SUMMARY
Regulatory T cells (Tregs) are required to establish immune tolerance to commensal microbes. Tregs accumulate abruptly in the skin during a defined window of postnatal tissue development. However, the mechanisms mediating Treg migration to neonatal skin are unknown. Here we show that hair follicle (HF) development facilitates the accumulation of Tregs in neonatal skin and that upon skin entry these cells localize to HFs, a primary reservoir for skin commensals. Further, germ-free neonates had reduced skin Tregs indicating that commensal microbes augment Treg accumulation. We identified Ccl20 as a HF-derived, microbiota-dependent chemokine and found its receptor, Ccr6, to be preferentially expressed by Tregs in neonatal skin. The Ccl20-Ccr6 pathway mediated Treg migration in vitro and in vivo. Thus, HF morphogenesis, commensal microbe colonization, and local chemokine production work in concert to recruit Tregs into neonatal skin, thereby establishing this tissue Treg niche early in life.
Graphical Abstract
INTRODUCTION
Regulatory T (Treg) cells play a critical role in establishing and maintaining immune homeostasis in peripheral tissues. Treg cells residing in these tissues demonstrate unique T cell receptor profiles and distinct functions when compared to cells found in lymphoid organs (Feuerer et al., 2009; Burzyn et al., 2013b). Although we are beginning to understand the fundamental biology of Treg cells in adult tissues, very little is known about the cellular and molecular mechanisms responsible for establishing tissue residence of these cells early in life.
Colonization by commensal microbes and formative shaping of the host-commensal relationship occurs in neonatal life (Dominguez-Bello et al., 2010), concurrently with the establishment of Treg cell populations at barrier sites where these commensals reside (Gollwitzer et al., 2014; Scharschmidt et al., 2015; Yang et al., 2015). Treg cells in barrier tissues, such as the skin and the intestine, are known to play an important role in promoting tolerance to commensal-derived antigens (Burzyn et al., 2013a; Scharschmidt et al., 2015). In the colon, commensals themselves facilitate generation of peripherally induced Treg cells that are critical for proper immune homeostasis (Atarashi et al., 2011; Tanoue et al., 2016). We recently demonstrated that establishment of immune tolerance to skin commensal bacteria is preferentially established early in neonatal life (Scharschmidt et al., 2015). In this work, we focused on defining the immunological mechanisms responsible for this process, namely a unique population of Treg cells in neonatal skin that is capable of establishing tolerance to skin commensals. However, perhaps the most striking result was the abrupt accumulation of Treg cells in skin during this defined window of postnatal tissue development. Factors that drive Treg cell accumulation into skin early in life and the relative role of commensal microbes in this process were not explored and are currently unknown.
Immune cell responses in tissues are highly influenced by tissue-specific cellular niches (Pasparakis et al., 2014). To date, the structure and function of these niches have been primarily studied in the intestine. Here, components of the gut-associated lymphoid tissue (GALT), including Peyer’s patches and isolated lymphoid follicles, are situated just deep to the intestinal mucosa, facilitating sampling of microbial antigens via microfold cells and coordination of appropriate B and T cell responses. Commensal microbes promote GALT development, and conversely, impaired GALT development leads to significant alterations in luminal bacterial composition, underlining the importance of these structures in intestinal host-commensal homeostasis (Bouskra et al., 2008; van de Pavert and Mebius, 2010; Maynard et al., 2012). In contrast, lymphoid structures are not found in healthy skin. Instead, emerging evidence suggests that adnexal structures, such as hair follicles (HFs), represent an important immune cell niche in this tissue. HFs serve as a high-traffic zone for antigen-presentation (Hansen and Lehr, 2014), a local source of cytokines and chemokines (Nagao et al., 2012), a primary reservoir for skin commensal microbes (Montes and Wilborn, 1969), and a site where multiple immune cells localize in the steady state, including Treg cells (Gratz et al., 2013; Sanchez Rodriguez et al., 2014; Collins et al., 2016).
Given that commensals preferentially colonize HFs (Montes and Wilborn, 1969), Treg cells localize to these structures (Gratz et al., 2013; Sanchez Rodriguez et al., 2014), and commensal microbes influence Treg cell biology at barrier sites (Tanoue et al., 2016), we sought to functionally dissect the relationship between HFs, the microbiota, and Treg cells in neonatal skin. We found that both HF development and commensal microbes play a major role in Treg cell migration to skin early in life. In addition, we define Ccl20-Ccr6 as a pathway that mechanistically links commensal colonization, HF development, and Treg cell migration to neonatal skin. Thus, coordinated interactions between commensal microbes and postnatal tissue development jointly facilitate recruitment of Treg cells to their niche in skin.
RESULTS
Hair Follicle Development Is Required for Treg Cell Accumulation in Neonatal Skin
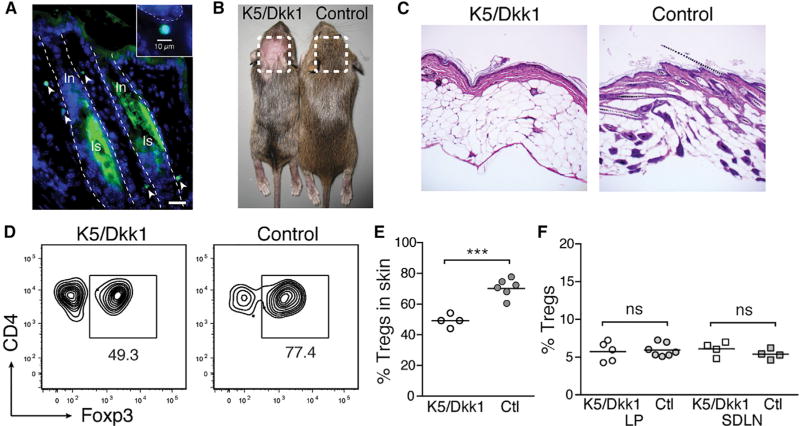
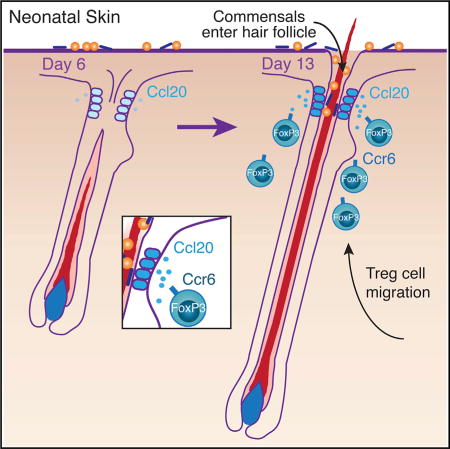
Treg cells accumulate in skin between days 6 and 13 of neonatal life, and this can be prevented by blocking lymphocyte migration from lymphoid organs during this window of time (Scharschmidt et al., 2015). We hypothesized that migration of neonatal Treg cells into skin was dependent on specific factors expressed during postnatal tissue development. In mice, hair follicle morphogenesis begins in utero and continues to progress through the first 2 weeks of life. Specifically, emergence and penetration of the hair shaft through the epidermis occurs after postnatal day 5 (Paus et al., 1999). Because HFs represent the primary niche for Treg cells in skin (Gratz et al., 2013; Sanchez Rodriguez et al., 2014) and have been demonstrated to produce chemokines capable of directing immune cell migration (Nagao et al., 2012), we hypothesized that signals produced by developing HFs facilitate Treg cell migration to neonatal skin. To test this hypothesis, we first characterized where Treg cells localize in skin early in neonatal life. Similar to their localization in adult skin (Gratz et al., 2013; Sanchez Rodriguez et al., 2014), Treg cells in neonatal skin primarily localized to HFs, with the majority found near the infundibulum and isthmus regions (Figure 1A).
Figure 1. Treg Cells Localize to HFs in Neonatal Skin and Are Reduced in the Absence of HF Morphogenesis.
(A) Immunohistochemical staining for GFP in skin from 13-day-old (D13) FoxP3GFP mice. Dashed outline, hair follicle; In, infundibulum, Is, isthmus. Scale bar, 20 µm. Inset: magnified image demonstrating GFP+ cell morphology and size.
(B) Pregnant K5+ females bred to K5/Dkk1 males were fed doxycycline starting at gestational date E13.5, preventing wnt-dependent hair follicle development in K5/Dkk1 but not single-transgenic (Ctl.) littermates.
(C) Representative histology of skin from D13 K5/Dkk1 and Ctl. littermates, taken from site-matched areas on the scalp.
(D) Flow cytometry plots of CD4+ T cells in D13 K5/Dkk1 versus Ctl. skin.
(E and F) Percentage of Treg cells of CD4+ cells in D13 K5/Dkk1 versus Ctl. skin (E), as well as in gut lamina propria (LP) and skin-draining lymph nodes (SDLN) (F).
Data were pooled from three replicate experiments on independent litters, each with ≥3 pups total.
To test whether HFs play a role in mediating Treg cell migration to neonatal skin, we employed an inducible model of abrogated HF development. Because chemokines are produced by HF epithelial cells and most “hairless” mouse models retain these cells but simply fail to develop hair shafts, we rationalized that a model in which the entire HF epithelium fails to develop would be the most appropriate system to test whether HFs play a role in facilitating Treg cell migration into neonatal skin. Initiation of HF morphogenesis is highly dependent on signaling through the wnt pathway during embryogenesis (Andl et al., 2002). In order to dissect whether accumulation of Treg cells was linked with HF development, we inducibly inhibited the wnt pathway in neonatal skin to block HF morphogenesis. In K5-rtTA tetO-Dkk1 (i.e., K5/Dkk1) double transgenic mice, administration of doxycycline induces overexpression of the pan-wnt inhibitor Dkk1 in cells expressing the keratin 5 promoter, thus enabling potent inhibition of the pathway in a tissue-specific fashion (Kistner et al., 1996; Chu et al., 2004; Liu et al., 2008). The skin of these mice demonstrates a specific defect in development of vibrissae and pelage HFs without any effect on proliferation or differentiation of keratinocytes in the interfollicular epidermis (Andl et al., 2002; Chu et al., 2004). We fed doxycycline to pregnant K5 single transgenic females starting on gestational day 13.5 after mating them to K5/Dkk1 males. Their double transgenic K5/Dkk1 offspring demonstrated defined regions of skin completely devoid of HFs, despite a normal appearing epidermis and normal interfollicular dermis (Figures 1B and 1C), consistent with previous reports (Andl et al., 2002; Jenkins et al., 2010). Examination of hairless K5/Dkk1 skin at postnatal day 13 revealed significantly reduced percentages of Treg cells compared with site-matched skin from single-transgenic (ctl) littermate controls (Figures 1D and 1E). Equivalent percentages of Treg cells were observed in intestinal lamina propria (LP) and skin-draining lymph nodes (SDLN) of 13-day-old double- and single-transgenic mice (Figure 1F), indicating that the reduction of neonatal Treg cells is skin specific and not the result of systemic or thymic wnt suppression. Thus, our results suggest that development of HFs facilitates the accumulation of Tregs in neonatal skin.
