Abstract
Exosomes are membrane-enclosed extracellular vesicles derived from cells, carrying biomolecules that include proteins and nucleic acids for intercellular communication. Owning to their advantages of size, structure, stability, and biocompatibility, exosomes have been used widely as natural nanocarriers for intracellular delivery of theranostic agents. Meanwhile, surface modifications needed to endow exosomes with additional functionalities remain challenging by their small size and the complexity of their membrane surfaces. Current methods have used genetic engineering and chemical conjugation, but these strategies require complex manipulations and have only limited applications. Herein, we present an aptamer-based DNA nanoassemblies on exosome surfaces. This in situ assembly method is based on molecular recognition between DNA aptamers and their exosome surface markers, as well as DNA hybridization chain reaction initiated by an aptamer-chimeric trigger. It further demonstrated selective assembly on target cell-derived exosomes, but not exosomes derived from nontarget cells. The present work shows that DNA nanostructures can successfully be assembled on a nanosized organelle. This approach is useful for exosome modification and functionalization, which is expected to have broad biomedical and bioanalytical applications.
Exosomes are nanosized (30–100 nm) membrane-enclosed extracellular vesicles that are derived from and secreted by many cell types, including leukocytes, epithelial cells, and cancerous cells.1,2 Serving an important role in cell communication, exosomes carry a full complement of biomolecules, including lipids, proteins, and nucleic acids.3–5 Owing to their high correlation with disease-related, especially cancer-related, alteration of physiological and pathological status, exosomes are widely postulated to be a potential source of biomarkers for bioanalytical applications and disease theranostics.6–9 Besides, as a type of naturally occurred nanocarrier, exosomes exhibit advantages in size, structure, stability and biocompatibility.10,11 For instance, nanosized exosomes can readily penetrate biological barriers, escape from phagocytosis, and accumulate in tissue microenvironments, such as tumors, via the enhanced permeability and retention effect. In addition, the bilayer membrane structure of exosomes is enhanced by deformable cytoskeleton and “gel-like” cytoplasm-derived content, making exosomes integral during trafficking and compatible for large quantities of soluble cargos. They are also nontoxic, immunologically inert, and resistant to nucleases in the bloodstream.12–14 Because of these and other unique characteristics, exosomes have been employed in the development of delivery systems for siRNAs, chemotherapeutics, and functional proteins.15,16
Although these intrinsic properties make exosomes excellent candidates as nanocarriers, their nanometer size range and biological complexity limit the further surface modifications and functionalizations, both prerequisites for endowing exosomal nanocarriers with targeting moieties, tracking markers, imaging agents or synergistic drugs. So far, surface modification strategies have mainly included two approaches: genetic engineering and chemical modifications.17,18 Genetic engineering transfects parent cells with encoded proteins of interest to generate exosomes with artificial receptors or other functional proteins.17,19,20 This method is effective in introducing recombinant proteins on exosomal surfaces, however suffering from such intrinsic drawbacks as complicated manipulations and limited range of applicable proteins. Chemical modifications, on the other hand, employed hydrophobic interactions or covalent ligations to introduce chemical ligands or functional molecules onto exosomal surfaces.21–23 However, hydrophobic probes are randomly inserted on exosome surfaces, and thus lack of precision and specificity. And that covalent ligations usually requires harsh conditions for chemical reactions, which could possibly affect the structure and function of exosomes and give rise to safety concerns. Thus, a mild, precise, but versatile method for exosomal surface modification is highly desired.
As an emerging type of immuno-affinity moiety, aptamers have been attractive in the field of cell labeling, cell surface modification, and cell–cell interaction.24–26 Aptamers are single-stranded oligonucleotides that bind to target molecules with great affinity and specificity. Generated via an in vitro selection process, they possess the advantages of easy chemical synthesis, biocompatibility and easy chemical modification with other functional agents.27,28 More importantly, aptamers can be combined with other DNA-based reactions and technologies, like Watson–Crick hybridization, polymerase chain reaction, rolling cycle reaction, and DNA-based nanotechnologies, to achieve a variety of biomedical applications on cell surfaces.29,30 Given the similarity between cell membrane and exosome membrane surfaces, aptamer-based molecular engineering should be useful in exosome surface modifications as well, to fulfill the demands of many biomedical applications.
