Abstract
Chromosome 17p13.3 is a gene rich region that when deleted is associated with the well-known Miller-Dieker syndrome. A recently described duplication syndrome involving this region has been associated with intellectual impairment, autism and occasional brain MRI abnormalities. We report 34 additional patients from 21 families to further delineate the clinical, neurological, behavioral, and brain imaging findings. We found a highly diverse phenotype with inter- and intrafamilial variability, especially in cognitive development. The most specific phenotype occurred in individuals with large duplications that include both the YWHAE and LIS1 genes. These patients had a relatively distinct facial phenotype and frequent structural brain abnormalities involving the corpus callosum, cerebellar vermis and cranial base. Autism spectrum disorders were seen in a third of duplication probands, most commonly in those with duplications of YWHAE and flanking genes such as CRK. The typical neurobehavioral phenotype was usually seen in those with the larger duplications. We did not confirm the association of early overgrowth with involvement of YWHAE and CRK, or growth failure with duplications of LIS1. Older patients were often overweight. Three variant phenotypes included cleft lip/palate (CLP), split hand/foot with long bone deficiency (SHFLD), and a connective tissue phenotype resembling Marfan syndrome. The duplications in patients with clefts appear to disrupt ABR, while the SHFLD phenotype was associated with duplication of BHLHA9 as noted in two recent reports. The connective tissue phenotype did not have a convincing critical region. Our experience with this large cohort expands knowledge of this diverse duplication syndrome.
Keywords: 17p13.3, Microarray, autism, Cleft lip/palate, marfanoid, split hand foot long bone deficiency, ABR, LIS 1, YWHAE, BHLHA9
INTRODUCTION
Human chromosome 17 is a small, gene rich chromosome associated with several well-known deletion and duplication syndromes. The first recognized was Miller-Dieker syndrome (MDS), caused by large deletions of 17p13.3 [Dobyns et al., 1983]. We previously showed that the severe brain phenotype is caused by deletion of both LIS1 (designated PAFAH1B1 in the UCSC browser and other genome databases) and the modifying gene YWHAE [Cardoso et al., 2003], but have not determined which genes are responsible for the MDS facial phenotype or Chiari malformation [Cardoso et al., 2003; Nagamani et al., 2009].
Several small series adding to 16 patients with dup 17p13.3 have been reported in the past 3 years that have begun to define the phenotypes associated with duplications of this region [Bi et al., 2009; Bruno et al., 2010; Hyon et al., 2011; Roos et al., 2009; Shimojima et al., 2010]. Duplications of the telomeric portion containing YWHAE were linked to high birth weight, large body size, dysmorphic facial appearance, mild developmental delay, autism, and behavior disorders such as increased aggression and preoccupation with food. Duplications of the centromeric region were associated with small body size, microcephaly, moderate to severe developmental delay and mild brain malformations – but not dysmorphic features – and duplication of LIS1 was suggested as the cause [Bi et al., 2009]. Brain-imaging studies were reported to show variable abnormalities of the corpus callosum and cerebellum in three of six imaged patients, with the remaining three scans reported as normal. Another approximately 70 individuals from 20 families with small duplications of distal 17p13.3 and split-hand/foot malformation and long bone deficiency (SHFLD) have been reported. However, none have had developmental delay, intellectual disability (ID) or autism [Armour et al., 2011; Klopocki et al., 2012].
Given the variable phenotype and genomic regions involved, we hypothesized that multiple genes were likely to contribute to the phenotype and that a larger group of patients would be required for accurate genotype-phenotype analysis. Our initial review of several patients with dup 17p13.3 suggested that the phenotypes were variable and that the association of specific genes with discrete phenotypes – including YWHAE and LIS1 – was probably premature. We have now ascertained 34 patients from 21 families with dup 17p13.3 by contacting colleagues directly or through two large clinical testing laboratories (Signature Genomic Laboratories and GeneDx), as well as from the DECIPHER database. All duplications included YWHAE, LIS1 or both, as well as varying combinations of 48 known genes located between the 17p telomere and LIS1 (data downloaded 11/25/2012 from the UCSC gene track in the UCSC genome browser). We critically assessed patients with respect to their developmental history, neurological and behavioral presentation, craniofacial phenotype, and medical complications. We also reviewed available brain imaging studies on 18 patients.
METHODS AND PATIENTS
Subject ascertainment
We ascertained 42 patients from 29 families personally, or by query of the Signature Genomics, GeneDx and DECIPHER databases for duplications in 17p13.3 that contained either YWHAE or LIS1. We eliminated six families from further analysis due to other genomic imbalances predicted to impact the phenotype. We eliminated another two families with chromosome rearrangements predicted to result in loss (rather than gain) of function. One had a small intragenic duplication within YWHAE, while the other had breakpoints in both YWHAE and CRK. We included two previously reported children (Patients 2 and 3 in their report), as unpublished brain imaging data became available [Roos et al., 2009]. We kept two unrelated patients with a second duplication in 17p13.2 consisting of 454 kb and 410 kb, as their phenotypes were typical of 17p13.3 duplication. The occurrence of more than one chromosome 17 rearrangement has been reported before and likely reflects the large number of segmental duplications and related propensity for instability on 17p [Shchelochkov et al., 2010]. We also kept one patient with the common dup 16p13.11, as it has been found in normal individuals although it is a risk factor for schizophrenia and other neuropsychiatric disorders [Hannes et al., 2009; Ingason et al., 2011; Ramalingam et al., 2011].
We thus included 34 patients from 21 families in our analysis. Clinicians (including geneticists, neurologists and behavioral pediatricians) supplied clinical data, brain imaging studies and photographs with parental consent. The Institutional Review Boards of Community Hospitals Central California, the University of Chicago, and Seattle Children’s Hospital approved this study.
Phenotype data
The clinical, brain magnetic resonance imaging (MRI), and chromosome microarray data were entered in a spreadsheet without identifying information. MRI’s were reviewed by WBD and re-reviewed without knowledge of clinical or array data by AJB. Facial photographs were analyzed for dysmorphic features by CJC and WBD, supplemented by clinical descriptions from referring physicians. We followed one subject and evaluated her facial features over 5 years; others were generally seen and described in one or two clinic visits. We specifically looked for evidence of facial hypotonia and an elongated face, as well as other features such as a prominent chin as this was mentioned as a frequent finding in other series [Bi et al., 2009; Bruno et al., 2010; Roos et al., 2009]. Growth was plotted on standard CDC growth charts, with data confined to a single time point for most patients. Developmental status was assessed from referring physician reports and, for a few patients, from detailed psychological studies outlined by the referring physician. The diagnosis of an autistic spectrum disorder was listed only if the referring physician noted this specifically in the records.
Chromosome microarrays
Arrays reported in this study utilized several different platforms as they were performed in different laboratories over several years during which time the resolution increased. The most common platforms used were oligonucleotide arrays: SignatureChipOS version 1 (105K- manufactured by Agilent Technologies, Santa Clara, CA) and SignatureChipOS version 2 (135K, manufactured by Roche NimbleGen, Madison WI).