Commensal Microbes Augment Accumulation of Treg Cells in Neonatal Skin
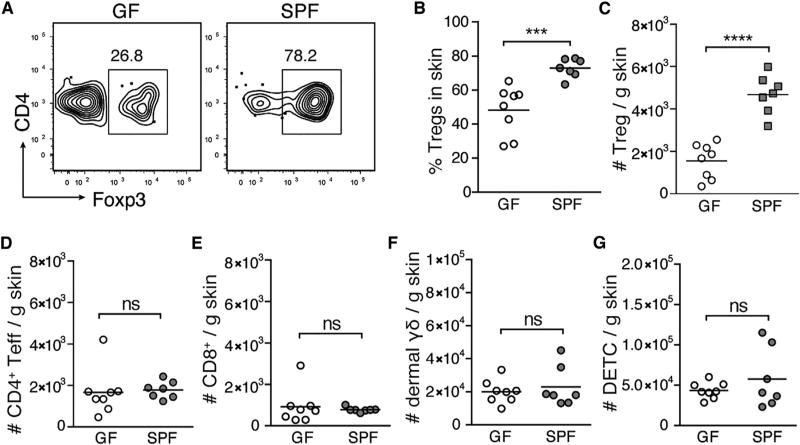
Commensal microbes colonize the skin surface immediately after birth (Dominguez-Bello et al., 2010). However, in neonatal mice colonization of the HF epithelium is delayed until follicular morphogenesis progresses to allow microbial access to deeper HF structures. Because HFs represent a key tissue niche for skin-resident microbiota (Montes and Wilborn, 1969; Lange-Asschenfeldt et al., 2011), we hypothesized that commensal microbes might influence signals produced by HFs to facilitate Treg cell accumulation. To test this, we examined T cell populations present in the skin of 13-day-old germ-free (GF) and specific-pathogen-free (SPF) mice. We found Treg cell percentages to be significantly reduced in the skin of GF neonates (Figures 2A and 2B). Treg cell numbers were also substantially lower in GF conditions (Figure 2C) approaching levels observed in 6-day-old SPF mice (Scharschmidt et al., 2015). In contrast, CD4+Foxp3neg T (CD4+ Teff) cells, CD8+ T cells, dermal γδ T cells, and dendritic epidermal T cells were present at equal numbers in 13-day-old SPF and GF skin (Figures 2D–2G). These results suggest a role for commensal microbes specifically in augmenting the accumulation of Treg cells in skin early in life.
Figure 2. Commensal Microbes Mediate Accumulation of Treg Cells in Neonatal Skin.
(A) Representative flow cytometry plots of CD4+ T cells from skin of D13 germ-free (GF) versus specific-pathogen-free (SPF) mice.
(B and C) Percentages (B) and absolute numbers (C) of Treg cells in skin of D13 GF versus SPF mice.
(D–G) Absolute numbers of FoxP3neg CD4+ effector T cells (Teff) (D), CD8+T cells (E), dermal γδ T cells (F), and dendritic epidermal T cells (DETC) (G) in skin of D13 GF versus SPF mice.
Data were pooled from two of three replicate experiments with ≥3 mice per group.
Identification of Ccl20 and Ccr6 as a Potential Pathway that Mediates Treg Cell Migration into Neonatal Skin
HF development and commensal microbes facilitate Treg cell migration into neonatal skin. Interestingly, commensal microbes begin to colonize HFs during the same window of time that Treg cells migrate to skin. Thus, we sought to determine if HFs and commensals jointly coordinate neonatal Treg cell migration and elucidate the molecular mechanisms underpinning this process. We have previously shown that accumulation of Treg cells in neonatal skin is largely migration dependent, as it can be blocked with FTY720, which prevents lymphocyte egress from lymphoid organs via functional antagonism of the sphingosine-1-phosphate receptor (Matloubian et al., 2004; Scharschmidt et al., 2015). We therefore hypothesized that the migration of Treg cells into neonatal skin occurs in response to a chemotactic signal originating from the tissue. To identify chemokine-chemokine receptor pairs that might be important in this process, we employed an unbiased discovery approach to select candidates that fit the following three criteria: (1) expression of the chemokine(s) is increased in skin concurrently with Treg cell influx into the tissue; (2) expression of the chemokine is augmented by the presence of commensal microbes; and (3) the corresponding chemokine receptor(s) is preferentially expressed by Treg cells in neonatal skin (outlined in Figure S1A).
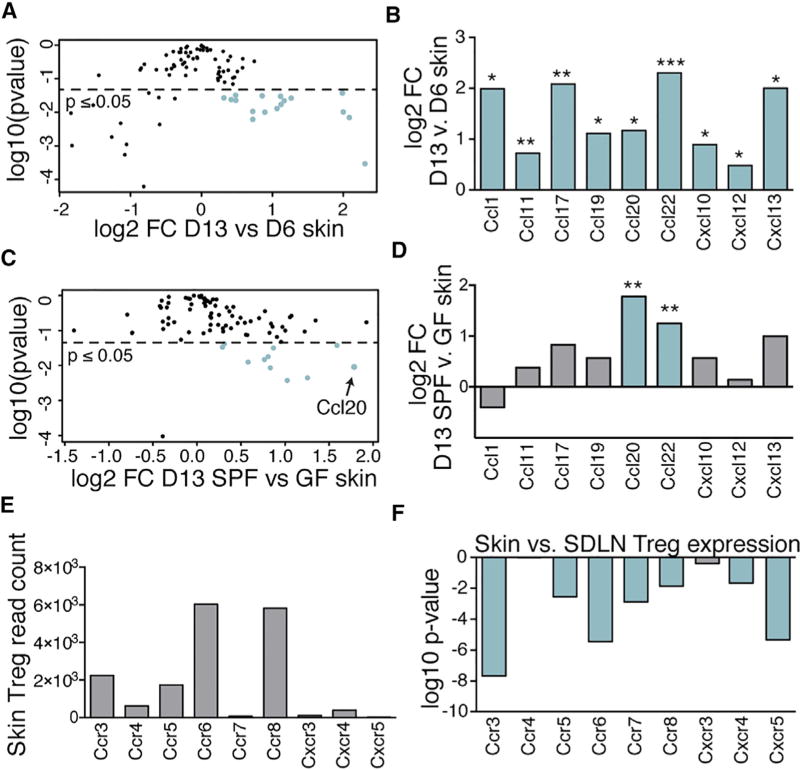
To determine which chemokines increase in skin early in life we employed a qRT-PCR chemokine array to compare levels of expression in 6-day-old (pre-Treg cell influx) versus 13-day-old skin (i.e., at the height of the Treg cell influx) (Figure 3A). We found 9 chemokines for which mRNA levels increased significantly in skin from postnatal day 6 to day 13 (Figure 3B). Of these, Ccl20 was a particularly intriguing candidate because keratinocytes in adult HFs produce this chemokine (Nagao et al., 2012). To dissect which of these chemokines have their production augmented by the presence of commensal microbes, we isolated RNA from whole skin of 13-day-old GF and SPF mice and performed qRT-PCR chemokine arrays (Figure 3C). Of the chemokines increased on postnatal day 13 relative to day 6, only Ccl20 and Ccl22 were significantly increased in the presence of commensals (Figure 3D).
Figure 3. Discovery Approach to Identify Chemokine-Chemokine Receptor Pathways Responsible for Treg Cell Migration to Neonatal Skin.
(A) Quantitative RT-PCR chemokine array was performed on RNA isolated from whole skin of three D6 and five D13 SPF mice. Volcano plot depicting log2FC versus log10 (p value) for all candidates tested.
(B) Log2FC for those chemokines significantly increased in D13 versus D6 skin.
(C) Identical qRT-PCR was performed on five D13 SPF versus three GF skin. Volcano plot depicting log2FC versus log10(p value) for all candidates tested.
(D) Log2FC in D13 SPF versus GF skin for chemokine subset also increased in D13 versus D6 skin.
(E) Treg cells were sorted from skin and LN from D13 SPF FoxP3DTR mice and subjected to RNA sequencing. Average raw gene counts in D13 skin Treg cells are shown for receptors corresponding to chemokines increased in D13 skin.
(F) Log10(p value) (corrected for multiple comparisons) of log2FC expression of receptors by skin versus SDLN Treg cells. Blue bar: p ≤ 0.05. See also Figure S1.
To determine which chemokine receptors were preferentially expressed on neonatal skin Treg cells, we isolated Treg cells from the skin and SDLNs of postnatal day 13 pups and subjected them to whole transcriptome RNA sequencing (Figure S1B). Of those receptors for which ligands increased in skin between postnatal day 6 and 13, Ccr6 (the receptor for Ccl20) had the highest expression levels in neonatal skin Treg cells (Figure 3E). Moreover, Ccr6 was preferentially expressed by skin Treg cells when compared to SDLN Treg cells (Figures 3F and S1C). Thus, based on our criteria, the Ccl20-Ccr6 pathway emerged as the top candidate warranting further validation and functional exploration.
