Abstract
Background
The diagnosis of asthma in children is challenging and relies on a combination of clinical factors and biomarkers including methacholine challenge, lung function, bronchodilator responsiveness, and presence of airway inflammation. No single test is diagnostic. We sought to identify a pattern of inflammatory biomarkers that was unique to asthma using a targeted metabolomics approach combined with data science methods.
Methods
We conducted a nested case-control study of 100 children living in a peri-urban community in Lima, Peru. We defined cases as children with current asthma, and controls as children with no prior history of asthma and normal lung function. We further categorized enrollment following a factorial design to enroll equal numbers of children as either overweight or not. We obtained a fasting venous blood sample to characterize a comprehensive panel of targeted markers using a metabolomics approach based on high performance liquid chromatography-mass spectrometry.
Results
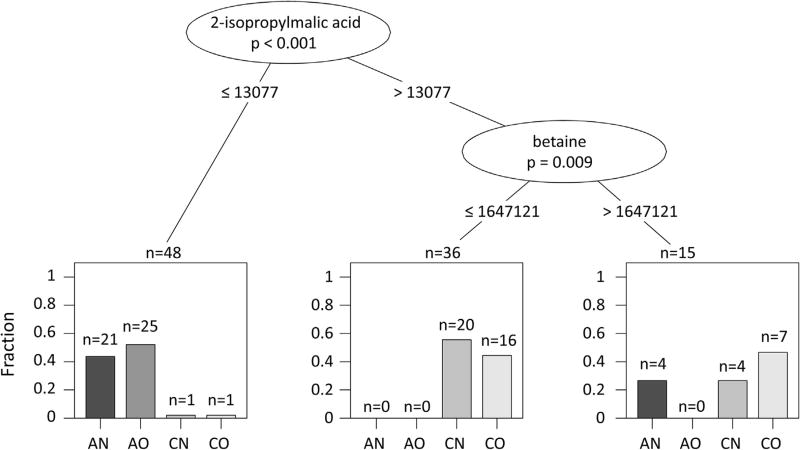
A statistical comparison of targeted metabolites between children with asthma (n = 50) and healthy controls (n = 49) revealed distinct patterns in relative concentrations of several metabolites: children with asthma had approximately 40–50% lower relative concentrations of ascorbic acid, 2-isopropylmalic acid, shikimate-3-phosphate, and 6-phospho-d-gluconate when compared to children without asthma, and 70% lower relative concentrations of reduced glutathione (all p < 0.001 after Bonferroni correction). Moreover, a combination of 2-isopropylmalic acid and betaine strongly discriminated between children with asthma (2-isopropylmalic acid ≤ 13 077 normalized counts/second) and controls (2-isopropylmalic acid > 13 077 normalized counts/second and betaine ≤ 16 47 121 normalized counts/second).
Conclusions
By using a metabolomics approach applied to serum, we were able to discriminate between children with and without asthma by revealing different metabolic patterns. These results suggest that serum metabolomics may represent a diagnostic tool for asthma and may be helpful for distinguishing asthma phenotypes.
1. Introduction
Asthma is the most common chronic disease in children. However, diagnosing asthma is challenging, as no single test is diagnostic. While methacholine challenge testing is considered the gold standard due to its high sensitivity, it has poor specificity, resulting in many false positives [1]. Bronchodilator reversibility is not uniform in children, and airway inflammation determined by fractional exhaled nitric oxide is unreliable and affected by the presence and degree of allergy and inflammation [2,3]. As such, most studies are based on a clinical definition based on symptom reporting and physician diagnosis. This complicates standardization of an asthma diagnosis and allows for subjectivity, especially among non-experts [4].
The search for airway or systemic biomarkers to help standardize the diagnosis of asthma has similarly met many challenges. Eosinophil count in induced sputum has been used to evaluate the presence of airway inflammation, but collection of an adequate sample and performing the test in a clinical setting can be difficult [5]. High serum immunoglobulin E is related to the presence and severity of asthma, but its use as a diagnostic tool is limited due to its lack of association with non-atopic asthma [6]. Serum periostin, a protein involved in the process of subepithelial fibrosis in asthma patients, was also proposed as a systemic biomarker of airways inflammation, but a recent external validation study showed an overall low diagnostic value [7]. The expression of galectins and their role in airway inflammation has also been studied, but recommendations on their use as biomarkers are still inconclusive [8].
Novel technologies have allowed for expansion of metabolomics, which includes a more comprehensive assessment of the metabolic pathways and the wide range of endogenous metabolites related to disease pathogenesis. Furthermore, the combination of metabolomics and data science may reveal insights into potential groups of biomarkers that might help identify individuals with asthma. We sought to utilize targeted metabolomics approaches to identify a pattern of serum biomarkers unique to children with asthma, leveraging a large case-control study performed in Lima, Peru. Additionally, given increased recognition of the association between obesity and asthma [9,10], we further categorized enrollment following a factorial design to enroll equal numbers of children as either overweight or not, based on both body mass index and body composition.
2. Materials and methods
2.1. Focused review of literature
We searched for articles in PubMed (www.pubmed.com) published in any language with the search terms “metabolomics OR metabolomic profiling”, “asthma”, “children” and “diagnosis”, with no date restrictions.
2.2. Study setting
Our study was conducted in Pampas de San Juan de Miraflores, a peri-urban community located 25 km south of the city center in Lima, Peru. For this analysis, serum samples were obtained from 100 individuals aged 9–19 years. We excluded children with a history of major surgery, or hospitalization for cardiac reasons in the past 3 months; a diagnosis of tuberculosis or currently receiving treatment for tuberculosis; a chronic respiratory condition other than asthma, or pregnancy at enrollment. The study was approved by the institutional review boards at the Johns Hopkins School of Medicine in Baltimore, USA and at A.B. PRISMA in Lima, Peru.
