Abstract
Although traditionally viewed as the “powerhouse” of the cell, an accruing body of evidence in the rapidly growing field of mitochondrial biology supports additional roles of mitochondria as key participants in a multitude of cellular functions. While it has been well established that mitochondria in different tissues have distinctive ultrastructural features consistent with differential bioenergetic demands, recent and emerging technical advances in flow cytometry, imaging, and “-omics”-based bioinformatics have only just begun to explore the complex and divergent properties of mitochondria within tissues and cell types. Moreover, contemporary studies evaluating the role of mitochondria in pluripotent stem cells, cellular reprogramming, and differentiation point to a potential importance of mitochondrial subpopulations and heterogeneity in the field of stem cell biology. This review assesses the current literature regarding mitochondrial subpopulations within cell and tissue types and evaluates the current understanding of how mitochondrial diversity and heterogeneity might impact cell fate specification in pluripotent stem cells.
1. Mitochondrial Structure and Diversity
Mitochondria are ubiquitous across eukaryotic organisms and are critical for cellular bioenergetics. The classical mitochondrial ultrastructural features, consisting of an outer membrane and an inner membrane containing invaginations that comprise the matrix-rich cristae, are specifically modified in a tissue-specific manner in order to meet cellular-explicit energy demands. The differences in mitochondrial morphology between cell and tissue types as well as in localization and distribution within the cell have been well documented [1, 2], and more recently, it has been demonstrated that even within a single cell, mitochondrial features fluctuate rapidly in response to alterations in a metabolic state [3, 4]. Although the exact molecular mechanisms underlying ultrastructural remodeling of mitochondria are a topic of investigation with much remaining to be discovered, the diversity observed in a mitochondrial phenotype includes a range in size (from 0.1 micron to 1.0 microns in diameter), shape (from spherical to elongated and tubular), and cristae density (from essentially devoid of cristae to dense cristae). As a mitochondrial form relates to function, early studies postulated that mitochondrial cristae density serves to increase the surface area, thereby enhancing oxidative phosphorylation [5]. With subsequent efforts identifying the inner membrane as containing the components of the electron transport chain and the capacity for ATP generation positively correlated with the cristae surface area (reviewed in [6]), the relationship between energy production and mitochondrial morphology, with specific modifications in the inner membrane and cristae structure, has become increasingly clear. For example, ATP synthase dimerization has been demonstrated to govern biogenesis of the inner membrane and cristae formation, and oligomerization of F1FO-ATP synthase has been proposed to mediate the formation of cristae by modulating inner membrane curvature [7–10]. Additionally, a number of candidate proteins involved in modulation of cristae formation and inner membrane organization have been identified across model organisms and specifically correlated with pathophysiological disease states in humans (reviewed in [5]). Mitochondrial morphological dynamics are also dependent upon fission and fusion of inner and outer membranes, mediated by large GTPases [11–13], including the cytosolic DRP1 [14], the mitofusin (MFN) proteins 1 and 2 (MFN1 and MFN2) [15], and optic atrophy 1 (OPA1) [16–19].
