Abstract
Background
Taenia solium, the cause of neurocysticercosis (NCC), has significant socioeconomic impacts on communities in developing countries. This disease, along with taeniasis is estimated to infect 2.5 to 5 million people globally. Control of T. solium NCC necessitates accurate diagnosis and treatment of T. solium taeniasis carriers. In areas where all three species of Taenia tapeworms (T. solium, Taenia saginata and Taenia asiatica) occur sympatrically, conventional microscope- and copro-antigen based diagnostic methods are unable to distinguish between these three Taenia species. Molecular diagnostic tools have been developed to overcome this limitation; however, conventional PCR-based techniques remain unsuitable for large-scale deployment in community-based surveys. Moreover, a real-time PCR (qPCR) for the discrimination of all three species of Taenia in human stool does not exist. This study describes the development and validation of a new triplex Taq-Man probe-based qPCR for the detection and discrimination of all three Taenia human tapeworms in human stools collected from communities in the Central Highlands of Vietnam. The diagnostic characteristics of the test are compared with conventional Kato Katz (KK) thick smear and copro-antigen ELISA (cAgELISA) method utilizing fecal samples from a community based cross-sectional study. Using this new multiplex real-time PCR we provide an estimate of the true prevalence of taeniasis in the source population for the community based cross-sectional study.
Methodology/Principal findings
Primers and TaqMan probes for the specific amplification of T. solium, T. saginata and T. asiatica were designed and successfully optimized to target the internal transcribed spacer I (ITS-1) gene of T. solium and the cytochrome oxidase subunit I (COX-1) gene of T. saginata and T. asiatica. The newly designed triplex qPCR (T3qPCR) was compared to KK and cAgELISA for the detection of Taenia eggs in stool samples collected from 342 individuals in Dak Lak province, Central Highlands of Vietnam. The overall apparent prevalence of taeniasis in Dak Lak province was 6.72% (95% confidence interval (CI) [3.94–9.50]) in which T. solium accounted for 1.17% (95% CI [0.37–3.17]), according to the T3qPCR. There was sympatric presence of T. solium, T. saginata and T. asiatica. The T3qPCR proved superior to KK and cAgELISA for the detection and differentiation of Taenia species in human feces. Diagnostic sensitivities of 0.94 (95% credible interval (CrI) [0.88–0.98]), 0.82 (95% CrI [0.58–0.95]) and 0.52 (95% CrI [0.07–0.94]), and diagnostic specificities of 0.98 (95% CrI [0.94–1.00]), 0.91 (95% CrI [0.85–0.96]) and 0.99 (95% CrI [0.96–1.00]) were estimated for the diagnosis of taeniasis for the T3qPCR, cAgELISA and KK thick smear in this study, respectively.
Conclusions
T3qPCR is not only superior to the KK thick smear and cAgELISA in terms of diagnostic sensitivity and specificity, but it also has the advantage of discriminating between species of Taenia eggs in stools. Application of this newly developed T3qPCR has identified the existence of all three human Taenia tapeworms in Dak Lak province and proves for the first time, the existence of T. asiatica in the Central Highlands and the south of Vietnam.
Author summary
Human taeniid tapeworms comprise three species, Taenia solium, Taenia saginata and Taenia asiatica. Taeniasis is a meat-borne zoonosis transmitted by the consumption of cysticerci in raw or undercooked pork for T. solium and T. asiatica (liver) and in beef for T. saginata. Accidental ingestion of T. solium eggs by humans also results in the formation of cysticerci, often in the brain, referred to as neurocysticercosis (NCC). T. solium NCC is a significant cause of morbidity and mortality owing to epilepsy in many resource-poor communities. In animals, ingestion of eggs passed by humans results in organ and/or carcass condemnation and suboptimal economic outcomes for farmers. The accurate diagnosis of T. solium tapeworm carriers is essential to monitor the success of control programs. In areas where all three species of Taenia tapeworms occur together, conventional diagnostic methods are unable to distinguish between the different species of Taenia. In this study, we develop and apply a T3qPCR capable of detecting and discriminating all three-tapeworm species in stools in a rapid and high-throughput fashion, suitable for large-scale community surveys. The newly developed T3qPCR proved superior to previously developed immunodiagnostic and conventional microscopic-based tests in terms of diagnostic sensitivity, specificity and the ability to identify and distinguish human Taenia species. This qPCR assay facilitated the identification of T. asiatica tapeworms in the Central Highlands of Vietnam, for the first time.
Introduction
Humans are infected with three species of Taenia—Taenia solium, Taenia saginata and Taenia asiatica [1,2]. While T. saginata has a global distribution, T. solium is distributed mostly in developing countries of Latin America, sub-Saharan Africa and Asia, and T. asiatica is restricted to certain Asian countries [3,4]. Taeniasis is estimated to infect 2.5 to 5 million people globally [5–7]. All three-tapeworm species utilize humans as the definitive hosts (adult tapeworm) due to the ingestion of undercooked and/or raw meat or liver [1,8]. The intermediate hosts of T. solium and T. asiatica are swine whereas the intermediate hosts of T. saginata are cattle (porcine/bovine cysticercosis) [3]. Humans may also become the accidental intermediate hosts of T. solium via the ingestion of food and water contaminated with T. solium eggs, which may result in neurocysticercosis (NCC) when the cysts lodge in the central nervous system. The clinical manifestations of T. solium NCC in humans are varied, ranging from being asymptomatic to severe neurological signs and symptoms such as epilepsy, paralysis, dementia, chronic headache, blindness or even death [9–11]. The symptoms of taeniasis in humans, on the other hand, are mostly subtle and mild, and include abdominal distension, abdominal pain, digestive disorders and anal pruritus (mostly for T. saginata and T. asiatica) [12,13].
Control of NCC necessitates accurate diagnosis and treatment of T. solium taeniasis carriers to break the cycle of transmission to pigs [14]. Although all cases of taeniasis necessitate treatment, species-specific identification from an epidemiological perspective will allow better targeted control programs to be implemented aimed at interrupting the lifecycle specific to each species of Taenia endemic within a community and region. Several diagnostic tools for detecting taeniasis, including microscopy, copro-antigen ELISAs, sero-antibody immunoblot [15–17], and copro-DNA tests have been developed[18,19]. Of these diagnostic tools, microscopy-based examinations of fecal samples have the limitation of poor diagnostic sensitivity owing to intermittent shedding of proglottids/eggs and low specificity owing to the inability to morphologically discriminate between the eggs of Taenia species [20]. Although self-detection of proglottid shedding is valuable for confirmation of individual (but not community-wide) taeniasis, it is however, not always reliable especially when deep pit latrines are being utilized. Moreover, in cases of infection with T. solium, the proglottids are immobile and may be misidentified as other worms [19]. Differentiation of T. saginata from T. asiatica based on proglottid morphology is onerous and challenging [21]. The cAgELISA utilizes polyclonal antigens and more sensitive (0.85, 95% CI [0.62–0.98]) than microscopy-based methods (0.53, 95% CI [0.11–0.97]) [22], is only capable of identification to a genus level [23–25]. More recently, a cAgELISA specific to T. solium was developed [20,26]. Another drawback of the cAgELISA is that it may miss cases of T. asiatica and T. saginata in areas where other human Taenia tapeworms co-exist [19]. Serum antibody detecting immunoblot assays using excretory/secretory or recombinant antigens have been developed for identification of antibodies to T. solium [15,17] and T. asiatica taeniasis [16]. Immunoblot assays were highly sensitive and specific when used in Taenia non-endemic areas, however resulted in a high proportion of false positive results in areas endemic for T. saginata [17]. Copro-PCRs are considered superior tools for the detection of Taenia eggs in fecal samples owing to their high diagnostic sensitivity and the ability to distinguish Taenia species. Currently, the only multiplex-PCR [27,28] and PCR-RFLP [21] that is able to differentiate all human Taenia tapeworms is unsuited for large-scale community surveys owing to labor intensive process of restriction digest and gel electrophoresis. A better suited real-time PCR [29] has been developed but it is only capable of discriminating T. solium and T. saginata. Loop-mediated isothermal amplification (LAMP) [30] has also shown promise as a potential point-of-care diagnostic technique capable of highly sensitive detection (one copy of target gene/reaction or at least five eggs/gram of feces) and differentiation of Taenia spp. in stool [30], however the technology is still far from point-of-care based and is considered equivocal to PCR [19].
