Abstract
Lysosomes are membrane-bound vesicles that contain hydrolases for the degradation and recycling of essential nutrients to maintain homeostasis within cells. Cancer cells have increased lysosomal function to proliferate, metabolize, and adapt to stressful environments. This has made cancer cells susceptible to lysosomal membrane permeabilization (LMP). There are many factors that mediate LMP such as Bcl-2 family member, p53; sphingosine; and oxidative stress which are often altered in cancer. Upon lysosomal disruption, reactive oxygen species (ROS) levels increase leading to lipid peroxidation, mitochondrial dysfunction, autophagy, and reactive iron. Cathepsins are also released causing degradation of macromolecules and cellular structures. This ultimately kills the cancer cell through different types of cell death (apoptosis, autosis, or ferroptosis). In this review, we will explore the contributions lysosomes play in inducing cell death, how this is regulated by ROS in cancer, and how lysosomotropic agents might be utilized to treat cancers.
1. Introduction
Lysosomes are membrane-enclosed vesicles that contain at least 60 hydrolases within an acidic environment. These hydrolases, which include the cathepsin family of proteases, are responsible for degradation, recycling, and disposal of cellular macromolecules [1]. Lysosomes are often termed the garbage disposal of the cell, but as our knowledge and understanding increase, the roles lysosomes play in other cellular functions expand [2]. The lysosomal degradation pathway regulates a variety of cellular functions such as autophagy, endocytosis, and phagocytosis to maintain cellular homeostasis [1]. In addition, this pathway directly or indirectly regulates cell signaling, metabolism, and degradation of protein aggregates and damaged organelles [3–5]. When the degradative pathway is dysregulated, diseases such as cancer can progress. This makes lysosomes a potential target for cancer therapy.
2. Lysosomal Biology
Lysosomes are the most acidic vesicles within the cell. This acidic pH is maintained by the action of a proton pump which hydrolyzes ATP to ADP in order to pump an H+ ion into the lumen of the lysosome [6]. The lysosomal membrane consists of a lipid bilayer and membrane proteins. The most abundant lysosomal membrane proteins are lysosome-associated membrane proteins 1 and 2 (LAMP-1 and LAMP-2). The inner lumen of these proteins is highly glycosylated and protects the lysosomal membrane from the digestive enzymes [7, 8]. These enzymes can digest DNA, RNA, sugars, lipids, and proteins. Among these enzymes is the diverse cathepsin protease family. Cathepsins A and G are serine proteases, meaning that their active site contains a vital serine. Cathepsins B, C, F, H, K, L, O, S, V, X, and W are cysteine proteases. Cathepsins D and E are aspartic proteases. Cysteine cathepsins are the most stable and active at an acidic pH. Like caspases, cathepsins have a wide range of cellular substrates. Cystatins, thyropins, and serpins prevent cathepsin substrates from binding and are thus endogenous inhibitors of cathepsins [9].
Lysosomal biogenesis is controlled by master regulators transcription factor EB (TFEB) and microphthalmia-associated transcription factor (MITF). These proteins receive cues in the cytoplasm and translocate into the nucleus to induce the transcription of lysosomal biogenesis network of genes [5, 10, 11]. TFEB and MITF are phosphorylated by mTOR in the cytoplasm and retained there by binding to 14-3-3 proteins [10]. Upon inhibition of the mTOR pathway under stress conditions, lysosomal biogenesis could be activated.
3. Lysosomes in Cancer
Lysosomes have been associated with diseases such as lysosomal storage disorders, neurodegenerative disorders, and cardiovascular disease [12, 13]. In cancer, lysosomal function is also altered. Many cancer cells have increased the number of lysosomes to maintain homeostasis by the increased degradation and recycling macromolecules to maintain cell proliferation and survive under stress condition in the microenvironment [4, 14, 15]. Indeed, increased expression of cathepsin B has been associated with increased cancer invasion [16]. Despite the ubiquitous nature of lysosomes in all mammalian cell types, cancer cells have been shown to increase lysosomal biogenesis [14, 17] and alter cellular biology [18, 19], thus affecting lysosomes. One such biological process that impacts lysosomes is sphingolipid metabolism. Altered sphingolipid metabolism has been found in many cancers [20–22]. Different cancer cell types overexpress sphingosine kinase (SK) [23–25] and downregulate acidic sphingomyelinase (ASM) [19]. These changes affect lysosomal membrane structure and function in cancer cells.