Although the specifics regarding how these nuclear-encoded proteins have been shown to participate in mitochondrial remodeling have been studied for almost two decades and extensively reviewed elsewhere [4, 20], far less is known regarding the cell signaling mechanisms that govern a mitochondrial phenotype and how this occurs on a tissue-specific basis during the process of differentiation. In undifferentiated embryonic stem cells (ESCs), mitochondria are characterized as having a globular shape, with few defined cristae and limited oxidative capacity [21]. This ovoid mitochondrial morphology and cristae arrangement are rather striking [21, 22] and also observed in oocytes (Figure 1), as well as in the inner cell mass of blastocysts—the originating source for ESCs [23]. The low matrix and cristae density are associated with slowly respiring state IV mitochondria, as compared to fast respiring state III mitochondria, which have a higher matrix and cristae density and a morphologically “condensed” ultrastructure [24–26]. Notably, the low cristae density and ovoid structure, along with distinctive perinuclear localization, are under consideration as features for cellular “stemness,” having been detected in adult somatic stem cells and reprogrammed stem cells (i.e., iPSCs [21, 27, 28]), in addition to numerous ESC lines. Intriguingly, observations of the modification of mitochondrial patterning in a spontaneously differentiating rhesus monkey adult mesenchymal cell (MSC) line (ATSC line) have demonstrated heterogeneity within stem cell populations and have led to the postulation that mitochondrial localization (deviation from a perinuclear region) may be a mechanism by which to monitor differentiation status, in vitro [28], although this remains to be firmly established, and applicability may be cell type- and status-dependent, as perinuclear localization can be induced in disease states and under conditions of hypoxia and apoptosis in nonstem cell lines [29–31]. A detailed analysis of several nonstem cell types, including HeLa, HUVECs, COS-7, cortical astrocytes, and primary hepatocytes, also demonstrated heterogeneity with respect to both distribution and functional properties [32]. Although in each cell type examined, mitochondria were distributed throughout the cell body, both cortical astrocytes and HUVECs demonstrated distinct perinuclear clustering, which was less pronounced in HeLa and COS-7 and absent in primary hepatocytes [32]. Functionally, it was noted that each cell type exhibited heterogeneity in mitochondrial membrane potential (potential (Δψm)), as each cell type examined contained distinct populations of mitochondria having both high or low Δψm, and no single mitochondrion contained regions of high and low Δψm. Quantitative analysis of Δψm in HeLa cells demonstrated that mitochondria localized to the perinuclear region had a greater percentage of mitochondria with low Δψm and were more closely associated with endoplasmic reticulum (ER) than their peripherally located counterparts [32].
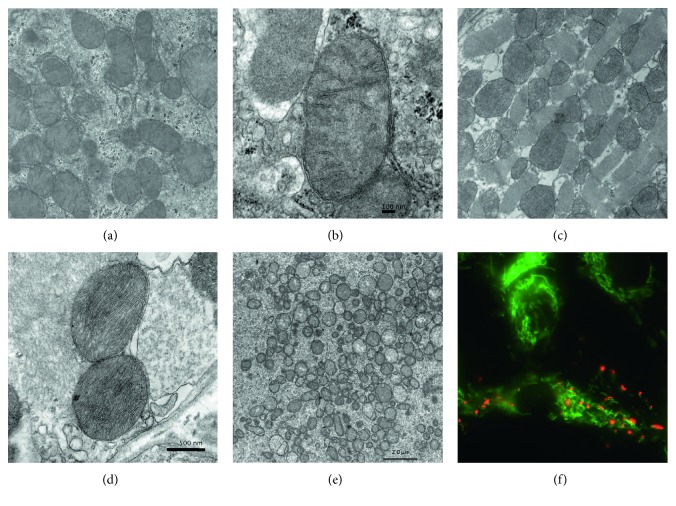
Figure 1.
Mitochondrial heterogeneity between tissue types and within cells. (a)–(e) Transmission electron micrographs depicting mitochondrial features in mouse liver ((a), enlarged in (b)), mouse heart ((c), enlarged in (d)), and a primary-stage human oocyte (e). (f) A fluorescent micrograph depicting JC-1-labeled KGN cells demonstrates heterogeneity in mitochondrial membrane potential between cells and between individual subcellular mitochondrial populations. JC-1 monomers (green) indicate low Δψm and JC-1 aggregates (red) indicate high Δψm. Scale bars as marked.
Morphological changes in the physical appearance of mitochondria during the process of differentiation are not well characterized, although it is well understood that mitochondrial ultrastructure varies dramatically between tissue types ([1], Figure 1). In a recent study evaluating the changes in mitochondrial features that accompany progressive states of MSC differentiation into the endothelial lineage, Shin et al. assessed mitochondrial numbers, length, resident area per cell, and morphology. Consistent with the high energy demands required for differentiation followed by a subsequent decline as cells approach a terminally differentiated phenotype, the authors determined that mitochondrial number, area per cell, length, and morphological complexity decreased corresponding with progression of differentiation [33]. To specifically address the mitochondrial reconfiguration that occurs during the differentiation process, Forni et al. monitored changes in mitochondrial mass, morphology, dynamics, and bioenergetics during MSC differentiation into osteocyte, chondrocyte, and adipocyte lineages. Data indicate that mitochondrial elongation and increases in MFN1 and MFN2 occurred during the early stages of adipocyte and osteogenic differentiation, whereas chondrogenesis was associated with a fragmented mitochondrial phenotype. Strikingly, the differentiation ability of MSCs was inhibited following knockdown of Mfn2 in adipogenesis and osteogenesis, while dominant-negative Drp1 impeded the chondrogenesis differentiation capability [34]. Together, these data provide strong supporting evidence for a fundamental role for mitochondrial dynamics, including the modulation of mitochondrial ultrastructure, in the differentiation process.