2.3. Study design
We conducted a cross-sectional analysis nested in a larger case-control study aimed at investigating the interaction between genetics and environmental exposures in children with asthma in Peru. We used a factorial design to enroll equal numbers of children categorized as asthma overweight (AO), asthma normal weight (AN), control overweight (CO), or control normal weight (CN). We defined overweight and obesity using BMI- for age-cutoffs outlined in Cole et al. [11] for the International Obesity Task Force and the age-and sex-specific curves for body fat published by McCarthy et al. for the Child Growth Foundation [12]. We used stratified random sampling of children in the parent study based on each of the above defined subgroups. We defined children with asthma as those with self- or parental-report of wheezing in the past 12 months, and who either had used asthma medications in the past 12 months or who have a physician diagnosis of asthma.
2.4. Clinical tests
Questionnaires were used to assess demographics, medication use and asthma status. Asthma control was assessed using the self-reported Asthma Control Test, a short questionnaire consisting of five questions about symptoms, medication use, control, and interruption of normal activities [13]. We also measured height and weight using a standardized approach, and calculated body mass index (BMI) in kg/m2. We measured body composition through bioimpedance using the TANITA TBF-300 body composition analyzer (TANITA Corporation, Inc., Arlington Heights, IL). We measured exhaled nitric oxide before spirometric testing using a portable analyzer (NIOXMINO, Aerocrine, Solna, Sweden) according to joint European Respiratory Society and the American Thoracic Society (ERS/ATS) recommendations [14]. We conducted spirometry in accordance with joint ERS/ATS guidelines using a flow-based portable spirometer (Jaeger/ERT, Hoechberg, Germany) to measure forced vital capacity (FVC) and forced expiratory volume at 1 s (FEV1) [15]. We obtained at least three acceptable and reproducible test and used a standard reference to obtain predicted values [16]. We tested all participants for reversibility with salbutamol (90 mcg/puff). Reversibility was defined as an increase of FEV1 by 12% or an increase of predicted FEV1 by 10%, consistent with National Asthma Education and Prevention Program (NAEPP-3) guidelines [17].
We collected a fasting blood sample for 100 participants at enrollment using standard phlebotomy techniques into nonheparinized 10-mL Vacutainer glass tubes (Becton-Dickinson, Franklin Lakes, NJ). Samples were allowed to coagulate and were centrifuged at 3500 RPM for 15–30 min within 1 h of sample collection. After separation, all samples were stored at −80 °C until ready to be analyzed. Atopy was defined as the presence of IgE antibodies to any of three allergen panels (animal epidermal and proteins mix, house dust mix, and a mold and yeast mix).
2.5. Targeted metabolomics analysis
We used a modified analytical platform published by Yuan et al. [18]. Briefly, serum samples were stored at −80 °C until analysis. Samples were thawed on ice and centrifuged at 16 000×g for 10 min at 4 °C prior to extraction. 200 µL of serum supernatant was transferred into a pre-labeled 1.5 mL microcentrifuge tube and 800 µL of precooled methanol (−80 °C) was added. Samples were vortexed gently for 1 min and placed into a −80 °C freezer. After incubation overnight, samples were centrifuged at 16 000µg for 10 min at 4 °C. 500 µL of the supernatant were transferred into another microcentrifuge tube, placed into a speed vacuum concentrator (Centrivap system, Labconco, Kansas City, MO) and vacuum was applied until samples were dry (approximately 2 h). Pellets were reconstituted in a solution containing 30% methanol and 70% water, vortexed briefly (1 min) and centrifuged at 16 000µg for 10 min at 4 °C. Supernatants were transferred into HPLC vials with 100 µL inserts and placed into a cooled (4 °C) HPLC autosampler (Ultimate 3000 WPS, Dionex Corp., Sunnyvale, CA). One sample was lost during this process. A detailed description of the chromatographic conditions, the mass spectrometer settings, and data processing parameters is included in the Appendix. Data were acquired using AB SCIEX Analyst software (version 1.6.2). Relative concentrations were normalized, i.e., the average response and standard deviation for each metabolite were calculated, and all values for that metabolite were then subtracted by the average and divided by the standard deviation. Initially, two study groups were assigned to the analytical runs in accordance with the study subjects asthma (n = 50) versus control (n = 49) group assignment and with the study subjects normal weight (n = 50) versus overweight (n = 49). This was followed by the assignment of the subgroups of AN (n = 25), AO (n = 25), CN (n = 25) and CO (n = 24).
2.6. Targeted lipid mediator analysis
We used a modified liquid chromatography (HPLC)-tandem mass spectrometry assay based on the analytical platform published by Masoodi et al. [19] to assess the pattern of 30 lipid mediators in serum, including several key prostaglandins, resolvins, hydroxylated eicosapentaenoic acids (HEPEs), hydroxyeicosatetraenoic acids (HETEs), and hydroxyoctadecadienoic acid (HODEs). We did not evaluate leukotriene concentrations because serum is not an ideal medium for analysis. A detailed description of the sample processing parameters is included in the Appendix.