This study describes the development and validation of a new multiplex Taq-Man probe-based qPCR for the detection and discrimination of all three species of Taenia in human stools. The diagnostic characteristics of the test are compared with the test characteristics of conventional KK thick smear and the cAgELISA using hyper immune rabbit anti-Taenia IgG polyclonal antibody by applying all three diagnostic tests to detect Taenia spp. in fecal samples collected as part of a community-based cross-sectional study carried out in the Central Highlands of Vietnam.
Materials and methods
Ethics statement
This study was reviewed and approved by the Behavioural and Social Sciences Human Ethics Sub-committee, the University of Melbourne (reference number 1443512) and conducted under the supervision of the local Center for Public Health, Dak Lak, Vietnam. Participants positive for taeniasis on KK thick smear were treated immediately by local medical officers with praziquantel (Distocide, Shinpoong Daewoo Pharma CO.LTD) at 20 mg/kg as a single dose, whereas those positive by cELISA and qPCR were treated once results were known.
Production of polyclonal antibodies in rabbits used for cAgELISA were obtained from the Ethical Committee for animal experiments of the Institute of Tropical Medicine, Antwerp (Reference number DG012S).
Multiplex real-time PCR
Primers and probes
The internal transcribed spacer I (ITS-1) gene of T. solium (GenBank accession number LC004200) was utilized to design a T. solium specific primer and Taq-Man-based hydrolysis probe for the specific amplification of T. solium. Sequences spanning the cytochrome oxidase subunit I (COX-1) gene were utilized to design a primer pair for specific amplification of T. saginata and T. asiatica concurrently, and two different species-specific Locked Nucleic Acid (LNA) probes were used for detection of T. saginata, and T. asiatica. Primers and probe of equine herpesvirus 4 (EHV4) used as an internal control were based on a previously published paper [31] (Table 1).
Table 1. Primers and probes for multiplex qPCR.
| Target species | Oligonucleotide | GenBank accession # | Target gene | Sequence (5’ - 3’) |
|---|---|---|---|---|
| T. solium | Forward primer | LC004200 | ITS-1 | TGTTAGCAGCAGTTGTGATG |
| Reverse primer | CCAACTCCACCCAGATTGA | |||
| Probe | Cy5/CCAGCGCTG/TAO/CTGTTGACTGA/IAbRQSp | |||
| T. asiatica and T. saginata | Forward primer of T. asiatica and T. saginata | AB107234 and AB271695 | COX-1 | GAGTACCAACAGGAATAAAGGT |
| Reverse primer of T. asiatica and T. saginata | CAACACAATACCAGTCACACC | |||
| Probe of T. asiatica | FAM/AA+CTA+T+C+CA+CCA/IAbkFQ | |||
| Probe of T. saginata | HEX/AA+CTA+T+T+CA+C+CA/IAbkFQ | |||
| EHV4 | Forward primer | M26171 | Glycoprotein gB | GATGACACTAGCGACTTCGA |
| Reverse primer | CAGGGCAGAAACCATAGACA | |||
| Probe | ROX/TTTCGCGTGCCTCCTCCAG/IAbRQSp |
“+” symbols found in the probe sequences indicate the presence of a Locked Nucleic Acid Base (LNA); IAbRQSp: Iowa Black RQ-Sp; IAbkFQ: Iowa Black FQ
Multiplex qPCR conditions
DNA of EHV4 was spiked into T3qPCR prepared reaction mixtures as an internal reaction control. The multiplex qPCR assay was run as a 20 μl reaction containing 10 μl of GoTaq Probe qPCR Master Mix (Promega Corporation, Madison, WI, USA), 2.44 μl of H2O, 350 nM of each primer of T. solium, T. saginata and T. asiatica, 40 nM of each primer of EHV, 200 nM probe of T. solium, 250 nM each probe of T. saginata and T. asiatica, 100 nM probe of EHV, 1 μl of EHV4 DNA diluted 20 times and 2 μl of DNA template.
The T3qPCR amplification was carried out in a Magnetic Induction Cycler, MIC (Bio Molecular Systems). The cycling conditions consisted of an initial denaturation step at 95°C for 2 minutes, followed by 40 amplification cycles, each comprising a denaturation step at 95°C for 30 seconds and annealing at 66°C for 60 seconds. All samples were tested in duplicate with positive and negative control samples were included in each amplification assay.
Multiplex qPCR controls
Synthesized gBlock gene fragments of T. solium, T. saginata and T. asiatica were used as the positive controls for the T3qPCR assays. The gBlocks Gene Fragments (Integrated DNA Technologies (IDT), Coralville, USA) are double-stranded and sequence-verified DNA molecules that vouch for the fidelity of the T3qPCR assays. The gBlock fragments of T. solium and T. asiatica were synthetically combined together to an amplicon size of 235 bp in length while the T. saginata gene fragment was synthesized as an individual amplicon with a length of 140 bp (Table 2).
Table 2. gBlocks positive controls.
| Organisms | GenBank accession number | Gene | Star-end position1 | Amplicon length |
|---|---|---|---|---|
| T. solium and T. asiatica | LC004200 | ITS-1 | 92–183 | 235 bp |
| AB107234 | COX-1 | 938–1077 | ||
| T. saginata | AB271695 | COX-1 | 938–1077 | 140 bp |
1 Original position in GenBank
In addition, gDNA from proglottids sourced from individuals in this study (following treatment and recovery) belonging to T. asiatica and T. saginata as well as T. solium from eggs sourced from the Centers of Disease Control and Prevention (Atlanta, GA, US) were utilized as positive controls for this study. T. asiatica and T. saginata proglottids were morphologically identified and confirmed using a previously published PCR [27] or singleplex PCR followed by DNA sequencing.
The individual qPCR reactions were optimized by subjecting individual gBlocks for each Taenia species to serial 10-fold dilutions ranging from 5 × 10−7 to 5 × 101 pg/μl and then mixed together to assess the diagnostic sensitivity, specificity and efficiency of the multiplex assay. EHV4 DNA templates were also included in optimization of the multiplex qPCR. Normal melt curves and absolute quantification analyses were used to determine the positive status of individual samples. Threshold and fluorescence cut off level were set up at 0.1 and 16%, respectively, except for fluorescence cut off level of EHV at 20%, and the first cycle of qPCR amplification was ignored from the analysis. The T3qPCR results were considered negative if cycle threshold (Ct) values were > 35. This value (5 × 10−5 pg/μl) was the limit of our standard curves (additional gBlocks dilutions were undetectable). Each sample was checked for fidelity/inhibition by comparing the Ct-value of the EHV4 in the sample compared to the EHV4 Ct-value in the negative control. An assay was deemed a failure when its EHV4 Ct-value was greater than two cycles different in comparison to the negative control EHV4 Ct-value.
The diagnostic specificity of the T3qPCR was confirmed utilizing DNA templates from 12 different human intestinal parasites and one canine tapeworm, including Cryptosporidium parvum, Giardia duodenalis, Haplorchis taichui, Hymenolepis nana, Opisthorchis viverrini, Ancylostoma ceylanicum, Necator americanus, Trichuris trichiura, Ascaris lumbricoides, Strongyloides stercoralis, Blastocystis hominis, Clonorchis sinensis and Taenia hydatigena. These parasitic DNA templates were sourced either from feces or adult worms, and confirmed using conventional PCR and DNA sequencing.
