Abstract
Through the one-bead two-compound (OB2C) ultra-high throughput screening method, we discovered a new small molecule compound LLS2 that can kill a variety of cancer cells. Pull-down assay and LC MS/MS indicated that galectin-1 is the target protein of LLS2. Galectin-1 is known to be involved in the regulation of proliferation, apoptosis, cell cycle, and angiogenesis. Binding of LLS2 to galectin-1 decreased membrane-associated H-Ras and K-Ras, and contributed to the suppression of pErk pathway. Importantly, combination of LLS2 with paclitaxel (a very important clinical chemotherapeutic agent) was found to exhibit synergistic activity against several human cancer cell lines (ovarian cancer, pancreatic cancer, and breast cancer cells) in vitro. Furthermore, in vivo therapeutic study indicated that combination treatment with paclitaxel and LLS2 significantly inhibits the growth of ovarian cancer xenografts in athymic mice. Our results presented here indicate that the OB2C combinatorial technology is a highly efficient drug-screening platform and LLS2 discovered through this method can be further optimized for anti-cancer drug development.
Keywords: One-Bead-Two-Compounds Library, Galectin-1 inhibitor
Introduction
Plasma membrane proteins exhibit various functions involving ion transport, cell adhesion, and sensory stimuli transduction(1). Some of them even play a pivotal role in human diseases such as death receptor, G protein-coupled receptor (GPCR), epidermal growth factor receptor (EGFR) and tumor necrosis factor receptor (TNF-R) in cancer, toll-like receptors (TLR) associates with immune response(2–5). In particular, death receptor present on the plasma membrane of cancer cells can be activated by chemical compounds to induce cell apoptosis. Therefore, membrane protein are attractive therapeutic targets for anti-cancer drug development(6). Among the pharmaceutically active heterocyclic compounds, the benzimidazole ring is an important pharmacophore(7). It might be fruitful to develop methods to synthesize large number of benzimidazole derivatives and screen them efficiently against cell surface receptors for biologic properties such as anti-cancer activities.
The one-bead one-compound (OBOC) combinatorial library method was developed as an ultra-high throughput method for synthesis and screening of chemical compounds, including peptides, peptidomimetics, small molecules, and macrocyclic natural product like molecules(8), for specific biological properties. Using “split-mix” synthesis method, millions of compounds can be rapidly synthesized on-bead. Functional compounds can then be screened by directly mixing the compound beads with the target protein. Beads interacting with the target are isolated and chemically decoded by Edman degradation or mass spectrometry(9, 10). The OBOC method has widespread applications in the search for new bioactive compounds. For example, Peng et al used this approach to identify a novel high-affinity peptidomimetics against α4β1 integrin(11). One-bead two-compounds (OB2C) method was first introduced by Meldal et al. They constructed fluorescence quenched substrate libraries for investigation of protease activity and successfully identify protease inhibitors(12). We later applied the OB2C concept to facilitate the discovery of pro-apoptotic peptides that target cell surface(13). In such OB2C libraries, each bead displays two compounds: a hexapeptide library compound (OBOC format) and a fixed well-defined cell adhesion ligand (second compound). When live cells are incubated with the OB2C bead library, each bead will be covered with a layer of cells by the action of the cell capturing or adhesion ligand, thus exposing their cell membrane proteins to the OB2C library compounds. Screening was performed by propidium iodide (PI) staining that can be used to identify dead cells. With this approach, two peptidic death ligands against T-cell leukemia cells (Molt-4) were identified(13).
Galectin-1, a 14 kDa lectin, is one of a galectin family with an affinity for β-galactosides. High expression of galectin-1 has been found in many human cancers including ovarian cancer(14), prostate cancer(15), lung cancer(16), breast cancer(17), kidney cancer(18) and pancreatic cancer(19). High expression of galectin-1 is directly implicated in the process of tumorigenesis(20, 21). Galectin-1 is involved in cancer progression and also associated with a poor prognosis in prostate, lung and ovarian cancers. Galectin-1 can localize to both intracellular and extracellular space. Intracellular galectin-1 binds to oncogenic H-Ras and activates pERK signaling pathway, resulting in cell transformation(22). Galectin-1 secreted by tumor cells exerts tumor immune suppression through the induction of apoptosis of activated T cells(23, 24), and stimulates tumor angiogenesis via the interaction of vascular endothelial growth factor receptor-2 (VEGFR2)(25). Recent papers reported that galectin-1 exhibits an oncogenic activity through the activation of H-Ras/Raf/ERK pathway, p21 and Bcl-2 in ovarian cancer(14), HIF/mTOR pathway in clear cell renal cell carcinoma (18) and hedgehog pathway in pancreatic cancer(19)
In this study, we adapted the powerful OB2C combinatorial library method for the discovery of synthetic death ligands against ovarian cancer in an ultra-high throughput fashion. In these OB2C benzimidazole libraries, each bead displays on its surface an ovarian cancer cell capturing molecule LXY30(26) (an α3β1 integrin binding ligand) and a benzimidazole library compound prepared by split and mix synthesis. When the bead library is incubated with SKOV-3 ovarian cancer cells which express high level of α3β1 integrin, each and every compound-bead are coated with cells. Cells that undergoing apoptosis can be readily detected by immunocytochemistry (ICC) using anti-cleaved caspase 3 antibody, and compound-beads coated with stained cells are considered candidate death ligands. Two death ligands against SKOV-3 cells have been identified from a OB2C benzimidazole-based small molecule library (74,088 permutations) and their pro-apoptotic effects have been confirmed. One of these ligands, LLS2, was active against SKOV-3 cells at low micromolar IC50. Pull-down assay followed by LC MS/MS indicated that galectin-1 is the target protein of LLS2. Molecular modeling studies suggested that LLS2 binds to the interface between the dimeric galectin-1 subunit, and is within 6 Å from the β-galactoside binding pocket. Binding of LLS2 to galectin-1 decreases membrane-associated Ras and contributes to the suppression of pERK pathway. In vivo study showed that LLS2 suppressed tumor growth in SKOV3 xenograft mice. Moreover, LLS2 was found to potentiate the anti-tumor activity of paclitaxel, both in vitro and in vivo.
Materials and methods
Construction of One-Bead Two-Compounds (OB2C) library
In OB2C library, a biotin molecule is co-displayed with library compound or testing molecule on the surface of the bead. Biotin is the site to introduce a cell-capturing ligand after biotinylated ligand-streptavidin (3:1) complex is incubated with the library beads. Here, we used biotinylated LXY30, a high affinity ligand against α3β1 integrin which is over-expressed in many epithelial cancers including SKOV3 ovarian cancer cells. The detail procedure was described in supplement material.
Synthesis of death ligands on beads and in soluble form
LLS1 and LLS2 were resynthesized on TentaGel resin in an OB2C format for validation of pro-apoptotic activity. For detail of synthesis, please refer to Supplementary Material. Soluble LLS2 and biotinylated LLS2 (LLS2-biotin) were synthesized on Rink amide resin using similar chemistry described in library synthesis. LLS2-biotin was designed to have biotin attached to the side chain of Lys, and two hydrophilic linkers between LLS2 and Lys(biotin). Detailed synthetic procedures of LLS2 and LLS2-biotin were described in Supplementary Material. The final products were cleaved off the beads with trifluoroacetic acid (TFA) cocktail, precipitated with cold ether and purified by reversed phase high-performance liquid chromatography (RP-HPLC). Although the starting scaffold used for synthesis of LLS2 and LLS2-biotin is racemic, by introducing another stereocenter from (S)-L-(3,4-diCl)phenylalanine, the final products formed two diastereoisomers which were successfully separated by HPLC with optimized conditions. LLS2 was found as the major diastereoisomer with (R, S) configuration at approximate ratio of 9:1 over its (S, S) isomer which was determined by HPLC peak integration.
Cell lines
The sources of cell lines including human ovarian cancer cell lines SKOV3, the lung cancer cell lines A549, the prostate cancer cell lines PC3, the pancreatic carcinoma cell line XPA3, the breast cancer cell line MCF7 and normal kidney cell line (MDCK) were obtained from the American Type Culture Collection (ATCC). Galectin-1 was examined by Western blotting after transfection with siGal-1 and pcDNA/Gal-1. Mycoplasma testing was routinely perform every month; no addition authentication was done by the authors.