RESULTS AND DISCUSSION
Subject groups
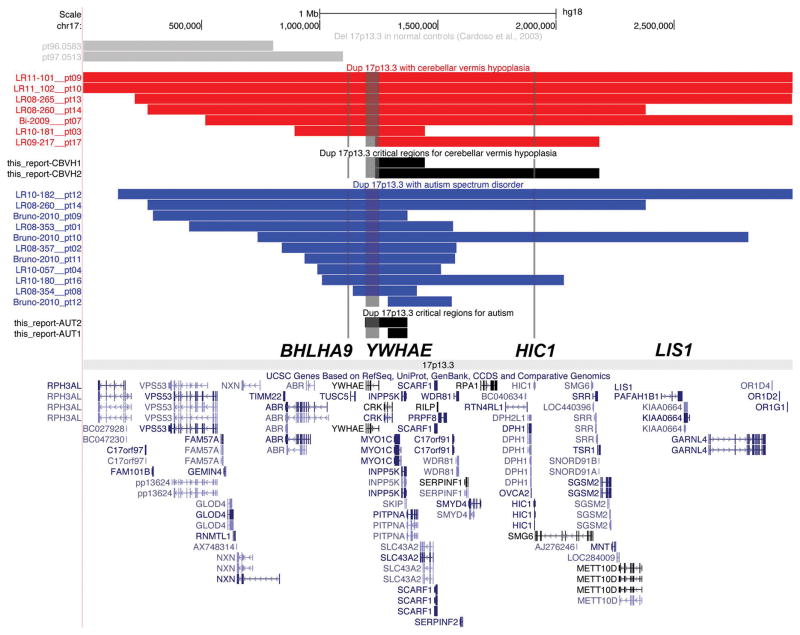
Due to heterogeneous duplication sizes and locations, we identified no subgroups with common breakpoints. However, our 34 subjects and all previously reported subjects fell into three size groups, and we divided our cohort into these 3 subgroups for genotype-phenotype analysis (Figure 1). These subsets included small telomeric duplications involving chr17:800,000-1,600,000 that include YWHAE and flanking genes (Group 1); larger duplications that involved most of 17p13.3 (Group 2), and small centromeric duplications involving chr17:2,000,000-3,000,000 that include LIS1 and flanking genes (Group 3).
Figure 1.
A map of chromosome band 17p13.3 showing the genome location and gene content of duplications in all 21 families in our study as well as 13 additional patients from the literature was made using the Custom Tracks function in the UCSC browser (http://genome.ucsc.edu). The map shows Group 1 dups in red, Group 2 dups in purple and Group 3 dups in blue so that the differences in duplication size and position are obvious. The map also shows other reported dups in gray [Bi et al., 2009; Bruno et al., 2010; Roos et al., 2009; Shimojima et al., 2010].
Prior reports implicated YWHAE and LIS1 as contributing to the duplication 17p13.3 phenotype [Bi et al., 2009; Bruno et al., 2010; Roos et al., 2009], and limited data from mouse (plus our unpublished data) suggest a possible role for HIC1 [Carter et al., 2000]. These genes are separated by 656 kb (YWHAE-HIC1) and 535 kb (HIC1-LIS1), and serve as sentinel genes for the telomeric (Group 1), large (Group 2) and centromeric (Group 3) regions. We recognize that these 3 sentinel genes may or may not be the pathogenic genes in these overlapping regions, so our data should not be interpreted to implicate these genes specifically. Given the 48 genes in the 17p13.3 region and evidence of incomplete penetrance, we hypothesized that multiple genes contribute to key components of the phenotype. After separating patients into these three groups, we examined sets of features emphasizing growth and development including autism, dysmorphic features and medical and neurological complications, and compared them between groups. For analysis of less common phenotypes including cerebellar vermis hypoplasia (CBVH), cleft lip/palate (CLP), SHFLD and marfanoid habitus, we evaluated all patients as a single group.
Overview and demographics
We analyzed data on 34 individuals from 21 families with dup 17p13.3, separated into groups as noted above. The probands presented with developmental disorders from infancy to 4–5 years of age (N=12), congenital anomalies (N=3), family history of a developmental disorder such as autism (N=1), failure-to-thrive (N=3), or abnormal fetal ultrasound (N=2). The demographics of these families did not differ substantially from the clinic populations where they were seen (Supplementary Table I). Multiple ethnicities were represented including African-American, Caucasian (16 families), Hispanic, East Indian and Asian. Parental ages were not increased. Our index patients were first evaluated between 6 months and 28 years of age. Several carrier parents were identified and included, although information regarding phenotype and behavior was limited for most. Data on the three affected fetuses in this cohort was also limited. With the exception of one child delivered prematurely at 33 weeks, all other index patients were delivered between 37 and 40 weeks gestation. Pregnancies were generally uncomplicated, except that one mother was in an automobile accident and another was diagnosed with hyperthyroidism during the pregnancy. Five of 17 patients for whom information was available were delivered by cesarean section, a rate similar to the general population.
Microarray data
Our microarray data (Table I and Figure 1) demonstrated very heterogeneous breakpoints, which we expected as 17p13.3 contains no important segmental duplications, although it does contain interspersed repetitive elements such as SINEs that can function as recombination substrates for non-homologous end-joining (NHEJ) or fork stalling and template switching (FoSTeS) events [Bi et al., 2009; Shchelochkov et al., 2010]. The duplications in this cohort ranged in size from a small 231.4 kb duplication involving only four genes including YWHAE to a large 4.0 Mb duplication encompassing ~87 genes from the 17p telomere past LIS1. In addition, three patients had second duplications in 17p13.2 (Patient 19, hereafter Pt19, and Pt21a1/a2), and one had a nested triplication within the dup (Pt9). These observations are consistent with the complex cytogenetic mechanisms previously reported with 17p rearrangements [Shchelochkov et al., 2010]. Another patient had a second duplication, dup 16p13.11 (pt17).
Table 1.