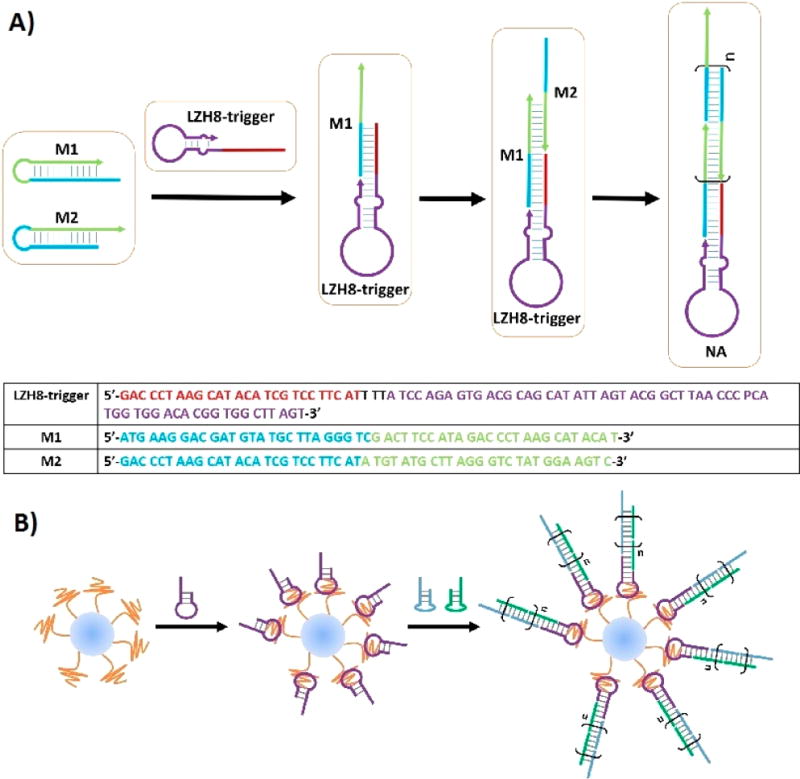
We herein report an aptamer-based nanoassembly (NA) method for exosome surface modifications (Figure 1). The aptamer used in this work was generated from a modified in vitro selection, wherein two artificial nucleotides were added in the initial library to deliver aptamers with better efficiency and affinity.31 The current approach combines the molecular recognition between an aptamer and its molecular target with the hybridization chain reaction (HCR) initiated by the aptamer-chimeric trigger (Figure 1A). Hence, a selective in situ self-assembly is achieved on the surface of exosomes without any complex manipulation or harsh reaction condition. Remarkably, by achieving what was designed, a functional DNA nanostructure is assembled on a nanosized organelle. This unprecedented strategy lays the scientific groundwork for further exosome-related biomedical applications.
Figure 1.
Schematic of aptamer-based DNA nanoassemblies on the surfaces of exosomes. (A) Schematic showing aptamer-chimeric triggered hybridization reaction. Sequences of chimeric aptamer trigger and building monomers are shown in the table. (B) Schematic showing aptamer-triggered in situ NAs on exosome surfaces.
To begin with, cancerous exosomes were isolated following the previously published protocol.32 Briefly, culture medium of HepG2 cell line was collected and centrifuged. The supernatant was filtered with a 0.22 µm filter, and ultracentrifuged to collect the exosomes. Nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM) imaging gave the size distribution of collected exosomes at between 50 and 100 nm (Figure S1, S2), corresponding to the reported size distribution range that distinguishes exosomes from other cell-derived vesicles.2,3
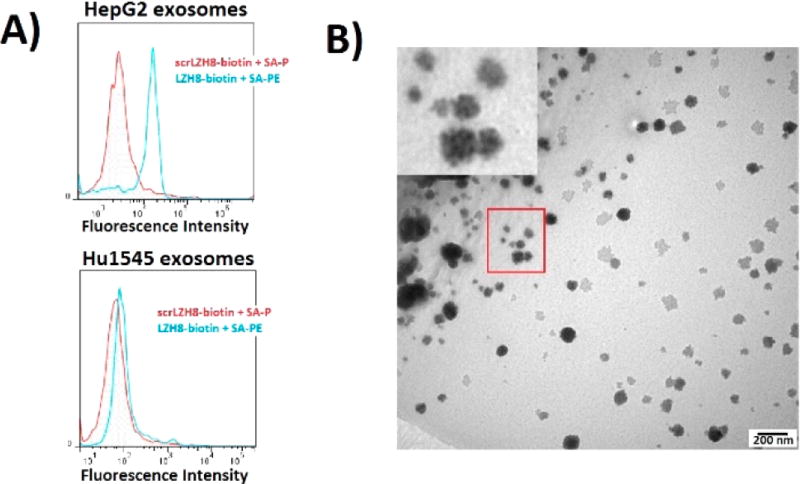
Aptamer binding on exosomes was screened from the artificial nucleotides-contained aptamers previously selected targeting HepG2 cells.31 Biotinylated aptamers were tested using exosomes immobilized on aldehyde/sulfate latex beads, followed by staining with streptavidin-phycoerythrin (SA-PE), and fluorescence determined by flow cytometry (Figure S3). Among aptamers selectively targeting liver cancer cells (HepG2) over non-cancerous liver cells (Hu1545), seven were found to bind HepG2 exosomes (Figure S4). But only aptamer LZH8, containing an expanded nucleotide P (Table S1), demonstrated excellent binding affinity and satisfactory specificity (Figure 2A). An immunogold experiment was further conducted to visualize the binding between LZH8 aptamer and HepG2 exosomes (Figure S5). Biotinylated aptamers were incubated with exosomes, followed by using streptavidin-conjugated gold nanoparticles (SA-AuNP, 10 nm) to further label the exosomes. The TEM image manifested attachment of AuNPs on the exosome surface, demonstrating the binding ability of aptamer LZH8 to HepG2 exosomes (Figure 2B). Notably, several aptamers showed excellent selectivity on HepG2 cells over Hu1545 cells,31 but they were not selective between these two types of exosomes. This indicates that the expression levels of these target proteins on cell surfaces differ from those on exosome surfaces. Although further identification of these proteins is outside the scope of the present paper, it is an ongoing subject of interest in our lab.