Immunocytoochemical (ICC) assay on beads
First, we reacted neutravidin (5 nmole) with LXY30-biotin in 1:2 molar ratio for 20 min. After gluing the OB2C beads onto the 12-well culture plate with 80% dimethylformamide, mixture of neutravidin and LXY30-biotin was added onto the beads-glued wells. After 20 min of incubation, SKOV3 cells (5 × 105) were then seeded onto the well and incubated at 37°C for 20 minutes. The unbound cells were removed by gentle washing with PBS. Cellbound beads were incubated at 37°C for another 24 hours, followed by fixation in 4% paraformaldehyde for 20 min. For ICC assay on beads, non-specific protein binding was blocked by adding 5% BSA and cell membrane was permeabilized with 0.5% Triton X-100. We used rabbit anti-human cleaved caspase 3 (Cell Signaling Technology) as the primary antibody. Beads were incubated with primary antibody (1:100 in PBS) for overnight at 4°C. After washing with PBS, beads were then incubated with the secondary antibody, an HRP-conjugated goat anti-rabbit IgG, for 1 hour at room temperature. HRP activity was finally detected using diaminobenzidine tetrahydrochloride (DAB) as a substrate for 3 min according to the manufacturer’s instructions (Biogenex).
Decoding of positive beads
Positive beads were isolated, followed by the treatment with 6M guanidine HCl (pH1.0), to remove the bound cells or any proteins or biomolecules produced by the cells, and washed with water thoroughly prior to chemical decoding using standard automatic Edman microsequencing (ABI Procise 494).
Cell survival assay
For immobilized LLS2, streptavidin coated plates were loaded with LLS2-biotin. 3 × 103 SKOV3 cells were seeded in these LLS2-coated plates. After 72 hours, cell survival assay was assessed using CellTiter-Glo Luminescent Cell Viability Assay kit (Promega). For solution phase cytotoxic assay, 3 × 103 SKOV3 cells were seeded into 96-well plates. After 24 hours, medium was removed and the cells were treated with the indicated concentrations of LLS2, and cell viability was determined after 72 hours. Briefly, the CellTiter-Glo reagent was diluted by CellTiter-Glo® Buffer. 100 μl of diluted reagent was added to plate well containing cells and culture medium. The plates were incubated at room temperature for 10 min, and the luminescence was quantified by plate reader (PerkinElmer).
Propidium Iodide (PI) staining on beads
After 48 hours, bead-bound cells were stained with PI 1 μg/mL for 10 min. After one wash with PBS, cells were photographed using a fluorescence microscope (Olympus IX81).
Analyses of combination index
The combination index (CI), computed with the multiple drug effect equation of Chou–Talalay has been widely used to quantify drug synergism. In our study, the CI values were determined for the drug combination in cell proliferation assays using CalcuSyn sofeware. CI < 0.9 indicates synergism, CI = 0.9–1.10 indicates additive interaction, and CI > 1.10 indicates antagonism(27).
Pull-down assay
2 × 106 SKOV3 cells were cultured for membrane protein extraction using using a commercial kit (ProteoExtract® Native Membrane Protein Extraction Kit, Millipore). 50μL streptavidin agarose gel slurry (Pierce) were mixed with 50μg biotunylated LLS2-biotin in the spin column. The mixture was incubated at 4°C for 2 hours followed by centrifugation at 1250 × g for 1 min at room temperature. After washing with TBS, agarose gel was incubated with 20μg membrane protein overnight at 4°C. Agarose gel beads were washed three times with TBS and then protein was eluted with 250 μl elution buffer (150 mM glycine-HCl, pH 1.5–2.5). The eluent was then neutralized by adding 10 μl neutralization buffer (Tris, pH 8.0). The eluted protein was digested by incubating overnight at 37°C with trypsin (Promega, Madison, WI; at 5 ng/mL) and subjected to MS analysis(28).
Protein identification by MS/MS
The eluted protein was then digested by incubating overnight at 37°C with trypsin at 5 ng/mL(Promega, Madison, WI) in 50mM NH4HCO3 buffer, pH 7.8. Tryptic peptides were extracted from the gel pieces in 1 volume of 0.1% trifluoroacetic acid, while vortexing for 5 min, followed by sonication for 5 min. Crude digest mixtures were concentrated and desalted using mC18 ZipTips (Millipore) followed by eluting in 1.5 mL of matrix (5mg of CHCA/mL in 50% ACN/0.1% TFA) for MALDI-TOF MS and MS/MS analyses. Both MS and MS/MS spectra were searched against the NCBI database, using MASCOT software from matrix science (www.matrixscience.com), to identify the proteins. The MALDI-TOF MS resolution for the peptides was around 20 000, and the mass accuracy was 0.01–0.02 Da. The MS/MS resolution was around 6000.
Plasmids and siRNA transfections
5 × 103 SKOV3 and HT29 cells were seeded on the 96 well plate and transfected with siRNA against galectin-1 (200 nM) (Ambion) and pcDNA/gal-1 (100 ng) (GenScript) respectively. 1 × 105 MDCK cells were seeded on the 6 well plate and transfected with H-Ras(G12V), K-Ras(G12V) and N-Ras(Q61K) plasmids (2 ug) (Addgene). Transfection was performed using Lipofectamine 3000 transfection reagent (Life Technologies) according to the manufacturer’s instructions. MDCK transfected cells were selected using 2 μg/ml puromycin after additional 7 days.
Ras activation assay
Stable K-Ras (G12V) and N-Ras (Q61K) transfected cells were serum starved overnight. The cells were stimulated with 100 ng/ml epidermal growth factor (EGF) for 20 mins, followed by treatment with 25 μM of LLS2 for 2 hours. Cells were lysed and Raf-1 Ras-binding domain (RBD)-agarose beads (Millipore) were added to cell lysates for 30 mins at 4°C followed by centrifugation at 14,000×g for 10 secs at 4°C. After washing, the agarose-bound Ras was incubated in 2X Laemmli reducing sample buffer (126 mM Tris/HCl, 20% glycerol, 4% SDS, 0.02% bromphenylblue) followed by electrophoresis and Western blotting with an anti-Ras antibody (Millipore).
Immunoblotting Analysis
SKOV3 and A549 cells were serum starved overnight and the cells were then treated with 100 ng/ml epidermal growth factor (EGF) for 20 mins following treatment with LLS2 (25 μM) for 2 hours. Cells were lysed in a RIPA lysis buffer (50 mM Tris-HCl pH 7.5, 0.5% sodium deoxycholate, 1% NP-40, 0.1% SDS, 150mM NaCl, 2mM EDTA, 50mM NaF, 1 mM DTT, 2 mg/mL aprotinin, and 2 mg/mL leupeptin) and incubated on ice for 20 min. After centrifugation at 12,000Xg for 20 min at 4°C, total cell lysates were collected. 20 μg of each lysate was boiled in 2X Laemmli SDS-PAGE sample buffer (126 mM Tris/HCl, 20% glycerol, 4% SDS, 0.02% Bromphenolblue) at 95°C for 10 mins followed by separation on 12% SDS-PAGE gels and transferred onto PVDF membrane (Bio-Rad). After blocking with 10% non-fat dried milk for 1 h, the membrane was incubated with the specific primary antibodies against phospho MEK, MEK, phosphor Erk, Erk, beta-actin (Cell Signaling) at 4°C overnight followed by incubation with corresponding secondary antibodies at 37°C for 1 hour. After addition of ECL substrate (Amersham), chemiluminescence signal was detected by CCD camera (Biorad).
Immunocytochemistry (ICC)
For light microscopy, cells were fixed in 4% para-formaldehyde. Non-specific protein binding was blocked by adding 5% BSA. We used rabbit antihuman galectin-1 (Abcam) or LLS2-biotin as primary antibody or primary ligand, respectively. Cells were incubated with primary antibody (1:200 in PBS) for overnight at 4°C. After washing with PBS, cells were then incubated with the secondary antibody, an rhodamine-conjugated goat anti-rabbit IgG (Jackson) or FITC-conjugated streptavidin (Sigma) in the case of primary ligand, for 1 hour at room temperature. Finally, the cells nuclear DNA were stained with DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride) (Invitrogen). After washing, the cells were mounted and photographed using a confocal microscope (Zeiss).
