Abstract
Hydrolytically degrading nano-polyplexes (HDG-NPs) that reverse charge through conversion of tertiary amines to carboxylic acids were investigated to improve intracellular un-packaging of siRNA and target gene silencing compared to a non-degradable analog (non-HDG-NPs). Both NP types comprised reversible addition-fragmentation chaintransfer (RAFT) synthesized diblock copolymers of a poly(ethylene glycol) (PEG) corona-forming block and a cationic block for nucleic acid packaging that incorporated butyl methacrylate (BMA) and either dimethylaminoethyl methacrylate (DMAEMA, non-HDG-NPs) or dimethylaminoethyl acrylate (DMAEA, HDG-NPs). HDG-NPs decreased significantly in size and released significantly more siRNA (~40%) than non-HDG-NPs after 24 h in aqueous solution. While both HDG-NPs and non-HDG-NPs had comparable uptake and cytotoxicity up to 150 nM siRNA doses, HDG-NPs achieved significantly higher target gene silencing of the model gene luciferase in vitro. High resolution FRET confocal microscopy was used to monitor the intracellular un-packaging of siRNA. Non-HDG-NPs had significantly higher FRET efficiency than HDG-NPs, indicating that siRNA delivered from HDG-NPs was more fully un-packaged and therefore had improved intracellular bioavailability.
Keywords: siRNA, polyplexes, RAFT, hydrolytically degrading, charge-reversal, FRET microscopy
INTRODUCTION
Small interfering RNA (siRNA) has emerged as a powerful research tool and potentially transformative clinical therapeutic.1 Due to its mode of action at the mRNA level, siRNA is a promising class of therapeutics for inhibiting targets considered to be hard-to-drug by conventional pharmacological approaches, such as intracellular enzymes, transcription factors, and protein–protein interactions.2,3 However, systemic and cellular delivery barriers currently limit the application of siRNAs as therapeutics.4 After systemic administration, siRNA molecules are rapidly cleared through filtration in the kidneys and do not have inherent properties that disposes them to distribute to tumors or other potential target sites.5,6 At the cellular level, siRNAs are unable to traverse the cellular membrane due to their size and negative charge and lack an inherent mechanism for endosomal escape.6,7 Thus, effective clinical use of siRNA is contingent upon the ability to safely and effectively deliver these molecules to the target tissue, cell type, and intracellular compartment of action.
The conventional approach for delivering siRNA is packaging with cationic polymers or lipids into nanosized polyplexes.8 The nanocarriers are typically formulated with an excess of cationic charge which enhances siRNA cellular uptake through interaction with the anionic cellular membrane.9 Strictly cationic lipoplexes and polyplexes can be effective as in vitro transfection reagents, but their highly positive zeta-potential and lack of stability preclude their application for systemic administration.10,11 Therefore, surface PEGylation has been widely employed as a strategy to shield the positive charge of siRNA polyplexes, which consequently reduces opsonization and increases stealth from the mononuclear phagocyte system (MPS) in vivo.12–14 For example, first-in-man class clinical trials have tested a cationic, PEGylated siRNA delivery vehicle for systemic administration.15–17
While PEGylated cationic polyplexes are feasible for in vivo systemic administration, they suffer from a lack of stability in vivo. This class of polyplexes, formed by electrostatic interaction, are disassembled at the glomerular basement membrane (GBM) and cleared primarily through the kidneys, resulting in only modest increases in circulation time and ability to distribute to target tissues relative to free siRNA.18,19 In previous work, we endeavored to improve the performance of cationic polyplexes through the incorporation of hydrophobicity into the polyplex core. In this study, we found that, relative to polyplexes formed with a purely cationic core, siRNA polyplexes formulated from diblock polymers of PEG and a copolymer block with a balanced composition of the cationic monomer dimethylaminoethyl methacrylate (DMAEMA) and the hydrophobic monomer butyl methacrylate (BMA) increased polyplex in vivo circulation time and also improved intracellular delivery by pH-dependent membrane disruption leading to enhanced endosomal escape.20
The current work builds upon our previous report by introducing a hydrolytic charge-reversing mechanism for increased un-packaging and bioavailability of siRNA within the cytosol (Scheme 1). Ideally, in vivo-ready polyplexes will possess characteristics which increase blood circulation time and stability (PEGylation and hydrophobicity). However, these characteristics can decrease gene silencing potency; PEGylation reduces cell uptake and core stabilization potentially hinders intracellular siRNA bioavailability. We have previously shown that reversible PEGylation promotes siRNA uptake and activity.21,22 To better understand the barrier of intracellular release for PEGylated polyplexes with balanced core hydrophobicity, the current project tested the impact of incorporating a hydrolytically degradable, charge-reversing cationic monomer and comparing the bioactivity and intracellular siRNA release relative to an analogous system made with a stable cationic monomer. Stimuli-responsive cues such as pH and ROS have been investigated for enhancing gene and drug-delivery potency.23–26 Specifically, charge-reversal has been investigated as a means to increase pDNA transfection27 and siRNA potency from polyplexes.28,29 Moreover, dimethylaminoethyl acrylate (DMAEA) has been utilized by Monteiro and coworkers for the timed release of siRNA from micelleplexes.28,30 Our report builds upon this work by utilizing hydrolytic charge-reversal in PEGylated polyplexes which have been optimized for systemic delivery in vivo, and this delivery system also effectively couples these functionalities with an active endosomal escape mechanism. Importantly, increased siRNA release from a charge-reversing carrier is measured in living cells using high resolution Förster Resonance Energy Transfer (FRET) confocal microscopy.
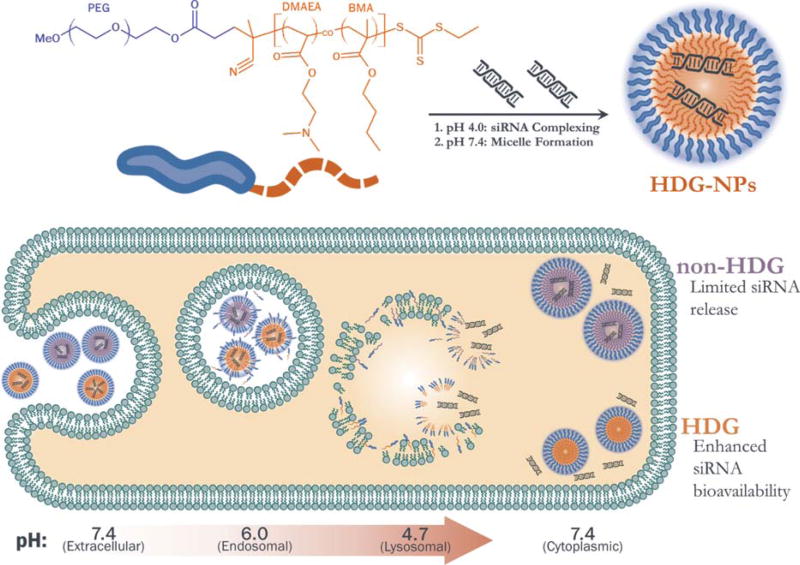
SCHEME 1.
Top: Polymer chemistry and siRNA loading protocol. Bottom: Hypothesis for enhanced intracellular release and activity of siRNA delivered by HDG-NPs.