Lysosomes also play an important role in drug resistance in cancer by sequestering weak-base chemotherapeutic drugs within the cell. This increases lysosomal biogenesis resulting in enlargement of the lysosomal compartment in cells [15]. The enlarged compartment allows significant concentration of chemotherapeutic drugs to be stored in lysosomes and blocks these drugs from reaching their cellular targets. In addition, lysosomes provide a mechanism for exocytosis of drugs from the cancer cells [15]. These mechanisms render cancer cells drug-resistant, thus highlighting lysosomes as a target for cancer therapy.
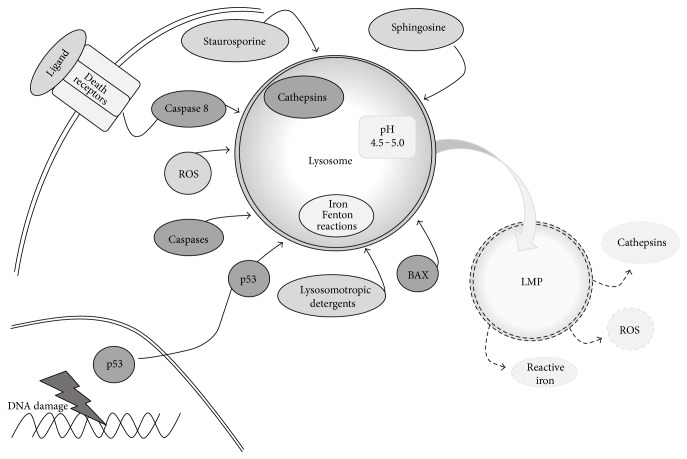
4. Lysosomal Membrane Permeabilization (Figure 1)
Figure 1.
Regulation of lysosomal membrane permeabilization (LMP). There are many factors that regulate lysosomal membrane permeabilization (LMP). These include increased levels of sphingosine, cathepsins, and ROS. The activation of caspase, Bax, and p53 and treatment with staurosporine, or lysosomotropic agents, also lead to LMP. This results in the release of ROS, cathepsins, and reactive iron from lysosomes.
Lysosomal membrane permeabilization (LMP) has been shown to be an effective therapeutic strategy in many cancer models [26]. LMP involves either the slight or the complete permeabilization of the lysosome. This permeabilization can cause lipid peroxidation and a partial or complete release of lysosomal contents. Cell death can be mediated by the reactive oxygen species (ROS) and/or lysosomal cathepsins [3, 4, 26]. In addition, sphingolipids can contribute to LMP [27]. Sphingosine has been shown to induce LMP when added to cells [27]. Upon TNFα, radiation, and DNA-damaging drug treatments, p53 is phosphorylated and translocates to lysosomes where it induces LMP [5]. Various cellular components can protect the lysosome from permeabilization such as cholesterol [28], lysosomal localization of heat shock protein 70 [29], and lipid peroxidation scavengers. Tocopherols are endogenous inhibitors of lipid peroxidation. Among tocopherols is α-tocopherol, otherwise known as vitamin E [30, 31]. Thus, there are many factors regulating LMP in cancer cells.
Cancer cells are sensitive to LMP by a variety of mechanisms. Cell lines transformed with oncogenic Src and Ras display altered lysosomal localization and decrease in LAMP-1 and LAMP-2 [18]. Decreases in the LAMP proteins prime cells for LMP. Other cancer cells increase lysosomal biogenesis [14, 17], increase lysosomal size, and alter heat shock protein 70 (HSP-70) localization creating destabilized lysosomes [29]. Cancer cells have altered sphingolipid metabolism which increases the amount of sphingosine and renders lysosomes sensitive to LMP [22, 27, 32]. Finally, many cancer cells have altered metabolism that increases ROS leading to destabilization of lysosomes leading to LMP [3, 23]. Thus, cancer cells might be sensitive to lysosome-mediated cell death.
5. Lysosome-Mediated Cell Death (LCD)
Since their discovery as the suicide bags of the cell, lysosomes have been explored as therapeutic targets in cancer. Due to these numerous alterations to this pathway, LMP is an effective way to kill many different cancer cell types. These include breast cancer [19, 30, 33], ovarian cancer [19], cervical cancer [19], colon cancer [18, 34, 35], prostate cancer [19], lung cancer [35–37], bone cancer [19], skin cancer [35], and AML [14]. Cancer cells are susceptible to lysosome-mediated cell death through increased ROS and lipid peroxidation leading to mitochondrial dysfunction and plasma membrane permeabilization [38]. Furthermore, the release of cathepsins caused cleavage and degradation of proteins leading to cell death [3]. The relations of lysosome-mediated cell death with other forms of cell death will be discussed below.
