Abstract
Cell division in Escherichia coli is mediated by a large protein complex called the divisome. Most of the divisome proteins have been identified, but how they assemble onto the Z ring scaffold to form the divisome and work together to synthesize the septum is not well understood. In this review, we summarize the latest findings on divisome assembly and activation as well as provide our perspective on how these two processes might be regulated.
An overview of the E. coli divisome
Cell division is a fundamental process that has to be executed precisely in all living organisms because an error in this process may result in cell death. To accomplish this task, E. coli employs a large protein complex called the divisome that consists of a protein network extending from the cytoplasm to all three layers of the cell envelope (de Boer, 2010, Lutkenhaus et al., 2012, Tsang & Bernhardt, 2015a). These proteins include a cytoskeletal protein that provides the scaffold for the assembly of the whole complex, peptidoglycan synthases and hydrolases that remodel the peptidoglycan network, a DNA translocase that coordinates cell division and chromosome segregation, proteins that coordinate peptidoglycan synthesis and invagination of the membranes and many other proteins with regulatory functions. Together, these proteins assemble into a tightly regulated peptidoglycan synthesis machine that builds a septum between the segregated chromosomes to safely separate the cell into two daughter cells while avoiding catastrophic consequences.
The past three decades have witnessed the identification of many divisome proteins in E. coli. Over 3 dozen proteins have been identified as components of the divisome and the number is still increasing. Among these proteins are a dozen that are essential or conditionally essential for division, including FtsZ, FtsA, ZipA, FtsE, FtsX, FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI and FtsN (Fig. 1). Most of these 12 proteins are highly conserved in the bacterial domain and constitute the basic components of the divisome. FtsZ is a bacterial tubulin homolog that polymerizes into filaments that coalesce into the Z ring at the future division site to provide a scaffold for the assembly of the entire divisome (Bi & Lutkenhaus, 1991, Lutkenhaus et al., 2012, de Boer, 2010, Erickson et al., 2010, Mukherjee et al., 1993, de Boer et al., 1992, RayChaudhuri & Park, 1992, Lowe & Amos, 1998). The dynamics of FtsZ filaments organize the synthesis of peptidoglycan at the septum and may also contribute to the driving force for constriction (Yang et al., 2017, Bisson-Filho et al., 2017, Erickson et al., 2010, Coltharp & Xiao, 2017). FtsA is an actin-like protein associated with the membrane through an amphipathic helix (Pichoff & Lutkenhaus, 2005, van den Ent & Lowe, 2000, Bork et al., 1992) and ZipA is a bitopic membrane protein with a large cytoplasmic domain that interacts with FtsZ (Hale & de Boer, 1997). Both FtsA and ZipA bind to a conserved C-terminal peptide (CCTP) of FtsZ to anchor FtsZ polymers to the membrane so that they can coalesce into the Z ring (Haney et al., 2001, Ma & Margolin, 1999, Mosyak et al., 2000, Szwedziak et al., 2012).
Fig. 1.
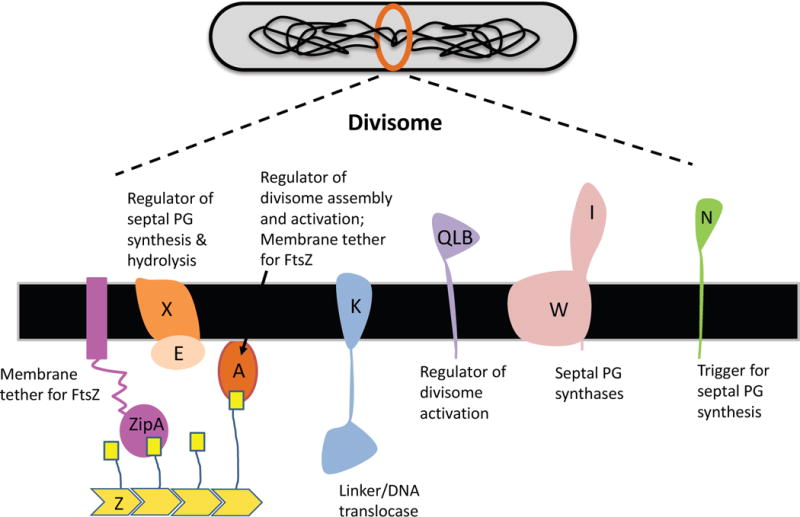
Components of the divisome. The twelve proteins that make up the essential components of the divisome in E. coli are indicate along with their functions. These include the Fts proteins, indicated by a capital letter, and ZipA. FtsZ assembles into a filament that is a scaffold for assembly of the complete divisome. FtsZ filaments are attached to the membrane by a short conserved C-termimal peptide (small yellow box) which binds to FtsA and ZipA.
Although FtsA or ZipA is sufficient for Z ring formation, both are necessary for the recruitment of downstream division proteins (Pichoff & Lutkenhaus, 2002). The latest findings indicate a critical role for FtsA in regulating the activity of the divisome (Liu et al., 2015, Tsang & Bernhardt, 2015b). FtsE and FtsX are the ATP-binding and membrane subunits, respectively, of an ATP-binding cassette transporter-like complex (Gill et al., 1986, Schmidt et al., 2004). FtsEX uses its ATPase cycle to control cell wall hydrolysis at the septum through regulation of amidases that cleave the stem peptide of peptidoglycan to promote cell separation (Yang et al., 2011). Recent findings suggest that FtsEX also acts on FtsA to regulate the assembly and activity of the divisome (Du et al., 2016). FtsK is a DNA translocase that helps resolve chromosome dimers and removes DNA from the constricting septum (Liu et al., 1998, Yu et al., 1998b, Steiner et al., 1999, Männik et al. 2017). Its essential function in cell division resides in its non-translocase transmembrane domain which links downstream divisome proteins to the Z ring (Yu et al., 1998a, Draper et al., 1998, Wang & Lutkenhaus, 1998, Chen & Beckwith, 2001). FtsK can be relatively easily bypassed under various conditions suggesting it was integrated into the pathway during evolution to improve the efficiency of division (Geissler & Margolin, 2005, Dubarry et al., 2010).