The hydrolytically degrading nano-polyplexes (HDG-NPs) were developed by replacing DMAEMA as the cationic monomer of non-hydrolytically degrading nano-polyplexes (non-HDG-NPs) with DMAEA. Like DMAEMA, DMAEA contains a tertiary amine that enables siRNA complexing and pH-dependent membrane disruption. However, the lack of a pendant methyl group off of the polymer backbone of DMAEA-containing polymers increases the rate of hydrolysis of the ester group, leaving behind a residual negatively-charged acrylic acid. Specifically, the increased hydrophilicity of the acrylate-based DMAEA monomer allows increased interaction of its ester with nucleophilic hydroxyls and leads to increased hydrolysis rates.31,32 Therefore, we hypothesized that the incorporation of DMAEA would enable a polymer charge-reversal-based siRNA release mechanism that would increase cytosolic availability and therefore, activity of siRNA delivered by PEGylated polyplexes with core chemistry comprising a balance of cationic and hydrophobic monomers. To test this hypothesis, we herein have compared the new HDG-NPs to the benchmark non-HDG-NPs for release kinetics in solution, cell uptake and viability, and target gene silencing. Finally, FRET confocal microscopy techniques were used to monitor the intracellular trafficking of nucleic acids delivered from both NPs.
EXPERIMENTAL METHODS
Materials
All chemicals were obtained from Sigma-Aldrich and used as received unless otherwise noted. DMAEMA, DMAEA, and BMA monomers were passed twice through an alumina column to remove inhibitors prior to polymerization. 2,2-Azobis(2-methylpropionitrile) (AIBN) was recrystallized twice from methanol prior to use. Dioxane was distilled under vacuum prior to use. Luciferase and scrambled siRNA sequences were designed and purchased through IDT. Fluorophore labeled dsDNAs were purchased through IDT and Sigma-Aldrich. Lipofectamine 2000 (LF2K) was obtained through Invitrogen. 21-mer dsDNA was used in DLS and fluorescent studies as a model of siRNA.
Synthesis of 4-cyano-4-(ethylsulfanylthiocarbonyl)sulfanylpentanoic acid
4-Cyano-4-(ethylsulfanylthiocarbonyl)sulfanylpentanoic acid (ECT) was synthesized according to previous protocols described by Convertine et al.33 and Moad et al.34 Briefly, ethanethiol (4.72 g, 76 mmol) was reacted with carbon disulfide (6.00 g, 79 mmol) in the presence of sodium hydride (3.15 g, 79 mmol) in diethyl ether for 1 h. The resulting sodium S-ethyl trithiocarbonate was further reacted with iodine (6.3 g, 25 mmol) to obtain bis(ethylsulfanylthiocarbonyl) disulfide, which was then refluxed with 4,4-azobis(4-cyanopentanoic acid) in ethyl acetate for 18 h to yield ECT. The crude ECT was purified by column chromatography using silica gel as the stationary phase and ethyl acetate:hexane (50:50) as the mobile phase. 1H-NMR (400 MHz, CDCl3): 3.35 (q, 2H, S—CH2—CH3), 2.69 (t, 2H, —CH2—CH2—COOH), 2.36–2.58 (m, 2H, —CH2—CH2—CH3), 1.88 (s, 3H, CH3—C—CN), 1.36 (t, 3H, S—CH2—CH3).
PEG RAFT macro-chain transfer agent generation and polymer synthesis
The chain transfer agent (CTA) ECT was conjugated to hydroxyl-functional PEG by a carbodiimide coupling strategy.35 Briefly, dicyclohexylcarbodiimide (4 mmol, 0.82 g) was added to the stirring solution of methoxy-poly(ethylene glycol)-hydroxyl (Mn = 5000, 2 mmol, 10 g), ECT (4 mmol, 1.045 g), and DMAP (10 mg) in 50 mL of dichloromethane. The reaction mixture was stirred for 48 h. The precipitated cyclohexyl urea was removed by filtration, and the dichloromethane layer was concentrated and precipitated into diethyl ether twice. The precipitated PEG-ECT was washed three times with diethyl ether and dried under vacuum (yield ~10 g). RAFT polymerization was then used to synthesize diblock copolymers from the PEG-ECT macroCTA. Either DMAEMA (0.236 g, 1.5 mmol) or DMAEA (0.415 g, 2.9 mmol) and BMA (0.213 g, 1.5 mmol or 0.333 g, 2.34 mmol) were added to separate solutions of PEG-ECT (100 mg, 0.02 mmol) in 3 mL dioxane. The solutions were purged with N2 for 30 min and then reacted at 70°C for 24 h using AIBN as an initiator at a 10:1 [CTA]:[Initiator] molar ratio. The reactions were quenched by exposure to air, and the resulting polymers were precipitated thrice into a mixture of pentane:diethyl ether (90:10) and vacuum dried. The reactivity ratios for the BMA and DMAEA monomers were determined using the Fineman-Ross method after polymerization under the conditions described above at three molar feed ratios [DMAEA:BMA]: [40:60], [50:50], and [60:40].
Polymer characterization
Polymers were characterized for composition and molecular weight by 1H-NMR (NMR, Bruker 400 MHz spectrometer equipped with a 9.4 T Oxford Magnet). Polydispersity index (PDI) of the polymers was determined using gel permeation chromatography (GPC) on a system running DMF + 0.1M LiBr as the mobile phase (Agilent Technologies, Santa Clara, CA) with inline Agilent refractive index and Wyatt mini-DAWN TREOS light scattering detectors (Wyatt Technology Corp., Santa Barbara, CA).
Polyplex preparation
For dsDNA- and siRNA-loaded polyplexes, the non-HDG and HDG polymers were initially dissolved in 100% ethanol (33.3 mg/mL), then diluted 10× with pH 4.0 citrate buffer (10 mM). Next, dsDNA/siRNA (50 µM, H2O) was added to the polymers at pH 4.0 at an N:P ratio of 5:1. After 30 min, 5× pH 8.0 phosphate buffer (10 mM) was used to raise the pH to ~7.4.
Hydrolysis of HDG-NPs
1H-NMR of HDG polymer hydrolysis
Both non-HDG and HDG micelles were initially dissolved in 100% ethanol (10 mg/mL), then diluted 10× with pH 7.4 PBS to give 1 mg/mL solutions of micelles. These solutions were frozen and lyophilized either immediately or after 24 and 48 h of incubation in PBS at 37°C. The lyophilized powder was then re-dissolved in CDCl3 at 10 mg/mL and analyzed by 1H-NMR (NMR, Bruker 400 MHz spectrometer equipped with a 9.4 T Oxford Magnet). The percent hydrolysis of each polymer was calculated by comparing the ratios of DMAEA/ DMAEMA (— O —CH2CH2—, δ 4.05s) to PEG (—OCH2CH2—, δ 3.65s) and normalizing to t = 0 h.
Dynamic light scattering
To make empty micelles, the non-HDG and HDG polymers were initially dissolved in 100% ethanol (10 mg/mL), then diluted 10× with pH 7.4 PBS to yield 1 mg/mL solutions. For dsDNA-loaded polyplexes, the non-HDG and HDG polymers were initially dissolved in 100% ethanol (33.3 mg/mL), then diluted 10× with pH 4.0 citrate buffer (10 mM). Next, dsDNA (50 µM, H2O) was added to the polymers at pH 4.0 at an N:P ratio of 5:1. After 30 min, 5× pH 8.0 phosphate buffer (10 mM) was used to raise the pH to ~7.4. The size of both loaded polyplexes and empty micelles was monitored by dynamic light scattering (DLS; Malvern Zetasizer Nano ZS, Malvern, UK) at 0 h and 24 h of incubation in aqueous solution.