2.7. Biostatistical methods
The primary objective of this analysis was to compare relative concentrations of an array of 308 selected metabolites by asthma status, and to identify interactions between these metabolites that may help uncover an asthma overweight phenotype. A secondary objective was to compare concentrations of 30 lipid mediators by asthma status. We performed t-tests to compare relative concentrations of metabolites between asthma and controls, and analysis of variance to compare relative concentrations of metabolites among the four study subgroups. We applied a Bonferroni correction to determine significance in the setting of multiple comparisons. For the targeted metabolomics analysis, a p < 0.00016 was considered statistically significant whereas for the targeted lipid mediator analysis, a p < 0.0017 was considered statistically significant. We then used statistical learning techniques [20] to determine combinations of metabolites that may help to visually and statistically discriminate between asthma and controls, and among the four study groups. First, we conducted a supervised principal component analysis - discriminant analysis (PCA-DA) using all metabolites among the four study groups. Second, we estimated the relative variable importance of single metabolites to discriminate between asthma and controls using a conditional random forest [21] that consisted of 3000 classification trees and five randomly sampled metabolites as candidates at each node. Third, we used a conditional classification tree analysis to identify combinations of metabolites that distinguished any of the four study subgroups [21]. We analyzed data using MarkerView software (version 1.2.1.1) and R (www.r-project.org).
3. Results
3.1. Findings of focused review
Of the twelve articles identified in our focused review, seven performed metabolomics analysis in exhaled breath condensate [22–28], two in urine [29,30], one in bronchoalveolar lavage fluid [31], one in plasma [32], and one in serum [33]. Five used nuclear magnetic resonance spectroscopy [22,24,28,29,33], three used liquid chromatography-mass spectrometry [27,30,32], three used gas chromatography-mass spectrometry [23,25,26], and one used both gas and liquid chromatography [31]. One study was performed in mice [31]. The aim of most studies was to find possible pathways associated with asthma pathogenesis, identify different phenotypes according to severity, and identify possible predictors of asthma control. Only two articles sought to discover biomarkers for asthma diagnosis. Most studies do not evaluate the effect of inhaled corticosteroid therapy on their results. We found no reports of the analysis of serum metabolomics on children, to identify possible biomarkers for asthma diagnosis.
3.2. Participant characteristics
We collected 100 samples, but 1 sample was lost during processing (50 children with asthma and 49 healthy controls). We did not observe differences in age (p = 0.20) or sex (p = 0.13) between study groups (Table 1). As expected, children with asthma had a higher prevalence of atopy (p = 0.007), exhaled nitric oxide (p < 0.001), serum IgE (p < 0.001), and a lower FEV1/FVC ratio (p < 0.001). Only 4 children (8%) reported using inhaled corticosteroids in the past 12 months. Mean ± SD for BMI was 28.4 ± 3.2 kg/m2 and 19.8 ± 2.2 kg/m2, and percentage of body fat was 38.1 ± 8.2% and 21.7 ± 5.8% in overweight and normal weight children, respectively.
Table 1.
Participant demographic and clinical characteristics.
| Asthma
|
Controls
|
|||
|---|---|---|---|---|
| Normal weight N = 25 |
Overweight N = 25 |
Normal weight N = 25 |
Overweight N = 25 |
|
| Age, mean (SD) | 13.9 (2.5) | 12.4 (1.8) | 14.3 (2.9) | 13.6 (2.9) |
| Male, n (%) | 16 (64.0) | 16 (64.0) | 13 (52.0) | 12 (48.0) |
| BMI in kg/m2, mean (SD) | 19.5 (2.2) | 26.9 (3.1) | 19.4 (2.5) | 27.3 (3.5) |
