Abstract
Golgins are a family of coiled-coil proteins located at the cytoplasmic surface of the Golgi apparatus and have been implicated in maintaining Golgi structural integrity through acting as tethering factors for retrograde vesicle transport. Whereas knockdown of several individual golgins in cultured cells caused Golgi fragmentation and disruption of vesicle trafficking, analysis of mutant mouse models lacking individual golgins have discovered tissue-specific developmental functions. Recently, homozygous loss of function of GOLGA2, of which previous in vitro studies suggested an essential role in maintenance of Golgi structure and in mitosis, has been associated with a neuromuscular disorder in human patients, which highlights the need for understanding the developmental roles of the golgins in vivo. We report here generation of Golga5-deficient mice using CRISPR/Cas9-mediated genome editing. Although knockdown studies in cultured cells have implicated Golga5 in maintenance of Golgi organization, we show that Golga5 is not required for mouse embryonic development, postnatal survival, or fertility. Moreover, whereas Golga5 is structurally closely related to Golgb1, we show that inactivation of Golga5 does not enhance the severity of developmental defects in Golgb1-deficient mice. The Golga5-deficient mice enable further investigation of the roles and functional specificity of golgins in development and diseases.
Keywords: cleft palate, CRISPR, Golgi, Golgb1, Golgin-84, mouse
Introduction
The mammalian Golgi apparatus consists of flattened membrane stacks, called cisternae, that are laterally interconnected by tubular membrane bridges to form the Golgi ribbon (Mellman and Warren 2000; Sütterlin and Colanzi 2010). Almost all secretory and membrane proteins made in mammalian cells are collected into transport vesicles from the ER, where they are synthesized, and transported across the Golgi membrane stacks in which they are subjected to sequential posttranslational modifications, such as glycosylation, before trafficking on to other cellular compartments or the plasma membrane (Gillingham and Munro 2016; Zhang and Wang 2015). To maintain the Golgi polarity and to ensure accurate sequential protein modifications, Golgi resident proteins are continuously recycled in retrograde coat protein I (COPI) vesicles through tethering to their target cisternae (Shorter and Warren 2002). A major class of membrane tethering proteins decorating the cytoplasmic surface of the Golgi are referred to as “golgins”, which consists of more than 10 proteins, with each containing multiple coiled-coil domains and a C-terminal transmembrane or membrane-tethering domain mediating anchorage to the Golgi (Gillingham and Munro 2016; Nozawa et al. 2005; Witkos and Lowe 2015). Different golgins are localized to distinct regions of the Golgi apparatus and the coiled-coil cytoplasmic domains form parallel homodimeric “rod-like” structures extending up to 300 nm away from the surface of the Golgi, making them ideal for capturing transport vesicles [reviewed by (Gillingham and Munro 2016; Ramirez and Lowe 2009)]. Recent studies have shown that the golgins are able to distinguish vesicles from different origins, suggesting that they provide specificity of vesicle traffic and make a major contribution to the maintenance of organization of the Golgi apparatus (Gillingham and Munro 2016; Wong et al. 2017; Wong and Munro 2014). However, whereas siRNA knockdown studies in cell culture systems have suggested an essential role for several golgins, including Golga2 (GM130), Golga5 (Golgin-84), and GMAP210, in vesicle trafficking and maintenance of Golgi structural integrity (Diao et al. 2003; Nakamura et al. 1995; Ríos et al. 2004), homozygous loss of function of either Golga2 or GMAP210 did not cause early embryonic lethality but led to distinct tissue-specific developmental defects in both mice and humans (Follit et al. 2008; Han et al. 2017; Liu et al. 2017; Smits et al. 2010). These data underscore the difficulty in explaining pathogenesis of developmental disorders from studies in cultured cells and the importance of investigating the physiological roles of the golgins through generation and analysis of mutant animal models.
Golga5 was first identified in a yeast two-hybrid screen using the Golgi-associated phosphatidylinositol(4,5)P2 5-phosphatase OCRL1 as bait (Bascom et al. 1999). Structurally, Golga5 is most similar to Golgb1, containing a coiled-coil cytoplasmic domain and a C-terminal transmembrane domain that anchors to the Golgi membrane (Bascom et al. 1999). Whereas studies of Golgb1-deficient rat and mouse models showed that Golgb1 plays crucial roles in craniofacial and skeletal development (Katayama et al. 2011; Lan et al. 2016), the physiological functions of Golga5 are unknown. We report here the generation and analysis of Golga5-deficent mice.