Immunohistochemistry (IHC)
Tissue sections were de-waxed using xylene twice and rehydrated with 100% ethanol for 5 minutes, 95% and 80% ethanol for 5 minute each. Then rinse in PBS. Antigen retrival was performed in 10 mM, pH 6.0 sodium citrate buffer at 95–100°C for 20 mins. After cooling down to room temperature, tissue sections were rinsed with PBS once followed by blocking endogenous peroxidase with 1% H2O2 and blocking non-specific binding site with Power Block (BioGenex) for 5 min at room temperature each. The tissue sections were then incubated with the specific antibody against cleaved-caspase 3 and Ki-67(Cell signaling) overnight, rinsed extensively three times with PBS. Nuclei were counterstained with hematoxylin. For secondary antibody labeling, same protocol used in ICC, was used in IHC.
In vivo xenograft tumor assays
Paclitaxel and LLS2 were prepared in 50% absolute alcohol and 50% Cremophor to make a 6 and 30 mg/ml solution respectively. Prior to administrations it was dilute with saline to produce 1 mg/ml and 5 mg/ml solution respectively. Female congenital athymic BALB/c nude (nu/nu) mice were purchased from the Jackson laboratory. 2.5 × 106 SKOV3 cells were subcutaneously injected to the right side of the mouse dorsal flank. The tumors were allowed to grow to about 100 mm3. Then, mice were randomly divided into control and treatment groups, and each group contained 5 mice. Mice were given a daily I.V. administartion for 5 successive days. Tumor size and body weight were measured every 3 days. In our previous study, the 10 mg/kg paclitaxel did not significantly inhibit tumor growth(29). Eighteen days after implantation, mice were sacrificed and tumor were excised, fixed in 10% formaldehyde, embedded in paraffin, and finally sectioned for histo-pathology examination.
Statistical analysis
All the in vitro experimental studies will be performed in triplicate in two different experiments. The x2 test or the Student’s t-test was used for comparison between variables. All results were expressed as the mean±s.d. and a value of P<0.05 was considered statistically significant.
Results
Construction of OB2C library
In this library, each bead displays on its surface a biotin and a benzimidazole library compound. Desirable cell capturing molecule such as LXY30 (an α3β1 integrin binding ligand) can be conveniently displayed on the bead surface, by premixing LXY30-biotin with streptavidin at 3:1 ratio, and then added to the bead library. The coding tag is a tri-peptide which resides inside the bead, such that it will not interfere with the screening. Figure 1 shows the synthetic scheme. The benzimidazole library has three diversities, constructed from 42 primary amines (R1NH2), 42 aldehydes (R2CHO) and 42 amino acids (X3, including L- and D-amino acids, natural and unnatural amino acids). The total permutation of the OB2C-S3 library is calculated to be 74,088 (42×42×42).
Figure 1. Synthetic scheme of library OB2C.
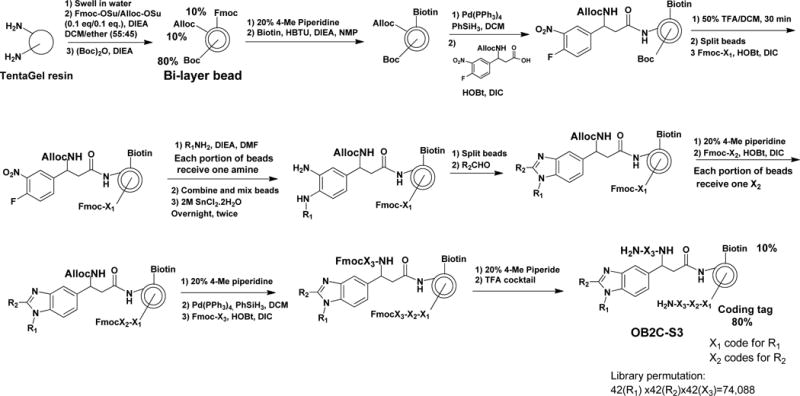
The library was synthesized on topologically segregated bi-layer TentaGel beads using split-mix synthesis method. The library has three diversities from 42 primary amines (R1NH2), 42 aldehydes (R2CHO) and 42 amino acids (X3) containing both L- and D-amino acids, natural and unnatural amino acids, respectively.
High-throughput screening using immunocytochemistry (ICC) assay on beads
Caspase-3, a pivotal executioner of apoptosis, is activated by cleavage into p20 and p12 fragments, and proteolytically cleaves several intracellular substrates(30). We believe an efficient colorimetric assay to detect cleaved caspase-3 in fixed cells will be an excellent approach to screen OB2C libraries for pro-apoptotic ligands. We used anti-cleaved caspase 3 antibody to perform ICC staining of cells bound on library beads. A positive cell bound bead is shown in Fig. 2a and 2b. For the discovery of death ligands against SKOV3 cells, a total of about 80,000 beads from OB2C-S3 library were screened; two positive beads were detected. These two positive beads were physically isolated by hand-held micropipette and their chemical identity determined by chemical decoding of the peptide tag with automatic Edman microsequencing. To validate the pro-apoptotic activity of these lead compounds, we first synthesized the death ligands on beads and then stained the bound cell for caspase 3 cleavage products. Our immunocytochemistry data indicated that LLS2 indeed has the ability to induce caspase 3, which is consistent with the screening results (Fig. 2c). In addition to confirming early apoptosis by caspase 3 staining on beads, we also validated cell death by propidium Iodide (PI), which stains late apoptotic cells with leaky cell membrane (31). After 48 hours of binding with beads displaying LLS2, SKOV3 cells were stained with PI. Many PI positive cells were detected (Fig. 2d).
Figure 2. Screening OB2C small molecule library for death ligands.
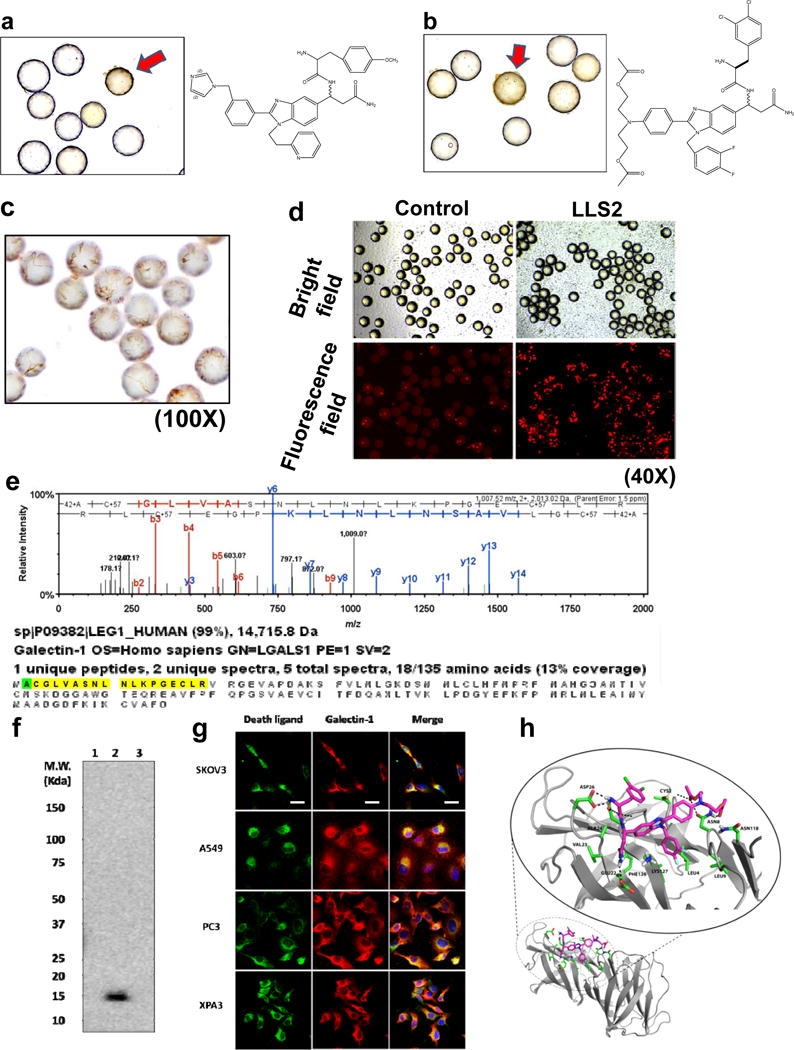
Cleaved caspase 3 positive beads (red arrow) in OB2C beads library. Positive beads were picked up for chemical decoding with automatic Edman microsequencing. (a) LLS1, (b) LLS2. Resynthesize death ligand LLS2 on beads for validation of death effect. (c) Cleaved caspase 3 staining.(100X) (d) Propidium Iodide staining for dead cells. (40X). Galectin-1 is a target protein of LLS2. (e) Eluted protein from pull-down assay was identified as galectin-1 by LC MS/MS. (f) Immunoblot analysis with anti-galectin-1 Ab, to validate the LC MS/MS result. 1: blank bead, 2: LLS2-beasd, 3: a irrelevant small molecule-bead control. (g) Immunostaining result reveals that LLS2 co-localizes with galectin-1. SKOV3, A549, PC3 and XPA3 cells were stained by biotinylated LLS2 (green), galectin-1 antibody (red), and nuclei were stained with DAPI (blue). (h) Computer modeling shows that LLS2 binds the interface of the galectin-1 dimer. Residues that are within 3.5 angstroms from LLS2 were shown. Scale bars: 50 μm.