Kinetics of dsDNA release
FRET monitoring of dsDNA release
The hydrolytically dependent release of dsDNA from NPs in aqueous solution was monitored by FRET-based measurements. FRET can be used to measure nucleic acid loading and release of FRET-labeled oligonucleotides, where a reduction in FRET efficiency over time is indicative of siRNA release.20,36 Two dsDNA molecules were co-loaded into polyplexes as described above. The two molecules were labeled with Alexa-488 and Cy5. These dsDNAs were each added at 50 mol % creating a FRET signal (Alexa-488 donor, Cy5 receptor) when concentrated in close proximity to one another within polyplexes. The FRET signal is not present when the molecules are simply co-dissolved in solution, and this approach therefore provides an optical readout of polyplex loading and release. Fluorescence intensity was measured at each time point using a plate reader (Tecan) at an excitation wavelength of 488 ± 5 nm. Alexa-488 (donor) emission was collected at 520 ± 5 nm, and Cy5 (acceptor) emission was collected at 670 ± 5 nm. The release of dsDNA was quantified as loss of %FRET signal which was calculated as follows:
Hemolysis assay
Whole blood was extracted from anonymous, consenting human donors, and red blood cells (RBCs) were isolated according to well-established protocols.37 RBCs were then incubated with the HDG- and non-HDG-NPs (concentration of 5 µg/mL) in buffers of pH 7.4, 6.8, and 6.2, which model the environments in the extracellular space and in the more acidic vesicles of the endolysosomal pathway. After 1 h of incubation, the RBCs were centrifuged and the supernatant was spectrophotometrically analyzed at 451 nm (Tecan Plate Reader) in order to determine percent hemolysis relative to 1% Triton X-100 detergent.
Cell culture
Human epithelial breast cancer cells (MDA-MB-231) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco Cell Culture, Carlsbad, CA) with 10% fetal bovine serum (FBS, Gibco) and 50 µg/mL gentamicin (Gibco). Some of the MDA-MB-231s used were lentivirally transduced to constitutively express firefly luciferase and green fluorescent protein (GFP). Luciferase-expressing MDA-MB-231s (L231) were used to assess cell viability and for screening efficiency of carrier delivery of luciferase siRNA.
Analysis of intracellular delivery by flow cytometry
MDA-MB-231 breast cancer cells were seeded in 24-well plates at 30,000 cells/well and allowed to adhere overnight. The cells were treated with polyplexes loaded with Alexa-488-labeled dsDNA at a final concentration of 100, 200, and 300 nM in DMEM supplemented with 10% FBS and 50 µg/ mL gentamicin. After 24 h, cells were washed with PBS and trypsinized. Trypsin was inactivated by adding serum containing media, and cells were centrifuged and resuspended in PBS containing 0.04% Trypan Blue to quench extracellular fluorescence. Relative cell fluorescence was quantified via flow cytometry to measure intracellular nucleic acid delivery (FACS Calibur, BD Biosciences, Franklin Lakes, NJ).
Cell viability
Viability of cells treated with polyplexes was assessed by measuring relative cell number based on luciferase activity. Luciferase-expressing MDA-MB-231 (L231) breast cancer cells were seeded at 2,000 cells/well in black, clear bottom 96-well plates and allowed to adhere overnight. Polyplexes containing a scrambled siRNA sequence were added to cells at 2:1, 5:1, 10:1, and 20:1 N:P ratios and final concentrations of 50, 100, 150, and 200 nM in DMEM supplemented with 10% FBS and 50 µg/mL gentamicin. After 24-h incubation, the cells were washed, and the media was replaced with luciferin-containing DMEM (150 µg/mL). Bioluminescence was quantified using a Xenogen Lumina III series IVIS (Caliper Sciences) to determine relative cell number compared to no treatment.
Target gene silencing
Luciferase-expressing L231 cells were seeded in black, clear bottom 96-well plates at 2,000 cells/well and allowed to adhere overnight. Cells were treated with polyplexes containing either anti-luciferase siRNA (100 and 150 nM) or a scrambled sequence (100 and 150 nM) as a control in DMEM supplemented with 10% FBS and 50 µg/mL gentamicin. After 24 h, media was replaced with luciferin-containing DMEM (150 µg/mL), and bioluminescence was quantified using a Xenogen Lumina III series IVIS (Caliper Sciences). In all cases, bioluminescent signal of treatment samples was normalized to the corresponding scrambled control to determine percent luciferase activity (n=5).
Analysis of intracellular oligonucleotide un-packaging by FRET confocal microscopy
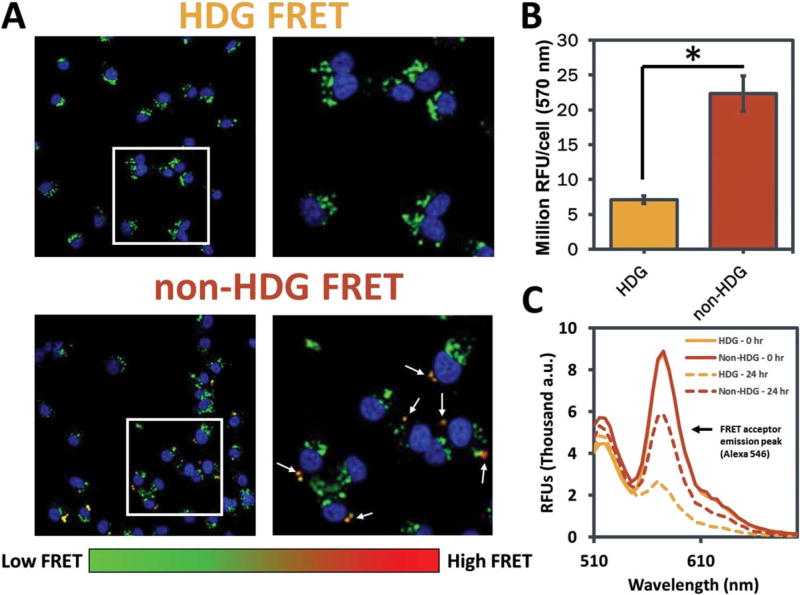
Intracellular un-packaging of dsDNA was monitored in live cells by FRET microscopy methods recently established.38 Briefly, MDA-MB-231 breast cancer cells were seeded in fibronectin-coated (50 µg/mL) 8-well chamber slides (Nunc, Thermo Fisher Scientific Inc., Waltham, MA) at a density of 10,000 cells/well. The cells were treated with polyplexes containing a 50:50 mix of Alexa-488- and Alexa-546-labeled dsDNA as a FRET pair at a total concentration of 100 nM in DMEM supplemented with 10% FBS and 50 µg/mL gentamicin. After 24 h, the cells were washed with PBS, and media was replaced with phenol red-free DMEM (10% FBS, 50 µg/mL gentamicin) containing DAPI nuclear stain. After 1-h incubation, cells were imaged using a Nikon C1si confocal microscope system (Nikon Instruments, Melville, NY) equipped with differential interference contrast transmitted light detector. A two-step imaging mode was performed; in the first pass, a 405 laser was used to acquire channel 1 (DAPI); in the second pass, the 488 laser was used for excitation while channels 2 and 3 were simultaneously acquired (Alexa 488/Alexa 546). The optical system was equipped with the 405/488/543 dichroic mirror, the 450/35 filter cube in the first position (Ch1), and the 515/30, 605/75 filter cube in the second position (Ch2/Ch3). Sensitized emission in the red channel (i.e., the increased fluorescence emission by the acceptor fluorophore due to FRET) was analyzed in a blinded, high throughput, whole-image analysis method using MATLAB (MathWorks, Natick, MA) code that calculated the average per cell sensitized emission fluorescence intensity by summing Ch3 and normalizing to cell number (algorithmically counted using DAPI staining). These data representing >9000 cells are presented in Figure 5(B).