Results and Discussion
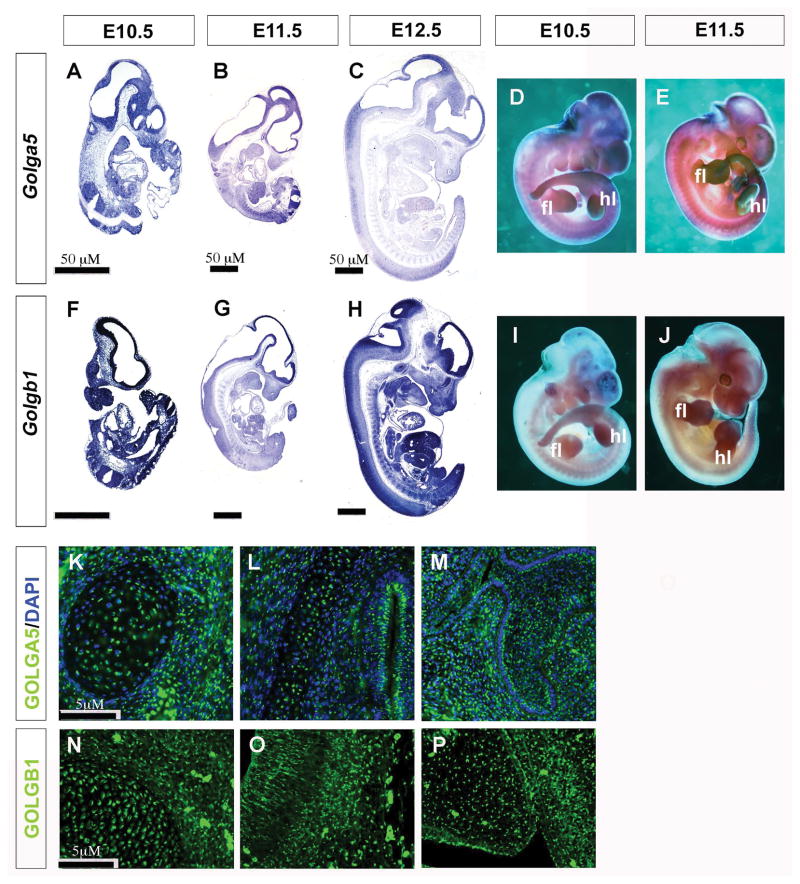
We first examined expression of Golga5 mRNAs in mouse embryos at several developmental stages using whole mount and section in situ hybridization assays. Whereas the developing limb buds and neural tube exhibit higher levels of Golga5 mRNA expression than in other tissues, section in situ hybridization showed that Golga5 mRNAs are expressed ubiquitously (Figure 1A – E). Similarly, Golgb1 mRNAs are expressed ubiquitously in developing mouse embryos, with higher levels in developing limbs and craniofacial tissues (Figure 1F – J). We further examined Golga5 and Golgb1 protein distribution using immunofluorescent staining and found that both proteins are distributed in similar punctate patterns characteristic of Golgi localization in all tissues examined, including neural tissues, developing palate, tooth germs, and Meckel’s cartilage (Figure 1K – P).
Figure 1. Overlapping expression of Golga5 and Golgb1 in mouse embryos.
(A – J) Detection of patterns of expression of Golga5 (A–E) and Golgb1 mRNAs (indicated by the purple stain) (F–J) in whole mount (D, E, I, J) and sagittal sections (A–C, and F–H) of mouse embryos at E10.5 (A, D, F, I), E11.5 (B, E, G, J) and E12.5 (C, H). Scale bar, 50 μm. fl, forelimb hl, hindlimb. (K–P) Distribution of Golga5 (K–M) and Golgb1 (N–P) proteins in mouse embryonic tissues detected by immunofluorscent staining (green). (K) E15.5 Meckle’s cartilage. (L) E15.5 brain tissue. (M) E15.5 maxillary molar tooth germ. (N) E13.5 Meckle’s cartilage. (O) E13.5 brain tissue. (P) E13.5 palatal shelf. Scale bar, 5 μm.