FtsQ, FtsL and FtsB are all bitopic membrane proteins that form a complex that acts as a scaffold for the recruitment of downstream divisome proteins (Carson et al., 1991, Guzman et al., 1997, Buddelmeijer & Beckwith, 2004). However, recent studies imply that this complex also plays a critical role in regulating the activity of the divisome (Liu et al., 2015, Tsang & Bernhardt, 2015b). FtsW is a polytypic membrane protein belonging to the widely conserved SEDS (shape, elongation, division and sporulation) family of membrane proteins (Ikeda et al., 1989). It was thought to be a flippase for lipid II (the peptidoglycan precursor)(Mohammadi et al., 2011, Mohammadi et al., 2014), however, the latest findings from Bernhardt’s and Rudner’s groups as well as Errington’s group indicate that the SEDS family proteins constitute a new family of peptidoglycan glycosyltransferases (Meeske et al., 2016, Cho et al., 2016, Emami et al., 2017). Thus, FtsW is a putative peptidoglycan glycosyltransferase dedicated to division working with its partner FtsI, also called penicillin binding protein 3 (PBP3), a peptidoglycan transpeptidase, to synthesize septal peptidoglycan (Botta & Park, 1981). FtsN is the last essential divisome protein recruited to the division site and is considered the trigger for cell constriction (Addinall et al., 1997, Lutkenhaus, 2009, Gerding et al., 2009). Recent studies from multiple groups reinforce this idea (Liu et al., 2015, Pichoff et al., 2015, Busiek & Margolin, 2014) but how exactly FtsN triggers cell constriction remains to be elucidated. The remaining 20 plus division proteins are not essential for division and are not discussed in this microreview.
Assembly of the divisome
Assembly of the divisome can be divided into two temporally distinct stages (Aarsman et al., 2005). The first stage involves formation of a Z ring at the future division site by FtsZ with the help of its membrane anchors FtsA and ZipA. Formation of the Z ring is temporally and spatially regulated to ensure that it is assembled precisely at the midcell so that cell division is coordinated with chromosome segregation, ensuring equal distribution of cellular contents to daughter cells (Lutkenhaus, 2007). A number of functionally redundant FtsZ associated proteins called Zaps (ZapA, ZapC and ZapD) enhance Z ring formation by cross-linking FtsZ polymers (Gueiros-Filho & Losick, 2002, Durand-Heredia et al., 2012, Durand-Heredia et al., 2011, Hale et al., 2011). In addition, ZapA (along with ZapB) has a unique role in linking the Z ring to the terminus region of the chromosome (Espeli et al., 2012, Buss et al., 2017, Buss et al., 2015). These proteins (except ZapB) localize to the Z ring through direct interaction with FtsZ. The ABC transporter-like complex FtsEX also localizes to the Z ring as it is forming (Schmidt et al., 2004). FtsE is reported to interact with FtsZ (Corbin et al., 2007), but how FtsEX localizes to the division site remains to be determined. Together, these proteins constitute the early components of the divisome and are referred to as the Z ring. After a temporary delay due to an unknown mechanism the second stage of divisome assembly occurs (Aarsman et al., 2005). During the second stage, 7 essential cell division proteins, FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI and FtsN, are recruited to the Z ring almost simultaneously. Although the determinants for midcell localization of a few of these proteins have been defined, such as FtsI and FtsN, the determinants of most of them still await identification. Once FtsN arrives at the Z ring, the divisome is activated to synthesize septal peptidoglycan to divide the cell.
The linear hierarchical assembly pathway
Studies of division protein localization following depletion of one of the components led to the idea that recruitment of the second cascade of divisome proteins to the Z ring proceeds in a linear hierarchical manner (Buddelmeijer & Beckwith, 2002). Take FtsQ for example, in its absence FtsZ, FtsA, ZipA, FtsEX and FtsK (upstream proteins) localize but the downstream proteins (FtsL, FtsB, FtsW, FtsI and FtsN) do not (Fig. 1). This approach has been used to determine the dependency relationship for localization of division proteins. However, it should be kept in mind that this dependence relationship is not necessarily a reflection of the order of assembly or a reflection of direct protein-protein interaction between adjacent proteins. For example, although FtsB can interact directly with FtsQ (Glas et al., 2015), its localization to the Z ring follows the localization of FtsL (Buddelmeijer et al., 2002), which interacts with both FtsQ and FtsB (Buddelmeijer & Beckwith, 2004). In fact, FtsQ, FtsL and FtsB form a subcomplex even before they localize to the Z ring (Buddelmeijer & Beckwith, 2004). It is also noteworthy that a direct protein-protein interaction between two division proteins does not necessarily mean that they would be adjacent in the pathway. FtsN interacts with FtsA directly (Busiek et al., 2012, Liu et al., 2015, Pichoff et al., 2015), but it does not localize to the Z ring as it forms. Instead, it is the last to localize to the Z ring and requires the presence of FtsA, FtsQ and FtsI (Addinall et al., 1997, Goehring et al., 2006). It should be noted that localization results using GFP-FtsN have to be interpreted with caution as overproduction of FtsN suppresses many division defects and can bypass some genes (Pichoff et al., 2015) Therefore, the hierarchical assembly pathway has to be interpreted with caution and there may exist multiple routes for division proteins to get to the division site.
The alternative assembly pathway
The first evidence for non-sequential assembly of the divisome came from studies that forced the localization of a late divisome protein to the Z ring in the absence of an upstream protein by fusing it to a protein that directly interacts with FtsZ (Goehring et al., 2005, Goehring & Beckwith, 2005). This method, called ‘premature targeting’, revealed that such a targeted late divisome protein not only recruits downstream division proteins but also back recruits upstream division proteins to the Z ring (Goehring et al., 2005). For example, a ZapA-FtsL fusion is able to recruit all downstream division proteins (except FtsN) and upstream proteins FtsK and FtsQ to the Z ring in the absence of FtsA (Goehring et al., 2005), which is normally required for localization of all these proteins. These findings suggest that most of the second stage divisome proteins except FtsN form a large protein complex that localizes to the Z ring with FtsK as a linker (Goehring & Beckwith, 2005).
