:224–229. doi: 10.1016/j.jchromb.2016.07.026. [DOI] [PubMed] [Google Scholar]
- 162.Hühner J, Jooß K, Christian N. ELECTROPHORESIS. 2016 doi: 10.1002/elps.201600457. n/a–n/a. [DOI] [Google Scholar]
- 163.Huhner J, Neususs C. Analytical and Bioanalytical Chemistry. 2016;408:4055–4061. doi: 10.1007/s00216-016-9498-8. [DOI] [PubMed] [Google Scholar]
- 164.Redman EA, Batz NG, Mellors JS, Ramsey JM. Analytical Chemistry. 2015;87:2264–2272. doi: 10.1021/ac503964j. [DOI] [PubMed] [Google Scholar]
- 165.Batz NG, Mellors JS, Alarie JP, Ramsey JM. Analytical Chemistry. 2014;86:3493–3500. doi: 10.1021/ac404106u. [DOI] [PubMed] [Google Scholar]
- 166.Redman EA, Mellors JS, Starkey JA, Ramsey JM. Analytical Chemistry. 2016;88:2220–2226. doi: 10.1021/acs.analchem.5b03866. [DOI] [PubMed] [Google Scholar]
- 167.Redman EA, Ramos-Payan M, Mellors JS, Ramsey JM. Analytical Chemistry. 2016;88:5324–5330. doi: 10.1021/acs.analchem.6b00622. [DOI] [PubMed] [Google Scholar]
- 168.Zhao YM, Sun LL, Knierman MD, Dovichi NJ. Talanta. 2016;148:529–533. doi: 10.1016/j.talanta.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 169.Engel N, Weiss VU, Wenz C, Rufer A, Kratzmeier M, Gluck S, Marchetti-Deschmann M, Allmaier G. Electrophoresis. 2015;36:1754–1758. doi: 10.1002/elps.201400510. [DOI] [PubMed] [Google Scholar]
- 170.Herwig E, Marchetti-Deschmann M, Wenz C, Rufer A, Redl H, Bahrami S, Allmaier G. Analytical Biochemistry. 2015;478:102–106. doi: 10.1016/j.ab.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 171.Park EJ, Kim MS, Lee HS, Lee KC, Na DH. Electrophoresis. 2015;36:918–923. doi: 10.1002/elps.201400539. [DOI] [PubMed] [Google Scholar]
- 172.Cai H, Song YL, Zhang J, Shi T, Fu Y, Li R, Mussa N, Li ZJ. Journal of Pharmaceutical and Biomedical Analysis. 2016;120:46–56. doi: 10.1016/j.jpba.2015.10.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.