Commensal Microbes Augment Production of Ccl20 in the Infundibulum of Developing Hair Follicles
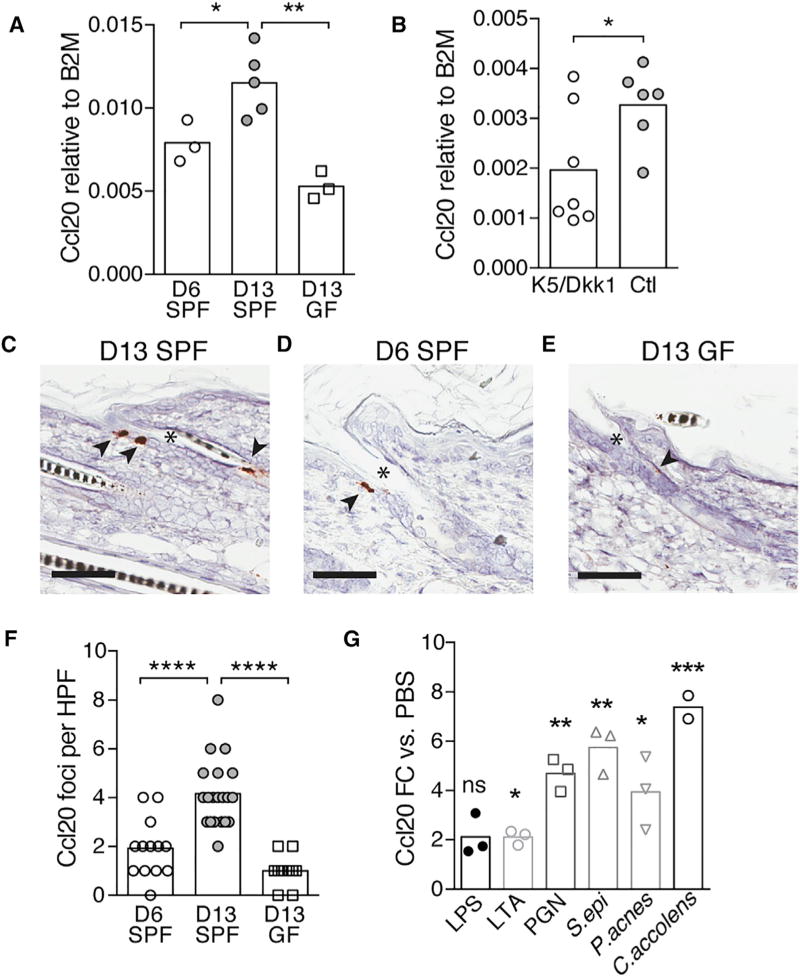
To validate findings from our discovery approach and localize the source of Ccl20 in neonatal skin, we performed qRT-PCR for Ccl20 on skin of 6-day-old SPF, 13-day-old SPF, and 13-day-old GF mice. Consistent with the results from our chemokine qRT-PCR array, we found that skin expression of Ccl20 increases between day 6 and day 13 of life and is reduced under GF conditions (Figure 4A). Because HF morphogenesis progresses significantly between postnatal day 6 and day 13 (Paus et al., 1999), we hypothesized that HFs represent the primary source of Ccl20 in neonatal skin. Consistent with this hypothesis, Ccl20 expression was significantly reduced when HF morphogenesis was abrogated, with lower transcript levels found in hairless skin from K5/Dkk1 mice as compared with normal hair-bearing skin of littermate controls (Figure 4B). In contrast, Ccl22, which was also elevated in D13 SPF skin versus D6 SPF or D13 GF skin, was not reduced in hairless skin from K5/Dkk1 mice (Figure S2A). To further verify that HFs in neonatal skin produce Ccl20, we performed RNA in situ hybridization for Ccl20 on skin tissue sections from 13-day-old SPF pups. Transcript signal was detected in keratinocytes located in the upper infundibular portion of HFs, but not elsewhere in the tissue (Figures 4C and S2B). This is the portion of the HF most densely populated by commensal microbes (Figure S2C) (Montes and Wilborn, 1969; Alexeyev et al., 2012) and around which Treg cells predominantly localize in neonatal skin (Figure 1A). Consistent with our qRT-PCR data, RNA in situ hybridization for Ccl20 in 6-day-old SPF skin and 13-day-old GF skin showed fewer foci of Ccl20 signal per high-powered field (Figures 4D–4F). To understand whether capacity for Ccl20 induction is limited to specific skin commensals or broadly conserved, we employed an in vitro human skin explant system in which we can selectively expose microbiallynaive fetal skin to commensal bacteria or bacterial surface molecules. Using this system, we found that overnight colonization with Staphylococcus epidermidis, Corynebacterium accolens, or Propionibacterium acnes was, in each case, sufficient to increase Ccl20 expression in skin as measured by qRT-PCR. Peptidoglycan and, to a lesser extent, lipoteichoic acid and lipopolysaccharide were also capable of augmenting Ccl20 in human fetal skin (Figure 4G). These results are consistent with a model in which conserved microbial epitopes activate innate immune pathways upstream of Ccl20 expression (Zhao et al., 2014).
Figure 4. Commensal Microbes Augment Ccl20 Expression in Developing HFs.
(A and B) Fold change expression of Ccl20 versus β 2-microglobulin mRNA transcripts in skin from D6 SPF, D13 SPF, and D13 GF mice (A) and D13 K5/Dkk1 versus Ctl littermates (B), pooled from three litters.
(C–E) Representative images of in situ hybridization (ISH) for Ccl20 mRNA transcript in the skin of D13 SPF (C), D6 SPF (D), and D13 GF (E) mice. Arrows, visualization of Ccl20 mRNA signal; *, hair follicle infundibulum. Scale bars, 60 µm.
(F) Quantification of Ccl20 ISH staining density (number of positive cells per 20× field).
(G) Fold change expression in Ccl20 (normalized to EIF3L) in human fetal skin after overnight exposure to skin commensal bacteria or bacterial surface molecules as compared with PBS-treated controls. LPS, lipopolysaccharide; LTA, lipoteichoic acid; PGN, peptidoglycan; S. epi, Staphylococcus epidermidis; P. acnes, Propionibacterium acnes; C. accolens, Corynebacterium accolens.
All data are representative of two independent experiments. See also Figure S2.
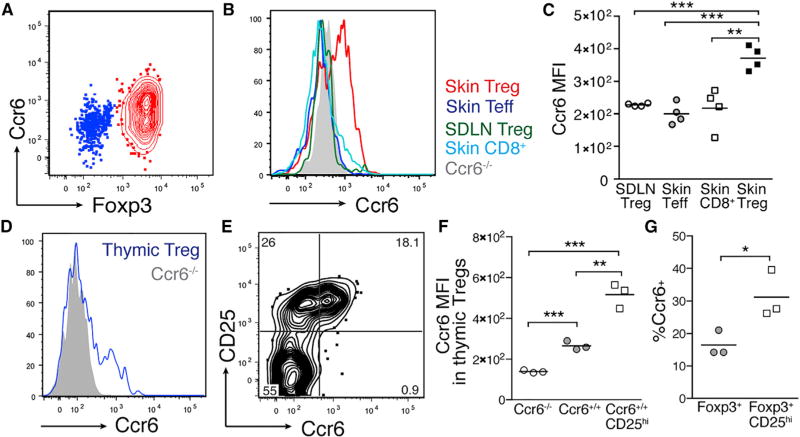
Ccr6 Is Preferentially Expressed by Treg Cells in Neonatal Skin and a CD25+ Subset of Thymic Treg Cells
After validating that Ccl20 expression is increased in skin from day 6 to day 13 of life, that this chemokine is almost exclusively expressed by the HF epithelium in neonatal skin, and that colonization with commensal microbes augments its expression, we next set out to quantify levels of the Ccl20 receptor, Ccr6, on neonatal skin Treg cells. Staining for Ccr6 by flow cytometry demonstrated significantly higher levels of expression by neonatal skin Treg cells as compared to Treg cells in the SDLN, skin Teff CD4+ cells, and skin CD8+ T cells (Figures 5A–5C). These results were consistent with our RNA sequencing data that showed elevated transcript levels for Ccr6 in neonatal skin compared to skin-draining LN Treg cells (Figures 3F and S1C). Moreover, these results demonstrate selective expression of Ccr6 on Treg cells versus other ab T cell subsets in neonatal skin, consistent with their preferential migration to skin during the postnatal period of increasing expression of Ccl20 in this tissue. Because Treg cells from skin-draining LN demonstrated minimal expression of Ccr6, we sought to quantify expression of Ccr6 on neonatal thymic Treg cells as the potential source of this skin-homing population. A discrete subset of neonatal thymic Treg cells expressed Ccr6 (Figure 5D). Notably, these Ccr6+ thymic Treg cells were enriched among CD25hi Treg cells (Figures 5E–5G), suggesting that these cells may constitute a phenotypically distinct subset in the developing thymus.
Figure 5. Neonatal Skin Treg Cells and a Subset of Thymic Treg Cells Preferentially Express Ccr6.
(A) Flow cytometry plot depicting Ccr6 expression on CD4+ T cells in neonatal skin.
(B) Histogram depicting relative mean fluorescent intensity (MFI) of Ccr6 by flow cytometry on Treg cells, CD4+ Teff cells, and CD8+ T cells from neonatal skin, as well as WT and Ccr6−/− Treg cells in SDLN.
(C) Average Ccr6 MFI on these same populations.
(D) Histogram depicting MFI of Ccr6 on neonatal thymic Treg cells (CD3+ CD4+ CD8neg FoxP3+).
(E) Flow cytometry plot depicting Ccr6 and CD25 co-staining by thymic Treg cells.
(F) Average MFI of Ccr6 on thymic Treg cells from Ccr6−/− mice versus wild-type mice versus CD25hi subset.
(G) Percent of all thymic Treg cells and CD25hi thymic Treg cells that stain positively for Ccr6.
All data are representative of three independent experiments with n ≥ 3 mice.