More direct evidence for non-sequential assembly of the divisome came from the surprising findings that several essential division proteins can be bypassed by mutations or overexpression of other division proteins without much of an effect on division (Fig. 2). ZipA, FtsEX and FtsK can all be bypassed individually by a single mutation in ftsA called ftsA* or by overexpression of FtsN (Geissler et al., 2003, Reddy, 2007, Geissler & Margolin, 2005, Pichoff et al., 2015) (Fig. 2B). Subsequent studies identified additional mutations in ftsA, ftsB, ftsL and ftsW that can bypass ZipA or FtsEX (Pichoff et al., 2012, Du et al., 2016, Tsang & Bernhardt, 2015b). Since ZipA, FtsEX and FtsK are normally necessary for the recruitment of downstream division proteins (Fig. 2A), it raises the question of how these divisome proteins are recruited in these bypass situations. With overexpression of FtsN, the N-terminal domain of FtsN, which interacts directly with FtsA, appears to be critical (Pichoff et al., 2015). This suggests that in the absence of an intermediate protein, overexpression of FtsN promotes the interaction between FtsN and FtsA and back-recruits the divisome proteins to the Z ring (Fig. 2B). In the case of bypass by activating mutations in division proteins (FtsL, B or W), the mechanism responsible for recruitment is less clear although the interaction between FtsN and FtsA is likely also the driving force, but this possibility needs to be verified. If it turns out to be true, we can conclude that there are two ways the second cascade of divisome proteins localize to the Z ring: one is through FtsK and the other is through FtsN. The FtsN-mediated pathway occurs when FtsN is overexpressed or the cells contain mutations in divisome proteins that can promote or strengthen FtsN’s interaction with FtsA. It is not clear why this assembly pathway does not occur under physiological conditions. One possibility is that the binding site for FtsN on FtsA is not available until all the other division proteins are present at the Z ring. Another possibility is that FtsN is somehow sequestered from the assembling divisome complex by an unknown mechanism. Results from a recent study favor the latter idea because the N-terminal domain of FtsN can localize to the Z ring in the absence of FtsQ (Busiek & Margolin, 2014), even though full length FtsN does not, as previously revealed by immunofluorescence microscopy (Goehring et al., 2006).
Fig. 2.
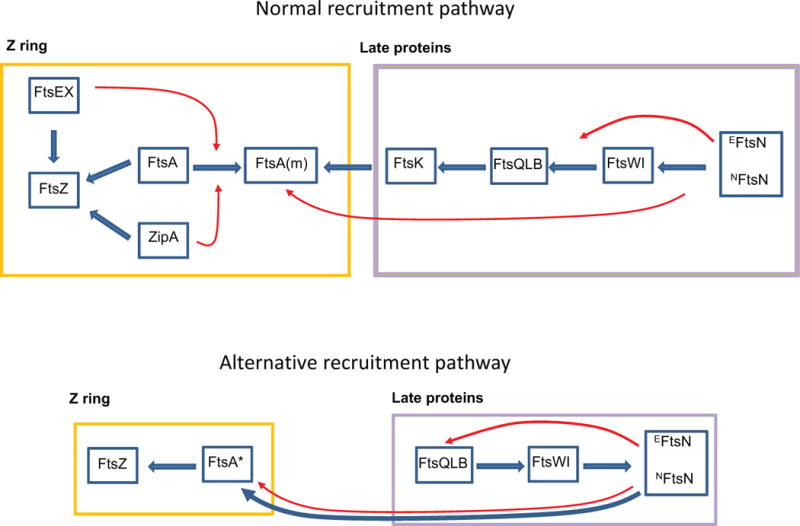
Assembly of the divisome. (A) Normal assembly pathway. The divisome assembles in two stages. In the first stage the Z ring forms when FtsZ filaments, attached to the membrane by ZipA and FtsA, coalesce into the ring. FtsEX acts on FtsA to convert it to a monomer (m) which recruits the late division proteins. ZipA also assists in this step. The last protein in the pathway FtsN activates septal PG synthesis with NFtsN interacting with FtsA in the cytoplasm and EFtsN acting in the periplasm. The blue arrows indicate known or suspected interactions and the red arrows indicate regulatory interactions. (B) Alternative assembly pathway. Some proteins can be bypassed by mutation or environmental conditions. ftsA* mutations, which make FtsA more monomeric, can bypass FtsEX, ZipA or FtsK individually. Under these conditions FtsN, normally the last protein recruited, is postulated to back recruit the other proteins. Overexpression of FtsN or activating mutations in ftsL and ftsB can also bypass FtsEX, ZipA or FtsK. It is likely that these conditions also favor the interaction between FtsA and FtsN.
A critical role for FtsA in divisome assembly
Although both FtsA and ZipA are necessary for recruitment of downstream divisome proteins to the Z ring, it is clear that FtsA plays a more important role: ZipA can be bypassed by many conditions as mentioned above and FtsA, but not ZipA, interacts with many divisome proteins. The fact that all mutations in FtsA that decrease self-interaction (but are functional) bypass ZipA lead to a model in which monomeric FtsA is responsible for recruiting downstream divisome proteins (Pichoff et al., 2012) (Fig. 2). In this model, ZipA modulates the polymerization state of FtsA at the Z ring, possibly by competing with FtsA for the CCTP of FtsZ. Importantly, the binding sites for downstream division proteins are also involved in FtsA’s self-interaction such that FtsA can only recruit downstream proteins when it becomes monomeric. Consistent with this model, the IC domain of FtsA, which is also involved in self-interaction, has been shown to interact with FtsN (Busiek et al., 2012). Overexpression of FtsN presumably bypasses ZipA because the interaction between FtsN and FtsA is favored allowing FtsN to back recruit other divisome proteins to the Z ring (Fig. 2B). Although this model is well supported by genetic data, there is a caveat: FtsN is the last recruit during divisome assembly and there must be another division protein that is earlier than FtsN and interacts with monomeric FtsA. This protein is likely FtsK because it is the first protein that localizes to the Z ring during the second stage of divisome assembly (Wang and Lutkenhaus, 1998) (Fig. 1), but direct evidence for interaction between FtsA and FtsK is missing so far.
FtsEX is also necessary for recruitment of downstream division proteins under physiological conditions (Schmidt et al., 2004). If FtsA directly recruits downstream proteins to the Z ring, then what is the role of FtsEX in recruitment? A recent study suggests that FtsEX directly regulates the polymerization state of FtsA to promote recruitment (Du et al., 2016) (Fig. 2A). ATP hydrolysis by FtsEX is not required but interaction between FtsX and a specific motif in FtsA is required as mutations in this motif that abrogate this interaction prevent the localization of the downstream protein FtsK to the Z ring (Du et al., 2016, Arends et al., 2009). How exactly the FtsEX-FtsA interaction leads to recruitment is not clear, but mutations that reduce FtsA self-interaction can bypass FtsEX and rescue FtsA mutants that are defective in interaction with FtsX. These genetic interactions imply that once FtsA is in the monomeric form FtsEX becomes dispensable for recruitment. Therefore, a model in which FtsEX interacts with FtsA to antagonize its polymerization or to maintain it in the monomeric form has been proposed (Du et al., 2016). However, additional biochemical evidence is required to support this model.