Genome data for duplication 17p13.3 patients
| Lisdb# | Pt# | Genome coordinates [hg18] | Size | Sentinel genes | Inheritance | Microarray |
|---|---|---|---|---|---|---|
| Group I | ||||||
| LR08-353 | 1 | (chr17:447,066-1,563,813)×3 | 1.12 Mb | YWHAE | paternal na* | SGL OS 135K |
| LR08-357 | 2 | (chr17:840,149-1,578,598)×3 | 738.4 kb | YWHAE | de novo | SGL OS 135K |
| LR10-181a1 | 3a1 | (chr17:894,983-1,443,730)×3 | 548.7 kb | YWHAE | maternal | OGT 105K |
| LR10-181a2 | 3a2 | same as 3a1 | “ | YWHAE | maternal | OGT 105K |
| LR10-057a1 | 4a1 | (chr17:991,473-1,513,936)×3 | 522.5 kb | YWHAE | germline | Genzyme Clarisure |
| LR10-057a2 | 4a2 | same as 4a1 | “ | YWHAE | mosaic | SGL OS 135K |
| LR10-179a1 | 5a1 | (chr17:1,007,000-1,619,962)×3 | 613.0 kb | YWHAE | paternal | SGL OS 135K |
| LR10-179a2 | 5a2 | same as 5a1 | “ | YWHAE | na | SGL OS 135K |
| LR09-270 | 6 | (chr17:1,039,126-1,491,785)×3 | 452.7 kb | YWHAE | de novo | SGL OS 105K |
| LR09-213a1 | 7a1 | (chr17:1,131,224-1,448,997)×3 | 317.8 kb | YWHAE | maternal | SGL OS 105K |
| LR09-213a2 | 7a2 | same as 7a1 | “ | YWHAE | na | SGL OS 105K |
| LR08-354a1 | 8a1 | (chr17:1,140,796-1,410,997)×3 | 270.2 kb | YWHAE | mat/pat | SGL OS 135K |
| LR08-354a2 | 8a2 | same as 8a1 | “ | YWHAE | mat/pat | SGL BAC |
| LR08-354a3 | 8a3 | na | na | na | na | |
| Group 2 | ||||||
| LR11-101 | 9 | (chr17:1-3,012,800)×3 and (chr17:1,448,942-1,503,503)×4 | 3.0 Mb | YWHAE-HIC1-LIS1 | de novo | Agilent 244K |
| LR11-102 | 10 | (chr17:1-4,052,847)×3 | 4 Mb | YWHAE-HIC1-LIS1 | de novo | Agilent 244K |
| LR10-068a1 | 11a1 | (chr17:52,934-2,501,198)×3 | 2.45 Mb | YWHAE-HIC1-LIS1 | maternal | SGL OS 135K |
| LR10-068a2 | 11a2 | dup confirmed by FISH | “ | YWHAE-HIC1-LIS1 | mosaic 72% | FISH |
| LR10-182 | 12 | (chr17:148,091-3,505,143)×3 | 3.4 Mb | YWHAE-HIC1-LIS1 | de novo | UNC SGLSelect 105K |
| LR08-265a1 | 13a1 | (chr17:219,072-2,996,360)×3 | 2.78 Mb | YWHAE-HIC1-LIS1 | paternal | Agilent 660K-quad |
| LR08-265a2 | 13a2 | same as13a1 | “ | YWHAE-HIC1-LIS1 | na | Agilent 660K-quad |
| LR08-260 | 14 | (chr17:271,999-2,277,555)×3 | 2.01 Mb | YWHAE-HIC1 | de novo | Genome Dx 105K |
| LR08-358 | 15 | (chr17:800,495-2,767,054)×3 | 2.16 Mb | YWHAE-HIC1-LIS1 | de novo | Agilent 105 K |
| LR10-180 | 16 | (chr17:1,010,661-2,032,333)×3 | 1.02 Mb | YWHAE-HIC1 | de novo | OGT 105K |
| LR09-217a1 | 17a1 | dup 17p13.3; dup 16p13.11 (chr17:1,235,314-2,183,796)×3 (chr16:14,957,300-16,195,404)×3 |
948.5 kb; 1.24 Mb | YWHAE-HIC1 | 17p maternal 16p de novo |
SGL OS 135K |
| LR09-217a2 | 17a2 | 17p same as 17a1;no 16p dup | “ | YWHAE-HIC1 | na | SGL OS 135K |
| Group 3 | ||||||
| LR10-114 | 18 | (chr17:1,963,450-2,816,939)×3 | 853.5 kb | LIS1 | de novo | Agilent 105K |
| LR09-065 | 19 | dup 17p13.3, dup 17p13.2 (chr17:2,165,727-4,237,227)×3 (chr17:4,014,003-4,468,074)×3 |
2.27 Mb, 454.0 kb | LIS1 | de novo | Genome Dx 105K |
| LR10-150a1 | 20a1 | (chr17:2,249,109-2,586,017)×3 | 336.9 kb | LIS1 | paternal | SGL OS 105K |
| LR10-150a2 | 20a2 | same as 20a1 | 336.9 kb | LIS1 | paternal | SGL OS 105K |
| LR10-150a3 | 20a3 | same as 20a1 | 336.9 kb | LIS1 | na | SGL OS 105K |
| LR10-056a1 | 21a1 | dup 17p13.3, dup 17p13.2 (chr17:2,260,944-2,560,922)×3 (chr17:4,028,199-4,436,327)×3 |
300.0 kb 408.1 kb |
LIS1 | maternal | SGL OS 135K |
| LR10-056a2 | 21a2 | same as 21a1 | “ | LIS1 | maternal | SGL OS 135K |
| LR10-056a3 | 21a3 | same as 21a1 | “ | LIS1 | na | SGL OS 135K |
Abbreviations: mat/pat, inherited but parent-of-origin unknown; mosaic, mosaicism implying de novo inheritance; na, not available; Pt#, patient #; *, maternal array normal. Chromosome microarrays: Agilent 105K, Agilent Human Genome CGH 105K; Agilent 244K, Agilent Human Genome CGH 244K (Agilent Technologies, Santa Clara, CA); Illumina 660K, Infinium HD Human660W-Quad BeadChip SNP array (Illumina, San Diego CA); SGL OS 105K and SGL OS135K, Signature Genomics Laboratories oligonucleotide arrays; OGT; Oxford Gene Technology; UNC University North Carolina.
Inheritance
The duplication was de novo in 10/21 and familial in 10/21 probands (Table I). Only one parent was tested in Family 1. Generally, parents were less affected than their child. Reports of less affected carrier parents are common for several well-known microdeletion and microduplication syndromes such as deletion 15q13.3 [Hoppman-Chaney et al., 2012] and duplication 16p13.11 [Hannes et al., 2009]. This most likely reflects ascertainment bias, variable expressivity, or incomplete penetrance. The mother of an affected fetus diagnosed by array on amniotic fluid cells was mosaic for the duplication in 72% of her peripheral blood cells (Pt11a2). In another family, one parent was an obligate mosaic carrier based on two affected offspring (Pt4a1/4a2) and normal array and FISH results in both parents. One family with two affected siblings (Pt8a1/a2) had no parental testing, although one parent must carry the duplication.
We also recognize that phenotypes such as Kabuki syndrome have been erroneously attributed to copy number variants and acknowledge this possibility [Vermeesch et al., 2011], especially for our patients with the variant CLP, SHFLD and marfanoid phenotypes. However, the constancy of a core clinical and brain phenotype in individuals with 17p13.3 duplication leads us to conclude that the duplication is indeed causative. Although carrier parents were seldom evaluated clinically, the limited information available suggests that few were functioning entirely normally. This would support a relatively subtle phenotype with significant variability, or the effects of modifier genes and environment. In addition, a few probands had near normal intelligence on formal testing but had significant problems with behavior that interfered with school performance.
Duplication 17p13.3 – General Characteristics
A wide range of developmental and health abnormalities occur with dup 17p13.3 including prenatal and growth abnormalities, neurodevelopmental disorders including autism, dysmorphic facial features and other congenital anomalies, and other medical and neurologic problems. These are summarized in Tables II-V and Supplementary Table II.
Table 2.
Prenatal findings in isolated 17p13.3 duplications
| Patient # | Indication | Prenatal US | CVS/AMNIO | Inheritance | Comments |
|---|---|---|---|---|---|
| 6 | Routine | dilated renal pelves | Not done | Not tested | Diagnosis in infancy |
| 4a1 | Recurrence dup 17p13, MA | Not available | CVS microarray dup 17p13.3 | Both parents normal; two affected sibs | Germline mosaicism in one parent; fetus terminated |
| 10 | Routine | Probable ACC; VMEG; oligo at 33 weeks | Amnio karyotype 46, XY | Not tested | Diagnosis in infancy |
| 11a1 | Routine | SUA; renal anomalies | Amnio karyotype 46, XY, microarray dup 17p13.3 | Maternal mosaicism 72% | Unilateral multi-cystic dysplastic kidney |
| 14 | MA, abnormal AFP | None | Amnio karyotype 46, XX | Not tested | Diagnosis at 2.5 years |
| 16 | Routine 1st trimester | Nuchal trans-lucency 7.5 mm | Not done | Not tested | Diagnosis at 2 years |
| 20a1 | Routine | Probable IVH, resolved before birth | Not done | Not tested | Diagnosis at 4 years |
| 21a1 | Routine 2nd trimester | VMEG, CP cysts, endocardial fibroelastosis | Amnio micoarray 17p13.3 dup | Maternal | Terminated with no autopsy; previous SB with hydrops |
Abbreviations: ACC, agenesis of the corpus callosum; AFP, alpha-fetoprotein; Amnio, amniocentesis; CP, choroid plexus; CVS, chorionic villus sample; dup, duplication; IVH, intraventricular hemorrhage; MA, maternal age; oligo, oligohydramnios; SB, stillbirth; SUA, single umbilical artery; US, ultrasound; VMEG, ventriculomegaly.