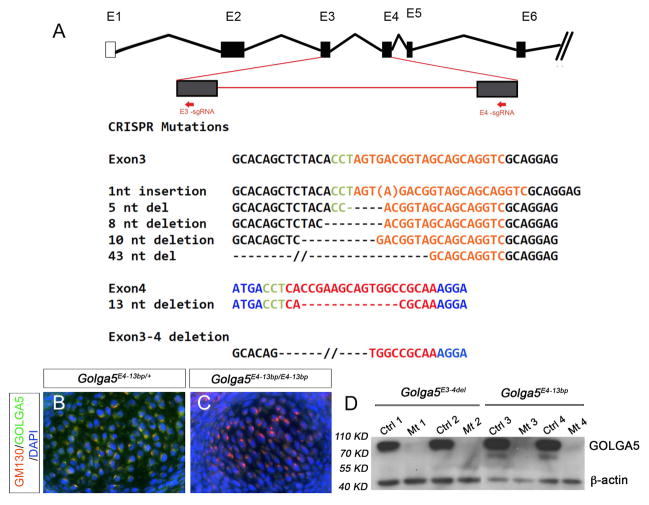
To generate Golga5-deficient mice, we designed and tested two guide RNAs targeting exon-3 and exon-4, respectively, of the Golga5 gene for CRISPR/Cas9-mediated genome editing in mouse embryonic kidney MK-4 cells. Subsequently, the guide RNAs were co-injected with in vitro transcribed mRNAs encoding humanized Cas9 protein into zygotes of FVB/NJ inbred mice to generate Golga5-targeted mice. Of 11 founder mice, three transmitted the same 1-bp insertion in exon-3 (Golga5E3ins1bp) to the G1 generation (Figure 2A). The Golga5E3ins1bp mutation causes frame shift and is predicted to cause a premature termination of translation of the mRNA product 6 nucleotides downstream of the insertion site. Four founder mice carried distinct frame-shifting small deletions in exon-3 (Figure 2A). Interestingly, one founder male mouse transmitted two distinct Golga5 mutant alleles affecting exon-4, a 13-bp deletion at the target site in exon-4 (Golga5E4-13bp) and a 1644-bp deletion from the middle of exon-3 to the middle of exon-4 (Golga5E3-4del) causing deletion of 248 nucleotides in the mRNA product (Figure 2A). The Golga5E4-13bp and Golga5E3-4del alleles are predicted to result in mutant mRNAs with premature termination of translation 59 and 52 nucleotides downstream of the deletion, respectively. Whereas alternative splicing from skipping exon-3 sequence would maintain the normal reading frame, the Golga5E4-13bp and Golga5E3-4del alleles are predicted to produce severely truncated peptides containing only the N-terminal one-third of the Golga5 protein and lacking the Golgi anchoring C-terminal domain. Thus, we established three distinct Golga5 mutant mouse lines, carrying the Golga5E3ins1bp, Golga5E4-13bp, and Golga5E3-4del mutations, respectively, for further analyses.
Figure 2. Generation of Golga5-deficient mice using CRISPR/Cas9-mediated genome editing.
(A) Top row shows schematic presentation of the genomic organization of part of the Golga5 gene. Boxes marked by E1 to E6 represent exons. Two red arrows mark the target sites of two guide RNAs (sgRNAs) designed to targeted exon-3 and exon-4, respectively. The sgRNA target sequences are shown in orange (E3-sgRNA) or red (E4-sgRNA) font with the PAM sequences shown in green. The frame-shift indel mutations found in founder mice are listed in comparison to the wildtype sequences corresponding to the two target sites. (B, C) Frontal sections through the Meckel’s cartilage region of E13.5 Golga5E4-13bp/+ heterozygous (B) and Golga5E4-13bp/E4-13bp homozygous (C) embryos showing co-immunofluorescent staining for GM130 (red) and GOLGA5 (green) proteins. DAPI counterstaining of nuclei was shown in blue. (D) Western blot detection of GOLGA5 and β-actin proteins in the control and Golga5E4-13bp/E4-13bp or Golga5E3-4del/E3-4del tissue lysates.
We carried out immunofluorescent staining and western blot analyses using the rabbit polyclonal anti-GOLGA5 antibody to evaluate effects of the mutations on the Golga5 protein product. As shown in Figure 2, whereas both Golga5E4-13bp homozygous mutant and heterozygous control embryos (n=6 each) showed similar punctate distribution of the GM130 protein, a widely used Golgi marker (Figure 2B, 2C), and the control embryos showed co-distribution of Golga5 and GM130 proteins (Figure 2B), the Golga5E4-13bp homozygous mutants had complete absence of Golga5 immunostaining (Figure 2C). Analysis of embryonic tissues from Golga5E3-4del homozygous mutant and heterozygous control embryos showed similar results (data not shown). Western blot analysis of tissue lysates from Golga5E4-13bp and Golga5E3-4del homozygous mutant embryos and their control littermates showed a clear Golga5-specific protein product of about 83 KD in the control samples that is absent in the homozygous mutant samples (Figure 2D, n=4). These results confirm that the Golga5E4-13bp and Golga5E3-4del mutations cause loss of the Golga5 protein.