Galectin-1 is target protein of LLS2
To clarify the molecular mechanism of LLS2 causing cell death, we used pull-down assay to determine the target protein of LLS2. SKOV3 cell membrane proteins bound to biotinylated LLS2 were first retained by streptavidin resin. Eluted proteins analyzed by LC MS/MS revealed galectin-1 as the putative target protein (Fig. 2e). The identity of galectin-1 as the target protein of LLS2 was further supported by immunoblot analysis of proteins (Fig. 2f) eluted from (i) blank streptavidin-beads, (ii) LLS2-biotin/streptavidin beads, and (iii) biotinylated unrelated small molecule/streptavidin beads, using anti-galectin-1 antibody as the western blot probe. As shown in Fig. 2f, a 14kd protein corresponding to galectin-1 was identified in lane 2 and lane 2 only, in which LLS2 was used as the affinity ligand, indicating that the target protein of LLS2 was indeed galectin-1. We then separately stained fixed and permeated SKOV3 cells with (i) LLS2-biotin followed by streptavidin-fluorophore, and (ii) anti-galectin-1 antibody, and demonstrated that fluorescent signals elicited by LLS2 and galectin-1 co-localized (Fig. 2g).
The putative binding site of LLS2 to dimeric galectin-1, predicted by computer modeling, is shown in Fig. 2h We performed multiple computational docking simulations to understand the binding of LLS compounds to galectin-1 monomer and dimer structures. Blind docking studies were first performed to rapidly scan the protein surface to identify putative binding surfaces. These initial studies were followed by the more accurate and fine-grained docking simulations across the previously identified binding interfaces. These docking studies of LLS2 with galectin-1 dimer suggest stable binding interactions that span both components of the homodimeric complex (Fig. 2h). LLS2 shows good shape complementarity with the dimer-binding pocket. The “R2” group of LLS2 forms hydrogen bond networks with the galectin-1 dimer, while the aromatic groups of LLS2 pack nicely against the hydrophobic core of the binding pocket.
LLS2 affects H-Ras(G12V) and K-Ras(G12V) localization
To elucidate the molecular mechanisms of LLS2 on tumor regression, the expression level and activity of signaling molecules associated with galectin-1 were analyzed. Galectin-1 located in cytoplasmic membrane(32) is known to cooperate with H-Ras in cell signaling and transformation (Fig. 3a). Since activated H-Ras(G12V) at the plasma membrane is essential for induction of oncogenic signaling pathway and its association with plasma membrane is stabilized by galectin-1,(22) we investigated whether membrane-associated H-Ras(G12V) can be affected by LLS2. Real time confocal images of EGF-treated cells, revealed that GFP-H-Ras(G12V) fusion resided on the plasma membrane migrated to intracellular compartments after treatment with LLS2 (25 μM) for 30 min (Fig. 3b). This is further supported by western blot analysis of plasma membrane, which showed that membrane-associated H-Ras level of MDCK cells was greatly diminished after treatment with LLS2 (Fig. 3c). LLS2 treatment was also found to (i) induce mis-colocalization of the EGF-stimulated K-Ras(G12V) fusion, but not the EGF-stimulated N-Ras(Q61K) fusion (Fig. 3d), and (ii) decreased the EGF-stimulated levels of K-Ras-GTP (Fig. 3d, 3e). An important downstream effect of Ras pathway is the activation of pErk, which is required for cell survival, transformation, proliferation and migration(33). We next examined the effect of LLS2 on MAPK/ERK pathway. Expression level of phospho-MEK and phospho-Erk was found to be down-regulated in SKOV3 and A549 cells after treatment with LLS2 for 24 hours (Fig. 3f). Together, the data support our notion that LLS2 induces cell death through suppression of the Ras-Erk pathway.
Figure 3. LLS2 causes membrane dissociation of H-and K-Ras.
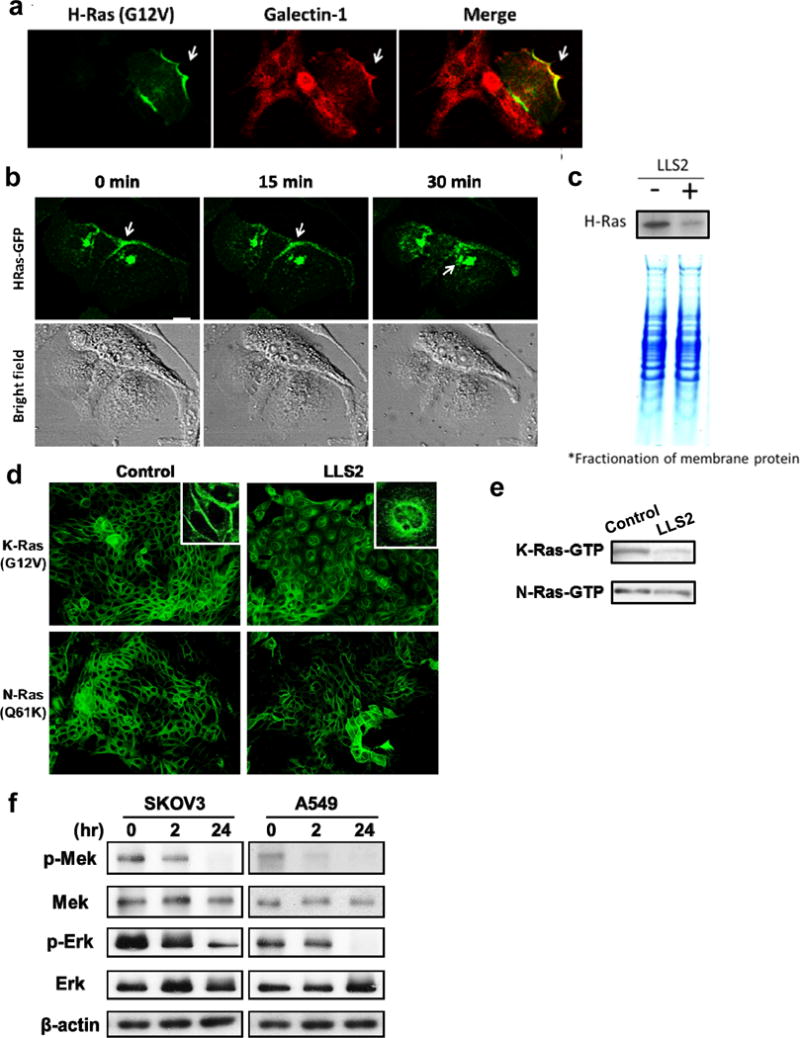
(a) Co-localization of galectin-1 and H-Ras. (b) Once activated by EGF, H-Ras will be partially trafficked to the plasma membrane leading to the activation of Erk pathway. After 15 mins treatment with LLS2(25 μM), there was a loss of membrane-localized H-Ras. Notably, intracellular H-Ras was augmented 30 mins after LLS2 treatment. Scale bars: 25 μm. (c) Quantification of membrane associated H-Ras (active form) by immunoblot analysis after treatment with buffer (−) or LLS2 (+), showing that LLS2 was able to lower the level of activated H- and K-Ras. (d) LLS2 also miscolocalized the EGF-stimulated K-Ras(G12V) but not N-Ras(Q61K). (e) Quantification of activated K- and N-Ras. (f) phospho-Mek and phospho-Erk were significantly down-regulated after treatment with LLS2 (25 μM) for 24 hours. Scale bars: 50 μm.