Table 5.
Brain imaging abnormalities in duplication 17p13.3 patients
| Pt# | Cerebral hemispheres | Corpus callosum | Lateral ventricles | Brainstem | Cerebellum | Posterior fossa, skull |
|---|---|---|---|---|---|---|
| Group 1 | ||||||
| 1 | N | N | N | N | N | N |
| 2 | N | Short | N | N | N | ±BOM |
| 3a1 | N | N | N | N | CBVH | MCM-BOM |
| 4a1 | N | N | N | N | N | N |
| 6 | N | Short | N | N | ±CBVH | ±BOM |
| 7a1 | Atrophy | Thin | VMEG | N | Atrophy | N |
| 7a2 | N | N | N | N | N | N |
| 8a1 | N | N | N | N | N | ±BOM |
| 8a2 | N | N | N | N | N | ±BOM |
| Group 2 | ||||||
| 9 | N | N | N | N | CBVH | BOM |
| 10 | N | Short | Fetal | N | CBVH | N |
| 13a1 | N | Thin | VMEG | N | DWM/CBVH | MCM-BOM |
| 14 | N | N | N | N | CBVH | MCM-BOM |
| 17a1 | N | Thin | VMEG | N | ±CBVH | CBTE, ±BOM |
| Group 3 | ||||||
| 18 | Atrophy | N | VMEG | N | N | N |
| 19 | N | N | N | N | N | N |
| 20a1 | N | N | VMEG | N | N | N |
| 21a2 | N | N | N | N | N | N |
Abbreviations: ±BOM, normal except for subtle basi-occipital changes such as downslanted foramen magnum or flat inferior occiput; CBTE, cerebellar tonsilar ectopia with Chiari 1 malformation; CBVH, cerebellar vermis hypoplasia; ±CBVH, probable or mild CBVH; DWM, Dandy-Walker malformation; MCM-BOM, mega-cisterna magna with associated basi-occipital skull anomalies; Pt#, patient number; Short, short and thin corpus callosum consistent with mild partial agenesis; VMEG, ventriculomegaly. Fetal, fetal VMEG described as normal on postnatal imaging.
Prenatal data
Eight patients had abnormal prenatal ultrasounds with findings summarized in Table II. All were referred for routine studies, except for one mother who had abnormal results on expanded AFP screening and another who was concerned about recurrence of duplication 17p13.3 diagnosed in a previous child. Potentially important findings included increased nuchal translucency (7.5 mm) in one fetus, fetal ventriculomegaly, renal anomalies and endocardial fibroelastosis.
Routine amniotic fluid karyotypes were all reported as normal, but subsequent microarrays revealed the duplication in three pregnancies. One mother was a heterozygous carrier of the duplication and another was a mosaic carrier. In a third family both parents had normal arrays and FISH consistent with germline mosaicism. Two mothers terminated affected pregnancies. In the remaining individuals with prenatal abnormalities the duplication was diagnosed sometime after birth.
Growth
Abnormal growth patterns have previously been reported in patients with dup 17p13.3 [Bi et al., 2009; Bruno et al., 2010; Roos et al., 2009; Shimojima et al., 2010]. We therefore reviewed available growth parameters in our cohort (Supplementary Table II). However, our data did not confirm previous reports of large size at birth or later tall stature with duplications involving YWHAE. We noticed a general tendency towards obesity from early childhood on in all three groups, but have insufficient data to define a critical region for this finding, or even separate our findings from those of the normal population. A recent report of a small deletion of CRK associated with postnatal growth deficiency suggests that this gene, located just centromeric to YWHAE, may be important in determining body size [Ostergaard et al., 2012]. Identification of further small duplications in this region will help clarify the genes important for the regulation of body size.
Dysmorphic facial features
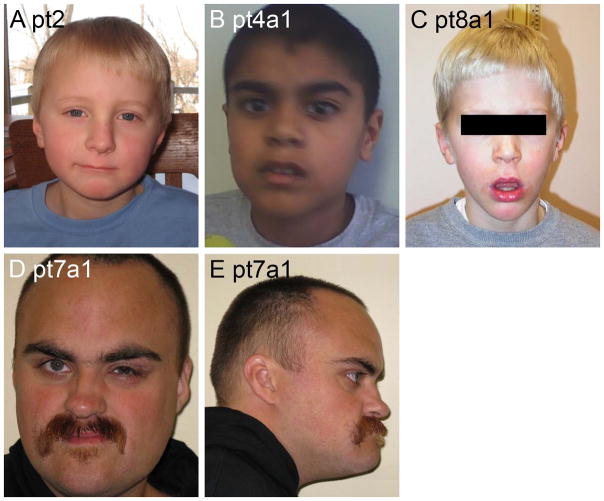
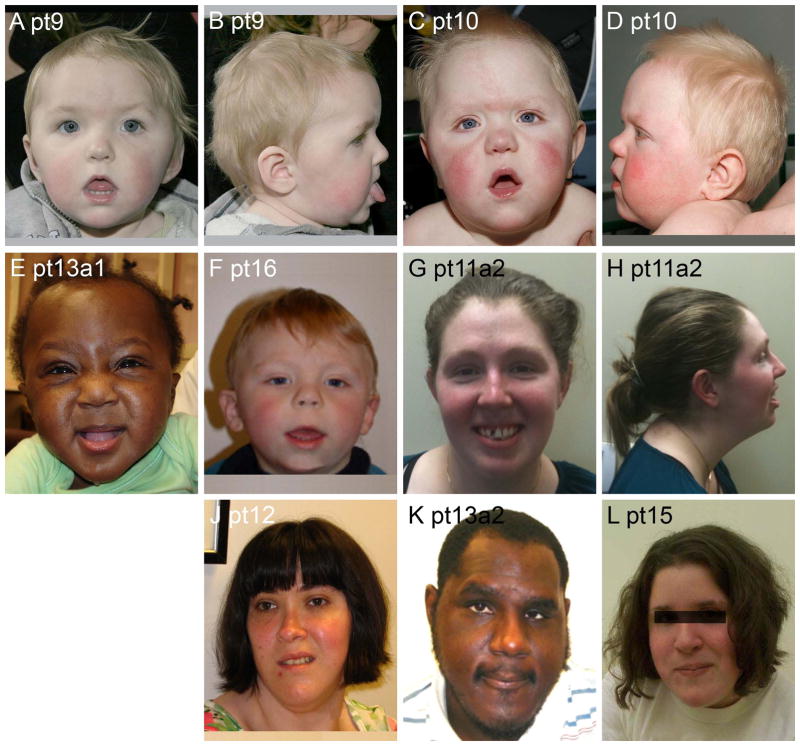
We examined facial photographs in 20 patients (Table III and Figures 2–8). Patients in all three groups were mildly dysmorphic, but with subtle and inconsistent features most consistent with the secondary effects of generalized hypotonia, such as mildly myopathic face, open mouthed expression and mildly increased facial length. In infants and young children, facial changes associated with hypotonia predominated. Several children had round faces as infants. With increasing age, evolving facial changes included lengthening of the face, an increasingly prominent jaw with pointed chin and small mouth. These features combined to form a generally consistent facial gestalt in 8/8 patients with photographs available in Group 2 (Table III). Serial photographs in pt14 of Group 2 document the evolution of the phenotype in the young child (Figure 4). In Group 1, many children had a hypotonic face, but facial characteristics were less distinctive than in Group 2. In Group 3, the five individuals with pictures were not convincingly dysmorphic beyond a hypotonic face seen in younger children.