FtsA is an actin-like protein and forms cytoplasmic filaments when its membrane targeting sequence (MTS) is removed (Pichoff et al., 2012). It can also polymerize into actin-like filaments on the membrane in vivo and in vitro (Szwedziak et al., 2012). Since regulation of the oligomerization state of FtsA has been suggested as a critical step in divisome assembly, one would expect that the ATPase cycle of FtsA would be critical for its function and some of the FtsA binding proteins might directly affect the polymerization dynamics of FtsA by regulating its ATPase cycle. However, the ATPase activity of FtsA and its polymerization dynamics have not been well characterized due to the difficulties in working with FtsA in vitro. One group has reported that purified E. coli FtsA binds to and hydrolyze ATP in the absence of membrane (Herricks et al., 2014). It would be interesting to see whether FtsEX and FtsN affect the ATPase activity and polymerization of FtsA.
Recently an FtsZ mutant was reported to bypass ZipA (Haeusser et al., 2015). Because this mutant displayed increased bundling in vitro, an “FtsZ centric” model was proposed to explain the bypass of ZipA (Haeusser et al., 2015). In this model, the essential function of ZipA is to bundle FtsZ polymers and the bundling of FtsZ polymers leads to the recruitment of downstream divisome proteins to the Z ring (Haeusser et al., 2015). It is not clear how FtsZ bundling can lead to recruitment and what the role of FtsA in recruitment is in this model. The authors also proposed that FtsA antagonizes FtsZ polymerization at the Z ring (Haeusser et al., 2015). However, it is hard to imagine how the Z ring is maintained in the absence ZipA and how FtsA mutants such as FtsA* bypass ZipA if FtsA antagonizes FtsZ polymerization. Nonetheless, this alternative model offers a different view of the recruitment process and the bypass of ZipA by the FtsZ mutant underscores the complexity of regulation of divisome assembly.
Activation of the divisome
Arrival of FtsN at the Z ring signals the completion of divisome core assembly and activates the divisome to start septal PG synthesis. The onset of PG synthesis results in the recruitment of additional cell wall remodeling enzymes and proteins that coordinate cell wall synthesis with invagination of the membranes, leading to cell constriction and separation. In the current self-enhancing model, the initial localization of FtsN to the Z ring is mediated by its N-terminal cytoplasmic domain, NFtsN, interacting with FtsA (Gerding et al., 2009, Liu et al., 2015). This interaction in the cytoplasm stabilizes FtsA in the on state (monomeric) for cell constriction but also brings the essential domain of FtsN, EFtsN, to the divisome to activate septal PG synthesis in the periplasm (Fig. 3)(Liu et al., 2015). Synthesis of septal PG leads to the recruitment and activation of amidases that generate denuded peptidoglycan strands at the septum, which are substrates for the C-terminal SPOR domain of FtsN (SFtsN) (Gerding et al., 2009, Yahashiri et al., 2015). SFtsN binding to denuded peptidoglycan strands in turn results in the recruitment of more FtsN to the divisome to activate septal PG synthesis, forming a positive feedback loop (Gerding et al., 2009).
Fig. 3.
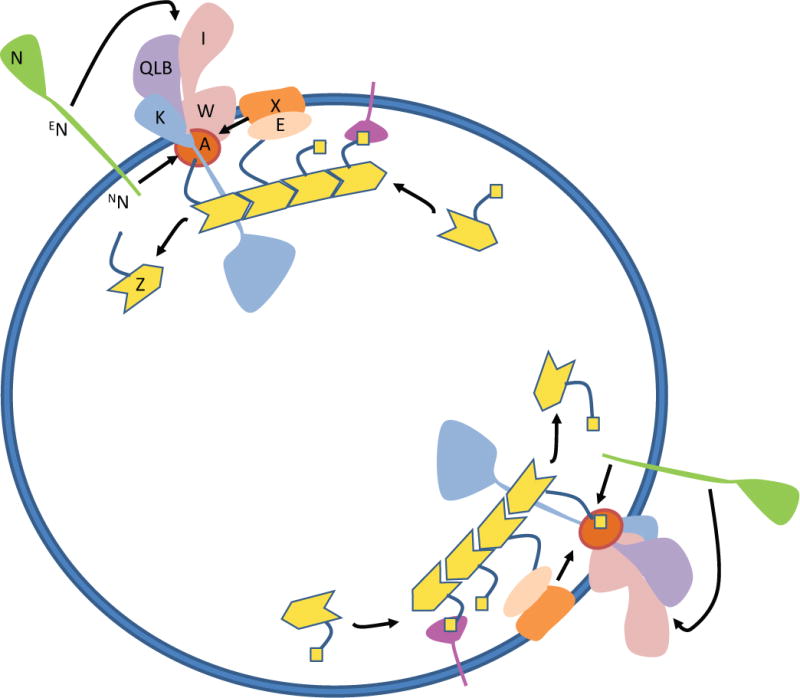
Activation of the divisome for septal PG synthesis. FtsEX acts on FtsA to make it monomeric such that FtsA recruits the downstream divisome proteins (FtsK, FtsQLB, and FtsWI). The arrival of FtsN triggers the activation of septal PG synthesis (FtsWI). Continuous ATP hydrolysis by FtsEX is required for PG synthesis and hydrolysis while the treadmilling of FtsZ filaments causes the PG synthesis machine to rotate around the septum resulting in symmetrical incorporation of PG.