Figure 2.
Aptamer with expanded nucleotide toward liver cancer cells recognizing liver cancer cell-derived exosomes. (A) Flow cytometry analysis of fluorescent labeling on immobilized exosomes. LZH8-biotin was used to label exosomes derived from HepG2 hepatocellular carcinoma cells (top) and Hu1545 immortalized liver cells (bottom), stained with SA-PE. Scrambled LZH8 (scrLZH8) was used as a negative control. (B) Transmission electron microscopy (TEM) imaging of immunogold labeling of exosomes using aptamer.
To build the proposed NA, the aptamer-chimeric trigger, or LZH8-trigger, was designed and tested its ability to trigger HCR when monomers (M1 and M2) are well designed (Figure 1A). Two annealed hairpin-structured monomers were inactive in buffer solution in the absence of the trigger, as each intermolecular complementary sequence was blocked by forming intracellular hairpins. When LZH8-trigger introduced, HCR was initiated that M1 was opened and hybridized by LZH8-trigger, followed by iteratively opening and hybridizing M2 and M1 monomers to form an elongated nanostructure. An agarose gel confirmation shows that LZH8-NA migrated much slower than single or mixed M1 and M2, and left a bright product signal band at the very top of the gel, indicating that a long and stable LZH8-NA had been formed in buffer solution after overnight incubation (Figure S6). The LZH8-NA channel containing bands with different sizes shows that different degrees of polymerization formed different lengths of the NAs.
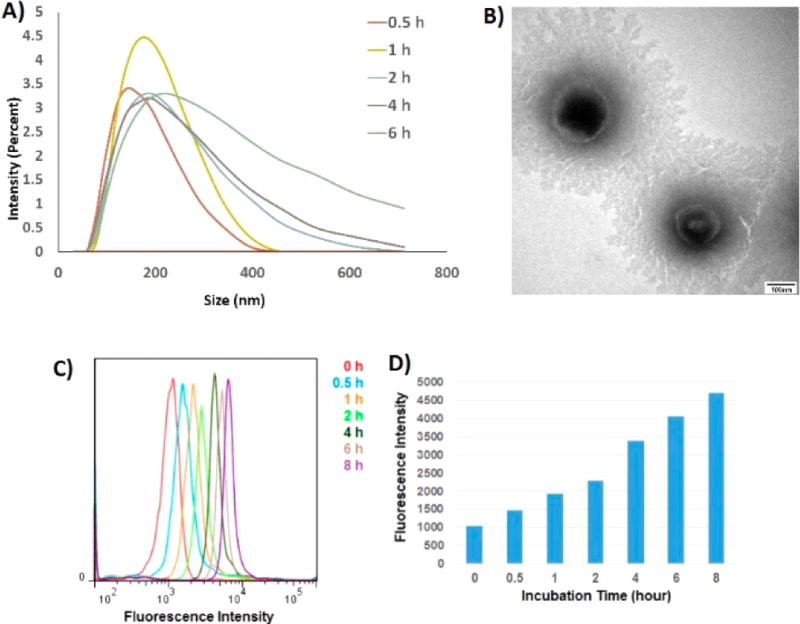
We then investigated if a NA could be built in situ on exosomal surfaces (Figure 1B). The LZH8-trigger was first incubated with exosomes to allow aptamer recognizing and binding on exosome surface targets. M1 and M2 monomers were then introduced to self-assemble the LZH8-NA. The time-dependent formation of LZH8-NA was monitored by dynamic light scattering (DLS). Figure 3A shows that the average size of exosome-LZH8-NA became larger, and the size distribution became broader with increasing incubation time. This result indicated that the lengths of LZH8-NA increased with time, forming longer, yet stable, NAs, while the different degrees of polymerization gave broader distribution range. TEM imaging shows similar results that overnight modified exosomes have sizes in the hundreds of nanometer scale (Figure 3B). Combining all these evidence, it was clearly proved that in situ assembly of LZH8-NA could be achieved on exosomal surfaces.