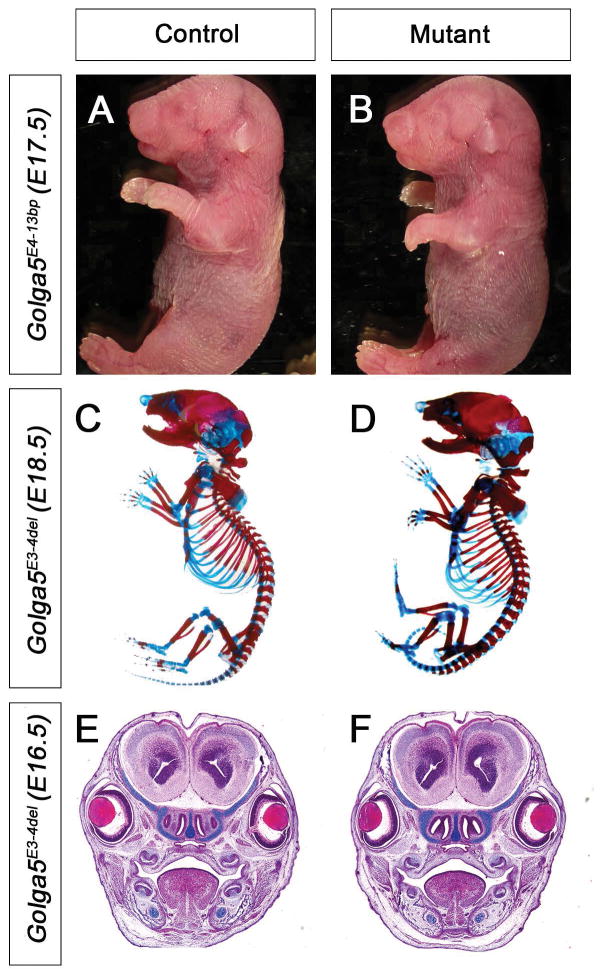
Genotyping of pups from heterozygous intercrosses in each line showed that the wildtype, heterozygous, and homozygous mutant mice are born at Mendelian ratios (1:2:1) (Table 1). Morphological and histological analyses of Golga5E3ins1bp, Golga5E4-13bp, and Golga5E3-4del homozygous embryos at distinct development stages from E13.5 to P0 were performed. No obvious morphological abnormality was consistently detected in any of the three mutant lines (Figure 3A, 3B, 3E, 3F). In addition, we made skeletal preparations of pups from E17 – P0 for at least three homozygous mutants for each of the three mutations. Skeletal abnormalities were not observed in the homozygous mutants (Figure 3C, 3D). We further monitored the mutant mice postnatally and grow at similar rates compared to their wildtype littermates. Breeding of at least three pairs of adult male and female homozygous mutant mice from each line showed that both male and female homozygous mice are fertile and that the females nursed their young similarly as control wildtype mice.
Table 1.
Summary of progeny analyzed for three independent Golga5 mutant mouse lines.
| Golga5 Mutations | # of litters | Total animals | Genotype | ||
|---|---|---|---|---|---|
| Wildtype | Heterozygous | Homozygous | |||
| Exon-3 1-bp insertion (Golga5E3ins1bp) | 7 | 66 | 16 (24%) | 35 (53%) | 15 (23%) |
| Exon-4 13-bp deletion (Golga5E4-13bp) | 8 | 74 | 19 (25.6%) | 33 (44.7%) | 22 (29.7%) |
| Exon-3 to 4 Deletion (Golga5E3-4del) | 13 | 233 | 42 (18.0%) | 129 (55.3%) | 62 (26.6%) |
Figure 3. Golga5-deficent mice have no obvious developmental abnormality.
(A, B) Whole mount views of representative E17.5 control (A) and Golga5E4-13bp homozygous (B) littermates. (C, D) Whole mount skeletal preparations of representative E18.5 control (C) and Golga5E3-4del homozygous mutant (D) embryos. (E, F) Frontal sections of E16.5 control (E) and Golga5E3-4del homozygous mutant (F) embryos. All organs and tissue structures in the Golga5 mutants appear normal.