FtsN was originally thought to activate septal PG synthesis by directly stimulating the transpeptidase activity of FtsI (PBP3) (Gerding et al., 2009). However, screening for suppressor mutations that bypass EFtsN function resulted in the identification of mutations in FtsA, FtsB and FtsL (Liu et al., 2015). Analysis of these mutants leads to a model in which FtsA and FtsQLB keep the divisome complex in the inactive state until FtsN arrives which then switches the complex to the active state by interacting with FtsA in the cytoplasm via NFtsN and relieving the suppression from FtsQLB in the periplasm via EFtsN (Liu et al., 2015). Study of a hyperactive FtsL* mutant by Bernhardt’s group also led to the conclusion that FtsQLB plays a critical role in activating the divisome complex for septal PG synthesis (Tsang & Bernhardt, 2015b). In their models, FtsA and FtsQLB exist in two different conformational states, OFF and ON, and the switch is controlled by FtsN (Liu et al., 2015, Tsang & Bernhardt, 2015b, Tsang & Bernhardt, 2015a). The activation signals in the cytoplasm (FtsA) and periplasm (FtsQLB) work synergistically to activate septal PG synthesis, but EFtsN alone is sufficient when expressed at a high level (and delivered to the periplasm)(Tsang & Bernhardt, 2015a, Liu et al., 2015). Presumably, FtsQLB in the OFF state suppresses the divisome activity by inhibiting the activity of the PG synthases FtsW and FtsI. The activation signal for FtsA correlates with its oligomerization state because many FtsA variants that are reduced for self-interaction are able to support cell division with reduced level of FtsN compared to wild type FtsA (Du et al., 2016). However, how the activation signal in FtsA is communicated to FtsQLB in the periplasm remains to be elucidated.
In addition to FtsA and FtsQLB, FtsEX has recently been shown to play a role in divisome activation as well (Du et al., 2016). FtsEX mutants predicted to be impaired in ATPase activity have been known to block cell constriction but not divisome assembly (Arends et al., 2009), a phenotype remarkably similar to inhibition of cell division by inhibitors which block the transpeptidase activity of FtsI. This similarity suggests that ATPase mutants of FtsEX prevent septal PG synthesis, but how is not clear. A recent study showed that this division inhibition by ATPase mutants of FtsEX depends on the interaction between FtsEX and FtsA, suggesting that FtsEX ATPase mutants lock FtsA in the inactive form or prevent FtsA from communicating with the FtsQLB complex (Du et al., 2016). Consistent with such a model, mutations in ftsB and ftsL that activate septal PG synthesis in the absence of EFtsN function provide resistance to the inhibition by the ATPase mutants of FtsEX, while mutations in ftsA that activate septal PG synthesis are still sensitive (Du et al., 2016). Also, reducing the interaction between FtsA and FtsEX renders cells resistant to inhibition caused by ATPase mutants of FtsEX (Du et al., 2016). Since the ATPase cycle of FtsEX has been known to govern cell wall hydrolysis at the septum by regulating amidase activity, these new findings indicate that the FtsEX complex uses its ATPase cycle to regulate PG synthesis as well as PG hydrolysis at the septum.
With the identification of FtsW as a putative PG glycosyltransferase, it is likely that FtsW and its cognate transpeptidase FtsI are the PG synthases primarily responsible for synthesizing septal PG. Thus, divisome activation means activating the enzymatic activities of these two PG synthases. In the current model, FtsW and FtsI would be active before they localize to the division site and FtsA and FtsQLB would suppress their activity at the Z ring (before FtsN arrives) (Liu et al., 2015). This is somewhat counterintuitive given that the bifunctional PG synthases, class A penicillin binding proteins, are kept inactive by an auto-inhibitory mechanism and are activated by cognate lipoproteins (Markovski et al., 2016, Paradis-Bleau et al., 2010, Typas et al., 2010). However, if the late divisome proteins (FtsK, FtsQLB, FtsWI but not FtsN) form a complex outside of the Z ring as suggested above, it is reasonable that the activity of FtsWI is suppressed by FtsQLB. An alternative possibility is that FtsW and FtsI, similar to the class A PBPs, are kept inactive by an auto-inhibitory mechanism at the Z ring before FtsN arrives. Arrival of FtsN at the Z ring might trigger a conformational change in FtsA and the FtsQLB complex such that they switch to the ON state. The ON signal in FtsA is transmitted across the membrane to the FtsQLB complex, which interacts with FtsW and FtsI to relieve inhibition. Mutations in ftsA, ftsB and ftsL that allow cells to divide with reduced FtsN function may switch FtsQLB to the ON state by causing a conformational change in these proteins. Mutations in ftsW or ftsI that enable the cells to start septal PG synthesis at a smaller cell size or require reduced FtsN function for division likely cause subtle conformational changes in the proteins that relieve the auto-inhibition. This model is slightly different than the one proposed by de Boer’s group due to the introduction of an auto-inhibitory mechanism for FtsW and FtsI instead of suppression by FtsQLB. However, both models are well supported by genetic data and future studies to understand the regulation of the catalytic activity of FtsW and FtsI would be critical to distinguish the two models.
FtsZ dynamics are coupled to activity of the divisome
The Z ring is a dynamic cytoskeletal element with a turnover time of less than 10 sec due to the intrinsic GTPase activity of FtsZ (Stricker et al., 2002). Ever since its discovery, the Z ring has been proposed to direct septal PG synthesis (Bi & Lutkenhaus, 1991), but how was not clear. We now know that it not only functions as a scaffold for the assembly of the entire division apparatus but that FtsZ filament dynamics are coupled to the synthesis of septal PG (Yang et al., 2017, Bisson-Filho et al., 2017). FtsZ mutants with reduced GTPase activity result in asymmetric constriction, producing daughter cells with contorted polar morphologies (Yang et al., 2017, Addinall & Lutkenhaus, 1996). This phenotype suggests that the GTPase activity of FtsZ is not essential for division or septal PG synthesis per se, but a dynamic Z ring is required for the distribution of PG synthesis so that symmetrical invagination of the septum occurs (Addinall & Lutkenhaus, 1996, Lutkenhaus et al., 2012, Yang et al., 2017).
Tracking the dynamics of FtsZ polymers at the Z ring revealed that they treadmill circumferentially at the division site (Bisson-Filho et al., 2017, Yang et al., 2017) (Fig. 3). The septal PG synthases FtsW and FtsI along with the other divisome proteins follow the movement of FtsZ polymers to distribute PG synthesis around the septum likely because they form a complex with FtsA which chases the movement of FtsZ polymers by binding to the CCTPs of FtsZ polymers. Reducing the GTPase activity of FtsZ slows down the movement of FtsZ polymers at the Z ring and leads to uneven incorporation of septal PG, resulting in the formation of distorted septa (Yang et al., 2017). One can imagine FtsZ polymers as trains running in shrinking circles and the PG synthases as workers that distribute the building blocks to form the septum. When the trains run at a constant speed, a smooth septum will be built overtime because septal PG material is incorporated evenly around the circle. However, when the trains are not running or are running too slowly, distribution of PG materials around the circle will be unequal. This uneven incorporation of PG changes the overall organization of PG at the division site, resulting in deformation of the septum. Interestingly, in the gram-positive bacterium Bacillus subtilis, the dynamics of FtsZ polymers at the Z ring not only control the distribution of PG synthesis but also the overall septal PG synthesis rate (Bisson-Filho et al., 2017). In other words, the faster FtsZ polymers treadmill, the higher the rate of septal PG incorporation, and vice versa. It is not clear why there is a difference between E. coli and B. subtilis, but it may reflect the different demands of septal PG synthesis in gram-positive bacteria which have a much thicker PG layer.