In vitro anticancer activity of LLS2
Immunoblots shows that high expression level of galectin-1 was found in colon cancer cell line HT29, prostate cancer cell line PC3, ovarian cancer cell line SKOV3, pancreatic cancer cell line XPA3, breast cancer cell line MCF7 and lung cancer cell line A549PC3 (Fig. 4a). We then tested the killing effect of the LLS2 compound in solution. Notably, we found that LLS2 killed SKOV3 cells with an IC50 value of 15.7 μM (Fig. 4b). In addition, LLS2 can kill HT29, PC3, XPA3, MCF7, and A549 with an IC50 value of 24.5 μM, 28.2 μM, 20.9 μM, 23.2 μM and 34.3 μM respectively. To further evaluate the specificity of galectin-1 as the target for LLS2 effect, we used siRNA to down-regulate galectin-1 in SKOV3 cells and pcDNA/galectin-1 to overexpress galectin-1 in HT29 cells (Fig. 4c). We found that SKOV3 cells transfected with negative control siRNA were more sensitive to LLS2 than those transfected with galectin-1 siRNA (Fig. 4d). Conversely, ectopic expression of galectin-1 increased sensitivity of HT29 cells to LLS2 (Fig. 4d). Although we cannot rule out the possibility of other therapeutic targets for LLS2, the result supports our notion that the major mechanisms of action of “death” function of LLS2 is through the inhibition of galectin-1 function, as evident by the difference in sensitivity to LLS2 with galectin-1 protein suppression and ectopic expression.
Figure 4. In vitro anticancer activity of LL2.
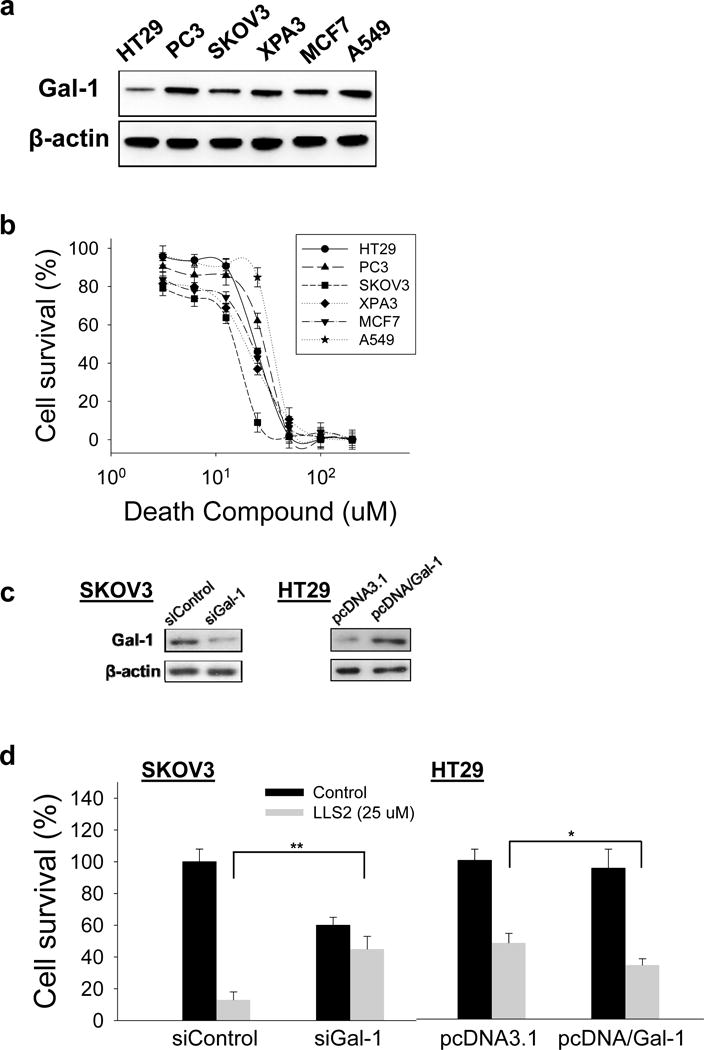
(a) Endogenous expression of galectin-1 in six different cancer cells. (b) Death ligand LLS2 (soluble form) kills a number of different cancer cell lines in solution. (c) Immunoblots demonstrate the suppression of endogenous galectin-1 expression by siRNA in SKOV3 cells, and ectopic over-expression of galectin-1 in HT-29 cells. (d) SKOV3 cells transfected with control siRNA, galectin-1 siRNA and HT29 cells transfected with control pcDNA3.1, pcDNA/Gal-1 were treated with/without LLS2 (25 uM) for 72 hours. *p<0.05; **p <0.01; ***p <0.001
LLS2 can potentiate the anti-tumor activity of paclitaxel on cancer cells
We examined the synergistic effects of LLS2 with some current chemotherapeutic drugs including docetaxel, paclitaxel, 5-fluorouracil (5-FU), oxaliplatin, carboplatin, doxorubicin and gemcitabine in vitro. Our combination studies showed that LLS2 displays synergistic effects with docetaxel, paclitaxel, 5-FU, oxaliplatin, carboplatin and doxorubicin. Among these chemotherapeutic drugs, paclitaxel is of particular interest, since it has synergistic effect with LLS2 on many cancer cell lines. Paclitaxel (Taxol), a very important chemotherapeutic agent, binds to microtubules to form the highly stable microtubules, causing cell cycle arrest at the G2/M phase. In addition, paclitaxel also exerts apoptotic activity through the mitotic block(34). In clinical cancer therapy, paclitaxel is a potent and toxic antineoplastic drug commonly used to treat a wide variety of human malignancies such as ovarian, breast and non-small cell lung cancers. Single agent and combination therapy have been extensively evaluated in clinical trials(35, 36). Because of its low solubility, paclitaxel is formulated in cremophor, which requires premedication of patients with benadryl and dexamethasone. The drug has also been nanoformulated with human serum albumin (Abraxane). Compounds that can potentiate the anti-tumor effects of paclitaxel will be of great clinical interest.
In our study, we observed that paclitaxel exhibited moderate cytotoxicity against PC3 and SKOV3 cancer cells at 2 nM. After the combined treatment with LLS2, much stronger anti-proliferative effect was achieved (Fig. 5a). This synergistic effect was confirmed by measuring the combination index (CI) (Fig. 5b) and it was further determined at various concentrations of LLS2 and paclitaxel against SKOV3 (Fig. 5c). Increased apoptosis and down-regulation of pERK were also observed after combined LLS2/paclitaxel treatment for 24 hours (Fig. 5d, 5e).
Figure 5. LLS2 can potentiate the anticancer effects of paclitaxel (PTX).
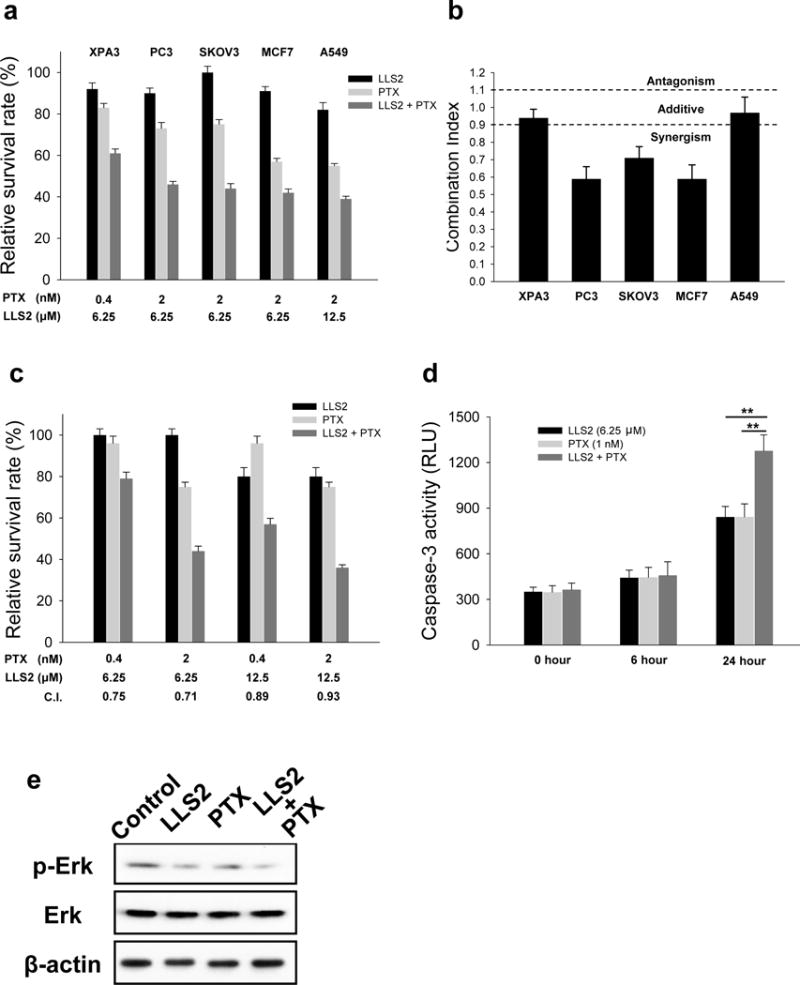
(a) The cytotoxicity of LLS2 alone, PTX alone, and the combination of LLS2 and PTX. (b) The combination index (C.I.) of the treatment of combination of LLS2 and PTX. C.I. < 0.9 indicates synergism, C.I. = 0.9–1.10 indicates additive interaction, and C.I. > 1.1 indicates antagonism. (c) LLS2 can potentiate the anticancer effects of PTX at various concentrations. (d) Apoptosis was increased and (e) phospho-ERK was down-regulated after combination treatment (LLS, 25 μM and PTX, 1nM) in SKOV3 cells for 24 hours.