6. Lysosomes and Apoptosis
Apoptosis is a form of program cell death involving mitochondrial dysfunction and activation of cysteine proteases called caspases. It leads to DNA condensation and membrane blebbing and eventually to the formation of apoptotic bodies that are phagocytosed by the surrounding cells. Mitochondrial dysfunction is triggered by the translocation of the Bcl-2 family member Bax to the mitochondria where it interacts with Bak and other BH3-only Bcl-2 family members such as Bid to form a pore allowing cytochrome c to be released and loss of membrane potential to occur. This leads to an increase in ROS and activation of caspase 9 and caspase 3 leading to cell death [39].
Lysosomes could play important roles in regulating apoptosis upstream of mitochondrial function and after caspase activation. Following oxidative stress, it was shown that low concentrations of hydrogen peroxide cause LMP before mitochondrial dysfunction and caspase activation [40]. Blocking cathepsin D activation also prevented the release of mitochondrial cytochrome c and caspase activation [41]. Moreover, ultraviolent radiation induces LMP under conditions of oxidative stress before mitochondrial release of cytochrome c [42]. Bax interacts with other BH3-only Bcl-2 family members such as BIM and BID at lysosomes contributing to LMP independent of its mitochondrial functions. BID is also a target of cathepsins allowing its translocation to the mitochondria to interact with Bax and Bak [42]. Similar to mitochondrial regulation, antiapoptotic Bcl-2 family members can prevent LMP [26]. This suggests that lysosomal disruption can lead to mitochondrial dysfunction.
Lysosomal disruption can also occur after mitochondrial dysfunction. Following loss of membrane potential, ROS production is increased. ROS destabilizes lysosomal membranes through lipid peroxidation leading to rupture [14, 27]. Activation of caspase 8 by death receptors or activation of caspase 9 has been associated with LMP [36, 43]. Overall lysosomes can play a role in either initiating or executing apoptosis.
7. Lysosome and Autophagy
Lysosomes fuse with autophagosomes forming an autolysosome to degrade extracellular or intracellular material [44]. Autophagy plays important roles in cancer cell adaptation to stress where it protects cancer cells from death during development and where its induction is limited to further progression of the disease [45]. Lysosomes function in autophagy regulation in three main areas: (i) lysosomal restoration, (ii) lysophagy, and (iii) autolysosomal degradation. Under normal conditions, lysosomal biogenesis occurs through biosynthesis and endocytic pathways to maintain homeostasis. Under stress conditions, the number of lysosomes decreased due to their role in degrading macromolecules for recycling or removing damaged organelles. Lysosomal levels are restored through a process called autophagic lysosomal reformation (ALR) [46]. This process can be prevented by autophagy inhibitors such as rapamycin and cathepsin inhibitors [46]. The second way autophagy regulates lysosomes is when lysosomes themselves become damaged such as through LMP. The damaged lysosomes are engulfed by autophagosomes which then fuse with functional lysosomes to remove them from the cells [47]. The levels of lysosomes are then restored by lysosomal biogenesis. Finally, the fusion of lysosomes and autophagosomes provides essential amino acids and nutrients to the cell and degrades damaged organelles [48]. If this process was left unchecked, the destruction of intracellular structures will lead to cellular collapse and a form of cell death called autosis [49]. This is dependent on functional lysosomes.
8. Lysosome and Ferroptosis
Ferroptotic cell death is a type of cell death that is distinct from apoptosis and autophagy [50, 51]. It is characterized by iron-dependent accumulation of ROS. Several proteins responsible for the regulation of iron such as ferritin and transferrin and the cysteine antiporter receptor have implicated the regulation of ferroptosis [52, 53]. One of the major storage sites for iron is lysosomes. In the presence of hydrogen peroxide, free iron undergoes a Fenton reaction creating reactive iron and increasing ROS [38]. The lysosomal disruptor siramesine induces a rapid rise in the lysosomal pH followed by lysosomal leakage mediated in part by inhibiting sphingomyelinase (ASM) [19]. This destabilization of lysosomal membranes leads to increased reactive iron and increased ROS causing cell death [30]. We found that the combination with a dual tyrosine kinase inhibitor of ErbB1 and ErbB2 tyrosine kinase receptors called lapatinib with siramesine could induce ferroptosis through blocking iron transport allowing the iron released by lysosomal disruption to accumulate and increase ROS [54]. The role lysosomes play in regulating ferroptosis through increased active iron and ROS requires future investigations.