Ccl20 Drives Migration of Neonatal Treg Cells In Vitro
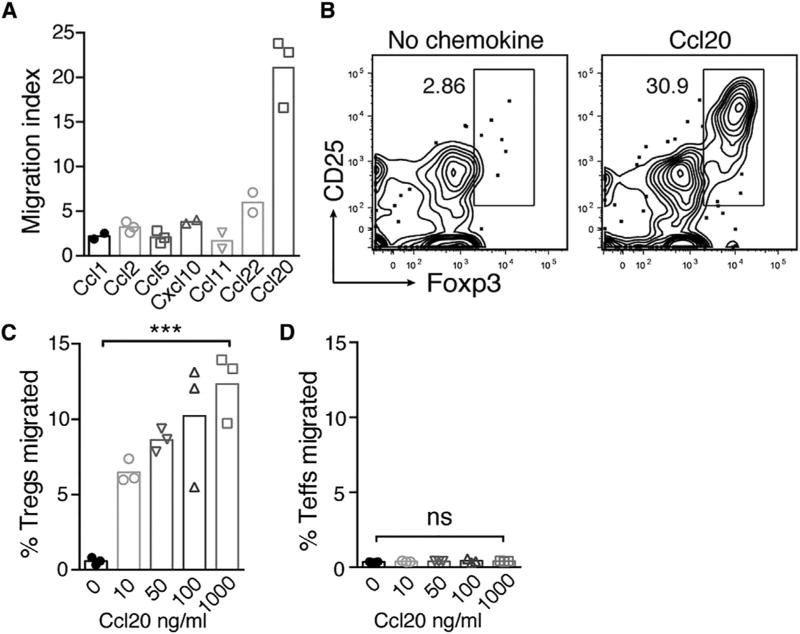
The HF and commensal-dependent increase in Ccl20 expression in neonatal skin, in conjunction with the preferential expression of Ccr6 by neonatal skin Treg cells, suggested a role for Ccl20 in driving Treg cell migration. We therefore sought to evaluate Ccl20’s capacity to drive migration of neonatal Treg cells compared to other chemokines expressed in skin during the neonatal period. We have previously shown that FTY720 treatment between postnatal days 5 and 11 results in accumulation of Treg cells in the thymus rather than SDLN (Scharschmidt et al., 2015). This observation, together with evidence for direct skin-homing by thymic αβ T cells (Washington et al., 1995) and the expression of Ccr6 by a subset of thymic Tregs cells (Figures 5D–5G), suggested that the Treg cell influx into neonatal skin might reflect Ccl20-mediated migration of Treg cells originating from the thymus. To test this, we incubated CD4+ single-positive thymocytes isolated from postnatal day 9 mice in a transwell system in the presence of various chemokines to evaluate their relative capacity to drive Treg cell migration. Of all the chemokines tested, including several observed to be increased in skin between day 6 and day 13, Ccl20 almost exclusively induced Treg cell migration, displaying a 5- to 10-fold higher migration index versus other chemokines (Figure 6A). Flow cytometry staining of cells in the lower transwell after 3 hr demonstrated significant enrichment for Treg cells in the presence of Ccl20 (Figure 6B), suggesting a preferential effect on Treg cell versus Teff cell migration. The percentage of Treg cells migrating in this assay increased in parallel with the concentration of Ccl20 (Figure 6C). In contrast, thymic CD4+ Teff cells did not demonstrate chemotaxis to Ccl20 at any of the doses tested (Figure 6D). Treg cells isolated from neonatal skin but not SDLN demonstrated Ccl20-induced migration comparable to thymic Treg cells (Figure S3). These results are consistent with the preferential expression of Ccr6 by neonatal skin and thymic Treg cells, as compared to LN Treg cells or CD4+ Teff cells (Figure 5). Taken together, Ccl20 was preferentially capable of driving migration of neonatal thymic and skin Treg cells in vitro.
Figure 6. Ccl20 Preferentially Drives Neonatal Treg Cell Migration In Vitro.
(A) CD4+ enriched D9 thymocytes were incubated in a transwell system with various chemokines, and cell migration to the lower transwell was assessed at 3 hr by flow cytometry. Treg cell migration index for various chemokines; index calculated as fold increase in Treg cells migrated in presence of chemokine (at 1,000 ng/ml) relative to no chemokine control. Each data point depicts a technical replicate.
(B) Representative flow cytometry plot of CD4+ population in lower transwell in the presence of Ccl20 at 100 ng/ml versus no chemokine control.
(C) Percentage of Treg cells migrated to lower transwell at varying doses of Ccl20. Calculated as number of Treg cells in lower transwell / number of Treg cells input to assay.
(D) Percentage of FoxP3neg CD4+ Teff cells migrated at varying doses of Ccl20.
Data in (A) are representative of two independent experiments; (B)–(D) are representative of three independent experiments. See also Figure S3.
Ccr6 Facilitates Migration of Treg Cells into Neonatal Skin In Vivo
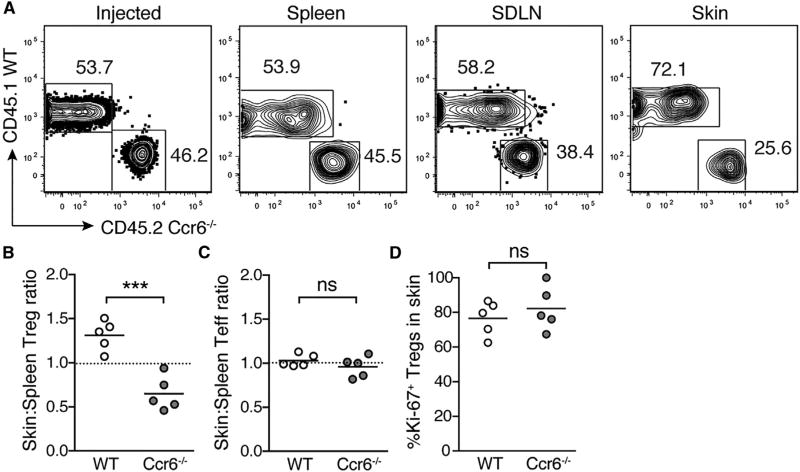
The capacity for Ccl20 to drive migration of neonatal Treg cells, in conjunction with its increased expression in HF epithelium, led us to hypothesize that expression of Ccr6 by neonatal Treg cells helps mediate their migration to skin early in life. Because competition between populations with distinct receptor profiles has been shown to be important in controlling lymphocyte entry into tissues (Campbell et al., 2007), we chose to test the role of Ccr6 in skin-homing of neonatal Treg cells using a neonatal adoptive transfer model. Populations of congenically labeled wild-type (WT) and Ccr6-deficient (Ccr6−/−) CD4+ single-positive thymocytes were transferred into RAG2-deficient recipients on postnatal day 9. Spleen, SDLN, and skin were harvested 2 weeks post-transfer, and flow cytometry was performed to assess the relative capacity of WT and Ccr6−/− Treg and Teff cells to migrate to these tissues. The ratio of Ccr6−/− to WT Treg cells was roughly equivalent upon transfer and unchanged in the spleen and SDLN upon harvest, suggesting that Ccr6 is not required for migration to these secondary lymphoid organs (Figure 7A). In contrast, the proportion of Ccr6−/− Treg cells was significantly reduced in the skin relative to both spleen (Figures 7A and 7B) and SDLN (Figures 7A, S4A, and S4B). Notably, CD4+ Teff cells distributed equally across skin, spleen, and SDLN irrespective of Ccr6 status (Figures 7C, S4C, and S4D). Ccr6−/− and WT Treg cells in skin post-transfer demonstrated equivalent Ki-67 expression (Figure 7D), suggesting that the absence of Ccr6 on Treg cells results in a diminished capacity for migration to skin rather than reduced local proliferation in the tissue. These results suggest that Ccr6-Ccl20 interactions play an important role in facilitating Treg cell migration to skin in neonatal life.
Figure 7. Ccr6 Expression on Treg Cells Facilitates Their Migration into Neonatal Skin.
(A) CD4+ enriched thymocytes from CD45.1 WT and CD45.2 Ccr6−/− mice were injected intraperitoneally into D9 rag2−/− neonates. Flow cytometry plots depicting percentage WT versus Ccr6−/− Treg cells in injected population on D0 and in spleen, skin-draining lymph nodes (SDLN), and skin of recipients 13 days post-transfer.
(B–D) Skin:spleen ratio for WT and Ccr6−/− Treg cells (i.e., percentage of WT Treg cells in skin divided by percentage of WT Treg cells in spleen) (B), skin:spleen ratio for WT and Ccr6−/− CD4+ Teffs (C), and percentage of Ki-67+ WT and Ccr6−/− Treg cells in skin (D).
Data are representative of two independent experiments with n ≥ 5 mice. See also Figure S4.
DISCUSSION
Elucidating the molecular mechanisms governing immune cell recruitment to barrier sites early in life is of fundamental importance in our attempts to understand how tissue immune cell niches are established. Because commensal microbes colonize barrier sites during this critical period of tissue and immune development, it is also paramount to explore how they cooperate with the host to facilitate an environment that lends to a productive and “mutualistic” adaptive immune response to commensal antigens. Here, we have shown that molecular signals jointly coordinated by commensal microbes and developing HFs direct migration of Treg cells into skin early in life. We found Treg cells to be reduced in the absence of either HF morphogenesis or commensal colonization, suggesting an independent and critical role for each of these factors in the accumulation and enrichment of skin Treg cells during this developmental window. HFs secrete chemokines, which can direct migration of immune cells into skin (Nagao et al., 2012). Likewise, bacteria, which reside in HFs, are capable of modulating chemokine expression by host epithelia (Maynard et al., 2012). To further dissect the molecular mechanisms mediating Treg cell migration, we utilized an unbiased discovery approach to determine a list of candidate ligands and corresponding receptors that might be involved. Ccl20 was increased in skin both during the window of Treg cell accumulation and in the presence of commensal microbes, was reduced in the hairless K5/Dkk1 model, and was expressed by keratinocytes in the infundibulum of developing HFs, the site of Treg accumulation in neonatal skin. Moreover, its receptor, Ccr6, was highly expressed on skin Treg cells relative to other T cell subsets in neonatal skin and Treg cells in SDLNs. Accordingly, Ccl20 demonstrated preferential capacity to induce neonatal Treg cell migration in vitro, and expression of Ccr6 facilitated Treg cell migration into neonatal skin in vivo. Together, these results support a model whereby colonization of developing HFs by commensal microbes helps to augment expression of Ccl20 by infundibular keratinocytes, thereby promoting migration of Ccr6+ Treg cells into neonatal skin.