Figure 3.
Time-course of in situ nanoassembly on membrane surface of exosomes. (A) DLS measuring size distributions of exosomes upon different times of NA formation. (B) TEM images showing exosomes with NA built overnight. (C) Flow cytometry monitoring fluorescence on exosomes upon different times of fluorescent NA formation. (D) Bar graph showing mean fluorescence intensity of FITC on exosomes versus different fluorescent NA formation times.
Next, we determined if NAs could be fluorescently functionalized delete on exosomal surfaces. To this end, fluorescein (FITC) was labeled on LZH8-trigger and two monomers as fluorescent reporters. 1011 exosomes were reacted with 10 µL aldehyde/sulfate latex beads (40 mg/mL) to immobilize exosomes. The time-course of fluorescence augmentation was investigated by flow cytometry (Figure S7). With different formation times, from 0.5 to 8 h, fluorescence intensity kept increasing, with nearly 5-fold increase, when comparing the fluorescence signal of 8 h with that of the beginning (Figure 3C,D). It was also found that fluorescence increased steadily under 4 °C, whereas diminished as formation time increased under 37 °C (Figure S8). This can be explained by the fact that exosomes under 37 °C are not as stable as those under 4 °C, given longer formation time.33
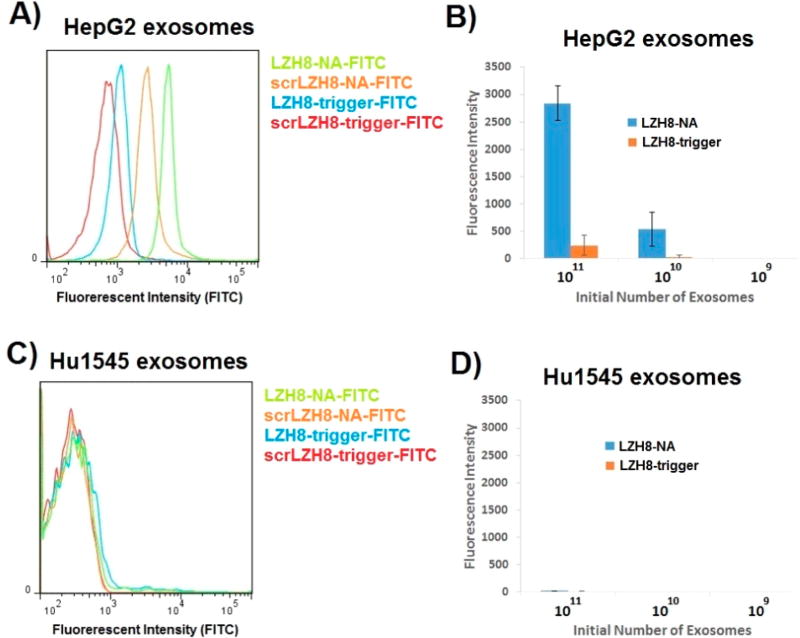
One advantage of using this method for exosome surface modification is that the aptamer-triggered NA is selectively assembled on target exosomes, but not on the nontarget exosomes, and this directly results from specific molecular recognition between aptamer and its target protein. To verify this point, we tested the fluorescent NA for fluorescence enhancement on surfaces of target HepG2 exosomes and nontarget Hu1545 exosomes (Figure 4A–D). 1011 exosomes of each type were reacted with 10 µL, 40 mg/mL aldehyde/sulfate latex beads for immobilization. The resultant immobilized exosomes were used to interact with LZH8-trigger, or scrLZH8-trigger to give the background, followed by NA assembly. This experiment showed that the fluorescence was increased 10-fold when LZH8-NA-FITC was assembled on HepG2 exosome surfaces, compared with that of LZH8-trigger-FITC. Aldehyde/sulfate latex beads immobilized with 1010 and 109 exosomes were also tested to explore the lower limit. Decreased fluorescence by nearly 5-fold enhancement, compared with LZH8-trigger-FITC, was detected from beads with 1010 initiating HepG2 exosomes. But no fluorescence enhancement was observed on beads with 109 initiating exosomes, showing the lower limit of detection. On the other hand, on Hu1545 exosomes, neither LZH8-NA nor LZH8-trigger gave any fluorescence for any of the groups. These results showed the selectivity of this strategy.