Although metabolic and structural remodeling during cell differentiation represents emerging fields of study in mitochondrial and stem cell biology, mitochondrial heterogeneity as it pertains to specific mitochondrial subpopulations within a single tissue or cell remains a considerably under-characterized component in cell biology. It is well understood that under physiological conditions, cells possess a heterogeneous mitochondrial population based on differences in membrane potential (Δψm [32, 35, 36]). Mitochondria with a low Δψm are generally regarded as metabolically quiescent (“resting” or damaged), whereas those with a high Δψm are viewed as metabolically active (“respiring”). However, there are caveats to this generalization, including that Δψm is maintained by the balance between the electron transport chain ATP synthases. Accordingly, even well-coupled (respiring) mitochondria can have a decreased Δψm. The heterogeneity in Δψm both within individual cells and between neighboring cells can be observed in vitro using fluorometric compounds, such as JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide). JC-1 undergoes a shift in spectral fluorescence from green (in mitochondria that are metabolically inactive) to red-orange upon conversion from monomeric (green) to aggregate (red-orange) form in actively respiring mitochondria. Utilizing the aggregate fluorescent properties of JC-1 to detect differences in mitochondrial respiration status in undifferentiated human ESCs (H9), Kumagai et al. have recently demonstrated that shifts in Δψm (e.g., cells with more aggregate-state JC-1) within undifferentiated ESC colonies may serve as a simple visual predictive indicator of cells that are undergoing the earliest stages of differentiation [37]. Due to the predominant metabolic reliance on glycolysis during the undifferentiated state [38–41], the observed increase in Δψm associated with oxidative phosphorylation is one of the first markers to distinguish differentiating cells in a mixed culture. Accordingly, mitochondrial metabolic changes have been proposed as early markers of stem cell differentiation [38–41], although how these changes occur on a cell type-specific basis and the relationship of metabolic shifts to cell fate specification remains less well defined. However, it has been shown that mitochondrial activity and stem cell function are intimately linked. In isolating undifferentiated mouse ESCs based on differences in Δψm, Schieke et al. obtained two distinct populations of ESCs which were indistinguishable based on morphology, yet had remarkably different resting oxygen consumption rates (low Δψm, ΔψmL; high Δψm, ΔψmH). Analysis of differentiation potential revealed that in the presence of bone morphogenetic protein 4 (BMP-4) in the absence of leukemia inhibitory factor (LIF) to promote mesoderm specification, ΔψmL ESCs exhibited a markedly greater mesodermal differentiation capacity (10-fold) than ΔψmH ESCs, whereas ΔψmH ESCs had a greater propensity for teratoma formation than ΔψmL ESCs [42]. Furthermore, treatment with rapamycin, a potent inhibitor of mTOR, resulted in a decrease in Δψm and oxygen consumption in undifferentiated mouse ESCs and augmented BMP-4-induced mesodermal differentiation [42]. Together, these data suggest a strong link between intrinsic mitochondrial metabolic function and stem cell fate.
Alterations in Δψm in human ESCs have also been demonstrated as a consequence of in vitro age (e.g., passage number). In undifferentiated H9 and PKU1 cell lines, Δψm increased significantly in late-passage cells as compared to their younger counterparts. The elevated Δψm also correlated with increases in total mitochondrial volume and generation of reactive oxygen species (ROS) [43]. Late-passage human ESCs also exhibited a reduced capacity for differentiation. While early-passage cells differentiated evenly into ectodermal, mesodermal, and endodermal lineages, high-passage cells preferentially differentiated into ectodermal lineage, although the authors acknowledge that the impact of prolonged duration on culture cannot be ruled out, rather than directly related to the passage number of the cell line [43]. Given that additional established markers of stemness, such as telomerase activity and pluripotency markers, were unaffected by passage number, the impact of in vitro age on mitochondrial function is striking and may represent an additional factor when screening stem cells for potential therapeutics. This type of in vitro aging has a similar impact on the mitochondria of iPSCs. In a direct comparison between iPSCs cultured for 1 month postcellular reprogramming (young iPSCs) and iPSCs cultured for over 1 year (aged iPSCs), H2O2-dependent Δψm depolarization occurred at a faster rate in aged iPSCs than that observed in their “younger” counterparts, demonstrating a diminished ability to counteract oxidant exposure with in vitro age. Moreover, the capacity for in vitro neurogenesis was diminished in the aged iPSCs versus the young iPSCs [44].