These results add to a growing body of literature that implicates HFs as tissue-specific structures integral to skin immune cell homeostasis. CD4+ T cells have been shown to cluster around HFs, where they interact with antigen-presenting cells (Collins et al., 2016). HFs themselves direct immune cell populations in the tissue, producing Ccl2 to recruit Langerhans cells to skin during inflammation (Nagao et al., 2012) and IL-7 and IL-15 to maintain epidermotropism of CD4+ and CD8+ memory T cell populations (Adachi et al., 2015). We have previously shown that Treg cells preferentially localize to HFs in murine and human skin (Gratz et al., 2013; Sanchez Rodriguez et al., 2014). Thus, it is intriguing but perhaps not surprising that these structures are also involved in directing Treg cells into the tissue. HFs represent a high-traffic area for antigen presentation, a dense site of microbial colonization, and an important reservoir of tissue stem cells (Cotsarelis et al., 1990). In this context, mechanistically linking the recruitment of Treg cells with HF formation may serve to prevent or limit inflammatory responses that could damage the tissue and jeopardize its capacity for repair. As HFs are not found at mucosal barrier sites, the growing literature on HFs as a skin-specific immune niche indicates that understanding skin immune homeostasis with respect to both self and foreign antigens will require dedicated study of these structures and the immune cell and microbial populations that localize to them.
Our results highlight a primary role for commensal microbes in facilitating HF-directed recruitment of Treg cells into skin. Commensals are also instrumental in augmenting Treg cell numbers in the colonic lamina propria (Round and Mazmanian, 2010; Atarashi et al., 2011). However, the mechanisms by which commensals influence skin Treg cell numbers appear to be distinct from those in the colon, where peripheral induction of commensal-specific Treg cells by a subset of bacterial species is believed to play a significant role (Tanoue et al., 2016). In skin, microbes exert their influence early in life by augmenting HF-mediated recruitment of Treg cells into the developing tissue. Whereas colonic Treg cell numbers remain persistently low under germ-free conditions, skin Treg cell numbers are equivalent or higher in adult GF versus SPF animals (Belkaid and Naik, 2013). This suggests that other factors can facilitate skin Treg cell accumulation later in life in the absence of microbes. However, what these other factors are and whether they induce migration, local proliferation, or peripheral induction of Treg cells remains unknown. Our finding that various skin bacteria and bacterial molecules can induce Ccl20 expression in developing skin, suggests that, in contrast to the induction of colonic Treg cells by specific commensals, the capacity to recruit Treg cells into the skin early in life may be redundant across bacterial phyla. Whether these skin Treg cells harbor T cell receptors specific for commensal antigens is an intriguing question worthy of further investigation. Furthermore, recognizing that, in contrast to mice, human thymic and hair follicle development begins in utero prior to significant microbial exposure (Holbrook and Minami, 1991; Mold and McCune, 2012), it will be important to evaluate timing of Treg cell accumulation in prenatal and neonatal human skin and dissect the relative contributions of hair follicle and microbial signals in this context.
We found a specific role for Ccl20 and its receptor, Ccr6, in mediating migration of Treg cells into neonatal skin. Intriguingly, this same receptor-chemokine pair is known to play a role in microbe-induced maturation of isolated lymphoid follicles, an important immunologic niche in the developing intestine (Bouskra et al., 2008). Though Ccr6 is known to mediate skin-homing properties of specific dermal γδ T cell (Gray et al., 2011; Cai et al., 2014) and CD4+ memory T cell subsets (Trifari et al., 2009; Duhen et al., 2009), here we describe its role in skin-homing of Treg cells. Ccr4 and Ccr10 have been shown to mediate migration of Treg cells into skin in other biological contexts (Sather et al., 2007; Xia et al., 2014). Though Treg cell recruitment via receptors other than Ccr6 may contribute to their rapid accumulation in neonatal skin, our data suggest a central role for the Ccl20-Ccr6 pathway in this process. Roles for Ccr6 in Treg cell homing have been previously described, especially in the context of memory responses and Th17-mediated inflammation (Kleinewietfeld et al., 2005; Yamazaki et al., 2008; Kitamura et al., 2010). This is consistent with the finding that S. epidermidis and other skin commensal bacteria elicit IL-17 production by resident CD4+ T cells (Naik et al., 2015).
We have previously shown that immune tolerance to skin commensal bacteria is established preferentially during neonatal versus adult life and that the “wave” of Treg cells into neonatal skin plays a critical role in this process (Scharschmidt et al., 2015). Consistent with this, Treg cells accumulating in other tissues early in life demonstrate heightened activation and superior capacity to suppress inflammation and prevent autoimmunity (Yang et al., 2015). Accordingly, efforts to understand the biology of Treg cells in neonatal tissues will have implications for host-commensal immune interactions as well as self-tolerance throughout life. Here, we have focused on tissue signals that preferentially direct accumulation of Treg cells into neonatal skin. Future studies should address the potential contribution of extra-cutaneous processes that generate these cells or facilitate their tissue-licensing, as well as host pathways and commensal products that promote their activation and their function in skin. Taken together, our work highlights that HFs and commensals are instrumental in establishing the Treg cell niche in neonatal skin, a process required for optimal host-commensal immune dialog in this barrier tissue.
STAR*METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
- EXPERIMENTAL MODEL AND SUBJECT DETAILS
- Experimental Animals
- Human fetal skin
- Bacterial strains
- METHOD DETAILS
- Tissue processing and flow cytometry
- Histopathology and immunohistochemistry
- RNA in situ hybridization
- Neonatal Adoptive Transfer Model
- In vitro transwell T cell migration assay
- qRT-PCR
- In vitro human skin explant system
- RNA sequencing of neonatal Treg cells
QUANTIFICATION AND STATISTICAL ANALYSIS
DATA AND SOFTWARE AVAILABILITY
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD4 BV650 Clone RM4-5 | BioLegend | Cat#100546; RRID: AB_2562098 |
| Anti-mouse CD3e BV711 Clone 145-2C11 | BD Biosciences | Cat#563123 |
| Anti-mouse CD8a BV605 Clone 53-6.7 | BioLegend | Cat#100744; RRID: AB_2562609 |
| Anti-mouse CD25 A780 Clone PC61.5 | eBioscience | Cat#47-0251-82; RRID: AB_1272179 |
| Anti-mouse CD45 A700 Clone 30-F11 | eBioscience | Cat#56-0451-82; RRID: AB_891454 |
| Anti-mouse CD45.1 PE Clone A20 | BD Biosciences | Cat#553776; RRID: AB_395044 |
| Anti-mouse CD45.2 A780 Clone 104 | eBioscience | Cat#47-0454-82; RRID: AB_1272175 |
| Anti-mouse CD152/CTLA-4 PE Clone UC10-4F10-11 | BD Biosciences | Cat#553720; RRID: AB_395005 |
| Anti-mouse CD196/Ccr6 A647 Clone 140706 | BD Biosciences | Cat#561753; RRID: AB_10893404 |
| Anti-mouse CD278/ICOS FITC Clone 398.4A | eBioscience | Cat#11-9949-82; RRID: AB_465458 |
| Anti-mouse TCRβ PerCp-Cy5.5 Clone H57-597 | eBioscience | Cat#45-5961-80; RRID: AB_925764 |
| Anti-mouse γδ TCR FITC Clone eBioGl3 | eBioscience | Cat#11-5711-82; RRID: AB_465238 |
| Anti-mouse/human Ki-67-PeCy7 Clone B56 | BD Biosciences | Cat#561283; RRID: AB_10716060 |
| Anti-mouse FoxP3-e450 Clone FJK-16 s | eBioscience | Cat#48-5773-82; RRID: AB_1518812 |
| Rabbit anti-GPF | Invitrogen | Cat#A11122; RRID: AB_221569 |
| Goat anti-Rabbit Alexa 488 | Invitrogen | Cat#A11070; RRID: AB_142134 |
| Prolong Gold Antifade Mountant with DAPI | Life Technologies | Cat#P36931 |
| Bacterial and Virus Strains | ||
| Staphylococcus epidermidis Tü3298 wild-type | Provided by Michael Otto; Augustin and Gotz, 1990 | N/A |
| Staphylococcus epidermidis Tü3298 with plasmid pJL71-2W-mcherry | Scharschmidt et al., 2015 | |
| Propionibacterium acnes HL110PA4 HM-555 | Provided by Yug Varma, strain originated from BEI Resources, NIAID, NIH as part of the Human Microbiome Project | HMP ID 9578 |
| Corynebacterium accolens Neubauer et al. | ATCC | 49725 |
| Biological Samples | ||
| Fetal skin samples, discarded tissue from elective termination procedures | Women’s Option Center at Zuckerberg SFGH | https://irb.ucsf.edu/not-human-subjects-research |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase from Clostridium histolyticum, Type XI | Sigma-Aldrich | Cat#C9407 |
| DNase | Sigma-Aldrich | Cat#DN25 |
| Hyaluronidase from bovine testes | Sigma-Aldrich | Cat#H3506 |
| Lipopolysaccharide from Escherichia coli O111:B4 | Sigma-Aldrich | Cat#L4391 |