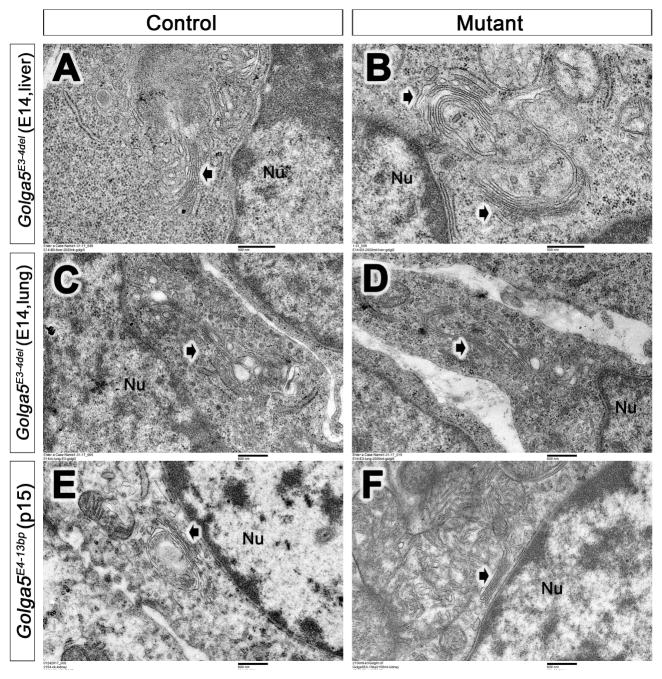
Since previous studies showed that siRNA-mediated knockdown of Golga5 caused Golgi fragmentation in several cell lines (Diao et al. 2003), we performed transmission electron microscopy (TEM) analysis of multiple tissues from Golga5 mutant and control littermates. TEM analysis of the heart, lung, and liver tissues of E14.5 Golga5E3-4del/E3-4del mutants and control littermates showed similar Golgi ribbon structure and organization (Figure 4A–D). We also examined the lung, liver, and kidney tissues from P15 Golga5E4-13bp/E4-13bp mutants and their littermates and no significant differences in Golgi structure and organization were detected between the mutant and control samples (Figure 4E, 4D).
Figure 4. Electron microscopy (TEM) analysis of Golgi structures in Golga5 mutant mouse tissues.
(A–D) Representative cells from E14.5 Golga5E3-4del/E3-4del homozygous embryonic liver (B) and lung (D) tissues exhibit similar Golgi (white arrows) structures as in their control littermate (A, C). (E, F) Representative kidney cells from P15 Golga5E4-13bp/E4-13bp mutant (F) and control (E) littermates. White arrow points to the Golgi. Nu, nucleus. Scale bar in A and B, 500 nm; scale bar in C–F, 600 nm.
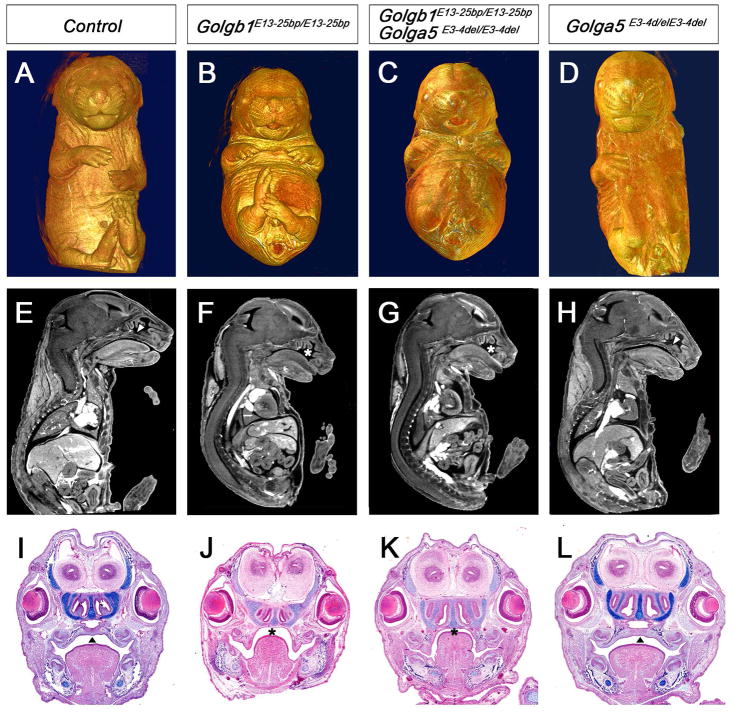
Since Golga5 is structurally closely related to Golgb1 and since we recently showed that mice lacking Golgb1 survive to birth and exhibit specific craniofacial developmental defects (Lan et al. 2016), we investigated whether the lack of obvious developmental defects in Golga5-deficient mice is due to functional complementation by Golgb1. We crossed Golga5E3-4del/E3-4del homozygous female mice with Golgb1E13-25bp/+ male mice, which are heterozygous for a 25-bp deletion in exon-13 of the Golgb1 gene that causes complete loss of Golgb1 function (Lan et al. 2016). We then crossed Golga5E3-4del/+Golgb1E13-25bp/+ double heterozygous progeny with Golga5E3-4del/E3-4del homozygous mice and found that the Golga5E3-4del/E3-4delGolgb1E13-25bp/+ mice (n=15) survive postnatally without obvious developmental abnormality and behaved similarly as the Golga5E3-4del/E3-4del and Golga5E3-4del/+Golgb1E13-25bp/+ littermates. We set up intercrosses of male and female Golga5E3-4del/+Golgb1E13-25bp/+ double heterozygous mice and analyzed the progeny at E14.5 to E18.5 by using Micro-CT and/or histology assays. We found that the Golga5E3-4del/E3-4delGolgb1E13-25bp/E13-25bp double homozygous pups were phenotypically similar to the Golgb1E13-25bp/E13-25bp mutant littermates, with both exhibiting domed head, shortened snout, cleft palate, and reduced alcian blue staining of cartilages (Figure 5A–L). No increase in severity or additional defects in embryonic tissues were found in Golga5E3-4del/E3-4delGolgb1E13-25bp/E13-25bp pups compared with Golgb1E13-25bp/E13-25bp single mutants. These results indicate that Golga5 does not complement Golgb1 function in vivo. The Golga5-deficient mice enable further studies of possible functional redundancy between Golga5 and other golgins.