9. Lysosomotropic Agents
LMP can be induced by numerous different stimuli that are collectively called lysosomotropic agents. Lysosomotropic agents are weak-base lipophilic or cationic amphiphilic drugs that accumulate in lysosomes. This occurs through diffusion across the lysosomal membrane where the agents become protonated and become trapped in the lysosome [26]. This causes damage to the lysosomal membranes leading to LMP. Lysosomotropic agents include metal nanoparticles [55], kinase inhibitor ML-9 [56], and numerous different types of pharmaceuticals. Pharmaceutical lysosomotropic agents include the antidepressants siramesine, nortriptyline, desipramine, imipramine, and clomipramine [19]. These have shown effectiveness in breast cancer, colon cancer, and CLL cells. Antimalarials mefloquine and chloroquine have shown effectiveness in breast cancer, lymphoma, and leukemia cells [14, 57–59]. Chloroquine has been investigated in clinical trials with only partial activity in lymphoma reported. There is, however, no evidence in these trails that chloroquine is acting through LMP. Antiallergy drugs terfenadine and loratadine [19] were effective at inducing cell death in breast and lung cancer cells. The treatments of stilbenoid antioxidant pterostilbene [35, 60] and antipsychotics chlorpromazine, thioridazine, and aripiprazole [19] showed efficacy in breast and leukemia cells. The use of these agents is summarized in Table 1. Many of these agents are FDA-approved or have been extensively studied in clinical trials but, with the exception of chloroquine, not in cancer patients [61]. This provides the foundation for many of these lysosomotropic agents to be clinically investigated for their efficacy in a variety of cancers in the near future.
Table 1.
The use of lysosomotropic agents as therapeutics in cancer.
| Lysosomotropic Agent | Model | Effective doses | Reference | |
|---|---|---|---|---|
| Siramesine | In vitro | Breast cancer lines: Mcf-7, Mcf-10A, and MDA-MB-468 | 1–10 μM | [18, 19, 30] |
| Cervix carcinoma cell lines: HeLa and ME-180 | 1–10 μM | |||
| Colorectal cancer cell lines: Hkh2 and HCT116 | 8 μM | |||
| Fibroblast cell line: NIH3&3-SrcY527F | 4–10 μM | |||
| Fibrosarcoma cell lines: WEHI-S and R4 | 5 μM | |||
| Mast cells (primary) | 2–20 μM | |||
| Osteosarcoma cell line: U2OS | 1–10 μM | |||
| Ovarian carcinoma cell line: SKOV3 | 8–10 μM | |||
| Prostate cancer cell lines: PC3 and Du145-P | 5–10 μM | |||
| In vivo | WEHI-R4 in BALB-c mice | 25–100 mg/kg/d | ||
| Mcf-7 in SCID mice | 30–100 mg/kg/d | |||
| PC3-MDR in SCID mice | 30 mg/kg | |||
|
| ||||
| Desipramine | In vitro | Breast cancer lines: Mcf-7 and Mcf-10A | 25 μM | [19] |
| Cervix carcinoma cell line: HeLa | 25–50 μM | |||
| Colorectal cancer cell lines: Hkh2 and HCT116 | 8 μM | |||
| Fibroblast cell line: NIH3&3-SrcY527F | 8–25 μM | |||
| Osteosarcoma cell line: U2OS | 25–50 μM | |||
| Ovarian carcinoma cell line: SKOV3 | 75–100 μM | |||
| Prostate cancer cell lines: PC3 and Du145-P | 5–10 μM | |||
| In vivo | Mcf-7 in SCID mice | 30 mg/kg, 2×/wk | ||
|
| ||||
| Nortriptyline | In vitro | Breast cancer line: Mcf-7 | 25–50 μM | [19] |
| Cervix carcinoma cell line: HeLa | 25–50 μM | |||
| Colorectal cancer cell lines: Hkh2 and HCT116 | 8 μM | |||