Although emerging evidence supports a role for Δψm in the maintenance of stemness, the impact of mitochondrial subcellular heterogeneity on cellular function, including differentiation, has not been evaluated. Based on a series of studies examining mitochondrial properties within individual cells [34, 35], Kuznetsov and Margreiter have neatly described mitochondrial heterogeneity as belonging to 4 major classifications (reviewed in [45]): (1) ultrastructure—mitochondria range in size (0.2 μm to 1.0 μm), shape (circular to elongated and tubular), and cristae density (no visible cristae/vacuole-like appearance to dense cristae) and can be found alone or physically networked; ultrastructure varies across tissue types, as well as within individual cells; and fission-fusion events, as well as respiratory status, also impact on morphology; (2) functional properties—mitochondria can differ in redox state, respiration, intramitochondrial Ca2+ and reactive oxygen species (ROS) levels, mitochondrial protein composition and content, and Δψm; (3) behavior—mitochondria respond differently to oxidative stress, starvation, apoptotic stimuli, and mitophagy signals and exhibit selective responses to toxins and substrates; (4) dynamics—cell type-specific intracellular localization, oscillatory movements, translocation events, filament extension and retraction, and fission and fusion events [45]. Additionally, new evidence also now demonstrates heteroplasmy in mitochondrial DNA (mtDNA) at the level of the single cell [46], bringing another layer into subcellular mitochondrial heterogeneity. However, despite the understanding that mitochondria within a cell may well serve different functions, precise characterization of subcellular mitochondrial subpopulations has proven challenging, due to difficulties in the isolation of specific mitochondrial subtypes for further analysis [45–47], and much remains to be discovered. In a discussion of mitochondrial heterogeneity pertaining to Δψm, Wikstrom et al. propose that low Δψm represents a mechanism by which mitochondria are selectively targeted for autophagy [36]. Although the dynamics that define mitophagy are unclear, it has been demonstrated that mitochondria depolarize prior to autophagy [48, 49] and join a preautophagic mitochondrial pool [49] characterized by small size and reduced levels of the mitochondrial fusion protein, OPA1 [49, 50]. It has also been proposed that mitochondrial heterogeneity may contribute to preservation, as reduced metabolic activity may serve to preserve genomic integrity [42].
2. Tissue- and Cell-Specific Mitochondrial Subpopulations
In addition to the differences in mitochondrial morphology mentioned above, mitochondria also differ functionally and are known to be involved in cellular processes beyond metabolism. Such functions are well characterized and ubiquitous, including cell death and differentiation, intracellular Ca2+ regulation, oxygen sensing, and ROS generation, while others are cell and tissue type-specific, such as steroid hormone biosynthesis, hormone signaling and responsiveness, thermogenesis, hemesynthesis, and processing of toxins [51]. These complex processes, in addition to those associated with metabolism, are carried out through bidirectional communication with the nucleus. In humans, 13 proteins, along with 2 rRNAs and 22 tRNAs, are encoded by the 37 genes contained within the small (16,569 bp) circular mitochondrial genome. The proteins serve as key constituents of mitochondrial electron transport chain (ETC) protein complexes I–IV that are embedded in the inner mitochondrial membrane. However, mitochondrial protein composition is estimated to approach 1500 proteins [52] encoded by the nuclear genome, presumably to execute the diverse array of functions performed by mitochondria. Of these, approximately 1000 proteins have been identified (although not functionally characterized) [53], with the vast majority predicted to localize to the mitochondrial inner membrane and the matrix [54, 55]. However, mitochondrial protein composition is not fixed, nor is it consistent between tissue types. A well-executed proteomic study using ultrapurified mitochondrial preparations revealed striking compositional differences between brain, kidney, liver, and heart mitochondria, determining that only 57% of mitochondrial proteins identified were consistently expressed between the examined tissues [55]. With nearly half of the protein composition of mitochondria differing between tissue types, the cell type-specific differences in mitochondrial function are also underscored. Although mitochondria are highly morphologically heterogeneous between tissues [1, 56], details regarding proteomic profiles on mitochondrial subpopulations within a single cell or tissue type are lacking, as is information regarding the ESC mitochondrial proteome [57]. Mitochondrial proteomic analyses in pluripotent stem cells as they undergo transformational changes related to reprogramming and differentiation would likely provide important information pertaining to the utility of stem cells in research and, as directed differentiation strategies improve, therapeutics and regenerative medicine.