| Lipoteichoic acid from Staphylococcus aureus | Sigma-Aldrich | Cat#L2515 |
| Peptidoglycan from Staphlococcus aureus | Sigma-Aldrich | Cat#77140 |
| Tryptic Soy Medium | BD-Bacto | Cat#211825 |
| Brain Heart Infusion Medium | BD-Bacto | Cat#237500 |
| Tween 80 | Fisher | Cat#T164-500 |
| Fatty acid-free Bovine Serum Albumin | Sigma-Aldrich | Cat#A8806 |
| Recombinant Murine Ccl1 | R&D Systems | Cat#845-TC-025 |
| Recombinant Murine Ccl2 | Peprotech | Cat#250-10 |
| Recombinant Murine Ccl5 | Peprotech | Cat#250-7 |
| Recombinant Murine Cxcl10 | Peprotech | Cat#250-16 |
| Recombinant Murine Ccl11 | Peprotech | Cat#250-1 |
| Recombinant Murine Ccl22 | Peprotech | Cat#250-23 |
| Recombinant Murine Ccl20 | Peprotech | Cat#250-27 |
| Critical Commercial Assays | ||
| RNeasy Fibrous Tissue Mini Kit | QIAGEN | Cat#74704 |
| gentleMACS M Tubes | Miltenyi Biotec | Cat#130-093-236 |
| Mouse Cytokines and Chemokines RT2 Profiler PCR Array | QIAGEN | Cat#PAMM-150Z |
| RNAlater Stabilization Solution | Life Technologies | Cat#AM7020 |
| iScript Kit Advanced cDNA Synthesis Kit for RT-qPCR | Bio-Rad | Cat#1725038 |
| Power SYBR Green Cells to CT Kit | Life Technologies | Cat#F410L |
| PureLink DNase Set | Life Technologies | Cat#12185010 |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | Cat#1725274 |
| EasySep Mouse CD4+ T Cell Isolation Kit | StemCell Technologies | Cat#19852 |
| EasySep Mouse PE Positive Selection Kit | StemCell Technologies | Cat#18554 |
| RNAscope 2.5 HD Detection Kit – Brown | Advanced Cell Diagnostics | Cat#322300 |
| Probe-Mm-Ccl20 | Advanced Cell Diagnostics | Cat#434051 |
| GasPak EZ Anaerobe Pouch System | BD Fisher Scientific | Cat#B260683 |
| Mouse FoxP3 Buffer Set | eBiosciences | Cat#00-5523-00 |
| Ghost Dye Violet 510 Live/Dead Stain | Tonbo Biosciences | Cat#13-0870-T100 |
| Deposited Data | ||
| RNA sequencing files | This paper | GEO: GSE87130; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87130 |
| Experimental Models: Organisms/Strains | ||
| SPF mice | Jackson Laboratory | C57BL/6J Cat#000664 |
| GF mice | Germ-free C57BL/6J from colony housed in Sonnenburg lab at Stanford University, originally obtained from Jeffrey Gordon’s lab at Wash U. | C57BL/6J |
| CD45.1 mice | Jackson Laboratory | B6.SJL-Ptprca Pepcb/BoyJ Cat#002014 |
| Ccr6 −/− mice | Jackson Laboratory | B6.129P2-Ccr6tm1Dgen/J Cat#005793 |
| FoxP3DTR mice | Jackson Laboratory | B6.129(Cg)-Foxp3tm3(DTR/GFP)Ayr/ J Cat#016958 |
| FoxP3GFP mice | Jackson Laboratory | B6.Cg-Foxp3tm2Tch/J Cat#006772 |
| Rag2−/− mice | Jackson Laboratory | B6(Cg)-Rag2tm1.1Cgn/J Cat#008449 |
| K5-rtTA x tetO-Dkk1 mice (mixed genetic background) | Provided by Sarah Millar; Chu et al., 2004; Kistner et al., 1996 | N/A |
| Oligonucleotides | ||
| Murine Ccl20 forward primer: TTGCTTTGGCATGGGTACTG | Nagao et al., 2012 | N/A |
| Murine Ccl20 reverse primer: TCGTAGTTGCTTGCTGCTTCTG | Nagao et al., 2012 | N/A |
| Murine Ccl22 forward primer: TCTGATGCAGGTCCCTATGGT | Kusumoto et al., 2007 | N/A |
| Murine Ccl22 reverse primer: TTATGGAGTAGCTTCTTCAC | Kusumoto et al., 2007 | N/A |
| Murine B2m forward primer: TTCTGGTGCTCTCACTGA | This paper | N/A |
| Murine B2m reverse primer: CAGTATGTTCGGCTTCCCATTC | This paper | N/A |
| Human Ccl20 forward primer: CTTTGATGTCAGTGCTGCTACT | Loiseau et al., 2016 | N/A |
| Human Ccl20 reverse primer: GATTTGCGCACACAGACAACT | Loiseau et al., 2016 | N/A |
| Human EIF3L forward primer: TGACCCCTACGCTTATCCCAG | Wang and Seed, 2003 | Primerbank ID 339275830c1 |
| Human EIF3L reverse primer: GTTTGCTGTTCATACTGACGTTC | Wang and Seed, 2003 | Primerbank ID 339275830c1 |
| Recombinant DNA | ||
| pJL71-2W-mcherry | Scharschmidt et al., 2015 | pJL71-2W-mcherry |
| Software and Algorithms | ||
| TopHat | Trapnell et al., 2009 | https://ccb.jhu.edu/software/tophat/index.shtml |
| SAMtools | Li et al., 2009 | http://www.htslib.org |
| DESeq2 | Anders and Huber, 2010 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| HTSeq | Anders et al., 2015 | http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html |
| R Statistical Computing Software | The R Foundation https://www.r-project.org/ | https://www.r-project.org/ |
| GraphPad Prism | GraphPad Software | http://www.graphpad.com/scientific-software/prism/ |
| FlowJo | FlowJo | https://www.flowjo.com/solutions/flowjo |
| Other | ||
| Mice Doxycycline Diet (1g/kg) | Bio-Serv | Cat#S4096 |
STAR*METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Rosenblum (michael.rosenblum@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Experimental Animals
C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME) then bred and maintained in the UCSF specific pathogen-free facility. K5/Dkk1 mice on a mixed genetic background were gifted by Sarah E. Millar. Dkk1 transgene expression was initiated with doxycycline chow (1g/kg, Bio-Serv) starting on gestational day E13.5 in pregnant K5+ females. Foxp3DTR mice, Foxp3GFP, CD45.1, Rag2−/− and Ccr6−/− (Ccr6tm1Dgen) mice were purchased from Jackson (Haribhai et al., 2007). C57BL/6 mice under germ-free conditions were bred, maintained and harvested at Stanford University in the colony of Justin Sonnenburg. All mice used in experiments were socially houses under a 12 hr light/dark cycle. Animals were 6 days to 10 weeks old at the time of experiments. Littermates of the same sex were randomly assigned to experimental groups, with both male and female mice included in experiments on neonatal animals. When littermates could not be used (e.g., SPF versus GF comparison), sex-matched animals born on identical days were harvested in parallel. All animals were used in scientific experiments for the first time. All animal experiments were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals and the guidelines of the Laboratory Animal Resource Center and Institutional Animal Care and Use Committee of the University of California, San Francisco.
Human fetal skin
Human fetal skin samples were obtained from second trimester pregnancy terminations (approximate gestational age 22–23 weeks) from the Women’s Options Clinic at Zuckerberg San Francisco General Hospital. Sex was unknown. As specimens were de-identified their use did not constitute human subjects research and IRB approval was not required.
Bacterial strains
For colonization of human fetal skin specimens, bacterial strains were grown for 24–48 hr at 37°C on agar plates. Staphylococcus epidermidis Tü3298 was grown on trytic soy agar, whereas Propionibacterium acnes HL110PA4 and Corynebacterium accolens ATCC 49725 were grown on brain heart infusion agar with 1% Tween™ 80. P. acnes was grown in anaerobic conditions using GasPak™ EZ Anaerobe Pouches (BD). For visualization of S. epi colonization in mouse skin, Staphylococcus epidermidis Tü3298 containing pJL71-2W-mcherry (Scharschmidt et al., 2015) was grown at 250 rpm for 48 hr in tryptic soy broth with 5 µg/ml erythromycin. Cellular mass from 2.5ml of saturated culture was pelleted and re-suspended in 100 µL PBS for skin colonization of each animal.
METHOD DETAILS
Tissue processing and flow cytometry
Isolation of cells from spleen, thymus and lymph nodes for flow cytometry was performed by mashing tissue over sterile wire mesh. For isolation of cells from skin, the entire trunk skin was harvested and lightly defatted. It was then minced with scissors and re-suspended in a 50ml conical with 1–3ml of digestion media comprised of 2mg/ml collagenase XI, 0.5mg/ml hyaluronidase and 0.1mg/ml DNase in RPMI with 1% HEPES, 1% penicillin-streptomycin and 10% fetal calf serum. This mixture was incubated in a shaking incubated at 37°C at 250 rpm for 45 min. An additional 15ml of RPMI/HEPES/P-S/FCS media was then added and the 50ml conical was shaken vigorously by hand for 30 s. Another 15ml of media was added and then the entire suspension was filtered through a sterile 100 µm cell strainer followed by a 40 µm cell strainer into a new 50ml conical. The suspension was then pelleted and the cell pellet was re-suspended in PBS for cell counting and staining.
Following isolation from the tissue, cells were labeled stained in PBS for 30 min at 4°C with surface antibodies and a live dead marker (Ghost Dye™ Violet 510, Tonbo Biosciences). For intracellular staining, cells were fixed and permeabilized using reagents and protocol found in the Foxp3 staining buffer kit (eBioscience). Fluorophore-conjugated antibodies specific for mouse cell surface antigens and intracellular transcription factors were purchased from eBioscience, BD Biosciences or BioLegend as detailed in the Key Resources Table. Samples were run on a Fortessa (BD Biosciences) in the UCSF Flow Cytometry Core. Treg cells from skin and SDLN of neonates were sorted on a FACS Aria2 (BD Biosciences). For longitudinal experiments comparing mice across time or ages, voltages were standardized using SPHERO Rainbow calibration particles (BD Biosciences). Accucheck counting beads (Invitrogen) were used calculate absolute numbers of cells. Flow cytometry data was analyzed using FlowJo software (FlowJo, LLC).