| Fibroblast cell line: NIH3&3-SrcY527F | 10–25 μM | |||
| Osteosarcoma cell line: U2OS | 25–50 μM | |||
| Ovarian carcinoma cell line: SKOV3 | 40–60 μM | |||
| Prostate cancer cell lines: PC3 and Du145-P | 40–80 μM | |||
|
| ||||
| Amlodipine | In vitro | Breast cancer line: Mcf-7 | 25–50 μM | [19] |
| Fibroblast cell line: NIH3&3-SrcY527F | 10–30 μM | |||
| Ovarian carcinoma cell line: SKOV3 | 37.5–50 μM | |||
| Prostate cancer cell lines: PC3 and Du145-P | 40–50 μM | |||
|
| ||||
| Terfenadine | In vitro | Breast cancer line: Mcf-7 | 25–50 μM | [19] |
| Colorectal cancer cell lines: Hkh2 and HCT116 | 8 μM | |||
| Fibroblast cell line: NIH3&3-SrcY527F | 2.5–5 μM | |||
| Ovarian carcinoma cell line: SKOV3 | 6–8 μM | |||
| Prostate cancer cell lines: PC3 and Du145-P | 1–10 μM | |||
| In vivo | Mcf-7 in SCID mice | 10 mg/kg, 2×/wk | ||
|
| ||||
| Mefloquine | In vitro | AML cells (primary) | 5–15 μM | [14] |
| AML cell lines: HL60, KG1A OCI-AML2, and TEX | 1–10 μM | |||
| APL cell line: NB4 | 5–7 μM | |||
| CML cell line: K562 | 6–10 μM | |||
| Dendritic cells (primary) | 25–50 μM | |||
| Erythroleukemic cell line: OCI-M2 | 7–9 μM | |||
| Gastric cancer cell lines: AGS, Hs746T, MKN45, MKN74,
NCI-N87, SNU1, SNU16, TCC1, YCC10, and YCC11 |
0.5–5 μM | |||
| Lymphosarcoma cell line: MDAY-D2 | 3–5 μM | |||
| Macrophage/monocyte cell lines: THP-1 and U937 | 5–18 μM | |||
| Oral cancer cell line: KVP20C | 5 μM | |||
| Prostate cancer cell line: PC3 | 5–40 μM | |||
| In vivo | K562, MDAY-D2, and OCI-AML2 in NOD-SCID mice | 50 mg/kg | ||
| Primary AML cells in NOD-SCID mice | 100 mg/kg/d | |||
| YCC or SNU1 in SCID mice | Unknown | |||
| PC3 in C57B1/6J mice | 200 μg/25 mg | |||
|
| ||||
| Primaquine | In vitro | Breast cancer cell line: Mcf-7 | 7 μM | [58] |
| Colon cancer cell lines: Caco-2 and HT-29 | 40–70 μM | |||
| Oral cancer cell line: KVB20C | 50–75 μM | |||
|
| ||||
| Atovaquone | In vitro | Oral cancer cell line: KVB20C | 2–12.5 μM | [59] |
|
| ||||
| Ciprofloxacin | In vitro | Cervix carcinoma cell line: HeLa | 10 μg/ml | [34] |
| Colorectal cancer cell line HCT116 | 1–5 μM | |||
|
| ||||
| Pterostilbene | In vitro | AML cell lines: HL-60, MV4-11, and OCI-AML2 | 25–75 μM | [58] |
| Macrophage cell lines: THP-1 and U937 | 25–75 μM | |||
| Melanoma cell line: A375 | 10–50 μM | |||
10. Conclusion
Lysosomes play a dynamic role in cells and are altered in cancer. The initiation of LMP in cancer cells is a novel mechanism to engage the different cell death mechanisms selective for cancer. LMP is induced by lysosomotropic agents through increased ROS, lipid peroxidation, and activation of cathepsins. Many of these lysosomotropic agents are FDA-approved and could be moved rapidly to the clinic. Targeting lysosomes to induce oxidative stress will be dependent on the context of other therapies and drug resistance mechanisms found in cancer cells. Further investigation is needed to understand the regulation of lysosome-mediated cell death and the use of lysosomotropic agents in combination with other standard chemotherapy drugs or novel targeted anticancer drugs. Nevertheless, targeting lysosomes provides hope that effective treatment against drug-resistant cancers could be developed.
Conflicts of Interest
There is no conflict of interest in publishing this manuscript.