| Body fat %, mean (SD) | 21.5 (6.5) | 35.6 (6.4) | 21.9 (5.1) | 40.6 (9.3) |
| Height in cm, mean (SD) | 149.5 (12.2) | 147.2 (10.0) | 148.5 (11.2) | 148.7 (11.3) |
| Atopy, n (%) (n = 93) | 24 (96.0) | 18 (72.0) | 13 (52.0) | 14 (56.0) |
| Serum IgE in kU/L, median (IQR) | 1331 (763.2–1971) | 1129 (331–2354) | 176.6 (83.2–557.2) | 296.6 (114.5–1159) |
| Pre-bronchodilator FVC, L (SD) | 3.21 (1.0) | 3.08 (0.7) | 3.07 (0.8) | 3.20 (0.8) |
| Pre-bronchodilator FEV1, L (SD) | 2.71 (0.8) | 2.59 (0.6) | 2.79 (0.7) | 2.85 (0.7) |
| Pre-bronchodilator FEV1/FVC %, mean (SD) | 85.5 (8.5) | 84.6 (7.3) | 91.1 (3.9) | 89.5 (3.1) |
| FEV1 reversibility, n (%) (n = 93) | 3 (12.0) | 5 (20.0) | 0 (0.0) | 0 (0.0) |
| FeNO ppb, median (IQR) | 34 (24–70) | 21 (11–46) | 13 (9–18) | 13 (8–18) |
| ACT score, mean (SD) | 22.9 (3.7) | 22.0 (3.4) | – | – |
| Use of β2-agonist inhalers for crisis control in the last 12 months, n (%) (n = 48) | 3 (12.0) | 9 (36.0) | – | – |
| Use of ICS in the last 12 months, n (%) (n = 48) | 1 (4.0) | 3 (12.0) | – | – |
3.3. Differences in the metabolome by asthma status and body composition
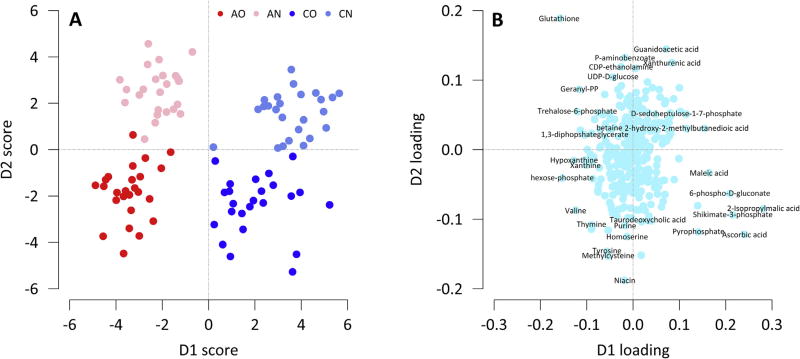
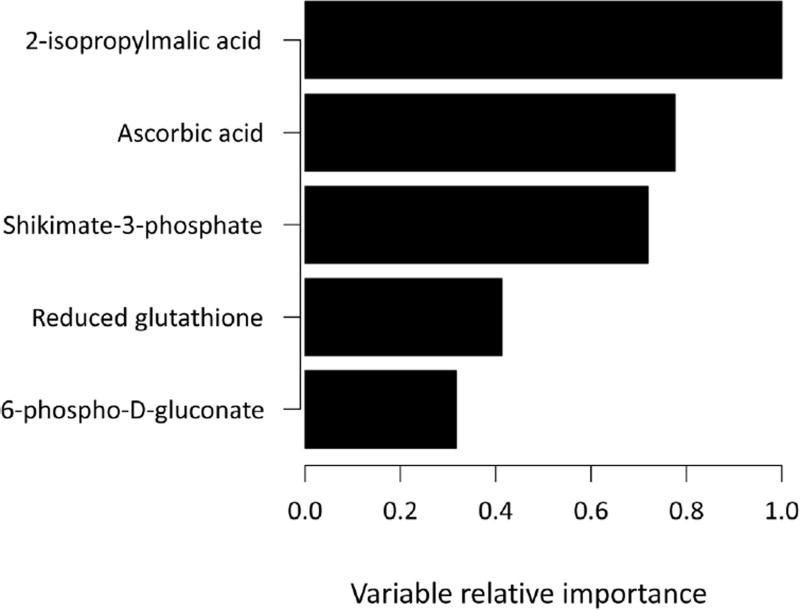
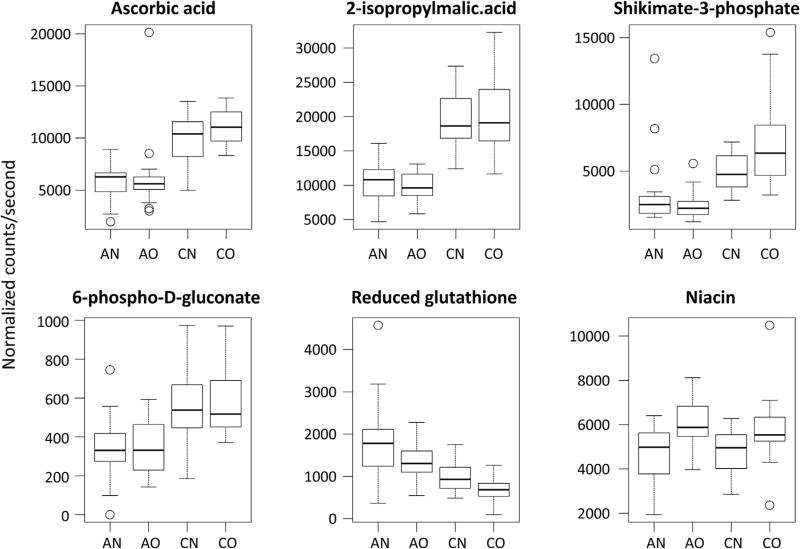
We first identified eighteen metabolites that were statistically different by asthma status after adjustment for multiple comparisons (Table 2). Their main functions can be found in the Appendix. We then plotted the results of the PCA-DA analysis to help uncover metabolites that could discriminate by asthma status and body composition (Fig. 1). There was a distinct separation between children with asthma and controls along the D1 score, and metabolites that contributed the most to this clustering were 2-isopropylmalic acid, ascorbic acid, shikimate-3-phosphate, 6-phospho-d-glutanate, and glutathione (Table 2). In fact, these five metabolites were also identified as the most important in classifying asthma status in random forest analysis (Fig. 2). Specifically, serum concentrations of 2-isopropylmalic acid, ascorbic acid, 6-phospho-d-gluconate and shikimate-3-phosphate were more than 40% lower in children with asthma compared to controls (Fig. 3). In contrast, the concentration of reduced glutathione was 70% higher among those with asthma compared to controls (Fig. 3). There was also good separation between overweight and normal weight children along the D2 score (Fig. 1), and the major contributor was niacin (Fig. 3).
Table 2.
Differences in metabolites between children with and without asthma.
| Proposed metabolite | Single reaction monitoring (SRM) |
Controls (normalized counts/second) |
Asthma (normalized counts/second) |
Fold difference | |||
|---|---|---|---|---|---|---|---|
| Q1/Q3 | p-value | Mean | SD | Mean | SD | ||
| 2-isopropylmalic acid | 175.0/115.0 | 3.28E-22 | 19 698 | 4731 | 10 055 | 2507 | 0.51 |
| ascorbic acid | 175.0/87.0 | 8.77E-16 | 10 465 | 2115 | 5985 | 2505 | 0.57 |
| 6-phospho-d-gluconate | 275.0/97.0 | 1.15E-10 | 568 | 175 | 333 | 147 | 0.59 |
| shikimate-3-phosphate | 253.1/97.0 | 1.18E-09 | 5844 | 2548 | 2756 | 1974 | 0.47 |
| Glutathione (red) | 308.1/162.0 | 2.43E-07 | 880 | 429 | 1559 | 748 | 1.77 |
| Pyrophosphate | 176.8/158.6 | 1.17E-06 | 2237 | 631 | 1575 | 637 | 0.70 |
| hexose-phosphate | 259.0/79.0 | 8.06E-06 | 11 224 | 3026 | 15 276 | 5251 | 1.36 |
| maleic acid | 115.0/71.0 | 8.08E-06 | 26 855 | 6292 | 21 191 | 5632 | 0.79 |
| 2-hydroxy-2-methylbutanedioic acid | 147.0/85.1 | 1.10E-05 | 28 085 | 5878 | 22 817 | 5409 | 0.81 |
| Hypoxanthine | 135.0/92.0 | 1.59E-05 | 49 000 | 23 081 | 70 564 | 24 139 | 1.44 |
| d-glucarate | 209.0/85.0 | 1.92E-05 | 2770 | 857 | 2129 | 517 | 0.77 |
| 2-oxobutanoate | 101.0/57.2 | 4.91E-05 | 11 197 | 3335 | 8631 | 2622 | 0.77 |
| Methylnicotinamide | 137.0/94.0 | 6.69E-05 | 9612 | 3277 | 12 546 | 3718 | 1.31 |
| trehalose-6-phosphate | 421.0/79.0 | 0.00016 | 218 | 107 | 294 | 84 | 1.35 |
Fig. 1.