FtsZ-dependent division
Francois Jacob once stated “the dream of a bacterium is to become two bacteria”. It is now clear that most bacteria achieve this dream using an FtsZ-dependent cell division mechanism. It is perhaps surprising that this mechanism is so widespread and conserved considering the diversity of bacteria and the evolutionary time involved. As discussed above, studies of this process in E. coli have revealed the basic components of the FtsZ-dependent division apparatus and the regulatory principles. However, as the process of division is explored in more bacterial species, we begin to see variations in this process despite conservation of many of the main components. Different proteins are employed to tether FtsZ polymers to the membrane. For example, B. subtilis employs SepF, EzrA and FtsA (Duman et al., 2013, Singh et al., 2007), while Caulobacter crescentus uses FzlC, FtsA and possibly a third protein (Meier et al., 2016). SepF and possibly other proteins are likely the membrane anchors in cyanobacteria and actinobacteria that lack FtsA (Marbouty et al. 2009, Gola et al. 2015). The assembly pathway is also altered compared to E. coli. For example, FtsN localizes to the Z ring earlier than FtsA in C. crescentus (Goley et al., 2011). Moreover, proteins that are highly conserved and critical for division in E. coli may be dispensable in other species. For example, FtsQ and FtsA can be bypassed in B. subtilis (Beall & Lutkenhaus, 1992, Beall & Lutkenhaus, 1989, Hamoen et al., 2006, Ishikawa et al., 2006) and deletion of FtsB has no significant effect on cell division in C. crescentus (Goley et al., 2011).
FtsN, which is highlighted here for its ability to trigger septation, is not as highly conserved (appears only in Proteobacteria) as the division proteins in the dcw cluster (FtsZ, A, I, W, L, and Q). As activation of the divisome is likely to be a conserved feature, it remains to be seen how this is accomplished in other bacteria. Those proteins encoded outside of the dcw region, ZipA, FtsK, FtsEX, FtsB and FtsN can be bypassed under certain conditions. These observations raise a question of what is absolutely required for FtsZ-dependent cell division. We suggest that there are only a few elements that are absolutely required: FtsZ, a membrane tether, PG synthetase (transglycoslyase and transpeptidase, FtsW and FtsI, respectively) and a protein that connects the PG synthases to the Z ring. In E. coli these activities are all encoded in the dcw cluster. All other proteins are encoded outside of this cluster and appear to be integrated into the system to facilitate and regulate the division process. This explains why certain proteins can be bypassed in E. coli and why some proteins can be substituted by functional homologs in other bacteria. What is clear though, is that a thorough understanding of this process in E. coli will serve as a useful guide and reference for studying this process in other bacteria.
Conclusion and outlook
The last few years have seen significant advancements in our understanding of the cytokinesis process in E. coli, including the elucidation of the role of FtsN in activating the divisome complex (PG synthases) for septal PG synthesis (Liu et al., 2015), the identification of factors that regulate divisome activation (Liu et al., 2015, Tsang & Bernhardt, 2015b, Du et al., 2016), the discovery of SEDS family proteins as PG glycosyltransferases (Cho et al., 2016, Emami et al., 2017, Meeske et al., 2016) and the revelation of the role of FtsZ dynamics in septal PG synthesis (Yang et al., 2017, Bisson-Filho et al., 2017). However, many challenges still remain. Two central questions about divisome assembly are how the late divisome proteins are connected to the Z ring and why there is a delay between Z ring formation and completion of divisome assembly. The current model suggests that the second cascade of divisome proteins (from FtsK to FtsN) form a complex that is recruited to the Z ring by interaction with FtsA monomers (Pichoff et al., 2015). Although FtsN interacts with FtsA, it is the last to be recruited, making it an unfavorable candidate to initiate the cascade. FtsK is most likely the one that links this sub-complex to FtsA, but how FtsK is recruited to the Z ring is not clear. There is some evidence that FtsK interacts with FtsZ (Dubarry et al., 2010) but its recruitment depends upon FtsA (Wang & Lutkenhaus, 1998) but there is no evidence for direct interaction between FtsA and FtsK. The model for the role of FtsA monomers in recruitment is well supported by genetic data, but suffers from the lack of biochemical data. Future characterization of FtsK localization and in vitro characterization of FtsA, although difficult, should provide some insight into these questions.
Another challenge is to understand how the divisome is activated for septal PG synthesis. With the identification of FtsW as a putative PG glycosyltransferase dedicated for division, it becomes apparent that most of the other divisome proteins are there to localize it and its cognate partner FtsI to the division site and to regulate their activities. Consistent with this, FtsA, FtsQLB, FtsN and FtsEX have been proposed to play critical roles in activating septal PG synthesis even though the mechanism is not clear (Du et al., 2016, Liu et al., 2015, Tsang & Bernhardt, 2015b). Perhaps the key to understanding this regulation is a detailed understanding of the function of FtsN, especially the function of NFtsN and EFtsN. Does NFtsN just capture FtsA in the monomer state or does it affect the conformation of FtsA and switch FtsA to the on state? Does EFtsN directly interact with FtsQLB? If it does, how does this interaction affect the conformation of the FtsQLB complex? If FtsN is not highly conserved how is septal PG synthesis regulated in other bacteria? Does the altered FtsQLB complex interact directly with FtsW and FtsI to activate their enzymatic activities? How does the ATPase cycle of FtsEX control the activation of FtsA or the signal transduction from FtsA to FtsQLB? How does increasing the osmolarity bypass FtsEX? Answers to all these questions require a combination of genetic, biochemical and imaging approaches to characterize these proteins and to reconstitute the whole process of PG synthesis by FtsW and FtsI. Despite the difficulties in working with these proteins, most of which are membrane proteins, we envision that answers to these questions will emerge soon with the development of biochemical assays to reconstitute PG synthesis, advancement in imaging and most importantly the collaboration between colleagues with different expertise.