Figure 5. Analysis of Golga5/Golgb1 double mutant mice.
(A–D) Whole mount ventral views of Micro-CT scanned and 3D reconstructed E17.5 control (A), Golgb1E13-25bp/E13-25bp homozygous (B), Golga5E3-4del/E3-4delGolgb1E13-25bp/E13-25bp double homozygous (C), and Golga5E3-4del/E3-4del mutant (D) embryos. (E – H) Representative mid-sagittal Micro-CT scans of E17.5 control (E), Golgb1E13-25bp/E13-25bp homozygous (F), Golga5E3-4del/E3-4delGolgb1E13-25bp/E13-25bp double homozygous (G), and Golga5E3-4del/E3-4del mutant (H) embryos. (I–J) Coronal histological sections of E16.5 embryos showing control (I), Golgb1E13-25bp/E13-25bp mutant (J), Golga5E3-4del/E3-4delGolgb1E13-25bp/E13-25bp double mutant (K), and Golga5E3-4del/E3-4del mutant (L) embryos. Arrowheads in E, H, I, and L point to normal fused secondary palate. Asterisk in F, G, J, and K marks the cleft palate defect.
Methods
Generation of Golga5-deficient mice using the CRISPR/Cas9 technology
Guide RNAs targeting exon-3 (target sequence 5′-GACCTGCTGCTACCGTCACT-3′ of the antisense strand) and exon-4 (target sequence 5′-TTGCGGCCACTGCTTCGGTG-3′ of the antisense strand), respectively, of the mouse Golga5 gene were designed using the CRISPR Design Tool at http://crispr.mit.edu. The target sequences were subcloned into the PX459 [pSpCas9(BB)-2A-Puro] mammalian Cas9-expression vector (Addgene #48139) and tested by transfection into cultured immortalized mouse embryonic kidney (MK4) cells as described previously (Lan et al. 2016). Both sgRNAs were synthesized in vitro, combined with humanized Cas9 mRNAs and co-injected into zygotes from FVB/NJ inbred mice at the Cincinnati Children’s Hospital Medical Center Transgenic Animal and Genome Editing Facility. Transgenic founder mice were identified by using Sanger sequencing analysis of PCR products containing the Golga5 exon-3 and exon-4 regions in tail biopsy genomic DNA samples. Founder mice were bred to FVB/NJ inbred mice to test for germline transmission and to establish independent mutant mouse lines. G1 mice heterozygous for frame-shifting mutations were intercrossed to analyze phenotype of homozygous mutants.
All animal studies were performed in accordance with the guidelines set by the Institutional Animal Care and Use Committee (IACUC) at the Cincinnati Children’s hospital Medical Center. The newly generated Golga5 mutant mouse lines will be shared with the research community upon request.
Other mouse strains
Golgb1E13-25bp mice (Lan et al. 2016) are maintained as heterozygotes by crossing with FVB/NJ wildtype animals. Golgb1E13-25bp/+ mice were crossed to newly generated Golga5E3-4del/+ mice to generate Golgb1E13-25bp/+Golga5E3-4del/+ mice, which were subsequently intercrossed to investigate whether the two mutations genetically interact.
Histology, in situ hybridization analyses, and immunofluorescent staining assays
Embryos were harvested at desired developmental stages (E13.5 to E18.5, noon of vaginal plug date was counted as day E0.5), fixed in 4% paraformaldehyde (PFA) in PBS overnight at 4°C, dehydrated through graded alcohols, embedded in paraffin, sectioned at 7μm thickness, and stained with alcian blue (A), hematoxylin (H) and eosin (E) for histology analysis. At least four pairs of control and mutant embryos were analyzed for each developmental stage.
Whole mount and section in situ hybridization assays were performed using established protocols (Lan and Jiang 2009; Lan et al. 2001; Liu et al. 2013; Zhang et al. 1999).