Multiplex qPCR confirmation
The positive results of T3qPCR were confirmed using either a conventional multiplex PCR targeting T. solium, T. saginata and T. asiatica developed by Jeon et al. (2009) [32] or a singleplex PCR for Taenia sp. following DNA sequencing.
The singleplex PCR utilized self-designed forward 5’-CATCATATGTTTACGGTTGG-3’ and reverse primer 5’-GACCCTAATGACATAACATAAT-3’ designed based on COX-1 gene and amplifying a gene of 350 bp. The assay comprised 2.5μl 10×CoralLoad PCR Buffer (Qiagen, Hiden, Germany), 12.5 pmol of each primer, 0.5 U of HotStar Taq DNA Polymerase, 2μl dNTPs (4 mM), 0.5 μl of MgCl2 (25 mM) and 1 μl of DNA, in a total volume of 25 μl. The cycling conditions consisted of an initial denaturation step at 95°C for 5 minutes, 52°C for 1 minute and 72°C for 2 minutes followed by 40 amplification cycles, each comprising a denaturation step at 95°C for 30 seconds, annealing at 52°C for 30 seconds and extension at 72°C for 30 seconds and final extension step at 72°C for 4 minutes. The singleplex PCR products were sent to Macrogen Inc. (Korea) for sequencing.
The multiplex PCR assay comprised 2.5 μl 10×CoralLoad PCR Buffer (Qiagen, Hiden, Germany), 12.5 pmol of each forward primer, 37.5 pmol reverse primer, 0.5 U of HotStar Taq DNA Polymerase, 2μl dNTPs (4 mM), 0.5 μl of MgCl2 (25 mM) and 1 μl of DNA, in a total volume of 25 μl. The cycling conditions consisted of an initial denaturation step at 95°C for 5 minutes, 56°C for 1 minute and 72°C for 2 minutes followed by 45 amplification cycles, each comprising a denaturation step at 95°C for 30 seconds, annealing at 56°C for 30 seconds and extension at 72°C for 30 seconds and final extension step at 72°C for 4 minutes.
DNA extraction of fecal samples
Samples stored in 5% potassium dichromate (w/v) were washed three times in distilled water and then subjected to DNA extraction. Approximately 250 mg washed stool was subsequently extracted using the PowerSoil DNA Isolation Kit (Mo Bio, Carlsbad, CA, USA). The protocol of extraction was in accordance with the manufacturer’s instructions, with the exception that provided beads were replaced with 1 g of Silica/Zirconia 0.5 mm beads (Daintree Scientific, Tasmania, Australia). DNA was eluted in a final volume of 100 μl of elution buffer. The quantity and quality of the extracted DNA was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE) and results were analyzed using the NanoDrop 1000 software. The DNA extracted was determined suitable when the yield was > = 2 ng/μl and the ratio of the absorbance at 260 and 280 nm and at 260 and 230 nm were > 1.70 and 2.0–2.2, respectively. The extracted DNA was stored at −20°C until use. All samples were subjected to the multiplex qPCR for the detection and differentiation of Taenia spp. as described earlier.
DNA extraction of proglottids
Proglottids were extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and eluted in 200 μl of AE Buffer.
Coproantigen enzyme-linked-immunosorbent assay
An in-house coproantigen detection ELISA [33] with slight modifications, as described by Mwape et al. (1990) [34], was performed on the stool samples. Briefly, a mixture of an equal amount of Phosphate Buffered Saline (PBS) and stool sample stored in 10% formalin was soaked for one hour with slight shaking and centrifuged at 2000 g for 30 minutes where after supernatant was collected. The Polystyrene ELISA plates (Nunc Maxisorp, Waltham, MA, USA) were coated with capturing hyper immune rabbit anti-Taenia IgG polyclonal antibody diluted at 2.5 mg/ml in carbonate-bicarbonate buffer (0.06 M, pH 9.6) and incubated for 1 hour at 37°C. The plates were washed once with PBS in 0.05% Tween 20 (PBS-T20). All wells were blocked by adding PBS-T20+2% New Born Calf Serum and incubated for 1 hour at 37°C. After washing the stool supernatant of 100 ml was added to all wells and incubated for 1 hour at 37°C followed by washing five times with PBS-T20. One hundred microliter of biotinylated hyper immune rabbit IgG polyclonal antibody diluted at 2.5 mg/ml in blocking buffer was added into each well. Between two cycles of incubating for 1 hour at 37°C followed by washing five times, 100 μl of streptavidin-horseradish peroxidase (Jackson Immunoresearch Lab, West Grove, PA, USA) diluted at 1/10,000 was added. Finally, 100 ml of ortho phenylenediamine (OPD) substrate, prepared by dissolving one tablet in 6 ml of distilled water and adding 2.5 ml of hydrogen peroxide, was added to all the wells, and incubated in a dark for 15 minutes at room temperature before stopping the reaction by adding 50 ml of sulphuric acid (4 N) to each well. The plates were read using an automated spectrophotometer at 490 nm with a reference of 655 nm. To determine the test result, the optical density (OD) of each stool sample was compared with the mean of a series of eight reference Taenia negative stool samples plus 3 standard deviations.
Microscopy examination
All fecal samples were microscopically examined for the presence of Taenia eggs using a duplicate KK thick-smear technique [35–38] and examined by trained parasitologists at Tay Nguyen University.
Study site and sampling
The fieldwork was conducted between May to October 2015 in Krong Nang, M’Drak and Buon Don districts in Dak Lak province, Vietnam (Fig 1). M’ Drak is located in the east of Dak Lak province with an average altitude of 400m to 500m above sea level and a tropical monsoon climate typical of the Vietnamese Central Coast. Krong Nang is situated to the north of Dak Lak at 800m above sea level. Buon Don, situated to the west of Dak Lak at approximately 330 m above sea level and had a hot and dry climate. Living standards in each of three districts is poor; open defecation using outdoor latrines is a common practice and livestock access to these latrine areas is, for the most part, unrestricted. Moreover, consumption of raw or undercooked blood, meat, organ tissues from domesticated and wild pigs and cattle is common. The practice of non-confinement of pigs and cattle is common in rural regions with slaughter activities commonly carried out in backyards without official meat inspection. Meat inspection is only carried out at abattoirs or slaughter-points, which serve the district level and/or clusters of large villages [32].
Fig 1. Map of study site.
Map of Dak Lak province showing districts where sampling was carried out.
The objective of this study was to estimate the true prevalence of human taeniasis in each of the three study districts. Based on the previous study of Van De at al. [13], the true prevalence of human taeniasis was assumed to be 7% and there was 95% certainty that this estimate was within 5% of the true population value (i.e. the true prevalence ranged from 2% to 12%). Ignoring the tendency for taeniasis to cluster within households we estimated that a total of 100 stool samples were required. We then assumed the average number of individuals eligible for the study per household was at least three and the between household (cluster) variance in taeniasis prevalence was around 1.14 times greater than the within household (cluster) variance, returning an intra-class correlation coefficient of 0.07 [39]. Our revised sample size, accounting for the tendency of taeniasis to cluster within households, was 114 for each district and 342 for the whole province.
Within each district, three villages were selected at random. Sixty-seven households were randomly sampled from within each of the selected villages. Within each household a maximum of three individuals over the age of seven years were selected at random and asked if they would like to take part in the study. Those that consented were then asked to provide informed written consent for stool donation. Participants under the age of 18 years were recruited with assent of themselves as well as written consent from their parents or legal guardians. During the participant recruitment phase investigators informed study participants that they could withdraw from the study at any stage.