FIGURE 5.
Analysis of intracellular un-packaging of oligonucleotides delivered from non-HDG- and HDG-NPs by FRET microscopy. A: Representative microscopy images of MDA-MB-231s after delivery of FRET-labeled dsDNA from non-HDG- and HDG-NPs. FRET signal is detected consistently within cells treated with non-HDG-NPs, whereas minimal FRET is detected within HDG-NP-treated cells. All images are presented at the same intensity scale. B: Microscopy image quantification of emission intensity at 570 nm (FRET acceptor emission; n=4, 9 images/n,>9000 cells total). C: Fluorescence spectra of FRET-labeled NPs at 0 and 24 h incubation in PBS.
Statistical analysis
All measurements are presented as mean ± standard error of the mean. One Way ANOVA coupled with post hoc Tukey means comparison test was used to determine statistical significance, and p<0.05 was considered significant.
RESULTS
Synthesis of non-HDG and HDG diblock copolymers
A previously reported RAFT synthesis scheme was used to synthesize two diblock copolymers (Supporting Information Fig. S2).20 In both cases, a carbodiimide coupling strategy using DCC and DMAP allowed the attachment of ECT to monohydroxyl-functional 5 kDa PEG, generating a PEG-ECT RAFT macroCTA. 1H-NMR (400 MHz CDCL3) revealed 91% substitution of the hydroxyl-PEG to PEG-ECT (data not shown).20 The second polymer block was then synthesized from the macroCTA and resulted in approximately equimolar (50:50) amounts of either DMAEMA and BMA (non-HDG) or DMAEA and BMA (HDG). 1H-NMR (400 MHz CDCL3) analysis of the products showed similar core compositions (non-HDG: 48.3% BMA, HDG: 55.9% BMA), overall degree of polymerizations (non-HDG: 120, HDG: 115), and molecular weights (non-HDG: 23 kDa, HDG: 21 kDa) (Table I and Supporting Information Fig. S3a). Both polymers were relatively monodispersed (non-HDG: 1.04, HDG: 1.35) as determined by GPC (Table I and Supporting Information Fig. S3b). The Fineman-Ross method was used to determine values of rBMA and rDMAEA. The reactivity ratios for the DMAEA/BMA system were rBMA = 1.21 and rDMAEA = 0.81, respectively indicating that there is the potential for compositional shifting, resulting in slightly longer segments of BMA (Supporting Information Fig. S4).
TABLE I.
Summary of the NMR and GPC Polymer Characterization of PEG-b-p(DMAEMA-co-BMA) [non-HDG] and PEG-b-p(DMAEA-co-BMA) [HDG] Diblock Copolymers
| Polymer | DPa | %BMAa | %DMAEMA/ DMAEAa |
Mna (Da) | PDIb |
|---|---|---|---|---|---|
| Non-HDG | 120 | 48.3 | 51.7 | 22,994 | 1.04 |
| HDG | 115 | 55.9 | 44.1 | 21,404 | 1.35 |
Determined by 1H-NMR.
Determined by GPC.
Analysis of HDG-NP hydrolysis
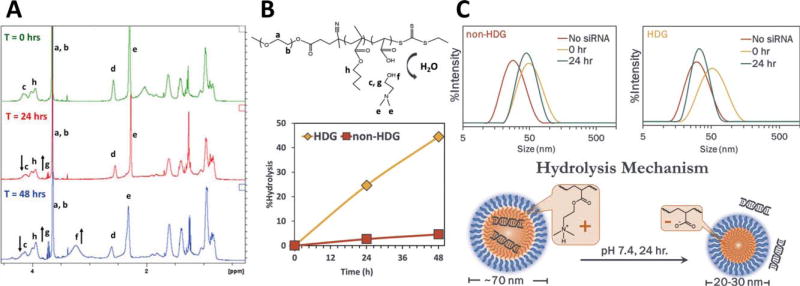
HDG-NPs were designed to hydrolyze in aqueous solutions by the incorporation of an unprotected ester bond adjacent to the polymer backbone. In contrast, the non-HDG polymer contains a pendant methyl group off the backbone, imparting hydrophobicity which reduces access of water and protects the ester from hydrolysis. 1H-NMR analysis confirmed HDG polymer hydrolysis upon incubation in PBS at 37°C. The decrease in signal from protons directly adjacent to the PDMAEA ester (δ 4.05s) and increase in signal from dimethylamino ethanol (DMAE) peaks (δ 3.3s and 3.7s) indicated significant polymer hydrolysis occurred over 24–48 h [Fig. 1(A)]. The extent of hydrolysis was quantified for both non-HDG and HDG polymers, and the HDG polymer showed ~25% and 45% hydrolysis at 24 and 48 h, respectively [Fig. 1(B)]. In contrast, the non-HDG polymer showed ~3% and 5% hydrolysis at 24 and 48 h. DLS measurement of NP size was used as an additional indicator that hydrolysis of the HDG polymer occurred within aqueous solutions [Fig. 1(C)]. For both polymers, the freshly-made dsDNA-loaded formulations were 60–70 nm. To provide an internal control for this experiment, the non-HDG and HDG polymers were assembled (without nucleic acid cargo) into empty micelles in PBS at pH 7.4. Both empty micelles were approximately 25 nm. After 24 h of incubation in aqueous solution, the nucleic acid-loaded HDG-NPs were reduced to the size of empty micelles (~25 nm) while non-HDG-NPs retained their initial size. These data suggest nucleic acid un-packaging from HDG-NPs over time as a result of exposure to an aqueous environment.
FIGURE 1.
Hydrolysis of siRNA-loaded polyplexes in aqueous solution. A: 1H-NMR analysis reveals HDG polymer hydrolysis at 24 and 48 h (arrows indicate either the appearance or disappearance of peaks over time due to hydrolysis). B: Structure of hydrolyzed HDG polymer (top) and quantification of polymer hydrolysis at 24 and 48 h (bottom). C: HDG-NPs (right) reduced in size after 24 h, indicating hydrolytically dependent nucleic acid release.
FRET measure of oligonucleotide release kinetics
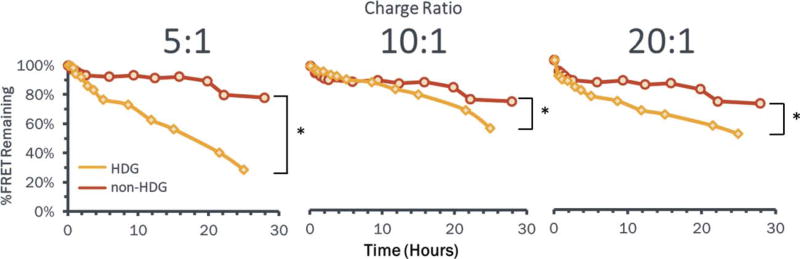
Increased release of dsDNA from HDG-NPs during incubation in aqueous solution was validated using a FRET-based readout (Fig. 2). The effect of hydrolysis of the HDG-NPs was apparent at extended time points based on the disappearance of FRET signal, indicating increased dsDNA release relative to the non-HDG-NPs. At 24 h, HDG-NPs released ~40% more dsDNA than non-HDG-NPs at a 5:1 N:P ratio (which was therefore used in functional in vitro studies). Increasing the N:P ratio partially mitigated the difference in dsDNA release, likely due to the excess of cationic charge present in higher N:P ratio formulations.