References
- 1.Saftig P., Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nature Reviews Molecular Cell Biology. 2009;10(9):623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 2.Settembre C., Ballabio A. Lysosomal adaptation: how the lysosome responds to external cues. Cold Spring Harbor Perspectives in Biology. 2014;6(6) doi: 10.1101/cshperspect.a016907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aits S., Jaattela M. Lysosomal cell death at a glance. Journal of Cell Science. 2013;126, Part 9:1905–1912. doi: 10.1242/jcs.091181. [DOI] [PubMed] [Google Scholar]
- 4.Kroemer G., Jaattela M. Lysosomes and autophagy in cell death control. Nature Reviews Cancer. 2005;5(11):886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 5.Settembre C., Fraldi A., Medina D. L., Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nature Reviews Molecular Cell Biology. 2013;14(5):283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouche V., Espinosa A. P., Leone L., Sardiello M., Ballabio A., Botas J. Drosophila Mitf regulates the V-ATPase and the lysosomal-autophagic pathway. Autophagy. 2016;12(3):484–498. doi: 10.1080/15548627.2015.1134081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J. W., Murphy T. L., Willingham M. C., Pastan I., August J. T. Identification of two lysosomal membrane glycoproteins. The Journal of Cell Biology. 1985;101(1):85–95. doi: 10.1083/jcb.101.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kundra R., Kornfeld S. Asparagine-linked oligosaccharides protect Lamp-1 and Lamp-2 from intracellular proteolysis. The Journal of Biological Chemistry. 1999;274(43):31039–31046. doi: 10.1074/jbc.274.43.31039. [DOI] [PubMed] [Google Scholar]
- 9.Stoka V., Turk V., Turk B. Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Research Reviews. 2016;32:22–37. doi: 10.1016/j.arr.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Roczniak-Ferguson A., Petit C. S., Froehlich F., et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Science Signaling. 2012;5(228, article ra42) doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sardiello M., Palmieri M., di Ronza A., et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 12.Maciejko J. J. Managing cardiovascular risk in lysosomal acid lipase deficiency. American Journal of Cardiovascular Drugs. 2017;17:217–231. doi: 10.1007/s40256-017-0216-5. [DOI] [PubMed] [Google Scholar]
- 13.Pereira C. S., Ribeiro H., Macedo M. F. From lysosomal storage diseases to NKT cell activation and back. International Journal of Molecular Sciences. 2017;18(3) doi: 10.3390/ijms18030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sukhai M. A., Prabha S., Hurren R., et al. Lysosomal disruption preferentially targets acute myeloid leukemia cells and progenitors. The Journal of Clinical Investigation. 2013;123(1):315–328. doi: 10.1172/JCI64180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhitomirsky B., Assaraf Y. G. Lysosomes as mediators of drug resistance in cancer. Drug Resistance Updates : Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy. 2016;24:23–33. doi: 10.1016/j.drup.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Ibrahim S. A., El-Ghonaimy E. A., Hassan H., et al. Hormonal-receptor positive breast cancer: IL-6 augments invasion and lymph node metastasis via stimulating cathepsin B expression. Journal of Advanced Research. 2016;7(5):661–670. doi: 10.1016/j.jare.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perera R. M., Stoykova S., Nicolay B. N., et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524(7565):361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fehrenbacher N., Bastholm L., Kirkegaard-Sorensen T., et al. Sensitization to the lysosomal cell death pathway by oncogene-induced down-regulation of lysosome-associated membrane proteins 1 and 2. Cancer Research. 2008;68(16):6623–6633. doi: 10.1158/0008-5472.CAN-08-0463. [DOI] [PubMed] [Google Scholar]
- 19.Petersen N. H., Olsen O. D., Groth-Pedersen L., et al. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell. 2013;24(3):379–393. doi: 10.1016/j.ccr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Don A. S., Hsiao J. H., Bleasel J. M., Couttas T. A., Halliday G. M., Kim W. S. Altered lipid levels provide evidence for myelin dysfunction in multiple system atrophy. Acta Neuropathologica Communications. 2014;2:p. 150. doi: 10.1186/s40478-014-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryland L. K., Fox T. E., Liu X., Loughran T. P., Kester M. Dysregulation of sphingolipid metabolism in cancer. Cancer Biology & Therapy. 2011;11(2):138–149. doi: 10.4161/cbt.11.2.14624. [DOI] [PubMed] [Google Scholar]