Table 3.
Facial features in duplication 17p13.3 patients
| Pt# | Age | OFC | Dysmorphic facial features | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (centile) | Gestalt | Hypo-tonic | Long | Malar hypo | Prom jaw | Pointed chin | Small mouth | Other | ||
| Group 1 | ||||||||||
| 2 | 4y | 50 | − | +/− | − | − | + | + | − | |
| 3a1 | 4y | NL | − | + | +/− | − | + | + | − | CLP, hypertelorism |
| 4a1 | 8y | <5 | − | − | − | + | − | − | − | |
| 5a1 | 3y | NA | − | − | − | − | − | + | + | CLP |
| 5a2 | adult | NA | + | − | + | + | + | + | + | CLP |
| 6 | 14mo | NA | − | − | −, rd | − | − | − | − | Blepharophimosis, CP |
| 7a1 | 27y | <3 | ± | − | + | − | + | + | + | Midface hypoplasia |
| 8a1 | 4y | 75–90 | ± | + | + | − | +/− | + | + | |
| Group 2 | ||||||||||
| 9 | 12mo | −1.5 SD | + | + | −, rd | − | + | + | + | Short nose |
| 10 | 13mo | NA | + | + | + | − | + | + | + | |
| 11a2 | 24y | NA | ++ | ? | + | − | + | + | − | |
| 12 | 28y | 50–75 | ++ | + | + | − | + | + | + | |
| 13a1 | 13mo | 25 | + | + | + | − | + | + | + | |
| 13a2 | adult | NA | ++ | NA | + | − | − | + | + | |
| 14 | 6y | 25–50 | + | + | + | − | − | + | + | Midface hypoplasia |
| 15 | 14y | 25–50 | ++ | NA | + | − | + | + | + | |
| 16 | 2y | NA | − | + | −, rd | − | − | + | − | Short nose; smooth philtrum |
| Group 3 | ||||||||||
| 18 | 15y | <3 | − | + | + | + | − | − | − | |
| 19 | 5.5y | 50 | − | − | − | + | − | − | − | |
| 20a1 | 3.9y | 50 | ± | + | + | + | − | + | + | |
| 20a2 | 18mo | 10 | ± | + | + | + | − | + | + | |
| 21a2 | 27y | NA | + | − | + | − | + | + | + | |
Abbreviations: CLP, cleft lip and palate; CP, cleft palate; hypo, hypoplasia; mo, months; NA, not available; prom, prominent; rd, round; SD, standard deviations; y, years.
Figure 2.
Facial photos of four patients from Group 1. Note the hypotonic face in Patient 8a1 (D).
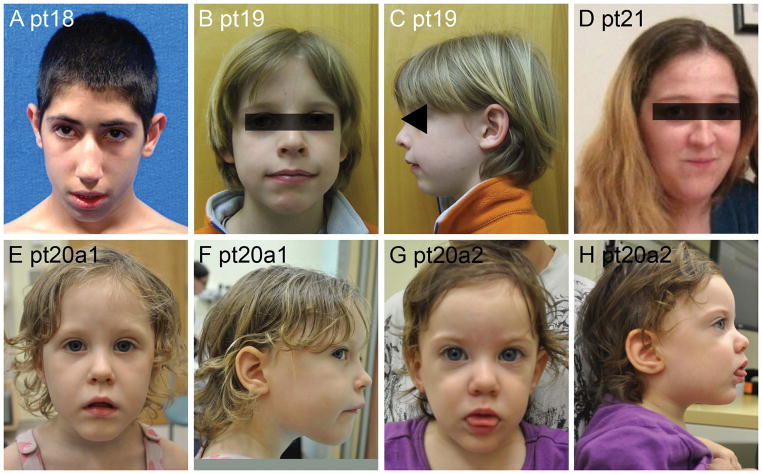
Figure 8.
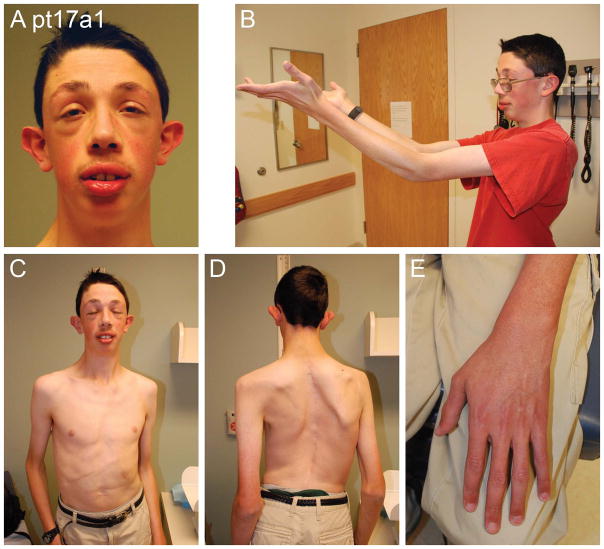
Facial and body photos of Patient 11 show dysmorphic facial features (A, C), and an unusual marfanoid habitus with scoliosis (D) and long limbs and fingers (B–E). He also has a dilated aortic root.
Figure 4.
Serial photos of Patient 14 from 2.5 to 7 years. Note increasing weight and facial lengthening with increasing age.
Medical and surgical complications
The medical problem most frequently reported in all groups was recurrent otitis media (Table VI), documented in 9 of 12 children with normal palates. Most were managed with tympanostomy tubes. Five children had early failure-to-thrive, including two given gastrostomy tubes for several months, and three who had gastroesophageal reflux. None had feeding issues persisting beyond infancy. Indeed, obesity and food foraging were reported in a few older children. Two patients had structural congenital heart disease, neither requiring surgery. Another two had urinary tract abnormalities, and two had significant spasticity that led to Achilles tendon lengthening procedures. One individual had precocious puberty at 7 years. Scoliosis requiring rod placement occurred in two older individuals in Groups 1 and 2, respectively. One adult in Group 2 had hypertension and depression.
Table 6.