FIGURE 2.
HDG-NPs release nucleic acid cargo more efficiently than non-HDG-NPs. FRET signal is used to track nucleic acid release kinetics where loss of FRET is indicative of nucleic acid release. At 5:1 N:P ratio, HDG-NPs released ~40% more dsDNA at 24 h than non-HDG-NPs. Increasing the N:P ratio reduced the impact of the HDG chemistry, likely due to the presence of excess cationic charge.
Comparison of non-HDG- and HDG-NP hemolysis, cell uptake, and viability
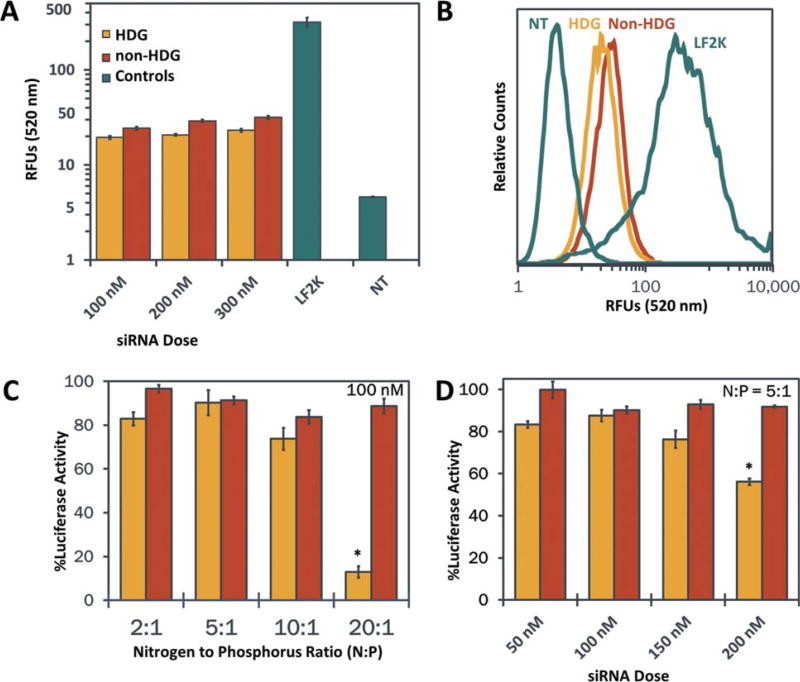
The hemolysis assay allows for the quantitative measure of pH-dependent membrane disruption and indicates the ability of the NPs to achieve endosomal escape.37 HDG- and non-HDG-NPs both exhibited switch-like, pH-dependent membrane disruption between pH 7.4 and 6.8 (Supporting Information Fig. S5). No significant difference in the hemolysis profiles of HDG- and non-HDG-NPs was observed, indicating that endosomal escape would not be a contributing factor to differences in silencing efficiency observed. Cell uptake was assessed in MDA-MB-231 breast cancer cells using flow cytometry to quantify the intracellular delivery of Alexa-488-labeled dsDNA from both non-HDG- and HDG-NPs. Both NPs exhibited cellular internalization of the nucleic acid cargo, with intracellular fluorescence significantly above no treatment [Fig. 3(A,B)]. The non-HDG- and HDG-NPs exhibited comparable levels of cell uptake at each dsDNA dose tested (100, 200, and 300 nM), but uptake in both groups was significantly lower than Lipofectamine 2000 uptake. There was a small dose-dependent increase in cell uptake within each group, but this effect was modest due to the inherently low uptake of this class of PEGylated, surface-shielded polyplexes that have ζ-potential of approximately 0 mV.
FIGURE 3.
Assessment of non-HDG- and HDG-NP cell uptake and cytocompatibility. A: Intracellular delivery of siRNA by polyplexes as quantified by flow cytometry (n=3). B: Flow cytometry histogram of cell uptake at 100 nM siRNA dose. C: Cell viability assessed at varying N:P ratios at 100 nM siRNA concentration (n=4). D: Cell viability assessed at varying siRNA concentrations at N:P ratio of 5:1 (n=4). LF2K=Lipofectamine 2000.
The effect of both polyplexes on cell viability was assessed in luciferase-expressing MDA-MB-231 breast cancer cells across a range of siRNA concentrations as well as N:P ratios. First, siRNA concentration was kept constant at 100 nM and the N:P ratio was varied between 2:1 and 20:1 [Fig. 3(C)]. Cell viability was not significantly different between either group up to 10:1 N:P ratios. At N:P ratio of 20:1, HDG-NPs were significantly more toxic to the cells than non-HDG-NPs. Next, the N:P ratio was held constant at 5:1 (where both polyplexes were most cytocompatible at 100 nM siRNA dose) and the siRNA concentration was varied between 50 and 200 nM [Fig. 3(D)]. There was decreased cytocompatibility of HDG-NPs at 200 nM, but there was no significant difference between non-HDG- and HDG-NPs up to 150 nM siRNA dose.
Comparison of target gene silencing by non-HDG- and HDG-NPs
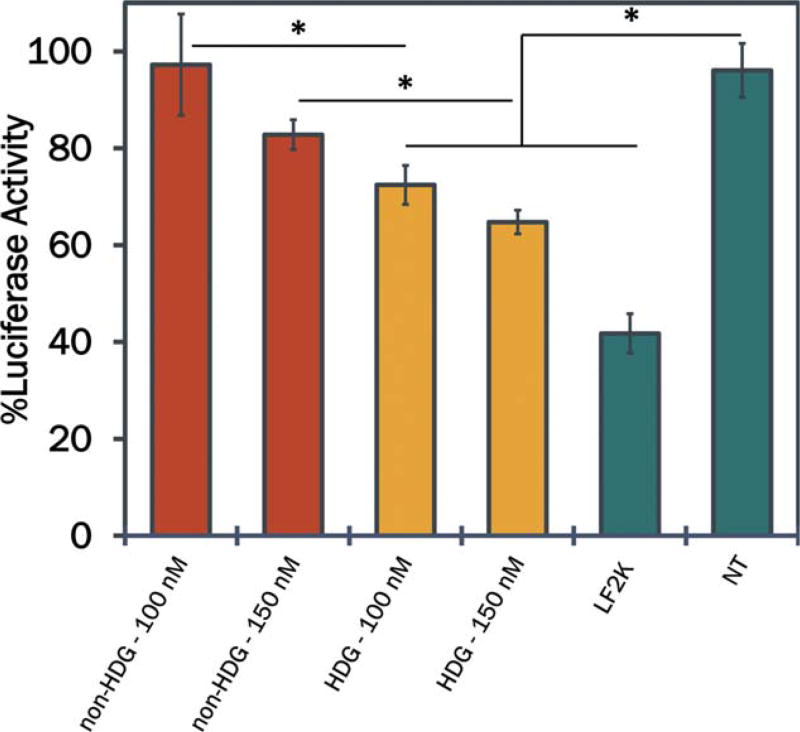
Potential efficacy of HDG-NPs for target gene silencing was assessed in vitro in MDA-MB-231 cells constitutively-expressing luciferase (L231s) as a model gene. Both non-HDG- and HDG-NPs, as well as the commercial transfection agent LF2K, were used to deliver anti-luciferase siRNA through incubation with L231s for 24 h, and gene silencing was determined by quantifying cellular bioluminescence. At each dose investigated, the HDG-NPs achieved significantly greater protein level knockdown of luciferase relative to non-HDG-NPs (Fig. 4). Moreover, the knockdown of luciferase was dose-dependent for both non-HDG- and HDG-NPs with up to ~40% knockdown observed by HDG-NPs at a 150 nM siRNA dose.