- 22.Truman J. P., Garcia-Barros M., Obeid L. M., Hannun Y. A. Evolving concepts in cancer therapy through targeting sphingolipid metabolism. Biochimica et Biophysica Acta. 2014;1841(8):1174–1188. doi: 10.1016/j.bbalip.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawamori T., Osta W., Johnson K. R., et al. Sphingosine kinase 1 is up-regulated in colon carcinogenesis. FASEB Journal. 2006;20(2):386–388. doi: 10.1096/fj.05-4331fje. [DOI] [PubMed] [Google Scholar]
- 24.Le Scolan E., Pchejetski D., Banno Y., et al. Overexpression of sphingosine kinase 1 is an oncogenic event in erythroleukemic progression. Blood. 2005;106(5):1808–1816. doi: 10.1182/blood-2004-12-4832. [DOI] [PubMed] [Google Scholar]
- 25.Nava V. E., Hobson J. P., Murthy S., Milstien S., Spiegel S. Sphingosine kinase type 1 promotes estrogen-dependent tumorigenesis of breast cancer MCF-7 cells. Experimental Cell Research. 2002;281(1):115–127. doi: 10.1006/excr.2002.5658. [DOI] [PubMed] [Google Scholar]
- 26.Boya P., Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27(50):6434–6451. doi: 10.1038/onc.2008.310. [DOI] [PubMed] [Google Scholar]
- 27.Dielschneider R. F., Eisenstat H., Mi S., et al. Lysosomotropic agents selectively target chronic lymphocytic leukemia cells due to altered sphingolipid metabolism. Leukemia. 2016;30(6):1290–1300. doi: 10.1038/leu.2016.4. [DOI] [PubMed] [Google Scholar]
- 28.Appelqvist H., Sandin L., Bjornstrom K., et al. Sensitivity to lysosome-dependent cell death is directly regulated by lysosomal cholesterol content. PloS One. 2012;7(11, article e50262) doi: 10.1371/journal.pone.0050262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyrd-Hansen M., Nylandsted J., Jaattela M. Heat shock protein 70 promotes cancer cell viability by safeguarding lysosomal integrity. Cell Cycle. 2004;3(12):1484–1485. doi: 10.4161/cc.3.12.1287. [DOI] [PubMed] [Google Scholar]
- 30.Ostenfeld M. S., Fehrenbacher N., Hoyer-Hansen M., Thomsen C., Farkas T., Jaattela M. Effective tumor cell death by sigma-2 receptor ligand siramesine involves lysosomal leakage and oxidative stress. Cancer Research. 2005;65(19):8975–8983. doi: 10.1158/0008-5472.CAN-05-0269. [DOI] [PubMed] [Google Scholar]
- 31.Zalkin H., Tappel A. L., Jordan J. P. Studies of the mechanism of vitamin E action. V. Selenite and tocopherol inhibition of lipid peroxidation in the chick. Archives of Biochemistry and Biophysics. 1960;91:117–122. doi: 10.1016/0003-9861(60)90463-X. [DOI] [PubMed] [Google Scholar]
- 32.Xia P., Gamble J. R., Wang L., et al. An oncogenic role of sphingosine kinase. Current Biology : CB. 2000;10(23):1527–1530. doi: 10.1016/S0960-9822(00)00834-4. [DOI] [PubMed] [Google Scholar]
- 33.Medina D. L., Fraldi A., Bouche V., et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Developmental Cell. 2011;21(3):421–430. doi: 10.1016/j.devcel.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erdal H., Berndtsson M., Castro J., Brunk U., Shoshan M. C., Linder S. Induction of lysosomal membrane permeabilization by compounds that activate p53-independent apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(1):192–197. doi: 10.1073/pnas.0408592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mena S., Rodriguez M. L., Ponsoda X., Estrela J. M., Jaattela M., Ortega A. L. Pterostilbene-induced tumor cytotoxicity: a lysosomal membrane permeabilization-dependent mechanism. PloS One. 2012;7(9, article e44524) doi: 10.1371/journal.pone.0044524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Q. Y., Shi J. G., Yao Q. H., et al. Lysosomal membrane permeabilization is involved in curcumin-induced apoptosis of A549 lung carcinoma cells. Molecular and Cellular Biochemistry. 2012;359(1-2):389–398. doi: 10.1007/s11010-011-1033-9. [DOI] [PubMed] [Google Scholar]
- 37.Mijatovic T., Mathieu V., Gaussin J. F., et al. Cardenolide-induced lysosomal membrane permeabilization demonstrates therapeutic benefits in experimental human non-small cell lung cancers. Neoplasia. 2006;8(5):402–412. doi: 10.1593/neo.05850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu F., Chen Z., Wang B., et al. The role of lysosome in cell death regulation. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(2):1427–1436. doi: 10.1007/s13277-015-4516-6. [DOI] [PubMed] [Google Scholar]
- 39.Green D. R., Llambi F. Cell death signaling. Cold Spring Harbor Perspectives in Biology. 2015;7(12) doi: 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eno C. O., Zhao G., Venkatanarayan A., Wang B., Flores E. R., Li C. Noxa couples lysosomal membrane permeabilization and apoptosis during oxidative stress. Free Radical Biology & Medicine. 2013;65:26–37. doi: 10.1016/j.freeradbiomed.2013.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Appelqvist H., Johansson A. C., Linderoth E., et al. Lysosome-mediated apoptosis is associated with cathepsin D-specific processing of bid at Phe24, Trp48, and Phe183. Annals of Clinical and Laboratory Science. 2012;42(3):231–242. [PubMed] [Google Scholar]