Given the complexity and diversity of mitochondrial proteomic profiles amongst mitochondrial subtypes, it is not surprising that mitochondria serve to perform a diverse spectrum of functions—with many more remaining to be discovered. Among the more well-characterized mitochondrial types within a single tissue are two cardiac mitochondrial subpopulations, subsarcolemmal mitochondria (SSM) and interfibrillar mitochondria (IFM). Initially characterized by ultrastructural differences observed between the two populations located beneath the sarcolemma and between the myofibrils, respectively, physical isolation of each mitochondrial subtype revealed distinct biochemical functional properties [58]. Specifically, succinate dehydrogenase and citrate synthase activities were elevated in the IFM population as compared to SSM, and oxidation of substrates was found to proceed 1.5× faster in IFM isolates than SSM preparations [58]. Subsequently, the Ca2+ uptake capability of IFM and SSM was comparatively demonstrated to differ, as a further indicator of mitochondrial heterogeneity between the two subpopulations [59], although differences in isolation strategies may have impacted some of these conclusions. Additionally, IFM demonstrated higher protein import rates for the precursors to malate dehydrogenase and ornithine carbamoyltransferase [60]. However, due to the technically difficult nature of the isolation procedures, in-depth comparative experimental characterization between the two subtypes has been impeded. More recently, Hatano et al. have developed a three-dimensional computational model, integrating electrophysiology, metabolism, and mechanics with subcellular structure. Using this intriguing multifaceted simulation approach, the authors demonstrate that the impact of the subcellular environment modulated mitochondrial function [61]. Although individual intrinsic functional differences between mitochondrial populations could not be examined in the study, this work highlights that mitochondria work within a subcellular “niche,” in which microenvironmental cues can govern function. It is intriguing to think of mitochondrial subpopulations as respondents to the microenvironment (similar to stem cells within a niche). As a first step toward evaluating “mitochondrial heteroplasmy” in a single mitochondrion, Pham et al. developed a microrespirometer to monitor mitochondrial respiration on individual organelles. Using this novel technological approach, the authors confirmed differences in respiration between coupled and uncoupled mitochondria [47]. Additionally, our own data utilizing a nanoparticle-sorting platform for the isolation of mitochondrial subpopulations similarly revealed differences in the ATP-generating capability between coupled and uncoupled mitochondria; however, a subset of uncoupled mitochondria could be induced to generate ATP when the microenvironment was altered [62]. As more information becomes available regarding intrinsic mitochondrial differences, computational models along with emerging technologies to evaluate mitochondrial subpopulations such as this will likely prove invaluable in the development of experimental paradigms and testable hypotheses.
3. Conclusion
The current understanding of mitochondrial function in stem cells is limited in scope as compared to the broader field of mitochondrial biology. Given the highly specialized features of mitochondria in differentiated cell types, it stands to reason that a separate field of study dedicated to mitochondrial fate specification and differentiation might coevolve with the stem cell field. In this way, stem cells serve as an excellent model to study “mitochondrial differentiation.” As each mitochondrion contains DNA encoding for only 13 proteins, yet contains a subtype-specific proteomic profile of up to 1500 nuclear-encoded proteins, the significance of nuclear communication in the regulation of mitochondrial function becomes increasingly clear. How the mitochondria and nucleus communicate on a per organelle basis (i.e., why some are activated while others remain resting, how cell type-specific functions are executed) remains to be determined. As nanoscaled technologies emerge for the study of subcellular organelles, the mechanisms that govern mitochondrial heterogeneity and function will be elucidated and perhaps provide additional platforms and metrics for stem cell reprogramming and differentiation.