LLS2 alone and LLS2/paclitaxel have anti-tumor activity in vivo
SKOV3 ovarian cancer xenograft model was used to examine the in vivo anti-tumor effect of LLS2. Both tumor size (Fig. 6a, 6b) and tumor weight (Fig. 6c) were smaller in LLS2-treated mice. Tumor response was found to be much more pronounced in the combination LLS2 and paclitaxel treatment group (Fig. 6a–c). Importantly, mice treated with the LLS2/paclitaxel combination regimen continued to gain weight and did not show any significant side effects (Fig. 6d). Excised tumors were analyzed for caspase 3 cleaved and ki-67 level. Increased cleaved caspase 3 positive cells (~10 fold) and decreased ki-67 positive cells were detected in LLS2-treated tumor, when compared with negative controls (Fig. 6e, 6f). Consistent with the in vitro and in vivo tumor response data, cleaved caspase 3 positive cells were dramatically increased and ki-67 positive cells were decreased in combination LLS2/paclitaxel treated group as compared to the other three groups (Fig. 6e, 7f). Together, these results indicate that LLS2 is an excellent drug lead for the development of a novel therapeutic combination regimen with paclitaxel, for the efficacious treatment of ovarian cancer and a number of other solid tumors.
Figure 6. LLS2 alone and LLS2/PTX have anti-tumor activity in SKOV3 xenograft model.
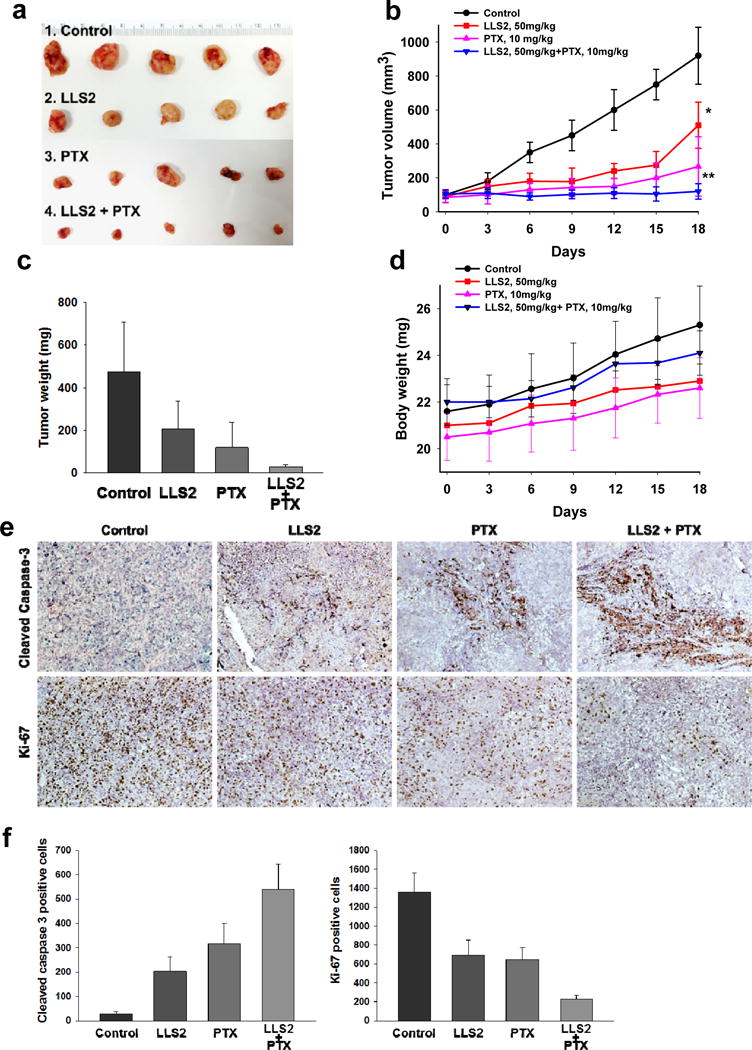
(a) Xenograft tumor. (b) Tumor growth curves, and (c) tumor weight of the xenografts in inoculated nude mice. (d) Body weight of nude mice. Briefly, 2.5 × 106 SKOV3 cells were subcutaneously injected to the right side of the dorsal flank of the female congenital athymic BALB/c nude mice. The tumors were allowed to grow to about 100 mm3. Then, mice were randomly divided into control and treatment groups (n=5). Mice were given a daily I.V. administartion for 5 successive days. (e) IHC detection of ki-67 and cleaved caspase-3 expression. (f) Quantification of immunostaining of ki-67 and cleaved caspase 3 positive cells. The cells were counted in 3 random chosen areas. *P< 0.05, **P < 0.01.
Discussion
In this study, we have established the utility of benzimidazole-based OB2C combinatorial library, in conjunction with a high throughput cell-based screening method to discover novel anti-cancer agents. Through screening a benzimidazole derivatives library (containing 74,088 discrete compounds), we have identified a novel pro-apoptotic compound LLS2. Further characterization of LLS2 revealed that galectin-1 is one of the target proteins and that LLS2 inhibits cell proliferation. Molecular modeling studies suggested that LLS2 binds to the interface between the dimeric galectin-1 subunits, and is within 6 Å from the β-galactoside binding pocket. We have also demonstrated one molecular mechanism of action of LLS2 on cell death. Binding of LLS2 to galectin-1 decreases membrane-associated H-Ras and K-Ras, and contributed to the suppression of pErk pathway. Moreover, we have found that LLS2 synergizes the anti-cancer effects of paclitaxel against several human cancer cell lines (ovarian cancer, pancreatic cancer, colon cancer, and breast cancer cells). In vivo study, LLS2 significantly suppressed tumor growth and led to significant tumor regression when used in combination with paclitaxel in SKOV3 xenograft model. Our present study suggests that cell-based OB2C combinatorial screening method is feasible for drug discovery and that LLS2 compound could potentially be used as a lead compound to develop a novel anticancer drug.
Compared with normal tissues, galectin-1 is overexpressed in cancer tissues and level of galectin-1 in tumors has been reported to be positively correlated with clinical staging, suggesting that galectin-1 participates in tumor progression. Moreover, a high level of galectin-1 expression has been found to correlate with poor prognosis in prostate, lung and ovarian cancers(15, 37, 38). Recent papers also reported that galectin-1 exerts an oncogenic activity through the activation of H-Ras/Raf/ERK pathway, p21 and Bcl-2 in ovarian cancer(14), HIF/mTOR pathway in clear cell renal cell carcinoma(18) and hedgehog pathway in pancreatic cancer(19). Galectin-1 secreted by tumor cells could suppresses tumor immune response through the induction of apoptosis of activated T cells(23, 24). Taken together, in addition to exerting direct inhibitory effect on ERK pathway, LLS2 might also suppresses other galectin-1 related pathway. In our preliminary results, LLS2 inhibited galectin-1 related angiogenesis. Work is currently underway in our laboratory on examining those galectin-1 related mechanisms.
It has been suggested that the hydrophobic pocket of galectin-1 interacts with the farnesyl group of H-Ras, stabilizing H-Ras-GTP interaction with the cell membrane(39). A single point mutation (L11A) in the galectin-1 hydrophobic pocket displaces H-RasG12V from the plasma membrane and inhibits biological activity of Ras(39). Since galectin-1’s hydrophobic pocket is essential for the binding to H-Ras, we hypothesize that LLS2/galectin-1 interaction may change the property or conformation of the hydrophobic pocket in galectin-1, and therefore attenuates the binding of galectin-1 to membrane associated H-Ras. This hypothesis, as well as how LLS2/galectin-1 interaction affects downstream targets, will need to be further confirmed.