Rare phenotypes and other health problems in duplication 17p13.3 patients
| Pt# | MARF | SHFLD | CL/P | Other health problems |
|---|---|---|---|---|
| Group 1 | ||||
| 1 | + | − | − | Flat feet, leg cramps, constipation |
| 2 | − | − | − | BOM |
| 3a1 | − | − | + | Pectus excavatum, hydrocele, pneumonia, BOM |
| 3a2 | − | − | + | BOM |
| 4a1 | − | − | − | |
| 5a1 | − | − | + | BOM |
| 5a2 | − | − | + | BOM |
| 6 | − | + | (+) | Cleft palate only, micrognathia, blepharophimosis, GER, bicuspid AoV, gastrostomy tube, orthopedic surgeries, BOM |
| 7a1 | − | − | − | Achilles lengthening |
| 8a1 | − | − | − | BOM |
| 8a2 | − | − | − | BOM |
| Group 2 | ||||
| 9 | − | − | − | FTT, BOM |
| 10 | − | − | − | Dislocated hip, inguinal hernia, recurrent URI, micropenis |
| 11a1 | − | − | − | Multicystic dysplastic kidney (unilateral) |
| 11a2 | − | − | − | |
| 12 | − | − | − | depression; hypertension; scoliosis with rods; incontinence, hydronephrosis; breast reduction, abdominoplasty, jaw reconstruction; Achilles lengthening |
| 13a1 | − | − | − | FTT, GER, gastrostomy tube, eczema |
| 13a2 | − | − | − | |
| 14 | − | − | − | FTT, GER; later overweight; precocious puberty |
| 15 | − | − | − | Hearing loss; UTI, ureteral reimplantation; sleep apnea; scoliosis; BOM |
| 16 | − | − | − | |
| 17a1 | + | − | − | Hiatal hernia; umbilical hernia; Morgagni hernia, VSD, hemivertebrae; GER, GT, BOM |
| Group 3 | ||||
| 18 | − | − | − | FTT |
| 19 | − | − | − | FTT, GER; gastrostomy tube; pneumonia; BOM |
| 20a1 | − | − | − | BOM |
| 20a2 | − | − | − | FTT |
| 21a1 | − | − | − | endocardial fibroelastosis |
| 21a2 | − | − | − | |
Abbreviations: AoV, aortic valve; CLP, cleft lip and palate; FTT, failure-to-thrive; F/U, follow-up; GER, gastroesophageal reflux; MARF, Marfanoid habitus; BOM, recurrent bilateral otitis media; Pt#, patient number; SHFLD, split hand-foot limb deficiency; URI, upper respiratory infections; UTI, serial urinary tract infections, VSD, ventricular septal defects.
Intellectual disability and neurologic problems
The most common developmental and neurologic problems were intellectual disability (ID) seen in 19 of 29 (66%) individuals, and hypotonia seen in 15 of 17 (88%) patients where this data was available (Table IV). Other health problems included seizures in four patients that were resistant to medical management in two, mild cerebral palsy in two, and a wide based, mildly ataxic gait attributed to hypotonia in several children. Many had mildly hyperextensible joints.
Table 4.
Developmental and neurologic status in duplication 17p13.3 patients
| Pt# | Hypotonia | Seizures | Cognition | ASD | Other |
|---|---|---|---|---|---|
| Group 1 | |||||
| 1 | + | − | Mild ID | + | Poor balance |
| 2 | + | − | Mild ID | + | |
| 3a1 | + | − | Mild ID | − | |
| 3a2 | na | − | Normal | − | |
| 4a1 | na | − | Mild ID | + | |
| 5a1 | + | − | Mild ID | − | |
| 5a2 | na | + | LD | − | ADD, unusual affect |
| 6 | + | − | Mod ID | − | |
| 7a1 | − | + | Severe ID | − | Mild spasticty, ataxia |
| 7a2 | na | − | LD | − | |
| 8a1 | + | − | Mild ID | − | |
| 8a2 | + | − | Mod ID | − | |
| 8a3 | na | − | Mild ID | + | Both parents have ASD |
| Group 2 | |||||
| 9 | + | − | Mild ID | − | |
| 10 | + | − | Mod ID | − | |
| 11a2 | na | − | LD | − | |
| 12 | na | + | LD | + | ADD, mild spasticity, ataxia, depression |
| 13a1 | − | − | Mild ID | − | |
| 13a2 | na | − | LD | − | |
| 14 | + | − | Mild ID | + | Poor balance, temper tantrums |
| 15 | + | − | Mild ID | − | Poor balance |
| 16 | + | − | Mild ID | + | Temper tantrums |
| 17a1 | + | − | Mild ID | − | |
| Group 3 | |||||
| 18 | na | − | Mod ID | − | ADHD |
| 19 | + | − | LD | − | ADHD, behavior problems |
| 20a1 | + | − | Mild ID | − | Temper tantrums |
| 20a2 | + | + | Mild ID | − | Temper tantrums |
| 20a3 | na | − | LD | − | |
| 21a2 | na | − | Normal | − | |
| 21a3 | na | − | Normal | − | |
Abbreviations: ADHD, attention deficity hyperactivity disorder; ASD, autistic spectrum disorder; ID, intellectual disability; LD, learning disability; Mod, moderate ID; na, not available; Pt#, patient number.
A wide spectrum of developmental and cognitive deficits was found in all three groups, with substantial inter- and intrafamilial variability (Table IV). In general, the size and location of the duplication correlated poorly with the degree of impairment. Combining groups, 14 of 29 had mild ID, and only 5 of 29 had moderate or severe ID, including two who had an affected but less disabled family member. Subjectively, the deficits in Group 3 seemed less severe than in Groups 1 and 2. For example, one Group 3 patient with LD (pt19) had a full scale IQ of 99, but showed significant learning and behavioral abnormalities without autism, while another (pt21a3) completed high school in regular classes. The four probands in Group 3 with centromeric duplication 17p13.3 have a less severe and less specific phenotype with normal brain imaging. The smallest region of overlap contains only three genes (METTL16, LIS1, KIAA0664 in Figure 1). While LIS1 duplication has been proposed as the cause of this phenotype [Bi et al., 2009], little is known about the function of the other two genes.
Across all groups aberrant behavior was frequent, but no consistent behavioral phenotype could be established. Attention deficit hyperactivity disorder, temper tantrums, occasional obsessive-compulsive disorder and food seeking behaviors were noted in all three groups, but we could not define critical regions for these. The presence of significant depression in the oldest proband suggests that psychiatric problems may occur in older individuals with this duplication. Neuropsychological testing was rarely reported in this cohort and would be of interest as there is a suggestion of a possibly distinct neurobehavioral phenotype associated with the duplication.
Autism spectrum disorder
A diagnosis of autism or autism spectrum disorder (ASD) was noted in 7 of 22 medical records of patients in Groups 1–2 (Table IV), but in none from Group 3. This suggests that one or more autism loci (contributory genes) are located in the telomeric portion of 17p13.3. We compared autism between Groups 1+2 and Group 3, but the difference was not statistically significant (p=0.29, Fisher’s exact test), probably because of small numbers. Among 11 patients with dup 17p13.3 and autism (all from Groups 1–2 as shown in Figure 1), the smallest region of overlap contains only two genes: MYO1C and INPP5K (Figure 10). Neither is a compelling autism candidate gene based on their known functions. Overexpression of Inpp5k in mice affects osmoregulation in kidney collecting ducts [Pernot et al., 2011]. Heterozygous missense mutations in the motor domain of MYO1C cause hearing loss in humans [Adamek et al., 2011]. However, MYO1C is a member of the myosin superfamily that binds phosphatidylinositol-4,5-bisphosphate (PIP2) and links the actin cytoskeleton to cellular membranes to affect mechano-signal transduction and membrane trafficking [Hokanson et al., 2006]. This function potentially links the protein to numerous intracellular signaling pathways including the PI3K-AKT pathway [Wickenden and Watson, 2010], so that further analysis will be important.
Figure 10.
Map of chromosome 17p13.3 shows possible critical regions for cerebellar vermis hypoplasia in red and autism in blue. The smaller critical regions were determined assuming a single locus for each disorder, while the larger regions allow for limited causal heterogeneity by removing the patient with the smallest duplication.