Conflicts of Interest
D. C. Woods declares no conflict of interest regarding the information contained herein.
References
- 1.Vafai S. B., Mootha V. K. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 2.Benard G., Bellance N., James D., et al. Mitochondrial bioenergetics and structural network organization. Journal of Cell Science. 2007;120(Part 5):838–848. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 3.Palmer C. S., Osellame L. D., Stojanovski D., Ryan M. T. The regulation of mitochondrial morphology: intricate mechanisms and dynamic machinery. Cellular Signalling. 2011;23(10):1534–1545. doi: 10.1016/j.cellsig.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 4.Wai T., Langer T. Mitochondrial dynamics and metabolic regulation. Trends in Endocrinology and Metabolism. 2016;27(2):105–117. doi: 10.1016/j.tem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Palade G. An electron microscope study of the mitochondrial structure. The Journal of Histochemistry and Cytochemistry. 1953;4(1):188–211. doi: 10.1177/1.4.188. [DOI] [PubMed] [Google Scholar]
- 6.Zick M., Rabl R., Reichert A. Cristae formation—linking ultrastructure and function of mitochondria. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2009;1793(1):5–19. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Paumard P., Vaillier J., Coulary B., et al. The ATP synthase is involved in generating mitochondrial cristae morphology. The EMBO Journal. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weimann T., Vaillier J., Salin B., Velours J. The intermembrane space loop of subunit b (4) is a major determinant of the stability of yeast oligomeric ATP synthases. Biochemistry. 2008;47:3556–3563. doi: 10.1021/bi702000g. [DOI] [PubMed] [Google Scholar]
- 9.Soubannier V., Vaillier J., Paumard P., Coulary B., Schaeffer J., Velours J. In the absence of the first membrane-spanning segment of subunit 4(b), the yeast ATP synthase is functional but does not dimerize or oligomerize. The Journal of Biological Chemistry. 2002;277:10739–10745. doi: 10.1074/jbc.M111882200. [DOI] [PubMed] [Google Scholar]
- 10.Giraud M. F., Paumard P., Soubannier V., et al. Is there a relationship between the supramolecular organization of the mitochondrial ATP synthase and the formation of cristae? Biochimica et Biophysica Acta. 2002;1555:174–180. doi: 10.1016/s0005-2728(02)00274-8. [DOI] [PubMed] [Google Scholar]
- 11.Chan D. C. Fusion and fission: interlinked processes critical for mitochondrial health. Annual Review of Genetics. 2002;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 12.Santel A., Fuller M. T. Control of mitochondrial morphology by a human mitofusin. Journal of Cell Science. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 13.Legros F., Lombes A., Frachon P., Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Molecular Biology of the Cell. 2002;13:4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Molecular Biology of the Cell. 2001;12(8):2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H., Detmer S. A., Ewald A. J., Griffin E. E., Fraser S. E., Chan D. C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. The Journal of Cell Biology. 2003;160(2):189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delettre C., Lenaers G., Griffoin J. M., et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nature Genetics. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 17.Lee H., Smith S. B., Yoon Y. The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure. The Journal of Biological Chemistry. 2017;292(17):7115–7130. doi: 10.1074/jbc.M116.762567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patten D. A., Wong J., Khacho M., et al. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. The EMBO Journal. 2014;33(22):2676–2691. doi: 10.15252/embj.201488349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander C., Votruba M., Pesch U. E., et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nature Genetics. 2000;26(2):211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 20.Kasahara A., Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends in Cell Biology. 2014;24(12):761–770. doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Baharvand H., Matthaei K. I. The ultrastructure of mouse embryonic stem cells. Reproductive Biomedicine Online. 2003;7(3):330–335. doi: 10.1016/s1472-6483(10)61873-1. [DOI] [PubMed] [Google Scholar]