Immunofluorescent staining was performed using paraffin or frozen sections following standard protocols (Lan et al. 2016; Xu et al. 2016; Zhou et al. 2011). Antibodies used include rabbit anti-Giantin (GOLGB1, Abcam ab24586; 1:300), mouse anti-GM130 (BD610823; 1:500), rabbit anti-GOLGA5 (Sigma-Aldrich HPA000992; 1:300).
Transmission electron microscopy (TEM) and micro-computed tomography (Micro-CT) imaging
For TEM, embryos were dissected at E14.5 from Golga5E3-4del/+ intercrosses. Lung, liver and heart tissues of each embryo were fixed with 3% glutaraldehyde individually. In addition, lung, liver and kidney tissues were harvested from Golga5E4-13bp/E4-13bp homozygous mutants and their littermates at postnatal day 15 (P15) and fixed in 3% glutaraldehyde solution. Samples were washed with sodium cacodylate buffer, post-fixed with 1% OsO4, processed through ethanol series, embedded in propylene oxide mixture, sectioned at 100 nm, and examined under a Hitachi Model 7650 transmission electron microscope as described previously (Lan et al. 2016).
For Micro-CT analysis, embryos from Golga5E3-4del/E3-4delGolgb1E13-25bp/+ intercrosses were harvested at desired developmental stages (E14.5, E16.5, E17.5 and E18.5) and fixed in 4% PFA for 48 hours, washed in PBS, and placed in 50% Lugol/PBS solution (Sigma-Aldrich, Catalog #L6146-1L) for 4 weeks, with frequent refreshing (Degenhardt et al. 2010). Embryos were scanned in the Cincinnati Children’s Hospital Medical Center Micro-CT core facility using MicroCAT II v. 1.9d (Imtek) with COBRA v.7.4 (Exxim Computing Corporation) software used for image reconstruction. Dicom files of Micro-CT scan were visualized, assembled and analyzed using Amira.
Western blot analysis
Four pairs of E11.5 Golga5E3-4del/E3-4del homozygous and wildtype control embryos and four pairs of Golga5E4-13bp/E4-13bp mutant and control lung and liver tissues from E13.5 embryos were lysed in RIPA buffer containing proteinase inhibitors (Santa Cruz, SC-24948). The supernatant was mixed with Laemmli buffer and separated by SDS-PAGE on 10 % Polyacrylamide gel. Western blots were incubated with rabbit anti-GOLGA5 (Sigma-Aldrich HPA000992; 1:1000) or the anti-β-Actin (Cell Signaling #3700, 1:3,000) antibody followed by washing and incubation with HRP-conjugated secondary antibodies. The signals were detected using Immobilon Western HRP peroxidase solution and Luminol reagents and exposed to X-ray film (Lan et al. 2016).
Skeletal preparations
Skeletal preparations of E18 and P0 pups were performed as described previously (Lan and Jiang 2009).
Acknowledgments
This work was supported by National Institutes of Health/National Institute of Dental and Craniofacial Research grant R03 DE023864 to YL. We thank Dr. Rulang Jiang for discussions. We thank the Cincinnati Children’s Hospital Medical Center Genome Editing and Transgenic Animal Facility for assistance in generation of the Golga5 mutant mice.
References
- Bascom RA, Srinivasan S, Nussbaum RL. Identification and characterization of golgin-84, a novel golgi integral membrane protein with a cytoplasmic coiled-coil domain. J Biol Chem. 1999;274(5):2953–2962. doi: 10.1074/jbc.274.5.2953. [DOI] [PubMed] [Google Scholar]
- Degenhardt K, Wright AC, Horng D, Padmanabhan A, Epstein JA. Rapid 3d phenotyping of cardiovascular development in mouse embryos by micro-ct with iodine staining. Circ Cardiovasc Imaging. 2010;3(3):314–322. doi: 10.1161/CIRCIMAGING.109.918482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao A, Rahman D, Pappin DJ, Lucocq J, Lowe M. The coiled-coil membrane protein golgin-84 is a novel rab effector required for golgi ribbon formation. J Cell Biol. 2003;160(2):201–212. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, San Agustin JT, Xu F, Jonassen JA, Samtani R, Lo CW, Pazour GJ. The golgin gmap210/trip11 anchors ift20 to the golgi complex. PLoS Genet. 2008;4(12):e1000315. doi: 10.1371/journal.pgen.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. Finding the golgi: Golgin coiled-coil proteins show the way. Trends Cell Biol. 2016;26(6):399–408. doi: 10.1016/j.tcb.2016.02.005. [DOI] [PubMed] [Google Scholar]