A labeled stool container was delivered to each individual. Fecal samples collected were divided into three aliquots. Approximately 5 g of each fecal sample was fixed in 5% potassium dichromate (w/v), and transported to the University of Melbourne, Australia for molecular analysis. The second and the third aliquots were stored in 5% formalin, shipped to the Institute of Tropical Medicine, Belgium, and kept at Tay Nguyen University, Vietnam, for cAgELISA testing and microscopy examination (KK), respectively.
Statistical analysis
Statistical analyses were carried out using R (R Development Core Team 2017) [40], Microsoft Excel 2007 and Microsoft Access 2007. Cohen’s kappa test statistic (K) was used for comparison of multiplex qPCR to Kato-Katz, and multiplex qPCR to Coproantigen ELISA.
The diagnostic sensitivity and specificity of multiplex qPCR, cAgELISA and KK were estimated using a Bayesian approach. The Bayesian approach utilised prior information based on both previous literature [22] and expert opinion (Table 3). Prior information for the diagnostic sensitivity and specificity for the T3PCR was elicited from six independent molecular parasitology diagnosticians. Prior information for the sensitivity and specificity of cAgELISA and KK derived from Praet et al. (2013) [22] (Table 3). Prior information for the true prevalence of taeniasis in the three study districts was obtained from previous studies carried out in the same region [41].
Table 3. Parameters of prior information.
| Parameter of interest | Mode | Lower limit 95% | |
|---|---|---|---|
| T3qPCR* | Se | 0.95 | 0.89 |
| Sp | 0.99 | 0.94 | |
| cAgELISA | Se | 0.85 | 0.62 |
| Sp | 0.92 | 0.90 | |
| KK | Se | 0.53 | 0.11 |
| SP | 1.00 | 1.00 | |
*Modes and lower limits for test characteristic of T3qPCR calculated as the average of the estimates elicited from each of the six experts
The model of estimation of diagnostic sensitivity and specificity for three tests applied to individuals from the same population with no gold standard test was based on Branscum et al. (2005) [42] with minor modification of the assumption that the three tests were independent [43]. The posterior distribution of diagnostic sensitivity and specificity were obtained using Markov chain Monte Carlo (MCMC) techniques. The MCMC sampler was run for 100,000 iteration and the first 1,000 ‘burn in’ samples were discarded. The point estimate and 95% credible interval (CrI) for diagnostic sensitivity and specificity were reported as the median and 2.5% and 97.5% quantiles of the posterior distribution of sensitivity and specificity. Parallel chains were run using diverse initial values to ensure that convergence was achieved to the same distribution [44]. Confirmation that the posterior estimates of the monitored parameters had converged to a stable distribution was achieved by plotting cumulative path plots for each variable [45,46] and quantified using the Raftery and Lewis convergence diagnostic [47,48].
Results
Optimization and validation of the multiplex qPCR
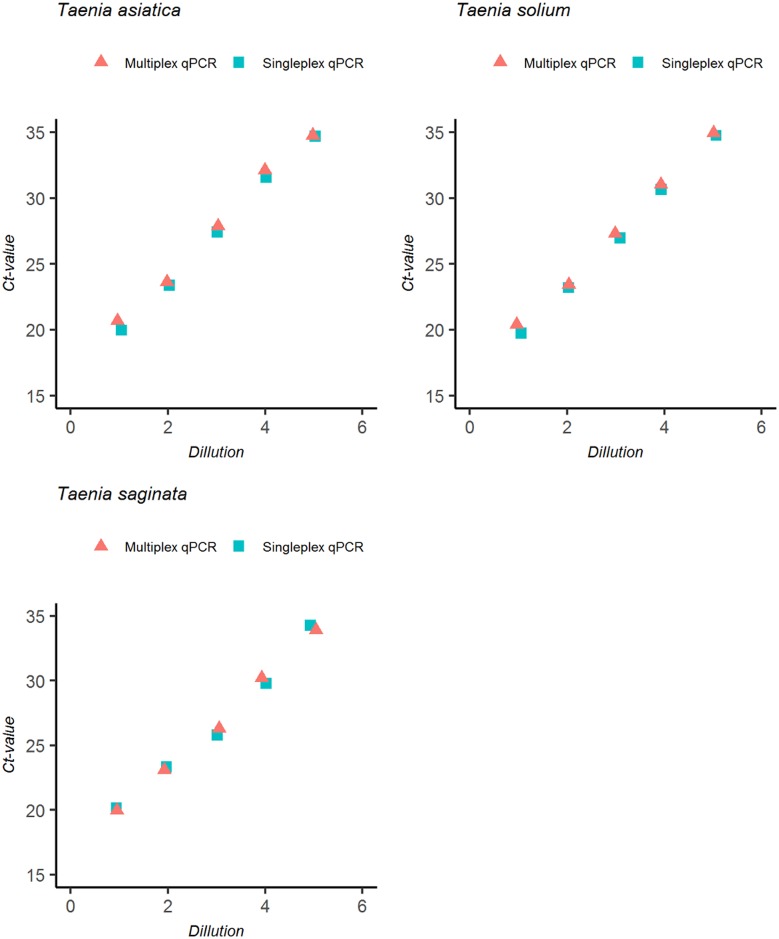
Designed primers/probes for the T3qPCR were highly species-specific and failed to cross-amplify non-target parasites including other species of Taenia. Multiplexing the assay had no effects on the sensitivity and efficiency of the qPCR (Fig 2).
Fig 2. Ct value comparison of multiplex to singleplex qPCR.
Assay optimization to determine effects of multiplex PCR set up on sensitivity and efficiency of PCRs compared to singleplex PCR using gBlock gene fragment (IDT Technologies) standard curve controls containing all PCR products.
Field study
Of 342 stool samples collected in Dak Lak province that were tested using T3qPCR and KK, 23 (6.7%, 95% CI [3.9–9.5]) and 7 (2%, 95% CI [0.9–4.4]) were positive for Taenia spp. For cAgELISA using a cut off of ≥ 0.2 OD, 21 (6.1%, 95% CI [3.9–9.4]) samples were classified positive for Taenia spp., whereas 3 (0.9%, 95% CI [0.2–2.7]) samples were classified positive for T. solium using a higher cut off of OD ≥ 0.55. The summarized test results are provided in Table 4.
Table 4. Results of three diagnostic tests for taeniasis.
| T3qPCR | KK | cAgELISA (OD>0.2) | cAgELISA (OD> = 0.55) | Individuals | ||||
|---|---|---|---|---|---|---|---|---|
| T. solium | T. saginata | T. asiatica | T. solium & T. saginata | T. saginata & T. asiatica | ||||
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 306 |
| 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 |
| 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 5 |
| 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 |
| 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 |
| 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 |
| 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 |
| 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 3 |
| 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
| 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 |
| 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 13 |
0: Negative; 1: Positive. Raw data available in S1 Data.
Seven individuals that were positive for taeniasis by KK that were administered praziquantel, eliminated at least one adult Taenia worm. In all cases, the species of Taenia identified was consistent with that detected by the T3qPCR, including one individual that harboured co-infection with T. saginata and T. asiatica. Amplification and DNA sequencing of the individual adult tapeworms recovered from this individual showed that one of them had 99% similarity with GenBank sequence LM146668 of T. asiatica and the other to have 100% similarity with GenBank sequence JN986712 of T. saginata.
Of the 23 samples positive for Taenia spp. using the T3qPCR, single infections of T. solium and T. saginata were identified in 3 (0.87%) and 14 (4.09%) individuals, respectively. No cases of a single infection with T. asiatica were identified. Mixed infections of T. solium and T. saginata occurred in one individual, and of T. saginata and T. asiatica in five individuals (Fig 3).
Fig 3. Infection status of human Taenia species.