PCA-DA analysis of metabolomics data. The data were processed as described in the methods section. The scores plot (A) shows a separated clustering of each of the study subgroups. Asthma and controls were separated along the D1-Scores axes. The loadings plot (B) shows the compounds responsible for the separation of asthma subgroups from control subgroups along the D1-loadings axes. Several of these compounds were previously identified during the statistical t-test group comparison and the random forest analysis (e.g. 2-isopropylmalic acid, ascorbic acid etc.).
Fig. 2.
Random forest analysis of metabolomics data. The bar plot shows the 5 most important metabolites responsible for separating the subjects by asthma status.
Fig. 3.
Boxplot of significantly changed metabolites according to subgroups (AN = Asthma Normal Weight; AO = Asthma Overweight; CN = Control Normal Weight; CO = Control Overweight).
In analysis of variance, niacin was the only significantly changed metabolite that was statistically different among study groups (Fig. 3; p = 0.00011); however, we could not identify a single metabolite that could clearly distinguish between overweight children with asthma and children in the other study groups (Appendix). Most of the observed differences between children with asthma and controls were still present in subgroup analyses, although they were less significant because of the lower group sizes (Appendix). In conditional tree analysis, we found that a combination of 2-isopropylmalic acid and betaine strongly discriminated between asthma and controls (Fig. 4). Moreover, the combination may be useful to discriminate between normal weight children with asthma and overweight children with asthma.
Fig. 4.
Classification tree analysis of metabolomics data. (AN = Asthma Normal Weight; AO = Asthma Overweight; CN = Control Normal Weight; CO = Control Overweight) The analysis shows the combination of the 2 molecules (isopropylmalic acid and betaine) responsible for the differentiation of asthma from controls, and asthma overweight from the other subgroups.
3.4. Differences in lipid mediators by asthma status and body composition
We did not identify important differences in lipid mediators by either asthma status or among study groups after Bonferroni correction. Nevertheless, several trends were apparent. For example, 12-HETE (mean 1702 and 864 pg/mL, respectively; p = 0.03) and 12-HEPE (87 and 22 pg/mL, respectively; p = 0.05) were higher in children with asthma than in controls, 12-HETE was higher in overweight children with asthma than in overweight controls (mean 2038 and 598 pg/mL; p = 0.007), and 20-HETE was higher in overweight children with asthma than in normal weight children with asthma (mean 296 pg/mL and 200 pg/mL; p = 0.009).
4. Discussion
To our knowledge, this is the first study of metabolomic profiling performed in blood samples of children with asthma, offering potentially non-invasive biomarkers of disease. Several metabolomic studies of lung disease have been performed using different fluids, such as exhaled breath condensate (EBC) [22,24,28], but some studies suggest that EBC may not give accurate results due to risk of contamination and the influence of endogenous and exogenous factors that complicate obtaining an adequate sample [34]. We applied targeted mass spectrometry-based metabolic profiling of sera in children with and without asthma, and in combination with data science methods we found important differences in concentrations of several metabolites that clearly discriminated asthma status.
Specifically, we found important differences in metabolites related to oxidative stress. Studies suggest that an imbalance in prooxidative and anti-oxidative processes in the airway could result in increased inflammation and tissue damage, and a lowered cellular reducing capacity has been linked with an increased risk of asthma and worse disease severity [35,36]. We found that hypoxantine, a metabolite involved in the superoxide anion generating system, was higher in children with asthma than in controls. We also found that reduced glutathione concentrations were higher in children with asthma than in controls. Reduced glutathione is an intracellular redox regulator capable of scavenging free radicals either directly, non-enzymatically or enzymatically by glutathione peroxidase, and has been associated with regulation of airway reactivity and inflammation [37,38]. Previous studies have found increased levels in the lungs and blood of subjects with asthma as part of an adaptive response to oxidative stress [39]. We also found higher levels of hexose-phosphate and lower levels of 6-phospho-d-gluconate, both intermediates in the pentose phosphate pathway and in the production of NADPH, which play a major role in the reducing capacity in all cells of the body and are involved in continuously replenishing the reduced glutathione pools [40]. Ascorbic acid concentrations were also lower in children with asthma when compared to controls. As one of the main components of the extracellular fluid lining of the alveolus, ascorbic acid plays a protective role against oxidative stress by directly scavenging oxide and hydroxide [40,41]. Low serum levels have been previously associated with pulmonary dysfunction and increased odds of developing asthma in children and adults [42].