- Han F, Liu C, Zhang L, Chen M, Zhou Y, Qin Y, Wang Y, Duo S, Cui X, Bao S, et al. Globozoospermia and lack of acrosome formation in gm130-deficient mice. Cell Death Dis. 2017;8(1):e2532. doi: 10.1038/cddis.2016.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama K, Sasaki T, Goto S, Ogasawara K, Maru H, Suzuki K, Suzuki H. Insertional mutation in the golgb1 gene is associated with osteochondrodysplasia and systemic edema in the ocd rat. Bone. 2011;49(5):1027–1036. doi: 10.1016/j.bone.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Lan Y, Jiang R. Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development. 2009;136(8):1387–1396. doi: 10.1242/dev.028167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Kingsley PD, Cho ES, Jiang R. Osr2, a new mouse gene related to drosophila odd-skipped, exhibits dynamic expression patterns during craniofacial, limb, and kidney development. Mech Dev. 2001;107(1–2):175–179. doi: 10.1016/s0925-4773(01)00457-9. [DOI] [PubMed] [Google Scholar]
- Lan Y, Zhang N, Liu H, Xu J, Jiang R. Golgb1 regulates protein glycosylation and is crucial for mammalian palate development. Development. 2016;143(13):2344–2355. doi: 10.1242/dev.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Mei M, Li Q, Roboti P, Pang Q, Ying Z, Gao F, Lowe M, Bao S. Loss of the golgin gm130 causes golgi disruption, purkinje neuron loss, and ataxia in mice. Proc Natl Acad Sci U S A. 2017;114(2):346–351. doi: 10.1073/pnas.1608576114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Lan Y, Xu J, Chang CF, Brugmann SA, Jiang R. Odd-skipped related-1 controls neural crest chondrogenesis during tongue development. Proc Natl Acad Sci U S A. 2013;110(46):18555–18560. doi: 10.1073/pnas.1306495110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Warren G. The road taken: Past and future foundations of membrane traffic. Cell. 2000;100(1):99–112. doi: 10.1016/s0092-8674(00)81687-6. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-gol matrix protein, gm130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa K, Fritzler MJ, Chan EK. Unique and shared features of golgi complex autoantigens. Autoimmun Rev. 2005;4(1):35–41. doi: 10.1016/j.autrev.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Ramirez IB, Lowe M. Golgins and grasps: Holding the golgi together. Semin Cell Dev Biol. 2009;20(7):770–779. doi: 10.1016/j.semcdb.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Ríos RM, Sanchís A, Tassin AM, Fedriani C, Bornens M. Gmap-210 recruits gamma-tubulin complexes to cis-golgi membranes and is required for golgi ribbon formation. Cell. 2004;118(3):323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Shorter J, Warren G. Golgi architecture and inheritance. Annu Rev Cell Dev Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- Smits P, Bolton AD, Funari V, Hong M, Boyden ED, Lu L, Manning DK, Dwyer ND, Moran JL, Prysak M, et al. Lethal skeletal dysplasia in mice and humans lacking the golgin gmap-210. N Engl J Med. 2010;362(3):206–216. doi: 10.1056/NEJMoa0900158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C, Colanzi A. The golgi and the centrosome: Building a functional partnership. J Cell Biol. 2010;188(5):621–628. doi: 10.1083/jcb.200910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkos TM, Lowe M. The golgin family of coiled-coil tethering proteins. Front Cell Dev Biol. 2015;3:86. doi: 10.3389/fcell.2015.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M, Gillingham AK, Munro S. The golgin coiled-coil proteins capture different types of transport carriers via distinct n-terminal motifs. BMC Biol. 2017;15(1):3. doi: 10.1186/s12915-016-0345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M, Munro S. Membrane trafficking. The specificity of vesicle traffic to the golgi is encoded in the golgin coiled-coil proteins. Science. 2014;346(6209):1256898. doi: 10.1126/science.1256898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liu H, Lan Y, Aronow BJ, Kalinichenko VV, Jiang R. A shh-foxf-fgf18-shh molecular circuit regulating palate development. PLoS Genet. 2016;12(1):e1005769. doi: 10.1371/journal.pgen.1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang Y. Grasps in golgi structure and function. Front Cell Dev Biol. 2015;3:84. doi: 10.3389/fcell.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao X, Hu Y, St Amand T, Zhang M, Ramamurthy R, Qiu M, Chen Y. Msx1 is required for the induction of patched by sonic hedgehog in the mammalian tooth germ. Dev Dyn. 1999;215(1):45–53. doi: 10.1002/(SICI)1097-0177(199905)215:1<45::AID-DVDY5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Zhou J, Gao Y, Zhang Z, Zhang Y, Maltby KM, Liu Z, Lan Y, Jiang R. Osr2 acts downstream of pax9 and interacts with both msx1 and pax9 to pattern the tooth developmental field. Dev Biol. 2011;353(2):344–353. doi: 10.1016/j.ydbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]