Histopathology and immunohistochemistry
For histopathology, skin tissue was fixed in 10% formalin and paraffin-embedded, sectioned and stained with hematoxylin and eosin by the UCSF Mouse Pathology Core. Histopathology images were acquired on a Leica microscope using a DS-Ri1 camera and NIS-Elements software (Nikon). For immunofluorescent tissue staining in Figure 1A, dorsal skin from Foxp3GFP mice was first fixed in 2% PFA for 6–8 hr, washed with PBS and left in 30% sucrose overnight before embedding in OCT and freezing in a isopentane solution cooled over liquid nitrogen. 12 µm thick sections were prepared on SuperFrost slides (VWR), stained with rabbit anti-GFP (Invitrogen) at 1:200 followed by Goat anti-Rabbit Alexa 488 (Invitrogen), washed in PBS and mounted with DAPI-containing medium. For images in Figure S2C, neonatal mice were colonized with S. epidermidis Tü3298 with pJL71-2w-mCherry, which expresses a 2W-Mcherry fusion peptide (Scharschmidt et al., 2015), and back skin was harvested 1 day after colonization. Tissue was fixed as above and counterstained with DAPI. All images were acquired on a Zeiss Imager M2 fluorescent microscope with Apotome. Representative images in Figures 1A and S2C are one of 4 biological replicates with staining done on two independent occasions.
RNA in situ hybridization
Mouse back skin was formalin-fixed and paraffin-embedded as above. 5 mm thick sections were freshly cut on SuperFrost slides, then de-paraffinized and subjected to hybridization with a probe specific for murine Ccl20 mRNA (Advanced Cell Diagnostics, #434051) prior to visualization with RNAscope® 2.5 HD Brown Detection Kit (ACD) following the manufacturer’s protocol. Slides were simultaneously subjected to hybridization with positive control Ppib probe and negative DapB control probe. Images in Figure 4 are representative of three biological replicates with technical replicates performed on two independent occasions. Quantification of staining density was performed by counting the number of positive cells per 20x magnification field across multiple slides per group.
Neonatal Adoptive Transfer Model
Thymuses were harvested and lymphocytes isolated from adult CD45.2 Ccr6−/− mice and congenically-marked age-matched CD45.1 C57BL/6 controls. Single-positive CD4+ T cells were isolated by EasySep™ magnetic bead enrichment (StemCell Technologies). Ccr6−/− and wt CD4+ cells were mixed in a 1:1 ratio in PBS, and 3 million cells were transferred into D9 Rag2−/− recipients via intraperitoneal injection. Spleen, SDLN and skin were harvested 13 days post-transfer.
In vitro transwell T cell migration assay
Transwell migration of lymphocytes was quantified as previously reported (Arnon et al., 2011). In brief, thymocytes were isolated at room temperature from D9 Foxp3DTR mice (to facilitate identification of Treg cells by GFP), subjected to EasySep™ CD4+ magnetic bead enrichment (StemCell Technologies) and re-suspended in migration media (RPMI containing 10mM HEPES and 2% fatty acid-free BSA). 600 µL of migration media containing either no chemokine or listed murine chemokines (PeproTech) in stated concentrations were transferred to the lower transwell of a 24-well plate. 250,000 cells were then transferred in 100 µL of migration media to upper portion of each 0.5µm transwell (Costar #3421). After 3 hr incubation at 37°C, cells from the lower transwell were collected and quantified by flow cytometry. Analogous experiments were performed with total un-enriched lymphocytes from skin-draining LN and with neonatal skin following CD4+ positive enrichment using PE-labeled CD4+ antibody and EasySep™ Mouse PE Positive Selection Kit (StemCell Technologies).
qRT-PCR
Circular 8 mm diameter pieces of mouse or human skin were placed in sterile eppendorf tubes in 1ml of RNAlater (ThermoFisher), stored overnight at 4°C and then maintained at −80°C until further processing. Total tissue RNA was isolated using RNAeasy fibrous tissue mini kit (QIAGEN) according to the manufacturer’s protocol with the following modification. Tissue was first transferred to 500 µL of RLT buffer in MACS M-tubes and physically disrupted using a gentleMACS™ Octo Dissociator (Miltenyi). The handbook protocol was followed thereafter including DNase treatment. For qRT-PCR chemokine array, cDNA was made using RT2 FirstStrand kit (QIAGEN) and then run on RT2 Profiler PCR Array Mouse Cytokines & Chemokines (QIAGEN). Alternatively for confirmatory qRT-PCR, 1 µg of RNA was converted to cDNA using iScript cDNA synthesis kit (Bio-Rad) and mixed with appropriate primers and SYBR® Green Supermix. All reactions were run on a StepOnePlus instrument (Applied Biosystems). Primers and sources can be found in the Key Resource Table (Wang and Seed, 2003; Kusumoto et al., 2007; Nagao et al., 2012; Loiseau et al., 2016).
In vitro human skin explant system
Multiple 8 mm punch biopsies were obtained from each human fetal skin specimen and placed individually into 0.4 µm PET-membrane transwells (Millipore) in a 12 well plate containing 1ml media per well of media (10% FBS, 1% HEPES, 1% penicillin-streptomycin in RPMI). A sterile cotton swab pre-wetted with PBS was used to colonize the skin surface with Staphylococcus epidermidis Tü3298, Propionibacterium acnes HL110PA4, or Corynebacterium accolens ATCC 49725 by touching the swab to a bacterial colony on an agar plate and then gently swabbing the skin surface. Alternatively 10 µL of a 5mg/ml solution of lipopolysaccharide (Sigma, L4391), lipoteichoic acid (Sigma, L2515) or peptidoglycan (Sigma, 77140) was applied to the skin surface. Control samples were treated with PBS only. Samples were incubated overnight at 37 degrees and then skin was transferred to RNAlater prior to RNA extraction and qRT-PCR as above.
RNA sequencing of neonatal Treg cells
Treg cells were sorted from the skin and SDLN of 13-day-old Foxp3DTR mice, gating on the live, CD3+, CD4+, CD25+ and GFP+ population. Cells from two animals were pooled for each sample, pelleted and flash frozen. RNA isolation was performed by Expression Analysis Q2 Solutions using QIAGEN RNeasy Spin columns and was quantified via Nanodrop ND-8000 spectrophotometer. RNA quality was checked by Agilent Bioanalyzer Pico Chip. Three SDLN and two skin samples met RNA quality criteria and were carried forward for sequencing. cDNA was created from 370 pg of input RNA with the SMARTer Ultra Low input kit and sequenced to a 25M read depth with Illumina RNASeq. Reads were aligned to UCSC GRCm38/mm10 reference genome with TopHat software (v. 2.0.12) (Trapnell et al., 2009). SAM files were generated with SAMtools from alignment results (Li et al., 2009). Read counts were obtained with htseq-count (0.6.1p1) with the union option (Anders et al., 2015). Differential expression was determined using the R/Bioconducter package DESeq2 (Anders and Huber, 2010).
QUANTIFICATION AND STATISTICAL ANALYSIS
The number of mice per group is annotated in corresponding figure legends. Data followed a Gaussian distribution and variation was similar between groups for each condition analyzed. Significance was assessed using the unpaired Student’s t test in GraphPad Prism software (GraphPad). In all figures, the mean value is visually depicted. P values correlate with symbols as follows: ns = not significant, p > 0.05, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001. Mice were allocated randomly into experimental groups after matching for age and gender. Specific numbers of animals can be found in corresponding figure legends. Investigators remained unblinded to group assignments throughout. No animals were excluded from statistical analysis.
DATA AND SOFTWARE AVAILABILITY
The accession number for RNA sequencing data is NCBI GEO: GSE87130.
Supplementary Material
Highlights.
Hair follicle development facilitates Treg accumulation in neonatal skin
Commensal microbes augment Treg accumulation in neonatal skin and induce Ccl20 expression
The Ccl20-specific receptor, Ccr6, is preferentially expressed by neonatal skin Tregs
The Ccl20-Ccr6 pathway mediates neonatal Treg migration and skin-homing
In Brief.
Regulatory T cells (Tregs) accumulate in the skin during a defined window of postnatal development, but the underlying mechanisms are unclear. Scharschmidt et al. demonstrate that microbial colonization of developing hair follicles is required for Treg migration and homing into neonatal skin and is mediated by the Ccl20-Ccr6 pathway.
Acknowledgments
We thank Jason G. Cyster and Abul K. Abbas for guidance on experimental design and critical review of the manuscript, Steven Higgenbottom for germ-free mouse husbandry, Gyula Szabo for assistance with RNA in situ hybridization experiments, and Carlos C. Benetiz for assistance with SPF animal husbandry. Flow cytometry data were generated in the UCSF Parnassus Flow Cytometry Core, which is supported by the Diabetes Research Center (DRC) grant, NIH P30 DK063720. Paraffin embedding of tissue, sectioning, and H&E staining were performed by the UCSF Mouse Pathology Core, which is supported by NIH 5P30CA082103-15. T.C.S. is supported by K08AR068409, Burroughs Wellcome Fund CAMS-1015631, and the UCSF Department of Dermatology. This work was primarily funded by grants to M.D.R.: K08AR062064, DP2AR068130, R21AR066821, and Burroughs Wellcome Fund CAMS-1010934.
Footnotes
Supplemental Information includes four figures and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2017.03.001.
AUTHOR CONTRIBUTIONS
T.C.S. designed the studies, performed the experiments, and analyzed the data. T.C.S. and M.D.R. wrote the manuscript. K.S.V. contributed to experiments, mouse husbandry, data collection, and analysis. M.L.P., E.G.L., K.C., N.A., and H.-A.T. assisted with mouse experiments. M.M.L. and R.S.R. assisted in RNA sequencing data analysis. M.D.R. oversaw all study design and data analysis. J.L.S., Z.G.L., and S.E.M. assisted in study design and contributed reagents. All authors discussed results and commented on the manuscript.