- 42.Oberle C., Huai J., Reinheckel T., et al. Lysosomal membrane permeabilization and cathepsin release is a Bax/Bak-dependent, amplifying event of apoptosis in fibroblasts and monocytes. Cell Death and Differentiation. 2010;17(7):1167–1178. doi: 10.1038/cdd.2009.214. [DOI] [PubMed] [Google Scholar]
- 43.Akazawa Y., Mott J. L., Bronk S. F., et al. Death receptor 5 internalization is required for lysosomal permeabilization by TRAIL in malignant liver cell lines. Gastroenterology. 2009;136(7):2365–2376. doi: 10.1053/j.gastro.2009.02.071. e2361-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Azad M. B., Chen Y., Gibson S. B. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxidants & Redox Signaling. 2009;11(4):777–790. doi: 10.1089/ars.2008.2270. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y., Henson E. S., Xiao W., et al. Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy. 2016;12(6):1029–1046. doi: 10.1080/15548627.2016.1164357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J., Zhou W., Lin J., et al. Autophagic lysosomal reformation depends on mTOR reactivation in H2O2-induced autophagy. The International Journal of Biochemistry & Cell Biology. 2016;70:76–81. doi: 10.1016/j.biocel.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 47.Hasegawa J., Maejima I., Iwamoto R., Yoshimori T. Selective autophagy: lysophagy. Methods. 2015;75:128–132. doi: 10.1016/j.ymeth.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 48.Klionsky D. J., Abdalla F. C., Abeliovich H., et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., Shoji-Kawata S., Sumpter R. M., Jr., et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(51):20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dixon S. J., Lemberg K. M., Lamprecht M. R., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yagoda N., von Rechenberg M., Zaganjor E., et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447(7146):864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mancias J. D., Wang X., Gygi S. P., Harper J. W., Kimmelman A. C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509(7498):105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schonberg D. L., Miller T. E., Wu Q., et al. Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell. 2015;28(4):441–455. doi: 10.1016/j.ccell.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood E. R., Truesdale A. T., McDonald O. B., et al. A unique structure for epidermal growth factor receptor bound to GW572016 (lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Research. 2004;64(18):6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 55.Sabella S., Carney R. P., Brunetti V., et al. A general mechanism for intracellular toxicity of metal-containing nanoparticles. Nanoscale. 2014;6(12):7052–7061. doi: 10.1039/c4nr01234h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kondratskyi A., Yassine M., Slomianny C., et al. Identification of ML-9 as a lysosomotropic agent targeting autophagy and cell death. Cell Death & Disease. 2014;5, article e1193 doi: 10.1038/cddis.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi A. R., Kim J. H., Woo Y. H., Kim H. S., Yoon S. Anti-malarial drugs primaquine and chloroquine have different sensitization effects with anti-mitotic drugs in resistant cancer cells. Anticancer Research. 2016;36(4):1641–1648. [PubMed] [Google Scholar]
- 58.Fernandes I., Vale N., de Freitas V., Moreira R., Mateus N., Gomes P. Anti-tumoral activity of imidazoquines, a new class of antimalarials derived from primaquine. Bioorganic & Medicinal Chemistry Letters. 2009;19(24):6914–6917. doi: 10.1016/j.bmcl.2009.10.081. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y., Li Y., Li Y., et al. Chloroquine inhibits MGC803 gastric cancer cell migration via the toll-like receptor 9/nuclear factor kappa B signaling pathway. Molecular Medicine Reports. 2015;11(2):1366–1371. doi: 10.3892/mmr.2014.2839. [DOI] [PubMed] [Google Scholar]
- 60.Hsiao P. C., Chou Y. E., Tan P., et al. Pterostilbene simultaneously induced G0/G1-phase arrest and MAPK-mediated mitochondrial-derived apoptosis in human acute myeloid leukemia cell lines. PloS One. 2014;9(8, article e105342) doi: 10.1371/journal.pone.0105342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y., Liao Z., Zhang L. J., Xiao H. T. The utility of chloroquine in cancer therapy. Current Medical Research and Opinion. 2015;31(5):1009–1013. doi: 10.1185/03007995.2015.1025731. [DOI] [PubMed] [Google Scholar]