FIGURE 4.
Target gene silencing of the model gene luciferase by non-HDG- and HDG-NPs at N:P ratio of 5:1 (n=5). HDG-NPs exhibited enhanced siRNA potency at both 100 and 150 nM doses. All gene silencing data are normalized to an analogous scrambled siRNA control with the same carrier (HDG-NP and non-HDG-NP) and at each dose (100 and 150 nM). LF2K=Lipofectamine 2000.
Intracellular un-packaging of oligonucleotides monitored by FRET confocal microscopy
The observed increase in gene silencing by the HDG-NPs, despite having similar hemolysis, cell uptake, and viability as non-HDG-NPs under the conditions tested, suggests that the addition of a hydrolytic, charge-reversal-based siRNA release mechanism improved carrier release and intracellular bioavailability of the siRNA. To test this in vitro, we monitored the FRET efficiency of paired dsDNAs, used as a model of siRNA, co-loaded into both particle types to investigate whether hydrolytically driven charge reversal increased intracellular un-packaging of oligonucleotides (Fig. 5 and Supporting Information Fig. S6). Non-HDG-NPs exhibited punctate regions of strong FRET efficiency within cells, suggesting that part of the dsDNA delivered into the cells remained packaged tightly within the NPs [Fig. 5(A)]. Quantification across all images acquired showed that non-HDG-NPs consistently retained significantly higher intracellular FRET signal than HDG-NPs after 24 h [Fig. 5(B)]. The HDG-NP and non-HDG-NP treatments administered to these cells were confirmed to have the same level of FRET efficiency at t=0 h [Fig. 5(C)]. Moreover, the increased loss of HDG-NP FRET was confirmed in solution at 24 h by re-measuring the same treatment aliquots after incubation in PBS [Fig. 5(C)].
DISCUSSION
The current work introduces a cytosolic release mechanism into in vivo-ready siRNA polyplexes which have been previously optimized for stability in the circulation and endosomal escape.20 The simple synthetic strategy utilized a one-step RAFT co-polymerization from a PEG macro-CTA precursor of a hydrophobic monomer (BMA) and a cationic monomer (DMAEA) which gradually reverses charge due to hydrolytic degradation. Increased nucleic acid release from the DMAEA-based HDG-NPs relative to DMAEMA-based non-HDG-NPs was confirmed by multiple methods in solution and to our knowledge, for the first time, intracellularly using high resolution FRET confocal microscopy techniques. The functional importance of cytosolic nucleic acid release was further confirmed as HDG-NPs achieved significantly higher gene silencing than non-HDG-NPs at all doses tested. The enhancement in gene silencing potency could prove important for systemic delivery applications, such as oncology, where repeated doses are needed for maintaining therapeutic efficacy and dose-limiting toxicities are common.
The charge reversing nature of HDG-NPs was confirmed by tracking both the polymer degradation and accelerated nucleic acid release when compared to the non-HDG-NP analogue. Chemical analysis through 1H NMR confirmed that DMAEA co-polymerized with BMA was gradually hydrolyzed to dimethylaminoethanol at physiologic pH and 37°C, whereas DMAEMA co-polymerized with BMA was stable against hydrolysis under the same conditions [Fig. 1(A,B)]. The rates of hydrolysis observed herein are comparable to previously reported homopolymers of DMAEA studied by Monteiro and coworkers,39 suggesting that co-polymerization of DMAEA with BMA did not significantly affect hydrolysis. Observable changes in the physicochemical characteristics of HDG-NPs relative to non-HDG-NPs further supported that hydrolysis occurred in the HDG-NPs [Fig. 1(C)]. After siRNA loading, non-HDG-NPs retained their size (~70 nm) when incubated for 24 h in aqueous solution. On the other hand, HDG-NPs were reduced to the size of unloaded micelles (~25 nm) during the same incubation period, suggesting release of the nucleic acid cargo. Tracking the nucleic acids directly through FRET measurements further corroborated these physicochemical results. FRET release kinetics revealed 40% lower FRET efficiency from HDG-NPs after 24–28 h incubation in aqueous solution (Fig. 2). The release quantified by measuring FRET kinetics from HDG-NPs at 5:1 N:P ratio was ~70%. For other comparable charge-reversal systems, cargo release has not been quantified28,40 or has been potentially underestimated at ~20% based on agarose gel retardation assays.29 The FRET-based release readout allowed quantification of release kinetics, which suggest that a significantly greater percentage of nucleic acid cargo is released from the HDG-NPs and is available for association with RISC machinery after 24–28 h. In summary, hydrolysis of the HDG-NPs was chemically confirmed, and the hydrolysis process proved to be functionally significant for increasing nucleic acid release.
In order to compare the non-HDG- and HDG-NPs directly for target gene silencing and intracellular un-packaging, we first endeavored to confirm that both formulations had comparable hemolysis, cell uptake, and effect on cell viability. No significant differences in hemolysis were observed between HDG- and non-HDG-NPs (Supporting Information Fig. S5). Importantly, both formulations exhibited switch-like, pH-dependent membrane disruption between extracellular (7.4) and early endosomal (6.8) pH, with no differences in endosomolytic potential between HDG and non-HDG formulations. HDG- and non-HDG-NPs also exhibited similar levels of cellular internalization, both being significantly lower than Lipofectamine 2000 due to surface PEGylation [Fig. 3(A,B)]. Cell viability was only significantly different at the highest dose (200 nM) and highest N:P ratio (20:1), where the HDG-NPs significantly reduced cell viability [Fig. 3(C,D)]. The decrease in cell viability at 20:1 N:P ratios is likely due to the increased mass of polymer necessary to deliver a constant dose (100 nM) at increasing N:P ratios. We have also observed that loading of these polymers with polyanionic cargo, such as siRNA and dsDNA, decreases their toxicity. It is likely that more free polymer is present in solution at the higher N:P ratio of 20:1 and that this is the cause of the cytotoxicity of this formulation. In seminal work performed by Kissel and coworkers, cytotoxicity of polycations in vitro was found to be highly dependent upon both polymer molecular weight and charge density.41 Moreover, Monteiro and coworkers reported cytotoxicity of large polymers (>5600 Da) containing the DMAEA monomer in the literature.42 Our observations confirm that cytotoxicity is of concern with the DMAEA-based polymers but suggest that toxicity can be mitigated for DMAEA copolymerized with BMA (lower charge density) when complexed with oligonucleotides under conditions where a minimum effective dose is used and there is not a significant excess of free polymer.
The level of target gene knockdown from HDG-NPs exceeded that of non-HDG-NPs in vitro (Fig. 4), proving the structure–function significance of increased siRNA cargo release from the HDG carrier. However, the level of knockdown achieved with either PEGylated, surface charge neutral formulation did not match that of LF2K. This is expected, as LF2K has a very high zeta potential that drives cell interactions and internalization in vitro. However, non-PEGylated, cationic transfection agents such as LF2K are not suitable for systemic delivery in vivo, whereas the HDG-NPs are more amenable to in vivo intravenous siRNA administration. The level of knockdown achieved in vitro does not reach that of some of the most optimized transfection reagents, but the performance is comparable to other PEGylated or decationized delivery systems.43,44 Overall, these data suggest that incorporation of mechanisms for un-packaging is a feasible route to improving the bioactivity of PEGylated polyplex delivery systems which notoriously are limited in their in vitro transfection potency relative to research-grade reagents that are optimized for in vitro use only.