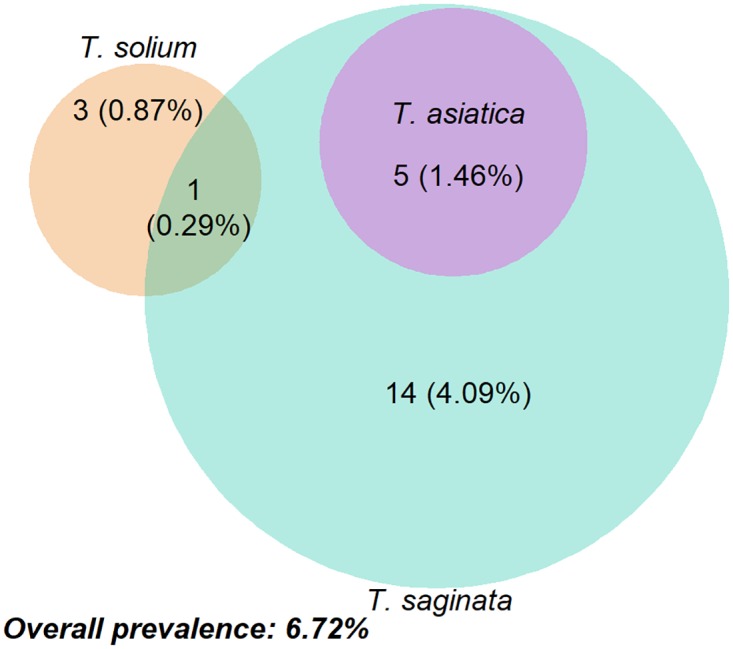
The venn diagram shows the proportional infection of Taenia spp. in Dak Lak province, Vietnam. Raw data available in S1 Data.
Comparison of T3qPCR with microscopy
The T3qPCR was more sensitive for detection of Taenia spp. eggs in human stools compared with KK. Twenty three of 342 individuals (6.72%, 95% CI [3.9–9.5]) were positive for Taenia egg using T3qPCR compared with seven of 342 individuals (2.05%, 95% CI [0.9–4.4]) positive using KK (Table 4). All the KK positive samples (7 of 342) were confirmed T. saginata and/or T. asiatica using the T3qPCR. No cases of T. solium detected by qPCR were positive by KK. Cohen’s Kappa test showed moderate agreement between the two methods (Table 5). The estimated diagnostic sensitivities of T3qPCR and KK for detecting Taenia spp. were 0.94 (95% CrI [0.88–0.98]) and 0.52 (95% CrI [0.07–0.94]), respectively, while the diagnostic specificity of the two tests were 0.98 (95% CrI [0.95–1.00]), and 0.99 (95% CrI [0.96–1.00]) (Table 6), respectively.
Table 5. The agreement statistics of T3qPCR, cAgELISA and KK for the detection of Taenia spp. in stool.
| T3qPCR | Kappa (95% CI)a | Observed agreement (%) | ||||
|---|---|---|---|---|---|---|
| Positive | Negative | Total | ||||
| cAgELISA (OD >0.2) | Positive | 8 | 13 | 21 | 0.32 (0.21–0.43) | 91.8 |
| Negative | 15 | 306 | 321 | |||
| Total | 23 | 319 | 342 | |||
| cAgELISA (OD ≥0.55) | Positive | 3 | 0 | 3 | 0.86 (0.75–0.96) | 99.7 |
| Negative | 1 | 338 | 339 | |||
| Total | 4 | 338 | 342 | |||
| KK | Positive | 7 | 0 | 7 | 0.45 (0.36–0.53) | 95.3 |
| Negative | 16 | 319 | 335 | |||
| Total | 23 | 319 | 342 | |||
a Kappa agreement level: K<0.2 Poor; 0.21–0.40 Fair; 0.41–0.60 Moderate; 0.61–0.80 Good; 0.81–1.00 Very good. Raw data available in S1 Data.
Table 6. The estimated characteristics of three diagnostic tests for the detection of Taenia spp. in stool.
| Test method | Parameter of interest | Median | 95% credible interval (CrI) |
|---|---|---|---|
| T3qPCR | Se | 0.94 | 0.88–0.98 |
| Sp | 0.98 | 0.95–1.00 | |
| cAgELISA(OD ≥0.55) | Se | 0.82 | 0.58–0.95 |
| Sp | 0.91 | 0.85–0.96 | |
| KK | Se | 0.52 | 0.07–0.94 |
| Sp | 0.94 | 0.89–0.98 |
Comparison of T3qPCR with cAgELISA
A cut off value for the cAgELISA of 0.2 was obtained by calculating the mean OD value of a series of 8-reference Taenia negative stool samples ± 3 SD. The cAgELISA identified 21 (6.14%) samples as positive for taeniasis however failed to detect 15 of 342 samples positive by T3qPCR. The agreement between the two diagnostic tests was fair with a Kappa value of 0.32 (95% CI [0.21–0.42]) (Table 5).
A cut off value of ≥0.55 OD on the cAgELISA was assumed to be more specific for the detection of T. solium antigens. Three (0.87%) of 342 individuals were positive for T. solium by this method in agreement with T3qPCR resulting in an apparent prevalence of 1.16% (95% CI [0.0–0.03]). The cAgELISA failed to detect any additional cases of T. solium detected by T3qPCR. There was very good agreement between the two diagnostic tests with Kappa of 0.86 (95% CI [0.75–0.96]) (Table 5). The diagnostic sensitivity and specificity of the cAgELISA was estimated to be 0.82 (95% CrI [0.58–0.95]) and 0.91 (95% CrI [0.85–0.96]), respectively (Table 6).
Discussion
The T3qPCR developed in this study demonstrated superior ability to both the KK thick smear and cAgELISA for the specific detection and discrimination of all three taeniid tapeworms found in humans, T. solium, T. asiatica and T. saginata. The assay proved highly specific with no cross-reactivity observed for 13 different intestinal parasites as well as with each other. The cross-reactivity of the T3qPCR with Echinococcus multilocularis and Echinococcus granulosus was not evaluated as DNA of these parasites are not expected to shed in stools. The innate advantage of this T3qPCR over other diagnostic methods is its ability to detect mixed cases of Taenia spp. infection, a necessary tool that provides a more comprehensive understanding of the epidemiology of taeniasis and Taenia cysticercosis in both humans and animals in regions where all three species are sympatric [49]. For example, in communities such as those in the Central Highlands of Vietnam, where taeniasis is highly endemic, use of the T3qPCR allows the true prevalence and risk factors for T. solium taeniasis carriers to be determined. Use of these diagnostic tools to inform taeniasis control programs will not only reduce the risk of T. solium NCC in humans, but will also assist in breaking the transmission cycle to backyard and free-roaming pigs, reducing economic losses to the small-holder farmers due to carcass condemnation [50–53]. Using the Bayesian approach described in this study, the T3qPCR had an estimated sensitivity of 0.94 (95% CrI [0.88–0.98]) compared to 0.83 (95% CrI [0.57–0.98]) for a similar Taq-Man probe based multiplex qPCR described by Praet et al. (2013) [22] for the specific detection and differentiation of T. solium and T. saginata. The specificity of the T3qPCR of 0.98, 95% CrI [0.95–1.00] was equivalent (0.99, 95% CrI [0.98–0.99]) to the multiplex qPCR developed by Praet et al. (2013) [22].