Other metabolites found in lower concentrations in children with asthma were 2-isopropylmalic acid, shikimate-3-phosphate, and 2-oxobutanoate, all of which are intermediates in amino acid synthesis. These molecules are usually found in fungi and bacteria but not in humans, and therefore the lower concentrations in children with asthma identified in our analysis could suggest a link between the human microbiome [29,30]. Other molecules that were significantly different and are usually found in bacteria and fungi were 2-hydroxy-2-methylbutadenoic acid and trehalose-6-phosphate. Although results from studies on the role of microbiome on asthma pathogenesis have been inconclusive, some suggest there could be an inverse relationship between reduced exposure and diversity of intestinal microbiota in infancy and allergic sensitization [43,44].
While no single metabolite could help discriminate by asthma status and body composition, our analyses suggest that a combination of 2-isopropylmalic acid and betaine may be helpful to discriminate not only by asthma status but may also help separate overweight asthma from other subgroups. Betaine is a methyl donor involved in a variety of biological processes [45]. Furthermore, increased levels of betaine have been linked to a worse prognosis in cardiovascular disease and enhanced atherosclerosis specially in the presence of the intestinal microbiota-dependent products [46]. Although we could not find other studies that have investigated the role of betaine on asthma pathogenesis, these preliminary findings could suggest an interaction between betaine and microbiota products in other pathophysiologic processes.
Several studies have demonstrated a positive association between obesity and asthma in children [9,10]. Obesity has a negative effect on lung mechanics, absolute airflows, and lung volumes, even in the absence of airway inflammation [47,48]. Moreover, obesity may be linked to a unique asthma phenotype, including a more severe clinical presentation and increased steroid resistance [49]. We found that niacin concentrations were higher in the overweight children with asthma compared to those with asthma and normal weight. Niacin is a potent stimulator of appetite and high niacin serum levels have been linked with increased insulin resistance [50,51]. Furthermore, current studies have found a significant association between niacin consumption and the increased prevalence of obesity in the United States [51]. Finally, there was a trend for a higher 20-HETE levels in overweight children with asthma compared to normal weight children with asthma, a potent vasoconstrictor whose upregulation is associated with oxidative stress [52].
Our study has some potential shortcomings, including a relatively small sample size and the exploratory nature of the metabolomics approach for inflammatory mediators. Nonetheless, despite the smaller sample size, we found important differences in metabolites between groups even after correction for multiple testing. Second, given that this was an exploratory study, further validation to investigate the potential use of differentiating biomarkers for asthma diagnosis in a larger population is warranted. Third, although the use of mass spectrometry for metabolite analysis allows for identification of a wide range of compounds, some metabolites might not be captured because of its destructive nature, given the requirement for separation, ionization, fragmentation and acceleration of the components through a magnetic field [53]. One of the strengths of this study was that less than 10% of the children were using inhaled corticosteroids, decreasing the risk of a difference in metabolic pathways resulting from medication use.
In summary, we provide evidence that a targeted metabolomics approach applied to serum could be used to discriminate among children with and without asthma by revealing different metabolic profiles. This could lead to an improvement in asthma diagnosis and may be further studied to discover new pathogenic pathways and possible therapeutic targets.
Supplementary Material
Acknowledgments
Funding
United States National Institutes of Health (R01ES018845).
Appendix A
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.rmed.2016.10.011.
Footnotes
Conflict of interest
The authors have no conflicts of interest to disclose.
References
- 1.Hewitt DJ. Interpretation of the “positive” methacholine challenge. Am. J. Ind. Med. 2008;51:769–781. doi: 10.1002/ajim.20631. [DOI] [PubMed] [Google Scholar]
- 2.Elmasri M, Romero KM, Gilman RH, Hansel NN, Robinson CL, et al. Pura study investigators. Longitudinal assessment of high versus low levels of fractional exhaled nitric oxide among children with asthma and atopy. Lung. 2014;192:305–312. doi: 10.1007/s00408-013-9551-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur. Respir. J. 2014;43:343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- 4.Yeatts K, Davis KJ, Sotir M, Herget C, Shy C. Who gets diagnosed with asthma? Frequent wheeze among adolescents with and without a diagnosis of asthma. Pediatrics. 2003;111:1046–1054. doi: 10.1542/peds.111.5.1046. [DOI] [PubMed] [Google Scholar]