Figure 4.
Selectivity of in situ fluorescent NA on membrane surface of exosomes and fluorescence enhancement on target exosomes. (A) Comparing fluorescence of LZH8-trigger-FITC and LZH8-NA-FITC upon binding on immobilized HepG2 exosome. (B) Comparing fluorescence of LZH8-trigger-FITC and LZH8-NA-FITC binding on HepG2 exosome with different initial amounts of exosomes immobilized on aldehyde/sulfate latex beads. (C) Comparing fluorescence of LZH8-trigger-FITC and LZH8-NA-FITC upon binding on immobilized Hu1545 exosome. (D) Comparing fluorescence of LZH8-trigger-FITC and LZH8-NA-FITC binding on Hu1545 exosome with different initial amounts of exosomes immobilized on aldehyde/sulfate latex beads.
To study further the selectivity of this fluorescent NA, exosomes were collected from another eight different types of cell lines, and 1011 exosomes of each type were used for immobilization and fluorescence investigation (Figure S9). Exosomes of HepG2 and Hep3B liver cancer cells provided similar fluorescence enhancement by introducing LZH8-NA-FITC, showing that these two liver cancer cells share the same target protein of LZH8. It was interesting to observe that a prostate cancer cell line, PC3, showed measurable fluorescence only upon the introduction of LZH8-NA-FITC. This demonstrates the strength of this method for detecting proteins at lower expression level. For other types of exosomes, LZH8-NA-FITC reported no detectable fluorescence enhancement.
To sum up, we have successfully developed a novel method of in situ selective DNA nanoassembly on membrane surfaces of nanosized exosomes. Other than fluorescent NAs demonstrated in this work, this strategy could also be used to build other NAs carrying various reporters and functionalities, such as tracking markers, imaging agents or therapeutics, taking advantage of properties of DNA molecules. These NAs showed potential for the functionalization of exosomes, thus extending applications of exosome-based disease theranostics.
Supplementary Material
Acknowledgments
The authors thank Dr. Kathryn R. Williams for manuscript review. The authors thank Dr. Chen Liu’s lab at Rutgers University for providing the Hu1545 cell lines, and Dr. Steven Benner’s lab at Foundation for Applied Molecular Evolution for providing artificial nucleotide phosphoramidites. We are indebted to the National Institutes of Health (GM079359, and CA133086), NSFC grants (21505032, 21325520 and 1327009).
Footnotes
ASSOCIATED CONTENT
The authors declare no competing financial interest.