FRET confocal microscopy studies were utilized to confirm that increased gene silencing bioactivity of HDG-NPs over non-HDG-NPs was mechanistically tied to increased siRNA release from the NPs within cells (Fig. 5). This was motivated by the concept that upon escape from the endolysosomal pathway, siRNA must release from its carrier and become bioavailable for loading into the RISC machinery to achieve gene silencing. The process of siRNA release from cationic carriers is likely catalyzed by the presence of competing intracellular counter ions such as RNA and proteins.45,46 However, we hypothesized that the incorporation of a charge-reversing cationic moiety would increase charge repulsion between the siRNA and polymer backbone of HDG-NPs, thus accelerating and increasing magnitude of siRNA release within the cytosol.32 Presumably, siRNA molecules tightly packaged within non-HDG-NPs would not be bioavailable for assembly into the RISC complex because of steric and electrostatic hindrance of the carrier polymer. Indeed, HDG-NP-treated cells had negligible FRET signal (indicating better nucleic acid un-packaging) while non-HDG-NP-treated cells consistently exhibited punctate intracellular FRET signal (indicating cargo that is still at least partially packaged) in microscopy studies (Fig. 5). Thus, our collective data indicate that the hydrolysis of the cationic siRNA-condensing moiety, and subsequent charge-reversal, of the HDG-NPs provided an effective mechanism for enhanced siRNA intracellular un-packaging and improved gene silencing bioactivity.
Cytosolic release of siRNA is often overlooked within non-viral gene delivery vector development. Here, a comparative study of HDG-NPs and non-HDG-NPs confirms that incorporation of a cytosolic release mechanism using the hydrolytically degradable, charge-reversing cationic monomer DMAEA improves intracellular release and bioactivity of endosomolytic, surface charge-neutral, PEGylated siRNA polyplexes. The HDG-NPs are stabilized upon initial formulation but have accelerated release of nucleic acid cargo in aqueous solutions relative to non-HDG-NPs; importantly, increased cargo release of the HDG-NPs was also confirmed intracellularly by FRET microscopy. Increased un-packaging of siRNA delivered by HDG-NPs correlated with significantly higher target gene silencing of the model gene luciferase in vitro compared to non-HDG-NPs, despite minimal differences in hemolysis, cell uptake, and viability under the test conditions. Overall, these results suggest that activity of endosomolytic, charge-neutral, PEGylated siRNA polyplexes can be increased by improving cytosolic release. This work motivates further exploration into charge-reversing systems, including carriers that respond to specific cues such as pH, oxidation/reduction, enzymatic cleavage, light activation, and so forth to leverage “smarter” intracellular un-packaging triggers.
Supplementary Material
Acknowledgments
Authors are grateful to the Vanderbilt Institute for Nanoscale Science and Engineering (VINSE) for use of facilities for DLS experiments.
Contract grant sponsor: National Institutes of Health; contract grant numbers: R21EB012750, R01EB019409
Contract grant sponsor: National Science Foundation; contract grant number: NSF#1005023, NSF#1445197
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Burnett JC, Rossi JJ, Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J. 2011;6:1130–1146. doi: 10.1002/biot.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dykxhoorn DM, Palliser D, Lieberman J. The silent treatment: siRNAs as small molecule drugs. Gene Ther. 2006;13:541–552. doi: 10.1038/sj.gt.3302703. [DOI] [PubMed] [Google Scholar]
- 3.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher H-P. Therapeutic silencing of an endogenous gene by systemic administration of modified siR-NAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. doi: http://www.nature.com/nature/journal/v432/n7014/suppinfo/nature03121_S1.html. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Lu Z, Wientjes MG, Au JS. Delivery of siRNA therapeutics: Barriers and carriers. AAPS J. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett DW, Davis ME. Effect of siRNA nuclease stability on the in vitro and in vivo kinetics of siRNA-mediated gene silencing. Biotechnol Bioeng. 2007;97:909–921. doi: 10.1002/bit.21285. [DOI] [PubMed] [Google Scholar]
- 6.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: Advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. NatMater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 8.Oh YK, Park TG. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev. 2009;61:850–862. doi: 10.1016/j.addr.2009.04.018. doi: http://dx.doi.org/10.1016/j.addr.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 9.Mislick KA, Baldeschwieler JD. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc Natl Acad Sci USA. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. doi: http://dx.doi.org/10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkataraman S, Ong WL, Ong ZY, Joachim Loo SC, Rachel Ee PL, Yang YY. The role of PEG architecture and molecular weight in the gene transfection performance of PEGylated poly(dimethylaminoethyl methacrylate) based cationic polymers. Biomaterials. 2011;32:2369–2378. doi: 10.1016/j.biomaterials.2010.11.070. doi: http://dx.doi.org/10.1016/j.biomaterials.2010.11.070. [DOI] [PubMed] [Google Scholar]
- 13.Mishra S, Webster P, Davis ME. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. Eur J Cell Biol. 2004;83:97–111. doi: 10.1078/0171-9335-00363. doi: http://dx.doi.org/10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 14.Sato A, Choi SW, Hirai M, Yamayoshi A, Moriyama R, Yamano T, Takagi M, Kano A, Shimamoto A, Maruyama A. Polymer brush-stabilized polyplex for a siRNA carrier with long circulatory half-life. J Control Release. 2007;122:209–216. doi: 10.1016/j.jconrel.2007.04.018. doi: http://dx.doi.org/10.1016/j.jconrel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 15.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: From concept to clinic. Mol Pharm. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 16.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. doi: http://www.nature.com/nature/journal/v464/n7291/suppinfo/nature08956_S1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuckerman JE, Gritli I, Tolcher A, Heidel JD, Lim D, Morgan R, Chmielowski B, Ribas A, Davis ME, Yen Y. Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proc Natl Acad Sci USA. 2014;111:11449–11454. doi: 10.1073/pnas.1411393111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuckerman JE, Choi CH, Han H, Davis ME. Polycation-siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proc Natl Acad Sci USA. 2012;109:3137–3142. doi: 10.1073/pnas.1200718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naeye B, Deschout H, Caveliers V, Descamps B, Braeckmans K, Vanhove C, Demeester J, Lahoutte T, De Smedt SC, Raemdonck K. In vivo disassembly of IV administered siRNA matrix nanoparticles at the renal filtration barrier. Biomaterials. 2013;34:2350–2358. doi: 10.1016/j.biomaterials.2012.11.058. doi: http://dx.doi.org/10.1016/j.biomaterials.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 20.