- 22.Chung S., Dzeja P. P., Faustino R. S., Perex-Terxic C., Behfar A., Terzic A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nature Clinical Practice. Cardiovascular Medicine. 2007;4(Supplement 1):S60–S67. doi: 10.1038/ncpcardio0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto S., Morimoto N., Yamanaka M., et al. Quantitative and qualitative changes of mitochondria in human preimplantation embryos. Journal of Assisted Reproduction and Genetics. 2017;34(5):573–580. doi: 10.1007/s10815-017-0886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hackenbrock C. R. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. I. Reversible ultrastructural changes with change in metabolic steady state in isolated liver mitochondria. The Journal of Cell Biology. 1966;30:269–297. doi: 10.1083/jcb.30.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hackenbrock C. R. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. II. Electron transport-linked ultrastructural transformations in mitochondria. The Journal of Cell Biology. 1968;37:345–369. doi: 10.1083/jcb.37.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goyer R. A., Krall R. Ultrastructural transformation in mitochondria isolated from kidneys of normal and lead-intoxicated rats. The Journal of Cell Biology. 1969;41:393–400. doi: 10.1083/jcb.41.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonergan T., Brenner C., Bavister B. Differentiation-related changes in mitochondrial properties as indicators of stem cell competence. Journal of Cellular Physiology. 2006;208(1):149–153. doi: 10.1002/jcp.20641. [DOI] [PubMed] [Google Scholar]
- 28.Lonergan T., Bavister B., Brenner C. Mitochondria in stem cells. Mitochondrion. 2007;7(5):289–296. doi: 10.1016/j.mito.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S., Kim H. Y., Lee S., et al. Hepatitis B virus X protein induces perinuclear mitochondrial clustering in microtubule- and dynein-dependent manners. Journal of Virology. 2007;81(4):1714–1726. doi: 10.1128/JVI.01863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Mehdi A. B., Pastukh V. M., Swiger B. M., et al. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Science Signaling. 2012;5(231, article ra47) doi: 10.1126/scisignal.2002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swiger B., Patel M., Pastukh V., Gillespie M. N., Al-Mehdi A. B. Hypoxia causes perinuclear mitochondrial clustering and nuclear oxidant stress in pulmonary artery endothelial cells (PAECS) via a dynein-dependent molecular motor. FASEB Journal. 2008;22:1174–1116. [Google Scholar]
- 32.Collins T. J., Berridge M. J., Lipp P., Bootman M. D. Mitochondria are morphologically and functionally heterogeneous within cells. The EMBO Journal. 2002;21(7):1616–1627. doi: 10.1093/emboj/21.7.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin J. W., Park S. E., Kang Y. G., Wu Y., Choi H. J., Shin J. W. Changes, and the relevance thereof, in mitochondrial morphology during differentiation into endothelial cells. PLoS One. 2016;11(8, article e0161015) doi: 10.1371/journal.pone.0161015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forni M. F., Peloggia J., Trudeau K., Shirihai O., Kowaltowski A. J. Murine mesenchymal stem cell commitment to differentiation is regulated by mitochondrial dynamics. Stem Cells. 2016;34(3):743–755. doi: 10.1002/stem.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuznetsov A. V., Troppmair J., Sucher R., Hermann M., Saks V., Margreiter R. Mitochondrial subpopulations and heterogeneity revealed by confocal imaging: possible physiological role? Biochimica et Biophysica Acta. 2006;1757(5-6):686–691. doi: 10.1016/j.bbabio.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Wikstrom J. D., Twig G., Shirihai O. S. What can mitochondrial heterogeneity tell us about mitochondrial dynamics and autophagy? The International Journal of Biochemistry & Cell Biology. 2009;41(10):1914–1927. doi: 10.1016/j.biocel.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Kumagai A., Suga M., Yanagihara K., Itoh Y., Takemori H., Furue M. K. A simple method for labeling human embryonic stem cells destined to lose undifferentiated potency. Stem Cells Translational Medicine. 2016;5(3):275–281. doi: 10.5966/sctm.2015-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Folmes C. D., Nelson T. J., Martinez-Fernandez A., et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metabolism. 2011;14(2):264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Folmes C. D., Dzeja P. P., Nelson T. J., Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11(5):596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Folmes C. D., Nelson T. J., Dzeja P. P., Terzic A. Energy metabolism plasticity enables stemness programs. Annals of the New York Academy of Sciences. 2012;1254:82–89. doi: 10.1111/j.1749-6632.2012.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi H. W., Kim J. H., Chung M. K., et al. Mitochondrial and metabolic remodeling during reprogramming and differentiation of the reprogrammed cells. Stem Cells and Development. 2015;24(11):1366–1373. doi: 10.1089/scd.2014.0561. [DOI] [PubMed] [Google Scholar]