References
- 1.Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, Laszlo V, Pallinger E, Pap E, Kittel A, Nagy G, Falus A, Buzas EI. Cell. Mol. Life Sci. 2011;68:2667. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Nat. Cell Biol. 2007;9:654. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 3.Fevrier B, Raposo G. Curr. Opin. Cell Biol. 2004;16:415. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Kooijmans SAA, Vader P, van Dommelen SM, van Solinge WW, Schiffelers RM. Int. J. Nanomed. 2012;7:1525. doi: 10.2147/IJN.S29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pisitkun T, Shen RF, Knepper MA. Proc. Natl. Acad. Sci. U. S. A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christianson HC, Svensson KJ, van Kuppevelt TH, Li JP, Belting M. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17380. doi: 10.1073/pnas.1304266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson RJ, Lim JWE, Moritz RL, Mathivanan S. Expert Rev. Proteomics. 2009;6:267. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 8.Tkach M, Thery C. Cell. 2016;164:1226. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 9.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, Mason MD, Clayton A. Mol. Cell. Proteomics. 2010;9:1324. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hood JL. Nanomedicine. 2016;11:1745. doi: 10.2217/nnm-2016-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakase I, Ueno N, Katayama M, Noguchi K, Takatani-Nakase T, Kobayashi NB, Yoshida T, Fujii I, Futaki S. Chem. Commun. 2017;53:317–320. doi: 10.1039/c6cc06719k. [DOI] [PubMed] [Google Scholar]
- 12.Azmi AS, Bao B, Sarkar FH. Cancer Metastasis Rev. 2013;32:623. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Andaloussi S, Lee Y, Lakhal-Littleton S, Li JH, Seow Y, Gardiner C, Alvarez-Erviti L, Sargent IL, Wood MJA. Nat. Protoc. 2012;7:2112. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 14.Lai RC, Yeo RWY, Tan KH, Lim SK. Biotechnol. Adv. 2013;31:543. doi: 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Nat. Biotechnol. 2011;29:341. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 16.Tian YH, Li SP, Song J, Ji TJ, Zhu MT, Anderson GJ, Wei JY, Nie GJ. Biomaterials. 2014;35:2383. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 17.Lai CP, Mardini O, Ericsson M, Prabhakar S, Maguire CA, Chen JW, Tannous BA, Breakefield XO. ACS Nano. 2014;8:483. doi: 10.1021/nn404945r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smyth T, Petrova K, Payton NM, Persaud I, Redzic JS, Graner MW, Smith-Jones P, Anchordoquy T. Bioconjugate Chem. 2014;25:1777. doi: 10.1021/bc500291r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morishita M, Takahashi Y, Nishikawa M, Sano K, Kato K, Yamashita T, Imai T, Saji H, Takakura Y. J. Pharm. Sci. 2015;104:705. doi: 10.1002/jps.24251. [DOI] [PubMed] [Google Scholar]
- 20.Morishita M, Takahashi Y, Matsumoto A, Nishikawa M, Takakura Y. Biomaterials. 2016;111:55. doi: 10.1016/j.biomaterials.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 21.Wang M, Altinoglu S, Takeda YS, Xu QB. PLoS One. 2015;10:e0141860. doi: 10.1371/journal.pone.0141860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakase I, Noguchi K, Fujii I, Futaki S. Sci. Rep. 2016;6:34937. doi: 10.1038/srep34937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cogoi S, Jakobsen U, Pedersen EB, Vogel S, Xodo LE. Sci. Rep. 2016;6:38468. doi: 10.1038/srep38468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu GZ, Zhang SF, Song EQ, Zheng J, Hu R, Fang XH, Tan WH. Angew. Chem., Int. Ed. 2013;52:5490. doi: 10.1002/anie.201301439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong X, Liu H, Zhao Z, Altman MB, Lopez-Colon D, Yang CJ, Chang LJ, Liu C, Tan W. Angew. Chem., Int. Ed. 2013;52:1472. doi: 10.1002/anie.201207063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan W, Donovan MJ, Jiang J. Chem. Rev. 2013;113:2842. doi: 10.1021/cr300468w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szeitner Z, Lautner G, Nagy SK, Gyurcsanyi RE, Meszaros T. Chem. Commun. 2014;50:6801. doi: 10.1039/c4cc00447g. [DOI] [PubMed] [Google Scholar]
- 28.Liang H, Zhang XB, Lv Y, Gong L, Wang R, Zhu X, Yang R, Tan W. Acc. Chem. Res. 2014;47:1891. doi: 10.1021/ar500078f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C, Wan S, Hou W, Zhang L, Xu J, Cui C, Wang Y, Hu J, Tan W. Chem. Commun. 2015;51:3723. doi: 10.1039/c4cc10047f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cansiz S, Zhang L, Wu C, Wu Y, Teng IT, Hou W, Wang Y, Wan S, Cai R, Jin C, Liu Q, Tan W. Chem. - Asian J. 2015;10:2084. doi: 10.1002/asia.201500434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Yang Z, Sefah K, Bradley KM, Hoshika S, Kim MJ, Kim HJ, Zhu G, Jimenez E, Cansiz S, Teng IT, Champanhac C, McLendon C, Liu C, Zhang W, Gerloff DL, Huang Z, Tan W, Benner SA. J. Am. Chem. Soc. 2015;137:6734. doi: 10.1021/jacs.5b02251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Nature. 2015;523:177. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokolova V, Ludwig AK, Hornung S, Rotan O, Horn PA, Epple M, Giebel B. Colloids Surf., B. 2011;87:146. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.