Paz(22) reported that membrane associated galectin-1 binds oncogenic H-Ras to mediate Ras membrane anchorage resulting in cell transformation. To date, only this study has addressed the membrane function of galectin-1, however, the larger role of membrane-associated galectin-1 in cancer is largely unknown. Our OB2C screening result demonstrates a new function of membrane galectin-1 as a death receptor, given that its interaction with LLS2 (as a proxy death ligand) leads to cell death. Therefore not only has our OB2C screening discovered a novel LLS2 death ligand, but it has also opened a window for the future study of a new membrane function featuring galectin-1’s role as a death receptor.
In contrast to biologicals, small molecules are orally bioavailable and their manufacturing cost is generally lower, which is an important advantage for patients and the healthcare system. Within the small molecule compounds, the benzimidazole scaffold is a common therapeutic pharmacophore found in several bioactive compounds(7). Its pharmacological activities including anti-viral(40), anti-microbial(41, 42), anti-parasitic(43), anti-inflammatory(44) and anti-hypertensive(45). Therefore, we chose the benzimidazole as the pharmacophore scaffold and utilized combinatorial chemistry to generate benzimidazole-based compounds.
MS analysis results of proteins pulled-down by LLS2 also revealed nucleophosmin, heterogenous nuclear ribonucleoprotein and filaggrin-2, making them potential therapeutic target proteins for LLS2. In fact, many FDA- approved and clinical effective drugs have been shown to act on multiple target proteins rather than a single target; examples are Imatinib (Novartis) and Sorafenib (Bayer). The main concern of off target effects is toxicity. Thus far, no obvious toxicities have been observed in the LLS2-treated mice (Fig 6d). This and other published work strongly suggest that galectin-1 is an excellent therapeutic target against tumor; we therefore would like to further improve the binding specificity and affinity of LLS2 to galectin-1. The “R2” group in LLS2 forms hydrogen bond with the galectin-1 dimer (Fig 2h), we could modify “R1” and the central scaffold to provide additional hydrogen bonding contacts (e.g., K127) with the dimer interface. Moreover, the binding pocket for LLS2 is also close enough (within 6 Å) to the known sugar-binding pocket of galectin-1. This warrants the extension our LLS2-virtual compound library to include sugar moieties attached by linkers of varying lengths to generate compounds that aim to target both the LLS2 and sugar binding pockets. This approach not only allows us to improve the binding affinity and specificity of the compound to galectin-1, but also inhibit the functional effect of glycan binding to galectin-1.
Considering the multitude of molecular entities and signaling pathways regulating the proliferation and cellular survival/cell death, the inhibition of a single target gene or transcriptional factor may not be sufficient to suppress neoplastic progression. In clinical oncology, cancer drugs with different mechanism of actions are often given in combination to maximize the therapeutic effects, decrease side effects and delay drug resistance(46). In this study, we have clearly demonstrated that LLS2 could synergize the anti-tumor efficacy of paclitaxel, both in vitro (Fig. 5) and in vivo (Fig. 6). We will further explore this synergy in xenograft model, patient-derived xenograft (PDX) model and canine spontaneous tumor model. Understanding the mechanism of this synergy may allow us to develop more efficacious drug combinations against human cancer.
In conclusion, through screening OB2C combinatorial small molecule libraries, we have identified LLS2 as the “death ligand”. Galectin-1 is the target of LLS2, and binding of LLS2 to galectin-1 decreases membrane-associated H-Ras and K-Ras, and contributes to the suppression of MAPK/ERK pathway. LLS2 exerts anti-tumor activity, and it also can potentiate the anti-cancer effects of paclitaxel in SKOV3 xenograft model. LLS2 is a promising lead compound to be developed into a potent anti-tumor drug, and LLS2/paclitaxel combination treatment is a potential new approach to enhance the therapeutic effects of paclitaxel, a very important cytotoxic chemotherapeutic drug clinically active against many different cancer types.
Supplementary Material
Acknowledgments
We thank Dr. A.W. Herren and Dr. B.S. Phinney (UC Davis Genome Center, University of California Davis, Davis, USA) for performing LC MS/MS. The authors would like to thank the Combinatorial Chemistry Shared Resource at University of California Davis for assistance with synthesis and screening of OB2C libraries, Utilization of this Shared Resource was supported by the UC Davis Comprehensive Cancer Center Support Grant (NCI P30CA093373)
Grant support: This work was supported by grants of the NIH/NCI (2R33 CA160132, to Dr. K. S. Lam).
Abbreviations list
- OB2C
one-bead two-compounds
- OBOC
one-bead one-compound
- GPCR
G protein-coupled receptor
- EGFR
epidermal growth factor receptor
- TNF-R
tumor necrosis factor receptor
- TLR
toll-like receptors
References
- 1.Engel A, Gaub HE. Structure and mechanics of membrane proteins. Annu Rev Biochem. 2008;77:127–48. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 3.Ritter SL, Hall RA. Fine-tuning of GPCR activity by receptor-interacting proteins. Nat Rev Mol Cell Biol. 2009;10:819–30. doi: 10.1038/nrm2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Croft M, Benedict CA, Ware CF. Clinical targeting of the TNF and TNFR superfamilies. Nat Rev Drug Discov. 2013;12:147–68. doi: 10.1038/nrd3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 6.Piechocki MP, Wu GS, Jones RF, Jacob JB, Gibson H, Ethier SP, et al. Induction of proapoptotic antibodies to triple-negative breast cancer by vaccination with TRAIL death receptor DR5 DNA. Int J Cancer. 2012;131:2562–72. doi: 10.1002/ijc.27534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sivakumar R, Pradeepchandran R, Jayaveera KN, Kumarnallasivan P, Vijaianand PR, Venkatnarayanan R. Benzimidazole: An Attractive Pharmacophore in Medicinal Chemistry. IJPR. 2011;3:19–31. [Google Scholar]
- 8.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–4. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 9.Sweeney MC, Pei D. An Improved Method for Rapid Sequencing of Support-Bound Peptides by Partial Edman Degradation and Mass Spectrometry. J Comb Chem. 2003;5:218–22. doi: 10.1021/cc020113+. [DOI] [PubMed] [Google Scholar]
- 10.Marani MM, Oliveira E, Cote S, Camperi SA, Albericio F, Cascone O. Identification of protein-binding peptides by direct matrix-assisted laser desorption ionization time-of-flight mass spectrometry analysis of peptide beads selected from the screening of one bead-one peptide combinatorial libraries. Anal Biochem. 2007;370:215–22. doi: 10.1016/j.ab.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 11.Peng L, Liu R, Marik J, Wang X, Takada Y, Lam KS. Combinatorial chemistry identifies high-affinity peptidomimetics against alpha4beta1 integrin for in vivo tumor imaging. Nat Chem Biol. 2006;2:381–9. doi: 10.1038/nchembio798. [DOI] [PubMed] [Google Scholar]
- 12.Meldal M. The one-bead two-compound assay for solid phase screening of combinatorial libraries. Biopolymers. 2002;66:93–100. doi: 10.1002/bip.10229. [DOI] [PubMed] [Google Scholar]