Treatment and recovery followed by morphological and genetic identification of two adult tapeworms from a single individual supported the T3qPCR results to confirm the presence of T. asiatica in the Central Highlands of Vietnam for the first time, extending the known distribution of this tapeworm to southern Vietnam. This assay also confirmed the co-existence of T. solium, T. saginata and T. asiatica in the Central Highlands region. The elimination of two adult tapeworms from a single individual also confirmed the ability of the T3qPCR to detect both species of worms as opposed to infection with hybrid tapeworm. In comparison to the KK thick smear, the T3qPCR offered a significantly greater sensitivity (0.94, 95% CrI [0.88–0.99]) for the detection of Taenia eggs in stool samples. There were 16 Taenia spp. T3qPCR-positive samples not detected by KK (Table 4). The innately lower sensitivity of KK to detect eggs in feces may explain this [54]. However, the unsuitability of microscopic-based techniques for the detection of Taenia spp. in faeces may also be compounded by the inability of Taenia spp. to actively shed/lay eggs through a uterine pore like Pseudophyllidians do so [55]. As opposed to microscopy-based techniques, the T3qPCR may be in addition to any released eggs, detecting sloughed tapeworm DNA present in the stool, however this needs to be confirmed. Most significantly, KK missed all four T. solium positive individuals confirmed by T3qPCR in this community. As a result, it is our opinion that microscopic based fecal examination (KK) is neither suitable nor recommended for screening for taeniasis. Therefore, real-time PCR or cAgELISA at a higher OD cut off is recommended for screening T. solium carriers in community-based surveys in South East Asia where sympatric infection of all three human Taenia tapeworms is common [26,4].
The estimated sensitivity of the cAgELISA assay with an OD cut off value at 0.55 using polyclonal antibodies from hyper-immunized rabbits against excretory/secretory tapeworm antigens in this study was estimated at 0.82 (95% CrI [0.58–0.95]), twofold higher than that of KK. There was a strong agreement with the T3qPCR, in which all three cAgELISA-positive samples were confirmed as T. solium infection by T3qPCR. The sensitivity of the cAgELISA in this study was lower than that reported by Allan et al. (1996) [56] who determined test sensitivity on purgation of a subset of coproantigen-positive cases. Using a Bayesian approach, Praet et al. (2013) [22] Showed that the diagnostic sensitivity and specificity of cAgELISA was 0.85 (95% CrI [0.62–0.98]) and 0.92 (95% CrI [0.85–0.96]), respectively, similar to that estimated for this study. In total, thirteen of 21 cAgELISA OD 0.2-positive samples (Table 5) were not confirmed to be positive for Taenia by T3qPCR and microcopy. This suggests that the cAgELISA may cross-react with other parasites rather than Taenia spp [56]. cAgELISA shows excellent potential for a large scale epidemiological community-based survey in regions where sympatric Taenia spp. exist as it is capable of specifically identifying T. solium carriers when the cut off OD is set at ≥0.55. Therefore, choosing an appropriate OD cut off value for cAgELISA seems to be an important diagnostic consideration when applying the assay for the diagnosis of T. solium taeniasis in different communities.
In conclusion, this study describes the development and test characteristics of a new T3qPCR for the detection and differentiation of all three human Taenia species in stool. The absence of a diagnostic gold standard necessitated the use of a Bayesian approach to estimate diagnostic test performance. In future, the usefulness of the T3qPCR for large-scaled epidemiological studies can be confirmed using a larger sample size of individuals, ideally in a community in which the prevalence of taeniasis is higher. The assay also has the potential use in anthelmintic efficacy trials targeting individual species of tapeworms, thereby directly allowing assessment of species-specific susceptibility to drugs as well as non-chemotherapeutic interventions.
Supporting information
(XLSX)
Acknowledgments
We are grateful to the Institute of Biotechnology and Environment Tay Nguyen University for providing resources and facilities for the fieldwork. We thank to Dr. Than Trong Quang, and village medical staff at Buon Don, Krong Nang and M’Drak district for assisting sample collection. The authors are most thankful to Ms. Nguyen Thi Ngoc Hien, Ms. Long Khanh Linh, and Ms. Nguyen Thi Lan Huong, who assisted with laboratory work.
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Ito A, Nakao M, Wandra T. Human taeniasis and cysticercosis in Asia. Lancet [Internet]. Lancet; 2003;362:1918–20. Available from: https://ezp.lib.unimelb.edu.au/login?url=https://search.ebscohost.com/login.aspx?direct=true&db=bth&AN=11611385&site=eds-live&scope=site [DOI] [PubMed] [Google Scholar]
- 2.Eom KS, Rim HJ. Morphologic descriptions of Taenia asiatica sp. n. Korean J. Parasitol. [Internet]. KOREA: Korean Society for Parasitology; 1993;31:1–6. Available from: https://ezp.lib.unimelb.edu.au/login?url=https://search.ebscohost.com/login.aspx?direct=true&db=cmedm&AN=8512894&site=eds-live&scope=site [DOI] [PubMed] [Google Scholar]
- 3.Eom KS, Jeon HK, Rim HJ. Geographical distribution of Taenia asiatica and related species. Korean J. Parasitol. [Internet]. 2009;47:S115–24. Available from: http://www.scopus.com/inward/record.url?eid=2-s2.0-73349089600&partnerID=40&md5=69f591284599d22b95e311ed24697a9d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ale A, Victor B, Praet N, Gabriël S, Speybroeck N, Dorny P, et al. Epidemiology and genetic diversity of Taenia asiatica: A systematic review. Parasites and Vectors [Internet]. 2014;7:45 doi: 10.1186/1756-3305-7-45 Available from: http://www.scopus.com/inward/record.url?eid=2-s2.0-84892662506&partnerID=40&md5=66a321ffff2fca8bb15ece08f3567071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murrell KD, Dorny P, Flisser A, Geerts S, Kyvsgaard NC, McManus DP, et al. WHO/FAO/OIE Guidelines for the surveillance, prevention and control of taeniosis/cysticercosis. OIE (World Organisation for Animal Health), WHO (World Health Organization) and FAO (Food and Agriculture Organization); 2005. [Google Scholar]
- 6.Schantz PM, Cruz M, Sarti E, Pawlowski ZS. Potential eradicability of taeniasis and cysticercosis. Bull. PAHO. 1993;27:397–403. [PubMed] [Google Scholar]
- 7.World Health Organization; Atlas: epilepsy care in the world. Geneva, Switzerland: World Health Organization; 2005. [Google Scholar]
- 8.Bowles J, McManus DP. Genetic characterization of the Asian Taenia, a newly described taeniid cestode of humans. Am. J. Trop. Med. Hyg. [Internet]. 1994;50:33–44. Available from: <Go to ISI>://CABI:19940804968 [PubMed] [Google Scholar]
- 9.Carabin H, Ndimubanzi PC, Budke CM, Nguyen H, Qian Y, Cowan LD, et al. Clinical manifestations associated with neurocysticercosis: a systematic review. PLoS Negl. Trop. Dis. [Internet]. 2011. [cited 2016 Jul 27];5:e1152 Available from: http://www.ncbi.nlm.nih.gov/pubmed/21629722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White AC. Neurocysticercosis: updates on epidemiology, pathogenesis, diagnosis, and management. Annu. Rev. Med. [Internet]. 2000. [cited 2016 Jul 27];51:187–206. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10774460 [DOI] [PubMed] [Google Scholar]