Acknowledgments
The work is supported by a grant from NIH to JL (GM29764).
References
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- Addinall SG, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- Addinall SG, Lutkenhaus J. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol. 1996;22:231–237. doi: 10.1046/j.1365-2958.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- Arends SJ, Kustusch RJ, Weiss DS. ATP-binding site lesions in FtsE impair cell division. J Bacteriol. 2009;191:3772–3784. doi: 10.1128/JB.00179-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. Nucleotide sequence and insertional inactivation of a Bacillus subtilis gene that affects cell division, sporulation, and temperature sensitivity. J Bacteriol. 1989;171:6821–6834. doi: 10.1128/jb.171.12.6821-6834.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J Bacteriol. 1992;174:2398–2403. doi: 10.1128/jb.174.7.2398-2403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bisson-Filho AW, Hsu YP, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, Brun YV, Garner EC. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 2017;355:739–743. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acady Sci U S A. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta GA, Park JT. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J Bacteriol. 1981;145:333–340. doi: 10.1128/jb.145.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddelmeijer N, Beckwith J. Assembly of cell division proteins at the E. coli cell center. Curr Opin Microbiol. 2002;5:553–557. doi: 10.1016/s1369-5274(02)00374-0. [DOI] [PubMed] [Google Scholar]
- Buddelmeijer N, Beckwith J. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol Microbiol. 2004;52:1315–1327. doi: 10.1111/j.1365-2958.2004.04044.x. [DOI] [PubMed] [Google Scholar]
- Buddelmeijer N, Judson N, Boyd D, Mekalanos JJ, Beckwith J. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc Natl Acady Sci U S A. 2002;99:6316–6321. doi: 10.1073/pnas.092128499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busiek KK, Eraso JM, Wang Y, Margolin W. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol. 2012;194:1989–2000. doi: 10.1128/JB.06683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busiek KK, Margolin W. A role for FtsA in SPOR-independent localization of the essential Escherichia coli cell division protein FtsN. Mol Microbiol. 2014;92:1212–1226. doi: 10.1111/mmi.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss J, Coltharp C, Shtengel G, Yang X, Hess H, Xiao J. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 2015;11:e1005128. doi: 10.1371/journal.pgen.1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss JA, Peters NT, Xiao J, Bernhardt TG. ZapA and ZapB form an FtsZ-independent structure at midcell. Mol Microbiol. 2017 doi: 10.1111/mmi.13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson MJ, Barondess J, Beckwith J. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J Bacteriol. 1991;173:2187–2195. doi: 10.1128/jb.173.7.2187-2195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol Microbiol. 2001;42:395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PD, Suh H, Marto JA, Garner EC, Bernhardt TG. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol. 2016;16172 doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltharp C, Xiao J. Beyond force generation: Why is a dynamic ring of FtsZ polymers essential for bacterial cytokinesis? BioEssays. 2017;39:1–11. doi: 10.1002/bies.201600179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Wang Y, Beuria TK, Margolin W. Interaction between cell division proteins FtsE and FtsZ. J Bacteriol. 2007;189:3026–3035. doi: 10.1128/JB.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- de Boer PA. Advances in understanding E. coli cell fission. Curr Opin Microbiol. 2010;13:730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper GC, McLennan N, Begg K, Masters M, Donachie WD. Only the N-terminal domain of FtsK functions in cell division. J Bacteriol. 1998;180:4621–4627. doi: 10.1128/jb.180.17.4621-4627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S, Pichoff S, Lutkenhaus J. FtsEX acts on FtsA to regulate divisome assembly and activity. Proc Natl Acady Sci U S A. 2016;113:E5052–5061. doi: 10.1073/pnas.1606656113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubarry N, Possoz C, Barre FX. Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Mol Microbiol. 2010;78:1088–1100. doi: 10.1111/j.1365-2958.2010.07412.x. [DOI] [PubMed] [Google Scholar]
- Duman R, Ishikawa S, Celik I, Strahl H, Ogasawara N, Troc P, Lowe J, Hamoen LW. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc Natl Acady Sci U S A. 2013;110:E4601–4610. doi: 10.1073/pnas.1313978110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J Bacteriol. 2012;194:3189–3198. doi: 10.1128/JB.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Heredia JM, Yu HH, De Carlo S, Lesser CF, Janakiraman A. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J Bacteriol. 2011;193:1405–1413. doi: 10.1128/JB.01258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami K, Guyet A, Kawai Y, Devi J, Wu LJ, Allenby N, Daniel RA, Errington J. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol. 2017;2:16253. doi: 10.1038/nmicrobiol.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biology Rev: MMBR. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Borne R, Dupaigne P, Thiel A, Gigant E, Mercier R, Boccard F. A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 2012;31:3198–3211. doi: 10.1038/emboj.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Elraheb D, Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acady Sci U S A. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol Microbiol. 2005;58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding MA, Liu B, Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. Self-enhanced accumulation of FtsN at Division Sites and Roles for Other Proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill DR, Hatfull GF, Salmond GP. A new cell division operon in Escherichia coli. Mol Gen Genet. 1986;205:134–145. doi: 10.1007/BF02428043. [DOI] [PubMed] [Google Scholar]
- Glas M, van den Berg van Saparoea HB, McLaughlin SH, Roseboom W, Liu F, Koningstein GM, Fish A, den Blaauwen T, Heck AJ, de Jong L, Bitter W, de Esch IJ, Luirink J. The soluble periplasmic domains of Escherichia coli cell division proteins FtsQ/FtsB/FtsL form a trimeric complex with submicromolar affinity. J Biol Chem. 2015;290:21498–21509. doi: 10.1074/jbc.M115.654756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Goehring NW, Gonzalez MD, Beckwith J. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol Microbiol. 2006;61:33–45. doi: 10.1111/j.1365-2958.2006.05206.x. [DOI] [PubMed] [Google Scholar]
- Goehring NW, Gueiros-Filho F, Beckwith J. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Genes Devel. 2005;19:127–137. doi: 10.1101/gad.1253805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gola S, Munder T, Casonato S, Manganelli R, Vicente M. The essential role of SepF in mycobacterial division. Mol Microbiol. 2015;97:560–576. doi: 10.1111/mmi.13050. [DOI] [PubMed] [Google Scholar]