- 13.Kumaresan PR, Wang Y, Saunders M, Maeda Y, Liu R, Wang X, et al. Rapid discovery of death ligands with one-bead-two-compound combinatorial library methods. ACS Comb Sci. 2011;13:259–64. doi: 10.1021/co100069t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P, Zhang P, Shi B, Zhou M, Jiang H, Zhang H, et al. Galectin-1 overexpression promotes progression and chemoresistance to cisplatin in epithelial ovarian cancer. Cell Death Dis. 2014;5:e991. doi: 10.1038/cddis.2013.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.A F, BruÃle vd, Castronovo DWaV. Increased expression of galectin-1 in carcinoma-associated stroma predicts poor outcome in prostate carcinoma patients. J Pathol. 2001;193:80–7. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH730>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 16.Carlini MJ, Roitman P, Nunez M, Pallotta MG, Boggio G, Smith D, et al. Clinical relevance of galectin-1 expression in non-small cell lung cancer patients. Lung Cancer. 2014;84:73–8. doi: 10.1016/j.lungcan.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Jung EJ, Moon HG, Cho BI, Jeong CY, Joo YT, Lee YJ, et al. Galectin-1 expression in cancer-associated stromal cells correlates tumor invasiveness and tumor progression in breast cancer. Int J Cancer. 2007;120:2331–8. doi: 10.1002/ijc.22434. [DOI] [PubMed] [Google Scholar]
- 18.White NM, Masui O, Newsted D, Scorilas A, Romaschin AD, Bjarnason GA, et al. Galectin-1 has potential prognostic significance and is implicated in clear cell renal cell carcinoma progression through the HIF/mTOR signaling axis. Br J Cancer. 2014;110:1250–9. doi: 10.1038/bjc.2013.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Bosch N, Fernandez-Barrena MG, Moreno M, Ortiz-Zapater E, Munne-Collado J, Iglesias M, et al. Galectin-1 Drives Pancreatic Carcinogenesis through Stroma Remodeling and Hedgehog Signaling Activation. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-13-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demydenko D, Berest I. Expression of Galectin-1 in malignant tumors. Exp Oncol. 2009;31:74–9. [PubMed] [Google Scholar]
- 21.Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 22.Paz A, Haklai R, Elad-Sfadia G, Ballan E, Kloog Y. Galectin-1 binds oncogenic H-Ras to mediate Ras membrane anchorage and cell transformation. Oncogene. 2001;20:7486–93. doi: 10.1038/sj.onc.1204950. [DOI] [PubMed] [Google Scholar]
- 23.Perillo NL, Pace KE, Seilhamer JJ, Baum LG. Apoptosis of T cells mediated by galectin-1. Nature. 1995;378:736–9. doi: 10.1038/378736a0. [DOI] [PubMed] [Google Scholar]
- 24.Rubinstein N, Alvarez M, Zwirner NW, Toscano MA, Ilarregui JM, Bravo A, et al. Targeted inhibition of galectin-1 gene expression in tumor cells results in heightened T cell-mediated rejection. Cancer Cell. 2004;5:241–51. doi: 10.1016/s1535-6108(04)00024-8. [DOI] [PubMed] [Google Scholar]
- 25.Stanley P. Galectin-1 Pulls the Strings on VEGFR2. Cell. 2014;156:625–6. doi: 10.1016/j.cell.2014.01.059. [DOI] [PubMed] [Google Scholar]
- 26.Xiao W, Li T, Bononi FC, Lac D, Kekessie IA, Liu Y, et al. Discovery and characterization of a high-affinity and high-specificity peptide ligand LXY30 for in vivo targeting of alpha3 integrin-expressing human tumors. EJNMMI research. 2016;6:18. doi: 10.1186/s13550-016-0165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou T-C, Talalay P. Generalized Equations for the Analysis of Inhibitions of Michaelis-Menten and Higher-Order Kinetic Systems with Two or More Mutually Exclusive and Nonexclusive Inhibitors. Eur J Biochem. 1981;115:207–16. doi: 10.1111/j.1432-1033.1981.tb06218.x. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh SY, Shih TC, Yeh CY, Lin CJ, Chou YY, Lee YS. Comparative proteomic studies on the pathogenesis of human ulcerative colitis. Proteomics. 2006;6:5322–31. doi: 10.1002/pmic.200500541. [DOI] [PubMed] [Google Scholar]
- 29.Xiao K, Li Y, Lee JS, Gonik AM, Dong T, Fung G, et al. “OA02” peptide facilitates the precise targeting of paclitaxel-loaded micellar nanoparticles to ovarian cancer in vivo. Cancer Res. 2012;72:2100–10. doi: 10.1158/0008-5472.CAN-11-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson DW, Thornberry NA. Caspases: killer proteases. Trends in Biochemical Sciences. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 31.Hug H, Los M, Hirt W, Debatin K-M. Rhodamine 110-Linked Amino Acids and Peptides as Substrates To Measure Caspase Activity upon Apoptosis Induction in Intact Cells. Biochemistry. 1999;38:13906–11. doi: 10.1021/bi9913395. [DOI] [PubMed] [Google Scholar]
- 32.Julie E, Thuy N, Douglas C, Lotan Dafna, Reuben L. Differential expression of endogenous galectin-1 and galectin-3 in human prostate cancer cell lines and effects of overexpressing galectin-1 on cell phenotype. Int J Oncol. 1999;14:217–24. [PubMed] [Google Scholar]
- 33.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008;9:747–58. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 34.Jordan MA, Wendell K, Gardiner S, Brent Derry W, Copp H, Wilson L. Mitotic Block Induced in HeLa Cells by Low Concentrations of Paclitaxel (Taxol) Results in Abnormal Mitotic Exit and Apoptotic Cell Death. Cancer Res. 1996;56:816–25. [PubMed] [Google Scholar]
- 35.Piccart MJ, Bertelsen K, James K, Cassidy J, Mangioni C, Simonsen E, et al. Randomized Intergroup Trial of Cisplatin–Paclitaxel Versus Cisplatin–Cyclophosphamide in Women With Advanced Epithelial Ovarian Cancer: Three-Year Results. J Natl Cancer Inst. 2000;92:699–708. doi: 10.1093/jnci/92.9.699. [DOI] [PubMed] [Google Scholar]
- 36.Malingré MM, Terwogt JMM, Beijnen JH, Rosing H, Koopman FJ, Tellingen Ov, et al. Phase I and Pharmacokinetic Study of Oral Paclitaxel. J Clin Oncol. 2000;18:2468–75. doi: 10.1200/JCO.2000.18.12.2468. [DOI] [PubMed] [Google Scholar]
- 37.Kim HJ, Jeon HK, Cho YJ, Park YA, Choi JJ, Do IG, et al. High galectin-1 expression correlates with poor prognosis and is involved in epithelial ovarian cancer proliferation and invasion. Eur J Cancer. 2012;48:1914–21. doi: 10.1016/j.ejca.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Szoke T, Kayser K, Baumhakel JD, Trojan I, Furak J, Tiszlavicz L, et al. Prognostic significance of endogenous adhesion/growth-regulatory lectins in lung cancer. Oncology. 2005;69:167–74. doi: 10.1159/000087841. [DOI] [PubMed] [Google Scholar]
- 39.Rotblat B, Niv H, André S, Kaltner H, Gabius H-J, Kloog Y. Galectin-1(L11A) Predicted from a Computed Galectin-1 Farnesyl-Binding Pocket Selectively Inhibits Ras-GTP. Cancer Res. 2004;64:3112–8. doi: 10.1158/0008-5472.can-04-0026. [DOI] [PubMed] [Google Scholar]
- 40.Tewari AK, Mishra A. Synthesis and antiviral activities of N-substituted-2-substituted-benzimidazole derivatives. Indian J Chem. 2006;45B:489–93. [Google Scholar]
- 41.Mavrova A, Anichina KK, Vuchev DI, Tsenov JA, Kondeva MS, Micheva MK. Synthesis and antitrichinellosis activity of some 2-substituted-[1,3]thiazolo[3,2-a]benzimidazol-3(2H)-ones. Biorg Med Chem. 2005;13:5550–9. doi: 10.1016/j.bmc.2005.06.046. [DOI] [PubMed] [Google Scholar]
- 42.Örena İ, Temiza Ö, Yalçına İ, Şenera E, Altanlarb N. Synthesis and antimicrobial activity of some novel 2,5- and/or 6-substituted benzoxazole and benzimidazole derivatives. Eur J Pharm Sci. 1998;7:153–60. doi: 10.1016/s0928-0987(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 43.Valdez J, Cedillo R, Hernández-Campos A, Yépez L, Hernández-Luis F, Navarrete-Vázquez G, et al. Synthesis and Antiparasitic Activity of 1H-Benzimidazole Derivatives. Bioorg Med Chem Lett. 2002;12:2221–4. doi: 10.1016/s0960-894x(02)00346-3. [DOI] [PubMed] [Google Scholar]
- 44.Sondhi SM, Rajvanshi S, Johar M, Bharti N, Azam A, Singh AK. Anti-inflammatory, analgesic and antiamoebic activity evaluation of pyrimido[1,6-a]benzimidazole derivatives synthesized by the reaction of ketoisothiocyanates with mono and diamines. Eur J Med Chem. 2002;37:835–43. doi: 10.1016/s0223-5234(02)01403-4. [DOI] [PubMed] [Google Scholar]
- 45.Kumar JR, Jawahar LJ, Pathak DP. Synthesis of Benzimidazole Derivatives: As Anti-hypertensive Agents. E J CHEM. 2006;3:278–85. [Google Scholar]
- 46.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