- 11.Teerasukjinda O, Wongjittraporn S, Tongma C, Chung H. Asymptomatic Giant Intraventricular Cysticercosis: A Case Report. Hawaii. J. Med. Public Health [Internet]. University Clinical, Education & Research Associates; 2016. [cited 2016 Jul 29];75:187–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27437162 [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzales I, Rivera JT, Garcia HH, Cysticercosis Working Group in Peru. Pathogenesis of Taenia solium taeniasis and cysticercosis. Parasite Immunol. [Internet]. 2016. [cited 2016 Sep 16];38:136–46. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26824681 [DOI] [PubMed] [Google Scholar]
- 13.Van De N, Le TH, Lien PTH, Eom KS. Current status of taeniasis and cysticercosis in Vietnam. Korean J. Parasitol. [Internet]. 2014;52:125–9. Available from: http://www.scopus.com/inward/record.url?eid=2-s2.0-84900338607&partnerID=40&md5=7bb85881ef5648db9bda21822618af53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayta H, Talley A, Gilman RH, Jimenez J, Verastegui M, Ruiz M, et al. Differentiating Taenia solium and Taenia saginata infections by simple hematoxylin-eosin staining and PCR-restriction enzyme analysis. J. Clin. Microbiol. [Internet]. UNITED STATES: American Society for Microbiology; 2000;38:133–7. Available from: https://ezp.lib.unimelb.edu.au/login?url=https://search.ebscohost.com/login.aspx?direct=true&db=cmedm&AN=10618076&site=eds-live&scope=site [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkins PP, Allan JC, Verastegui M, Acosta M, Eason AG, Hugo Garcia H, et al. Development of a serologic assay to detect Taenia solium taeniasis. Am. J. Trop. Med. Hyg. 1999;60:199–204. [DOI] [PubMed] [Google Scholar]
- 16.Jeon H-K, Eom KS. Immunoblot patterns of Taenia asiatica taeniasis. Korean J. Parasitol. [Internet]. 2009;47:73–7. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2655338&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine MZ, Lewis MM, Rodriquez S, Jimenez JA, Khan A, Lin S, et al. Development of an Enzyme-Linked Immunoelectrotransfer Blot (Eitb) Assay Using Two Baculovirus Expressed Recombinant Antigens for Diagnosis of Taenia Solium Taeniasis. J. Parasitol. 2007;93:409–17. doi: 10.1645/GE-938R.1 [DOI] [PubMed] [Google Scholar]
- 18.Dorny P, Brandt J, Zoli A, Geerts S. Immunodiagnostic tools for human and porcine cysticercosis. Acta Trop. [Internet]. 2003;87:79–86. Available from: http://www.sciencedirect.com/science/article/pii/S0001706X03000585 [DOI] [PubMed] [Google Scholar]
- 19.Raoul F, Li T, Sako Y, Chen X, Long C, Yanagida T, et al. Advances in diagnosis and spatial analysis of cysticercosis and taeniasis. Parasitology [Internet]. Cambridge University Press; 2013. [cited 2016 Jul 29];140:1578–88. Available from: http://www.journals.cambridge.org/abstract_S0031182013001303 [DOI] [PubMed] [Google Scholar]
- 20.Mayta H, Gilman RH, Prendergast E, Castillo JP, Tinoco YO, Garcia HH, et al. Nested PCR for Specific Diagnosis of Taenia solium Taeniasis. J. Clin. Microbiol. [Internet]. United States, AMERICAN SOCIETY FOR MICROBIOLOGY; 2008;46:286–9. Available from: https://ezp.lib.unimelb.edu.au/login?url=https://search.ebscohost.com/login.aspx?direct=true&db=edsbl&AN=RN225804162&scope=site [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somers R, Dorny P, Geysen D, Nguyen LA, Thach DC, Vercruysse J, et al. Human tapeworms in north Vietnam. Trans. R. Soc. Trop. Med. Hyg. [Internet]. 2007;101:275–7. Available from: http://www.scopus.com/inward/record.url?eid=2-s2.0-33845972668&partnerID=40&md5=0efd13a7fe328db479061db6a50da7f6 [DOI] [PubMed] [Google Scholar]
- 22.Praet N, Verweij JJ, Mwape KE, Phiri IK, Muma JB, Zulu G, et al. Bayesian modelling to estimate the test characteristics of coprology, coproantigen ELISA and a novel real-time PCR for the diagnosis of taeniasis. Trop. Med. Int. Heal. [Internet]. 2013;18:608–14. Available from: http://dx.doi.org/10.1111/tmi.12089 [DOI] [PubMed] [Google Scholar]
- 23.Guezala MC, Rodriguez S, Zamora H, Garcia HH, Gonzalez AE, Tembo A, et al. Development of a species-specific coproantigen ELISA for human Taenia solium taeniasis. Am J Trop Med Hyg. 2009/08/27. 2009;81:433–7. [PubMed] [Google Scholar]
- 24.Deplazes P, Eckert J, Pawlowski ZS, Machowska L, Gottstein B. An enzyme-linked immunosorbent assay for diagnostic detection of Taenia saginata copro-antigens in humans. Trans. R. Soc. Trop. Med. Hyg. [Internet]. 1991. [cited 2016 Aug 1];85:391–6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/1719664 [DOI] [PubMed] [Google Scholar]
- 25.Allan JC, Craig PS, Garcia Noval J, Mencos F, Liu D, Wang Y, et al. Coproantigen detection for immunodiagnosis of echinococcosis and taeniasis in dogs and humans. Parasitol. [Internet]. 1992; Available from: https://ezp.lib.unimelb.edu.au/login?url=https://search.ebscohost.com/login.aspx?direct=true&db=edsagr&AN=edsagr.US9300547&site=eds-live&scope=site [DOI] [PubMed] [Google Scholar]
- 26.Aung AK, Spelman DW. Taenia solium Taeniasis and Cysticercosis in Southeast Asia. Am. J. Trop. Med. Hyg. [Internet]. 2016. [cited 2016 Jul 28];94:947–54. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4856625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeon H-K, Chai J-Y, Kong Y, Waikagul J, Insisiengmay B, Rim H-J, et al. Differential diagnosis of Taenia asiatica using multiplex PCR. Exp. Parasitol. [Internet]. United States: Academic Press; 2009;121:151–6. Available from: https://ezp.lib.unimelb.edu.au/login?url=https://search.ebscohost.com/login.aspx?direct=true&db=cmedm&AN=19017531&site=eds-live&scope=site [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki H, Allan JC, Sato MO, Nakao M, Sako Y, Nakaya K, et al. DNA differential diagnosis of taeniasis and cysticercosis by multiplex PCR. J. Clin. Microbiol. [Internet]. United States: American Society for Microbiology; 2004;42:548–53. Available from: https://ezp.lib.unimelb.edu.au/login?url=https://search.ebscohost.com/login.aspx?direct=true&db=cmedm&AN=14766815&scope=site [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon CA, McManus DP, Acosta LP, Olveda RM, Williams GM, Ross AG, et al. Multiplex real-time PCR monitoring of intestinal helminths in humans reveals widespread polyparasitism in Northern Samar, the Philippines. Int. J. Parasitol. [Internet]. 2015. [cited 2016 May 11];45:477–83. Available from: http://www.sciencedirect.com/science/article/pii/S0020751915000752 [DOI] [PubMed] [Google Scholar]
- 30.Nkouawa A, Sako Y, Nakao M, Nakaya K, Ito A. Loop-mediated isothermal amplification method for differentiation and rapid detection of Taenia species. J Clin Microbiol [Internet]. 2008/11/14. 2009;47:168–74. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2620829/pdf/1573-08.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambert SB, Whiley DM, O’Neill NT, Andrews EC, Canavan FM, Bletchly C, et al. Comparing nose-throat swabs and nasopharyngeal aspirates collected from children with symptoms for respiratory virus identification using real-time polymerase chain reaction. Pediatrics [Internet]. MBBS, Queensland Paediatric Infectious Diseases Laboratory, Royal Children’s Hospital, Herston Queensland 4029, Australia. sblambert@uq.edu.au: American Academy of Pediatrics; 2008;122:e615–20. Available from: https://ezp.lib.unimelb.edu.au/login?url=https://search.ebscohost.com/login.aspx?direct=true&db=cmedm&AN=18725388&site=eds-live&scope=site [DOI] [PubMed] [Google Scholar]