- 5.Desai M, Oppenheimer J. Year in review: asthma. Ann. Allergy Asthma Immunol. 2015;114:170–172. doi: 10.1016/j.anai.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Checkley W, Robinson CL, Baumann LM, Romero K, Combe JM, Gilman RH, et al. Effect of urbanisation on the relationship between total serum IgE and asthma. Eur. Respir. J. 2013;41:1074–1081. doi: 10.1183/09031936.00025512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagener AH, de Nijs SB, Lutter R, Sousa AR, Weersink EJM, Bel EH, et al. External validation of blood eosinophils, FE NO and serum periostin as surrogates for sputum eosinophils in asthma. Thorax. 2015;70:115–120. doi: 10.1136/thoraxjnl-2014-205634. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez-Cuellar S, de la Fuente H, Cruz-Adalia A, Lamana A, Cibrian D, Giron RM, et al. Reduced expression of galectin-1 and galectin-9 by leucocytes in asthma patients: Gal-1 and Gal-9 in asthma. Clin. Exp. Immunol. 2012;170:365–374. doi: 10.1111/j.1365-2249.2012.04665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beuther DA, Sutherland ER. Overweight, obesity, and incident asthma: a meta-analysis of prospective epidemiologic studies. Am. J. Respir. Crit. Care Med. 2007;175:661–666. doi: 10.1164/rccm.200611-1717OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu P-C, Kieckhefer GM, Gau B-S. A systematic review of the association between obesity and asthma in children. J. Adv. Nurs. 2013;69:1446–1465. doi: 10.1111/jan.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole TJ, Bellizzi MC, Flegal KM, Dietz WH. Establishing a standard definition for child overweight and obesity worldwide: international survey. BMJ. 2000;320:1240. doi: 10.1136/bmj.320.7244.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy HD, Cole TJ, Fry T, Jebb SA, Prentice AM. Body fat reference curves for children. Int. J. Obes. 2006;30:598–602. doi: 10.1038/sj.ijo.0803232. [DOI] [PubMed] [Google Scholar]
- 13.Schatz M, Kosinski M, Yarlas AS, Hanlon J, Watson ME, Jhingran P. The minimally important difference of the Asthma Control Test. J. Allergy Clin. Immunol. 2009;124:719–723 e1. doi: 10.1016/j.jaci.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 14.Reddel HK, Taylor DR, Bateman ED, Boulet L-P, Boushey HA, Busse WW, et al. An official American Thoracic Society/European Respiratory Society statement: asthma control and exacerbations: standardizing endpoints for clinical asthma trials and clinical practice. Am. J. Respir. Crit. Care Med. 2009;180:59–99. doi: 10.1164/rccm.200801-060ST. [DOI] [PubMed] [Google Scholar]
- 15.Miller MR. Standardisation of spirometry. Eur. Respir. J. 2005;26:319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 16.Quanjer PH, Stanojevic S, Cole TJ, Baur X, Hall GL, Culver BH, et al. Multiethnic reference values for spirometry for the 3–95-yr age range: the global lung function 2012 equations. Eur. Respir. J. 2012;40:1324–1343. doi: 10.1183/09031936.00080312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Expert Panel Report 3 (EPR-3), Guidelines for the diagnosis and management of asthma–summary report 2007. J. Allergy Clin. Immunol. 2007;120:S94–S138. doi: 10.1016/j.jaci.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 18.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion–switching, targeted mass spectrometry–based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masoodi M, Mir AA, Petasis NA, Serhan CN, Nicolaou A. Simultaneous lipidomic analysis of three families of bioactive lipid mediators leukotrienes, resolvins, protectins and related hydroxy-fatty acids by liquid chromatography/electrospray ionisation tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2008;22:75–83. doi: 10.1002/rcm.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman J, Hastie T, Tibshirani R. Springer series in Statistics. Vol. 1. New York: Springer; 2001. The Elements of Statistical Learning [Internet] [Google Scholar]
- 21.Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: a conditional inference framework. J. Comput. Graph Stat. 2006;15:651–674. [Google Scholar]
- 22.Carraro S, Giordano G, Reniero F, Carpi D, Stocchero M, Sterk PJ, et al. Asthma severity in childhood and metabolomic profiling of breath condensate. Allergy. 2013;68:110–117. doi: 10.1111/all.12063. [DOI] [PubMed] [Google Scholar]
- 23.Gahleitner F, Guallar-Hoyas C, Beardsmore CS, Pandya HC, Thomas CP. Metabolomics pilot study to identify volatile organic compound markers of childhood asthma in exhaled breath. Bioanalysis. 2013;5:2239–2247. doi: 10.4155/bio.13.184. [DOI] [PubMed] [Google Scholar]
- 24.Ibrahim B, Marsden P, Smith JA, Custovic A, Nilsson M, Fowler SJ. Breath metabolomic profiling by nuclear magnetic resonance spectroscopy in asthma. Allergy. 2013;68:1050–1056. doi: 10.1111/all.12211. [DOI] [PubMed] [Google Scholar]
- 25.Dallinga J, Robroeks C, Van Berkel J, Moonen E, Godschalk R, Jobsis Q, et al. Volatile organic compounds in exhaled breath as a diagnostic tool for asthma in children. Clin. Exp. Allergy. 2009;40:68–76. doi: 10.1111/j.1365-2222.2009.03343.x. [DOI] [PubMed] [Google Scholar]
- 26.Cap P, Chladek J, Pehal F, Maly M, Petru V, Barnes P, et al. Gas chromatography/mass spectrometry analysis of exhaled leukotrienes in asthmatic patients. Thorax. 2004;59:465–470. doi: 10.1136/thx.2003.011866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montuschi P, Santini G, Valente S, Mondino C, Macagno F, Cattani P, et al. Liquid chromatography-mass spectrometry measurement of leukotrienes in asthma and other respiratory diseases. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014;964:12–25. doi: 10.1016/j.jchromb.2014.02.059. [DOI] [PubMed] [Google Scholar]
- 28.Santini G, Mores N, Penas A, Capuano R, Mondino C, Trové A, et al. Electronic nose and exhaled breath NMR-based metabolomics applications in airways disease. Curr. Top. Med. Chem. 2016;16:1610–1630. doi: 10.2174/1568026616666151223113540. [DOI] [PubMed] [Google Scholar]
- 29.Saude EJ, Skappak CD, Regush S, Cook K, Ben-Zvi A, Becker A, et al. Metabolomic profiling of asthma: diagnostic utility of urine nuclear magnetic resonance spectroscopy. J. Allergy Clin. Immunol. 2011;127:757–764 e6. doi: 10.1016/j.jaci.2010.12.1077. [DOI] [PubMed] [Google Scholar]