- 42.Schieke S. M., Ma M., Cao L., et al. Mitochondrial metabolism modulates differentiation and teratoma formation capacity in mouse embryonic stem cells. The Journal of Biological Chemistry. 2008;283(42):28506–28512. doi: 10.1074/jbc.M802763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie X., Hiona A., Lee A. S., et al. Effects of long-term culture on human embryonic stem cell aging. Stem Cells and Development. 2011;20(1):127–138. doi: 10.1089/scd.2009.0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masotti A., Celluzzi A., Petrini S., Bertini E., Zanni G., Compagnucci C. Aged iPSCs display an uncommon mitochondrial appearance and fail to undergo in vitro neurogenesis. Aging. 2014;6(12):1094–1108. doi: 10.18632/aging.100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuznetsov A. V., Margreiter R. Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. International Journal of Molecular Sciences. 2009;10(4):1911–1929. doi: 10.3390/ijms10041911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jayaprakash A. D., Benson E. K., Gone S., et al. Stable heteroplasmy at the single-cell level is facilitated by intercellular exchange of mtDNA. Nucleic Acids Research. 2015;43(4):2177–2187. doi: 10.1093/nar/gkv052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pham T. D., Wallace D. C., Burke P. J. Microchambers with solid-state phosphorescent sensor for measuring single mitochondrial respiration. Sensors (Basel) 2016;16(7) doi: 10.3390/s16071065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elmore S. P., Qian T., Grissom S. F., Lemasters J. J. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. The FASEB Journal. 2001;15(12):2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 49.Twig G., Graf S. A., Wikstrom J. D., et al. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. American Journal of Physiology. Cell Physiology. 2006;291(1):C176–C184. doi: 10.1152/ajpcell.00348.2005. [DOI] [PubMed] [Google Scholar]
- 50.Kim I., Rodriguez-Enriquez S., Lemasters J. J. Selective degradation of mitochondria by mitophagy. Archives of Biochemistry and Biophysics. 2007;462(2):245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McBride H. M., Neuspiel M., Wasiak S. Mitochondria: more than just a powerhouse. Current Biology. 2006;16(14):R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 52.Taylor S. W., Fahy E., Zhang B., et al. Characterization of the human heart mitochondrial proteome. Nature Biotechnology. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 53.Ryan M. T., Hoogenraad N. J. Mitochondrial-nuclear communications. Annual Review of Biochemistry. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- 54.Calvo S., Jain M., Xie X., et al. Systematic identification of human mitochondrial disease genes through integrative genomics. Nature Genetics. 2006;38(5):576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- 55.Mootha V. K., Bunkenborg J., Olsen J. V., et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115(5):629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 56.Fleischer S., Fouser G., Fleisher B., Casu A., Kritchevsky G. Lipid composition of mitochondria from bovine heart liver, and kidney. Journal of Lipid Research. 1967;8:170–180. [PubMed] [Google Scholar]
- 57.Shekari F., Baharvand H., Salekdeh G. H. Organellar proteomics of embryonic stem cells. Advances in Protein Chemistry and Structural Biology. 2014;95:215–230. doi: 10.1016/B978-0-12-800453-1.00007-5. [DOI] [PubMed] [Google Scholar]
- 58.Palmer J. W., Tandler B., Hoppel C. L. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. The Journal of Biological Chemistry. 1977;252(23):8731–8739. [PubMed] [Google Scholar]
- 59.Palmer J. W., Tandler B., Hoppel C. L. Heterogeneous response of subsarcolemmal heart mitochondria to calcium. The American Journal of Physiology. 1986;250:H741–H748. doi: 10.1152/ajpheart.1986.250.5.H741. [DOI] [PubMed] [Google Scholar]
- 60.Takahashi M., Hood D. A. Protein import into subsarcolemmal and intermyofibrillar skeletal muscle mitochondria. Differential import regulation in distinct subcellular regions. The Journal of Biological Chemistry. 1996;271(44):27285–27291. doi: 10.1074/jbc.271.44.27285. [DOI] [PubMed] [Google Scholar]
- 61.Hatano A., Okada J., Washio T., Hisada T., Sugiura S. Distinct functional roles of cardiac mitochondrial subpopulations revealed by a 3D simulation model. Biophysical Journal. 2015;108(11):2732–2739. doi: 10.1016/j.bpj.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chandrasekhar K., MacDonald J. A., Navaroli D. M., Tilly J. L., Woods D. C. Analysis of nanosorted oocyte mitochondrial subpopulations reveals active but reversible repression of non-respiring mitochondria. Reproductive Sciences. 2015;22(Supplement):p. 96A. [Google Scholar]