- 32.Ng-Nguyen D, Stevenson MA, Traub RJ. A systematic review of taeniasis, cysticercosis and trichinellosis in Vietnam. Parasit. Vectors [Internet]. Parasites & Vectors; 2017;10:150 Available from: http://parasitesandvectors.biomedcentral.com/articles/10.1186/s13071-017-2085-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allan JC, Avila G, Garcia Noval J, Flisser A, Craig PS. Immunodiagnosis of taeniasis by coproantigen detection. Parasitology [Internet]. ENGLAND: Cambridge University Press; 1990;101 Pt 3:473–7. Available from: https://ezp.lib.unimelb.edu.au/login?url=https://search.ebscohost.com/login.aspx?direct=true&db=cmedm&AN=2092303&site=eds-live&scope=site [DOI] [PubMed] [Google Scholar]
- 34.Mwape KE, Phiri IK, Praet N, Muma JB, Zulu G, van den Bossche P, et al. Taenia solium infections in a rural area of Eastern Zambia-A community based study. PLoS Negl. Trop. Dis. 2012;6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thienpont D. Diagnosing helminthiasis by coprological examination. Janssen Research Foundation; 1986. [Google Scholar]
- 36.Hoff R, Hoff R, Sleigh A, Mott K, Barreto M, de Paiva TM, et al. Comparison of filtration staining (bell) and thick smear (kato) for the detection and quantitation of schistosoma mansoni eggs in faeces. Trans. R. Soc. Trop. Med. Hyg. [Internet]. Oxford University Press; 1982. [cited 2016 Aug 10];76:403–6. Available from: http://trstmh.oxfordjournals.org/cgi/doi/10.1016/0035-9203(82)90201-2 [DOI] [PubMed] [Google Scholar]
- 37.Teesdale CH, Amin MA. A simple thick-smear technique for the diagnosis of Schistosoma mansoni infection. Bull. World Health Organ. [Internet]. 1976. [cited 2016 Aug 10];54:703–5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/1088516 [PMC free article] [PubMed] [Google Scholar]
- 38.Katz N, Chaves A, Pellegrino J. A simple device for quantitative stool thick-smear technique in Schistosomiasis mansoni. Rev. do Inst. Med. Trop. Saão Paulo. 1972;14:397–400. [PubMed] [Google Scholar]
- 39.Asaava LL, Kitala PM, Gathura PB, Nanyingi MO, Muchemi G, Schelling E. A survey of bovine cysticercosis/human taeniosis in Northern Turkana District, Kenya. Prev. Vet. Med. 2009;89:197–204. doi: 10.1016/j.prevetmed.2009.02.010 [DOI] [PubMed] [Google Scholar]
- 40.Team R Core. R: A Language and Environment for Statistical Computing [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2017. http://www.r-project.org [Google Scholar]
- 41.Van Chuong N. Status of helminthic infection in humans in 14 provinces spanning the Central and Central Highland regions of Vietnam [Internet]. IMPE-QN. 2005. [cited 2016 Mar 1]. Available from: http://www.impe-qn.org.vn/impe-qn/vn/portal/InfoDetail.jsp?area=58&cat=1068&ID=597 [Google Scholar]
- 42.Branscum AJ, Gardner IA, Johnson WO. Estimation of diagnostic-test sensitivity and specificity through Bayesian modeling. Prev. Vet. Med. 2005;68:145–63. doi: 10.1016/j.prevetmed.2004.12.005 [DOI] [PubMed] [Google Scholar]
- 43.Bouma A, Stegeman JA, Engel B, de Kluijver EP, Elbers ARW, de Jong MCM. Evaluation of Diagnostic Tests for the Detection of Classical Swine Fever in the Field without a Gold Standard. J. Vet. Diagnostic Investig. [Internet]. 2001. [cited 2016 Nov 1];13:383–8. Available from: http://vdi.sagepub.com/cgi/content/long/13/5/383 [DOI] [PubMed] [Google Scholar]
- 44.Gelman A. Inference and monitoring convergence In: Gilks W, Richardson S, Spiegelhalter D (Eds., editors. Markov Chain Monte Carlo Pract. [Internet]. Chapman & Hall, London; 1996. p. 131–143. https://ezp.lib.unimelb.edu.au/login?url=https://search.ebscohost.com/login.aspx?direct=true&db=msn&AN=MR1397966&site=eds-live&scope=site [Google Scholar]
- 45.Yu B, Mykland P. Looking at Markov samplers through cusum path plots: a simple diagnostic idea. Stat. Comput. [Internet]. 1998;8:275–286. Available from: http://www.springerlink.com/content/nm8811rt432jn287/ [Google Scholar]
- 46.Casella G, Fienberg S, Olkin I. Springer Texts in Statistics [Internet]. Design. 2006. http://books.google.com/books?id=9tv0taI8l6YC [Google Scholar]
- 47.Raftery AE, Lewis SM. One Long Run with Diagnostics: Implementation Strategies for Markov Chain Monte Carlo. Stat. Sci. 1992;7:493–7. [Google Scholar]
- 48.Raftery EA, Lewis MS. How many iterations in the Gibbs sampler? In: Benardo J, Berger J, Dawid A, Smith A (Eds., editors. Bayesian Stat. 4 [Internet]. The Clarendon Press, Oxford University Press, New York; 1992. p. 763–774. https://ezp.lib.unimelb.edu.au/login?url=https://search.ebscohost.com/login.aspx?direct=true&db=msn&AN=MR1380266&site=eds-live&scope=site [Google Scholar]
- 49.Ito A, Yanagida T, Nakao M. Recent advances and perspectives in molecular epidemiology of Taenia solium cysticercosis. Infect. Genet. Evol. 2015;40:357–67. doi: 10.1016/j.meegid.2015.06.022 [DOI] [PubMed] [Google Scholar]
- 50.Bhattarai R, Carabin H, Proaño J V., Flores-Rivera J, Corona T, Flisser A, et al. Cost of neurocysticercosis patients treated in two referral hospitals in Mexico City, Mexico. Trop. Med. Int. Heal. [Internet]. 2015. [cited 2015 Oct 4];20:1108–19. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25726958 [DOI] [PubMed] [Google Scholar]
- 51.Rajkotia Y, Lescano AG, Gilman RH, Cornejo C, Garcia HH, Cysticercosis Working Group of Peru for TCWG of. Economic burden of neurocysticercosis: results from Peru. Trans. R. Soc. Trop. Med. Hyg. [Internet]. Oxford University Press; 2007. [cited 2016 Nov 3];101:840–6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17507067 [DOI] [PubMed] [Google Scholar]
- 52.Praet N, Speybroeck N, Manzanedo R, Berkvens D, Nforninwe DN, Zoli A, et al. The Disease Burden of Taenia solium Cysticercosis in Cameroon. PLoS Negl. Trop. Dis. [Internet]. 2009;3(3):e406 doi: 10.1371/journal.pntd.0000406 Available from: <Go to ISI>://WOS:000265536800021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choudhury AA, Conlan J V, Racloz VN, Reid SA, Blacksell SD, Fenwick SG, et al. The economic impact of pig-associated parasitic zoonosis in Northern Lao PDR. Ecohealth. 2013/02/19. 2013;10:54–62. doi: 10.1007/s10393-013-0821-y [DOI] [PubMed] [Google Scholar]
- 54.Llewellyn S, Inpankaew T, Nery SV, Gray DJ, Verweij JJ, Clements ACA, et al. Application of a Multiplex Quantitative PCR to Assess Prevalence and Intensity Of Intestinal Parasite Infections in a Controlled Clinical Trial. Bethony JM, editor. PLoS Negl. Trop. Dis. [Internet]. Public Library of Science; 2016. [cited 2016 Aug 10];10:e0004380 Available from: http://dx.plos.org/10.1371/journal.pntd.0004380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price DL. Procedure Manual for the Diagnosis of Intestinal Parasites. CRC Press; 1994. [Google Scholar]
- 56.Allan JC, Velasquez-Tohom M, Torres-Alvarez R, Yurrita P, Garcia-Noval J. Field trial of the coproantigen-based diagnosis of Taenia solium taeniasis by enzyme-linked immunosorbent assay. Am J Trop Med Hyg. 1996/04/01. 1996;54:352–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.