The standard critical region mapping approach assumes that a single gene is responsible for the autism phenotype in all individuals with dup 17p13.3. While this is usually valid for rare phenotypes caused by one or a few genes, it could exclude strong candidate genes for more common disorders with known genetic heterogeneity. Prior reports identified YWHAE and the adjacent CRK as possible candidate genes for the duplication 17p13.3 syndrome. Despite one report [Bruno et al., 2010] that excludes both genes based on autism in a patient with a slightly more centromeric duplication (their Patient 12), we still consider YWHAE and CRK to be good candidates for autism spectrum disorders seen in this cohort (Figure 10). Two further patients recently evaluated and not included in this report have small duplications involving YWHAE and CRK, and both have an autism spectrum disorder (CRC, unpublished data).
Brain imaging
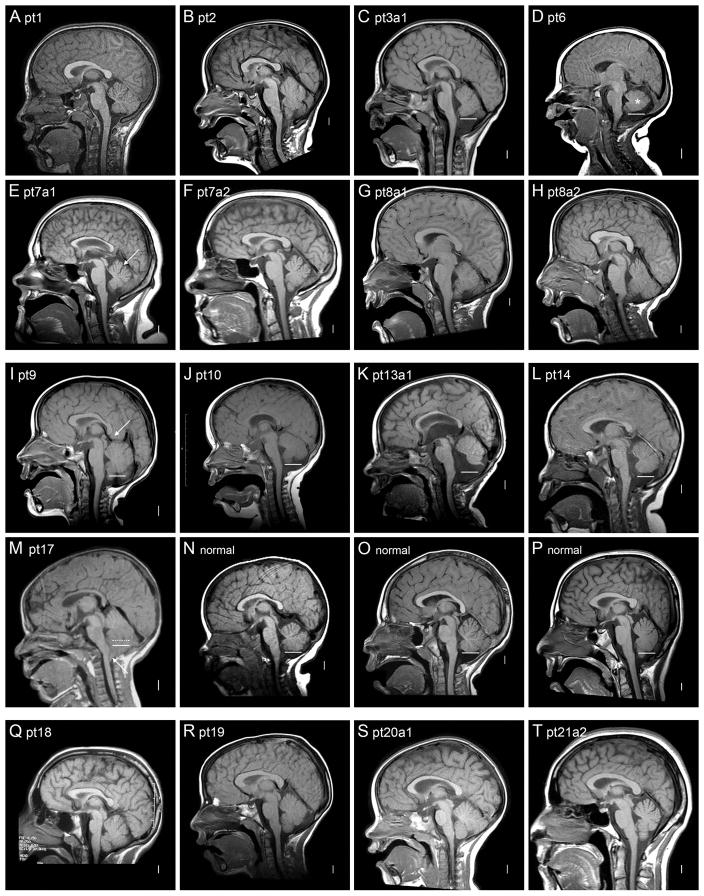
We reviewed brain-imaging studies performed for clinical care in 18 patients including 9 in Group 1 (2 affected sibs, both with imaging), 5 in Group 2, and 4 in Group 3 (Figure 9). The most consistent abnormalities involved the corpus callosum, cerebellum, posterior fossa and skull. We found mild hypogenesis of the corpus callosum in 2/9 patients in Group 1 and 1/5 in Group 2. Several others had mild, nonspecific thinning of the corpus callosum. The posterior fossa changes were more interesting, as we found mild but convincing CBVH in 5/5 patients in Group 2, but only 1/9 in Group 1 (Table V). Despite small numbers, the difference is statistically significant (p=0.003, Fisher’s exact test). If we recalculate after adding the borderline CBVH seen in patient 5, the difference remains significant (p=0.021, Fisher’s exact test).
Figure 9.
Brain imaging. T1–weighted midline sagittal brain magnetic resonance images are shown from individuals from Group 1 (A–H), Group 2 (I–M), and Group 3 (Q–T), as well as from three normal individuals (N–P). The solid white bars seen in multiple images show the level of the obex, which typically marks the level of the most inferior portion of the vermis as seen in the normal controls (N–P). The dotted white bar in M marks the lower limit of the vermis, which is difficult to see because of Chiari malformation in this patient. These images show a mildly small cerebellar vermis in two of eight patients in Group 1 (C and D), and in all five patients in Group 2 (I–M). The arrow in E points to a mildly prominent primary fissure, which suggests mild atrophy. The asterisk in D marks the smallest vermis, with normal vermian anatomy not well seen, probably because the image is just off midline. The arrow in image I points to mild protrusion or ectopia of the mesial parietal lobes into the tentorial notch between the corpus callosum and superior vermis. The vermis appears normal in all patients in Group 3 (Q–T). The images also show steeply angled foramen magnum (B, C, G, H, I, K, L) and flat inferior occiput (D, G, H, I, M) in most patients in Groups 1 and 2.
Several abnormalities involved the posterior fossa and/or posterior skull including mega-cisterna magna or Dandy-Walker malformation (DWM), sharply angled (downward anterior to posterior) foramen magnum, and flat inferior occiput (Table V). Combining these anomalies, we found mild posterior skull changes in 4/5 patients in Group 2, and in more subtle changes in 5/8 patients in Group 1 (striking in only 1/8). We found none of these changes in the four imaged patients in Group 3.
Among seven patients with definite CBVH or DWM (shown in Figure 9), the smallest region of overlap contains only four genes (CRK, MYO1C, INPP5K, PITPNA; Figure 10), none compelling candidate genes for a cerebellar developmental disorder. Further, the frequency of CBVH is higher in Group 2 than Group 1 patients, and the malformations are more severe. The higher penetrance of CBVH in Group 2 patients suggests that the region between YWHAE and LIS1 contains an important cerebellar developmental gene, although it may not be the only cerebellar developmental gene in the region.
If we exclude the smallest duplication with CBVH (pt3), the region encompasses a larger genomic region that includes HIC1 and other genes in our Group 2 (Figure 10). Hic1 is a tumor suppressor in mice and haploinsuffiency produces gross developmental defects including exencephaly, cleft palate, craniofacial defects and limb abnormalities suggesting an important developmental role [Carter et al., 2000; Grimm et al., 1999]. Also, Hic1 is expressed in cranial mesenchyme during brain development in mice (Kathleen Millen, PhD, unpublished data). We previously showed that haploinsufficiency of Foxc1 (and FOXC1 in humans) results in cerebellar hypoplasia in both mouse and human [Aldinger et al., 2009]. However, this gene is expressed in cranial mesenchyme but not the cerebellum itself, which implicates defects in signaling from the mesenchyme into the developing cerebellum as a cause of cerebellar hypoplasia. Thus, loss of HIC1 might also affect mesenchymal to cerebellar signaling leading to CBVH.
Unusual variant phenotypes
Our data demonstrate three distinct rare variant phenotypes associated with duplication 17p13.3, including CLP (or cleft palate alone in one girl), SHFLD, and marfanoid habitus. The first two phenotypes were observed only in individuals with small distal duplications, while data in the third phenotype is difficult to interpret (Figure 11). These finding suggest complex genomic regulation of the region. Alternatively, although less likely, these phenotypes could be unrelated to the duplication.
Figure 11.