- Goley ED, Yeh YC, Hong SH, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. Assembly of the Caulobacter cell division machine. Mol Microbiol. 2011;80:1680–1698. doi: 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Devel. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Weiss DS, Beckwith J. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J Bacteriol. 1997;179:5094–5103. doi: 10.1128/jb.179.16.5094-5103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusser DP, Rowlett VW, Margolin W. A mutation in Escherichia coli ftsZ bypasses the requirement for the essential division gene zipA and confers resistance to FtsZ assembly inhibitors by stabilizing protofilament bundling. Mol Microbiol. 2015;97:988–1005. doi: 10.1111/mmi.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Hale CA, Shiomi D, Liu B, Bernhardt TG, Margolin W, Niki H, de Boer PA. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J Bacteriol. 2011;193:1393–1404. doi: 10.1128/JB.01245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen LW, Meile JC, de Jong W, Noirot P, Errington J. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol Microbiol. 2006;59:989–999. doi: 10.1111/j.1365-2958.2005.04987.x. [DOI] [PubMed] [Google Scholar]
- Haney SA, Glasfeld E, Hale C, Keeney, He Z, de Boer P. Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J Biol Chem. 2001;276:11980–11987. doi: 10.1074/jbc.M009810200. [DOI] [PubMed] [Google Scholar]
- Herricks JR, Nguye D, Margolin W. A thermosensitive defect in the ATP binding pocket of FtsA can be suppressed by allosteric changes in the dimer interface. Mol Microbiol. 2014;94:713–727. doi: 10.1111/mmi.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Sato T, Wachi M, Jung HK, Ishino F, Kobayashi Y, Matsuhashi M. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J Bacteriol. 1989;171:6375–6378. doi: 10.1128/jb.171.11.6375-6378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Kawai Y, Hiramatsu K, Kuwano M, Ogasawara N. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol Microbiol. 2006;60:1364–1380. doi: 10.1111/j.1365-2958.2006.05184.x. [DOI] [PubMed] [Google Scholar]
- Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. 2005;187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Persons L, Lee L, de Boer PA. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol Microbiol. 2015;95:945–970. doi: 10.1111/mmi.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Draper GC, Donachie WD. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol Microbiol. 1998;29:893–903. doi: 10.1046/j.1365-2958.1998.00986.x. [DOI] [PubMed] [Google Scholar]
- Lowe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annual Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. FtsN–trigger for septation. J Bacteriol. 2009;191:7381–7382. doi: 10.1128/JB.01100-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton. 2012;69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J Bacteriol. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Männik J, Bailey MW, O’Neill JC, Männik J. Kinetics of large-scale chromosomal movement during asymmetric cell division Escherichia coli. PLoS Genet. 13:e1006638. doi: 10.1371/journal.pgen.1006638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbouty M, Saguez C, Cassier-Chauvat C, Chauvat F. Characterization of the FtsZ- interacting septal proteins SepF and Ftn6 in the spherical-celled cyanobacterium Synechocystis strain PCC 6803. J Bacteriol. 2009;91:6178–85. doi: 10.1128/JB.00723-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovski M, Bohrhunter JL, Lupoli TJ, Uehara T, Walker S, Kahne DE, Bernhardt TG. Cofactor bypass variants reveal a conformational control mechanism governing cell wall polymerase activity. Proc Natl Acady Sci U S A. 2016;113:4788–4793. doi: 10.1073/pnas.1524538113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, Rudner DZ. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier EL, Razavi S, Inoue T, Goley ED. A novel membrane anchor for FtsZ is linked to cell wall hydrolysis in Caulobacter crescentus. Mol Microbiol. 2016;101:265–280. doi: 10.1111/mmi.13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, Sijbrandi R, Lutters M, Verheul J, Martin NI, den Blaauwen T, de Kruijf B, Breukink E. Specificity of the transport of lipid II by FtsW in Escherichia coli. J Biol Chem. 2014;289:14707–14718. doi: 10.1074/jbc.M114.557371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, van Dam V, Sijbrandi R, Vernet T, Zapun A, Bouhss A, Diepeveen-de Bruin M, Nguyen-Disteche M, de Kruijff B, Breukink E. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, Seehra J, Somers WS. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19:3179–3191. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acady Sci U S A. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Du S, Lutkenhaus J. The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol Microbiol. 2015;95:971–987. doi: 10.1111/mmi.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBOJl. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Shen B, Sullivan B, Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol. 2012;83:151–167. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D, Park JT. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- Reddy M. Role of FtsEX in cell division of Escherichia coli: viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J Bacteriol. 2007;189:98–108. doi: 10.1128/JB.01347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KL, Peterson ND, Kustusch RJ, Wissel MC, Graham B, Phillips GJ, Weiss DS. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J Bacteriol. 2004;186:785–793. doi: 10.1128/JB.186.3.785-793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh JK, Makde RD, Kumar V, Panda D. A membrane protein, EzrA, regulates assembly dynamics of FtsZ by interacting with the C-terminal tail of FtsZ. Biochemistry. 2007;46:11013–11022. doi: 10.1021/bi700710j. [DOI] [PubMed] [Google Scholar]
- Steiner W, Liu G, Donachie WD, Kuempel P. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acady Sci U S A. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Freund SM, Lowe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang MJ, Bernhardt TG. Guiding divisome assembly and controlling its activity. Curr Opin Microbiol. 2015a;124:60–65. doi: 10.1016/j.mib.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang MJ, Bernhardt TG. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol Microbiol. 2015b;95:925–944. doi: 10.1111/mmi.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Bernhardt TG, Vollmer W. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Lowe J. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 2000;19:5300–5307. doi: 10.1093/emboj/19.20.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- Yahashiri A, Jorgenson MA, Weiss DS. Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc Natl Acady Sci U S A. 2015;112:11347–11352. doi: 10.1073/pnas.1508536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acady Sci U S A. 2011;108:E1052–1060. doi: 10.1073/pnas.1107780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science. 2017;355:744–747. doi: 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Tran AH, Sun A, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998a;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Weihe EK, Margolin W. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J Bacteriol. 1998b;180:6424–6428. doi: 10.1128/jb.180.23.6424-6428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]