- 30.Mattarucchi E, Baraldi E, Guillou C. Metabolomics applied to urine samples in childhood asthma; differentiation between asthma phenotypes and identification of relevant metabolites: metabolomics in childhood asthma. Biomed. Chromatogr. 2012;26:89–94. doi: 10.1002/bmc.1631. [DOI] [PubMed] [Google Scholar]
- 31.Ho WE, Xu Y-J, Xu F, Cheng C, Peh HY, Tannenbaum SR, et al. Metabolomics reveals altered metabolic pathways in experimental asthma. Am. J. Respir. Cell Mol. Biol. 2013;48:204–211. doi: 10.1165/rcmb.2012-0246OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGeachie MJ, Dahlin A, Qiu W, Croteau-Chonka DC, Savage J, Wu AC, et al. The metabolomics of asthma control: a promising link between genetics and disease: integrative metabolomics of asthma control. Immun. Inflamm. Dis. 2015;3:224–238. doi: 10.1002/iid3.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung J, Kim S-H, Lee H-S, Choi GS, Jung Y-S, Ryu DH, et al. Serum metabolomics reveals pathways and biomarkers associated with asthma pathogenesis. Clin. Exp. Allergy. 2013;43:425–433. doi: 10.1111/cea.12089. [DOI] [PubMed] [Google Scholar]
- 34.Izquierdo-García JL, Peces-Barba G, Heili S, Diaz R, Want E, Ruiz-Cabello J. Is NMR-based metabolomic analysis of exhaled breath condensate accurate? Eur. Respir. J. 2011;37:468–470. doi: 10.1183/09031936.00094010. [DOI] [PubMed] [Google Scholar]
- 35.Owen S, Pearson D, Suarez-Mendez V, O'Driscoll R, Woodcock A. Evidence of free-radical activity in asthma. N. Engl. J. Med. 1991;325:586–587. doi: 10.1056/NEJM199108223250816. [DOI] [PubMed] [Google Scholar]
- 36.Comhair SA, Bhathena PR, Dweik RA, Kavuru M, Erzurum SC. Rapid loss of superoxide dismutase activity during antigen-induced asthmatic response. Lancet. 2000;355:624. doi: 10.1016/S0140-6736(99)04736-4. [DOI] [PubMed] [Google Scholar]
- 37.Nadeem A, Siddiqui N, Alharbi NO, Alharbi MM, Imam F, Sayed-Ahmed MM. Glutathione modulation during sensitization as well as challenge phase regulates airway reactivity and inflammation in mouse model of allergic asthma. Biochimie. 2014;103:61–70. doi: 10.1016/j.biochi.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Comhair SA, Bhathena PR, Farver C, Thunissen FB, Erzurum SC. Extracellular glutathione peroxidase induction in asthmatic lungs: evidence for redox regulation of expression in human airway epithelial cells. FASEB J. 200(15):70–78. doi: 10.1096/fj.00-0085com. [DOI] [PubMed] [Google Scholar]
- 39.Reynaert NL. Glutathione biochemistry in asthma. Biochim. Biophys. Acta. 2011;1810:1045–1051. doi: 10.1016/j.bbagen.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Kuehne A, Emmert H, Soehle J, Winnefeld M, Fischer F, Wenck H, et al. Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol. Cell. 2015;59(3):359–371. doi: 10.1016/j.molcel.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 41.Harris JR. Subcellular Biochemistry: Ascorbic Acid: Biochemistry and Biomedical Cell Biology. 25. London: Plenum Press; 1996. p. 464. [Google Scholar]
- 42.Allen S, Britton JR, Leonardi-Bee JA. Association between antioxidant vitamins and asthma outcome measures: systematic review and meta-analysis. Thorax. 2009;64:610–619. doi: 10.1136/thx.2008.101469. [DOI] [PubMed] [Google Scholar]
- 43.Huang YJ. Asthma microbiome studies and the potential for new therapeutic strategies. Curr. Allergy Asthma Rep. 2013;13:453–461. doi: 10.1007/s11882-013-0355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bisgaard H, Li N, Bonnelykke K, Chawes BLK, Skov T, Paludan-Müller G, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 2011;128:646–652. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 45.Sharma S, Litonjua A. Asthma, allergy, and responses to methyl donor supplements and nutrients. J. Allergy Clin. Immunol. 2014;133:1246–1254. doi: 10.1016/j.jaci.2013.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Tang WHW, Buffa JA, Fu X, Britt EB, Koeth RA, et al. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur. Heart J. 2014;35:904–910. doi: 10.1093/eurheartj/ehu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beuther DA, Sutherland ER. Obesity and pulmonary function testing. J. Allergy Clin. Immunol. 2005;115:1100–1101. doi: 10.1016/j.jaci.2004.12.1141. [DOI] [PubMed] [Google Scholar]
- 48.Biring MS, Lewis MI, Liu JT, Mohsenifar Z. Pulmonary physiologic changes of morbid obesity. Am. J. Med. Sci. 1999;318:293–297. doi: 10.1097/00000441-199911000-00002. [DOI] [PubMed] [Google Scholar]
- 49.Farzan S. The asthma phenotype in the obese: distinct or otherwise? J. Allergy. 2013;2013:1–8. doi: 10.1155/2013/602908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koh Y, Bidstrup H, Nichols DL. Niacin increased glucose, insulin, and C-peptide levels in sedentary nondiabetic postmenopausal women. Int. J. Womens Health. 2014;6:913–920. doi: 10.2147/IJWH.S69908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li D. Chronic niacin overload may be involved in the increased prevalence of obesity in US children. World J. Gastroenterol. 2010;16:2378. doi: 10.3748/wjg.v16.i19.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams JM, Murphy S, Burke M, Roman RJ. 20-hydroxyeicosatetraeonic acid: a new target for the treatment of hypertension. J. Cardiovasc Pharmacol. 2010;56:336–344. doi: 10.1097/FJC.0b013e3181f04b1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fearnley LG, Inouye M. Metabolomics in epidemiology: from metabolite concentrations to integrative reaction networks. Int. J. Epidemiol. 2016 doi: 10.1093/ije/dyw046. http://dx.doi.org/10.1093/ije/dyw046. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.