References
- Adachi T, Kobayashi T, Sugihara E, Yamada T, Ikuta K, Pittaluga S, Saya H, Amagai M, Nagao K. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat. Med. 2015;21:1272–1279. doi: 10.1038/nm.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyev OA, Lundskog B, Ganceviciene R, Palmer RH, McDowell A, Patrick S, Zouboulis C, Golovleva I. Pattern of tissue invasion by Propionibacterium acnes in acne vulgaris. J. Dermatol. Sci. 2012;67:63–66. doi: 10.1016/j.jdermsci.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dev. Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- Arnon TI, Xu Y, Lo C, Pham T, An J, Coughlin S, Dorn GW, Cyster JG. GRK2-dependent S1PR1 desensitization is required for lymphocytes to overcome their attraction to blood. Science. 2011;333:1898–1903. doi: 10.1126/science.1208248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin J, Gotz F. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiology Letters. 1990;54:203–207. doi: 10.1016/0378-1097(90)90283-v. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat. Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouskra D, Brézillon C, Bérard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nat. Immunol. 2013a;14:1007–1013. doi: 10.1038/ni.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, Sefik E, Tan TG, Wagers AJ, Benoist C, Mathis D. A special population of regulatory T cells potentiates muscle repair. Cell. 2013b;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Xue F, Fleming C, Yang J, Ding C, Ma Y, Liu M, Zhang H-G, Zheng J, Xiong N, Yan J. Differential developmental requirement and peripheral regulation for dermal Vγ4 and Vγ6T17 cells in health and inflammation. Nat. Commun. 2014;5:3986. doi: 10.1038/ncomms4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JJ, O’Connell DJ, Wurbel MA. Cutting Edge: Chemokine receptor CCR4 is necessary for antigen-driven cutaneous accumulation of CD4 T cells under physiological conditions. J. Immunol. 2007;178:3358–3362. doi: 10.4049/jimmunol.178.6.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu EY, Hens J, Andl T, Kairo A, Yamaguchi TP, Brisken C, Glick A, Wysolmerski JJ, Millar SE. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development. 2004;131:4819–4829. doi: 10.1242/dev.01347. [DOI] [PubMed] [Google Scholar]
- Collins N, Jiang X, Zaid A, Macleod BL, Li J, Park CO, Haque A, Bedoui S, Heath WR, Mueller SN, et al. Skin CD4(+) memory T cells exhibit combined cluster-mediated retention and equilibration with the circulation. Nat. Commun. 2016;7:11514. doi: 10.1038/ncomms11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 2009;10:857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, Lee J, Goldfine AB, Benoist C, Shoelson S, Mathis D. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, Nicod LP, Lloyd CM, Marsland BJ. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 2014;20:642–647. doi: 10.1038/nm.3568. [DOI] [PubMed] [Google Scholar]
- Gratz IK, Truong H-A, Yang SH-Y, Maurano MM, Lee K, Abbas AK, Rosenblum MD. Cutting Edge: memory regulatory t cells require IL-7 and not IL-2 for their maintenance in peripheral tissues. J. Immunol. 2013;190:4483–4487. doi: 10.4049/jimmunol.1300212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Suzuki K, Cyster JG. Cutting edge: Identification of a motile IL-17-producing gammadelta T cell population in the dermis. J. Immunol. 2011;186:6091–6095. doi: 10.4049/jimmunol.1100427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S, Lehr C-M. Transfollicular delivery takes root: the future for vaccine design? Expert Rev. Vaccines. 2014;13:5–7. doi: 10.1586/14760584.2014.862500. [DOI] [PubMed] [Google Scholar]
- Haribhai D, Lin W, Relland LM, Truong N, Williams CB, Chatila TA. Regulatory T cells dynamically control the primary immune response to foreign antigen. J. Immunol. 2007;178:2961–2972. doi: 10.4049/jimmunol.178.5.2961. [DOI] [PubMed] [Google Scholar]
- Holbrook KA, Minami SI. Hair follicle embryogenesis in the human. Characterization of events in vivo and in vitro. Ann. N Y Acad. Sci. 1991;642:167–196. [PubMed] [Google Scholar]
- Jenkins MK, Chu HH, McLachlan JB, Moon JJ. On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu. Rev. Immunol. 2010;28:275–294. doi: 10.1146/annurev-immunol-030409-101253. [DOI] [PubMed] [Google Scholar]
- Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lübbert H, Bujard H. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc. Natl. Acad. Sci. USA. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Farber JM, Kelsall BL. CCR6 marks regulatory T cells as a colon-tropic, IL-10-producing phenotype. J. Immunol. 2010;185:3295–3304. doi: 10.4049/jimmunol.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinewietfeld M, Puentes F, Borsellino G, Battistini L, Rötzschke O, Falk K. CCR6 expression defines regulatory effector/memory-like cells within the CD25(+)CD4+ T-cell subset. Blood. 2005;105:2877–2886. doi: 10.1182/blood-2004-07-2505. [DOI] [PubMed] [Google Scholar]
- Kusumoto M, Xu B, Shi M, Matsuyama T, Aoyama K, Takeuchi T. Expression of chemokine receptor CCR4 and its ligands (CCL17 and CCL22) in murine contact hypersensitivity. J. Interferon Cytokine Res. 2007;27:901–910. doi: 10.1089/jir.2006.0064. [DOI] [PubMed] [Google Scholar]
- Lange-Asschenfeldt B, Marenbach D, Lang C, Patzelt A, Ulrich M, Maltusch A, Terhorst D, Stockfleth E, Sterry W, Lademann J. Distribution of bacteria in the epidermal layers and hair follicles of the human skin. Skin Pharmacol. Physiol. 2011;24:305–311. doi: 10.1159/000328728. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Chu EY, Watt B, Zhang Y, Gallant NM, Andl T, Yang SH, Lu M-M, Piccolo S, Schmidt-Ullrich R, et al. Wnt/β-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol. 2008;313:210–224. doi: 10.1016/j.ydbio.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau C, Requena M, Mavigner M, Cazabat M, Carrere N, Suc B, Barange K, Alric L, Marchou B, Massip P, et al. CCR6(−) regulatory T cells blunt the restoration of gut Th17 cells along the CCR6-CCL20 axis in treated HIV-1-infected individuals. Mucosal Immunol. 2016;9:1137–1150. doi: 10.1038/mi.2016.7. [DOI] [PubMed] [Google Scholar]
- Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mold JE, McCune JM. Immunological tolerance during fetal development: from mouse to man. Adv. Immunol. 2012;115:73–111. doi: 10.1016/B978-0-12-394299-9.00003-5. [DOI] [PubMed] [Google Scholar]
- Montes LF, Wilborn WH. Location of bacterial skin flora. Br. J. Dermatol. 1969;81(s1):1–23. doi: 10.1111/j.1365-2133.1969.tb12829.x. [DOI] [PubMed] [Google Scholar]
- Nagao K, Kobayashi T, Moro K, Ohyama M, Adachi T, Kitashima DY, Ueha S, Horiuchi K, Tanizaki H, Kabashima K, et al. Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat. Immunol. 2012;13:744–752. doi: 10.1038/ni.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Linehan JL, Han S-J, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M, Haase I, Nestle FO. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 2014;14:289–301. doi: 10.1038/nri3646. [DOI] [PubMed] [Google Scholar]
- Paus R, Müller-Röver S, Van Der Veen C, Maurer M, Eichmüller S, Ling G, Hofmann U, Foitzik K, Mecklenburg L, Handjiski B. A comprehensive guide for the recognition and classification of distinct stages of hair follicle morphogenesis. J. Invest. Dermatol. 1999;113:523–532. doi: 10.1046/j.1523-1747.1999.00740.x. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Rodriguez R, Pauli ML, Neuhaus IM, Yu SS, Arron ST, Harris HW, Yang SH-Y, Anthony BA, Sverdrup FM, Krow-Lucal E, et al. Memory regulatory T cells reside in human skin. J. Clin. Invest. 2014;124:1027–1036. doi: 10.1172/JCI72932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather BD, Treuting P, Perdue N, Miazgowicz M, Fontenot JD, Rudensky AY, Campbell DJ. Altering the distribution of Foxp3(+) regulatory T cells results in tissue-specific inflammatory disease. J. Exp. Med. 2007;204:1335–1347. doi: 10.1084/jem.20070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharschmidt TC, Vasquez KS, Truong H-A, Gearty SV, Pauli ML, Nosbaum A, Gratz IK, Otto M, Moon JJ, Liese J, et al. A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity. 2015;43:1011–1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T, Atarashi K, Honda K. Development and maintenance of intestinal regulatory T cells. Nat. Rev. Immunol. 2016;16:295–309. doi: 10.1038/nri.2016.36. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat. Immunol. 2009;10:864–871. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- van de Pavert SA, Mebius RE. New insights into the development of lymphoid tissues. Nat. Rev. Immunol. 2010;10:664–674. doi: 10.1038/nri2832. [DOI] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154–e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington EA, Kimpton WG, Holder JE, Cahill RN. Role of the thymus in the generation of skin-homing alpha beta and gamma delta virgin T cells. Eur. J. Immunol. 1995;25:723–727. doi: 10.1002/eji.1830250314. [DOI] [PubMed] [Google Scholar]
- Xia M, Hu S, Fu Y, Jin W, Yi Q, Matsui Y, Yang J, McDowell MA, Sarkar S, Kalia V, Xiong N. CCR10 regulates balanced maintenance and function of resident regulatory and effector T cells to promote immune homeostasis in the skin. J. Allergy Clin. Immunol. 2014;134:634–644.e10. doi: 10.1016/j.jaci.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Yang XO, Chung Y, Fukunaga A, Nurieva R, Pappu B, Martin-Orozco N, Kang HS, Ma L, Panopoulos AD, et al. CCR6 regulates the migration of inflammatory and regulatory T cells. J. Immunol. 2008;181:8391–8401. doi: 10.4049/jimmunol.181.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 2015;348:589–594. doi: 10.1126/science.aaa7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Xia J, Wang X, Xu F. Transcriptional regulation of CCL20 expression. Microbes Infect. 2014;16:864–870. doi: 10.1016/j.micinf.2014.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.