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. Balancing cationic and hydrophobic content of PEGylated siRNA polyplexes enhances endosome escape, stability, blood circulation time, and bioactivity in vivo. ACS Nano. 2013;7:8870–8880. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Yu SS, Miteva M, Nelson CE, Werfel T, Giorgio TD, Duvall CL. Matrix metalloproteinase responsive, proximity-activated polymeric nanoparticles for siRNA delivery. Adv Funct Mater. 2013;23:3040–3052. doi: 10.1002/adfm.201202215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Miteva M, Kirkbride KC, Cheng MJ, Nelson CE, Simpson EM, Gupta MK, Duvall CL, Giorgio TD. Dual MMP7-proximityactivated and folate receptor-targeted nanoparticles for siRNA delivery. Biomacromolecules. 2014;16:192–201. doi: 10.1021/bm501394m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun H, Guo B, Cheng R, Meng F, Liu H, Zhong Z. Biodegradable micelles with sheddable poly(ethylene glycol) shells for triggered intracellular release of doxorubicin. Biomaterials. 2009;30:6358–6366. doi: 10.1016/j.biomaterials.2009.07.051. doi: http://dx.doi.org/10.1016/j.biomaterials.2009.07.051. [DOI] [PubMed] [Google Scholar]
- 24.Cheng R, Meng F, Deng C, Klok HA, Zhong Z. Dual and multistimuli responsive polymeric nanoparticles for programmed site-specific drug delivery. Biomaterials. 2013;34:3647–3657. doi: 10.1016/j.biomaterials.2013.01.084. doi: http://dx.doi.org/10.1016/j.biomaterials.2013.01.084. [DOI] [PubMed] [Google Scholar]
- 25.Meng F, Zhong Z, Feijen J. Stimuli-responsive polymersomes for programmed drug delivery. Biomacromolecules. 2009;10:197–209. doi: 10.1021/bm801127d. [DOI] [PubMed] [Google Scholar]
- 26.Zhu C, Jung S, Luo S, Meng F, Zhu X, Park TG, Zhong Z. Co-delivery of siRNA and paclitaxel into cancer cells by biodegradable cationic micelles based on PDMAEMA-PCL-PDMAEMA triblock copolymers. Biomaterials. 2010;31:2408–2416. doi: 10.1016/j.biomaterials.2009.11.077. doi: http://dx.doi.org/10.1016/j.biomaterials.2009.11.077. [DOI] [PubMed] [Google Scholar]
- 27.Prata CAH, Zhao Y, Barthelemy P, Li Y, Luo D, McIntosh TJ, Lee SJ, Grinstaff MW. Charge-reversal amphiphiles for gene delivery. J Am Chem Soc. 2004;126:12196–12197. doi: 10.1021/ja0474906. [DOI] [PubMed] [Google Scholar]
- 28.Gu W, Jia Z, Truong NP, Prasadam I, Xiao Y, Monteiro MJ. Polymer nanocarrier system for endosome escape and timed release of siRNA with complete gene silencing and cell death in cancer cells. Biomacromolecules. 2013;14:3386–3389. doi: 10.1021/bm401139e. [DOI] [PubMed] [Google Scholar]
- 29.Foster AA, Greco CT, Green MD, Epps TH, Sullivan MO. Light-mediated activation of siRNA release in diblock copolymer assemblies for controlled gene silencing. Adv Healthcare Mater. 2015;4:760–770. doi: 10.1002/adhm.201400671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truong NP, Gu W, Prasadam I, Jia Z, Crawford R, Xiao Y, Monteiro MJ. An influenza virus-inspired polymer system for the timed release of siRNA. Nat Commun. 2013;4:1902. doi: 10.1038/ncomms2905. [DOI] [PubMed] [Google Scholar]
- 31.van de Wetering P, Zuidam NJ, van Steenbergen MJ, van der Houwen OAGJ, Underberg WJM, Hennink WE. A mechanistic study of the hydrolytic stability of poly(2-(dimethylamino)ethyl methacrylate) Macromolecules. 1998;31:8063–8068. [Google Scholar]
- 32.Funhoff A, van Nostrum C, Janssen ACA, Fens MAM, Crommelin DA, Hennink W. Polymer side-chain degradation as a tool to control the destabilization of polyplexes. Pharm Res. 2004;21:170–176. doi: 10.1023/b:pham.0000012165.68765.e6. [DOI] [PubMed] [Google Scholar]
- 33.Convertine AJ, Benoit DSW, Duvall CL, Hoffman AS, Stayton PS. Development of a novel endosomolytic diblock copolymer for siRNA delivery. J Control Release. 2009;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. doi: http://dx.doi.org/10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moad G, Chong YK, Postma A, Rizzardo E, Thang SH. Advances in RAFT polymerization: The synthesis of polymers with defined end-groups. Polymer. 2005;46:8458–8468. doi: http://dx.doi.org/10.1016/j.polymer.2004.12.061. [Google Scholar]
- 35.Chong YK, Le TPT, Moad G, Rizzardo E, Thang SH. A more versatile route to block copolymers and other polymers of complex architecture by living radical polymerization: The RAFT process. Macromolecules. 1999;32:2071–2074. [Google Scholar]
- 36.Alabi CA, Love KT, Sahay G, Stutzman T, Young WT, Langer R, Anderson DG. FRET-labeled siRNA probes for tracking assembly and disassembly of siRNA nanocomplexes. ACS Nano. 2012;6:6133–6141. doi: 10.1021/nn3013838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans BC, Nelson CE, Yu SS, Beavers KR, Kim AJ, Li H, Nelson HM, Giorgio TD, Duvall CL. Ex vivo red blood cell hemolysis assay for the evaluation of pH-responsive endosomolytic agents for cytosolic delivery of biomacromolecular drugs. Forthcoming. 2013 doi: 10.3791/50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miteva M, Kirkbride KC, Kilchrist KV, Werfel TA, Li H, Nelson CE, Gupta MK, Giorgio TD, Duvall CL. Tuning PEGylation of mixed micelles to overcome intracellular and systemic siRNA delivery barriers. Biomaterials. 2015;38:97–107. doi: 10.1016/j.biomaterials.2014.10.036. doi: http://dx.doi.org/10.1016/j.biomaterials.2014.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran NTD, Truong NP, Gu W, Jia Z, Cooper MA, Monteiro MJ. Timed-release polymer nanoparticles. Biomacromolecules. 2013;14:495–502. doi: 10.1021/bm301721k. [DOI] [PubMed] [Google Scholar]
- 40.Truong NP, Jia Z, Burgess M, Payne L, McMillan NAJ, Monteiro MJ. Self-catalyzed degradable cationic polymer for release of DNA. Biomacromolecules. 2011;12:3540–3548. doi: 10.1021/bm2007423. [DOI] [PubMed] [Google Scholar]
- 41.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: Influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24:1121–1131. doi: 10.1016/s0142-9612(02)00445-3. doi: http://dx.doi.org/10.1016/S0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 42.Truong NP, Jia Z, Burges M, McMillan NAJ, Monteiro MJ. Self-catalyzed degradation of linear cationic poly(2-dimethylaminoethyl acrylate) in water. Biomacromolecules. 2011;12:1876–1882. doi: 10.1021/bm200219e. [DOI] [PubMed] [Google Scholar]
- 43.Novo L, Takeda KM, Petteta T, Dakwar GR, van den Dikkenberg JB, Remaut K, Braeckmans K, van Nostrum CF, Mastrobattista E, Hennink WE. Targeted decationized polyplexes for siRNA delivery. Mol Pharm. 2014;12:150–161. doi: 10.1021/mp500499x. [DOI] [PubMed] [Google Scholar]
- 44.Oe Y, Christie RJ, Naito M, Low SA, Fukushima S, Toh K, Miura Y, Matsumoto Y, Nishiyama N, Miyata K, Kataoka K. Actively-targeted polyion complex micelles stabilized by cholesterol and disulfide cross-linking for systemic delivery of siRNA to solid tumors. Biomaterials. 2014;35:7887–7895. doi: 10.1016/j.biomaterials.2014.05.041. doi: http://dx.doi.org/10.1016/j.biomaterials.2014.05.041. [DOI] [PubMed] [Google Scholar]
- 45.Arigita C, Zuidam N, Crommelin DA, Hennink W. association and dissociation characteristics of polymer/DNA complexes used for gene delivery. Pharm Res. 1999;16:1534–1541. doi: 10.1023/a:1015096302720. [DOI] [PubMed] [Google Scholar]
- 46.De Smedt S, Demeester J, Hennink W. Cationic polymer based gene delivery systems. Pharm Res. 2000;17:113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.