Map of chromosome 17p13.3 shows possible critical regions for rare abnormalities including cleft lip and palate (CLP) in blue, split hand/foot with long bone deficiency (SHFLD) in purple, and marfanoid habitus in red. The rearrangements associated with CLP and SHFLD suggest a more complex mechanism than simple duplication, as explained in the text. All three dups associated with CLP have one breakpoint located within the ABR gene. All 4 dups associated with SHFLD shown here as well as 16/17 additional dups associated with SHFLD from another recent report [Klopocki et al., 2012] have one breakpoint located within or between the ABR and TUSC5 genes, suggesting disruption of regulatory elements.
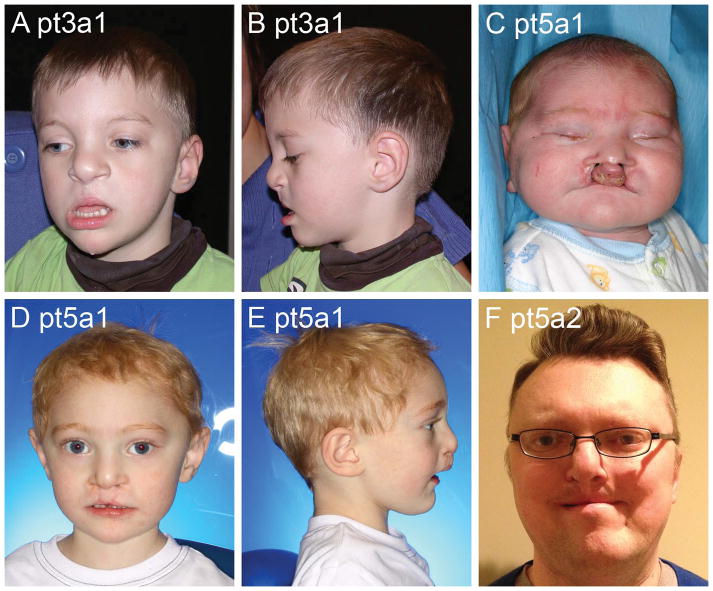
Cleft lip and palate
Probands in two families had CLP and surgical repair obscuring the usual dup 17p13 facial phenotype (Figure 6). In one family the proband (pt3a1) and his intellectually normal mother both had CLP, while the duplication carrier maternal grandmother did not have CLP or ID. In a second family, the carrier father (Pt5a2) had learning disabilities and CLP. At age 2 years, the affected infant had mild global delay on the Bayley Scales of Infant Development. A third child, described below, had cleft palate and micrognathia consistent with Robin sequence.
Figure 6.
Facial photos of three individuals with cleft lip and palate including one pre-surgical photo (C) and the remainder with repaired cleft lip (A, B and D–F). Also note the typical dup 17p13 adult face in Patient 5a2 (F).
Our observation of CLP or cleft palate alone in five patients from three families is novel. All three duplications contained BHLHA9 and several centromeric genes (BHLHA9, TUSC5, YWHAE, CRK, MYO1C, INPP5K, PITPNA), and had at least one breakpoint in the SHFLD region (chr17:853509-1151031). Indeed, all three duplications associated with CLP or cleft palate alone had one breakpoint located within ABR (Figure 11), so that CLP or cleft palate could result from haploinsufficiency of ABR rather than from duplication of surrounding genes. Cleft palate was reported in 3/8 patients with deletions of distal 17p13.3 that included ABR but not LIS1 [Bruno et al., 2010; Mignon-Ravix et al., 2010; Nagamani et al., 2009], and in 3/27 patients with Miller-Dieker syndrome [Dobyns et al., 1991] using older technology that could not define the molecular breakpoints (but 2/3 had visible chromosome deletions).
Heterozygous and homozygous Abr null mouse mutants as well as Abr−/−;Bcr−/− double mutants were reported to have a normal phenotype [Kaartinen et al., 2001]. Further, the CLP and cleft palate phenotypes are unlikely to be caused by duplication of BHLHA9, as expression in both mouse and human is restricted to the distal limb bud mesenchyme underlying the apical ectodermal ridge [Klopocki et al., 2012]. Thus, the molecular pathogenesis of CLP associated with dup 17p13.3 is not clear.
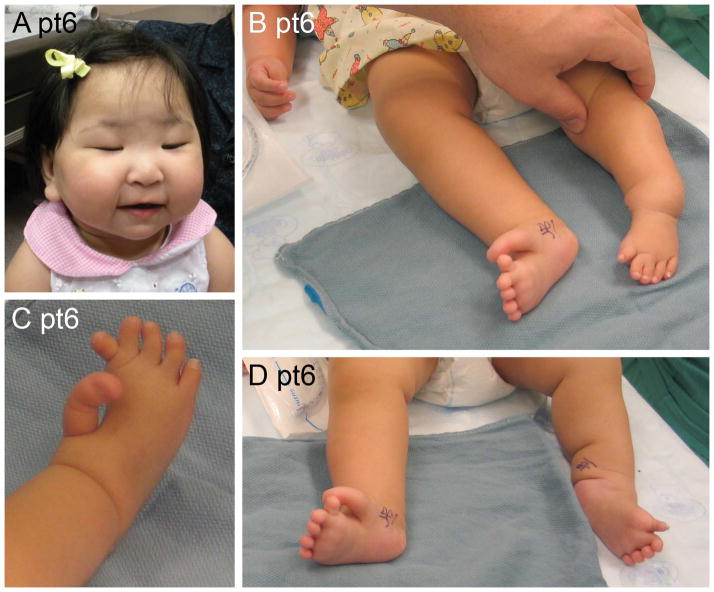
SHFLD and cleft palate
This single remarkable child (pt6) has distinctive dysmorphic facial features that include severe blepharophimosis, round cheeks, small jaw and cleft palate (Figure 7), with the latter two consistent with Pierre Robin sequence. Her lower limb defects are consistent with those described in SHFLD. Her left leg has a short tibia that diverges from the fibula at the ankle consistent with a Jones type IV tibial hemimelia, as well as a distally broad fibula distally lacking normal articulation at the ankle. She also has a left equinus deformity, hypoplastic first metatarsal, hypoplastic proximally-placed hallux, and left 2–3 toe syndactyly. Her right foot has a hypoplastic metatarsal with a deep cleft between the first and second toes, greatly enlarged and proximally placed hallux, and intercalary polydactyly attached to the second toe. We initially considered that this girl had a syndrome unrelated to 17p13.3 duplication. However, the publications related to duplication of BHLHA9 on 17p13.3 describe the SHFLD part of her phenotype [Armour et al., 2011; Klopocki et al., 2012].
Figure 7.
Facial and limb photos of a girl with split hand/foot with long bone deficiency show blepharophimosis, full cheeks and Robin sequence (A). She also has tibial deficiency with intercalary polydactyly and syndactyly (B–D).
These two reports described 58 patients from 20 families with SHFLD associated with small distal dup 17p13.3, and in addition noted incomplete penetrance and highly variable expressivity [Armour et al., 2011; Klopocki et al., 2012]. The critical region derived from Pt6 and the first paper [Armour et al., 2011], and the smaller critical region from the Klopocki et al. paper [ 2012] are shown in Figure 11. The latter study described SHFLD in 42/80 dup 17p13.3 carriers and attributed the phenotype to duplication of the small BHLHA9 gene. They also reported higher penetrance in males than females, with 30/42 male duplication carriers affected with SHFLD, but only 12/40 female carriers affected (P=0.0003, Fisher’s Exact Test). Our calculation of the P value includes probands, as they were not designated in the paper.
The authors attributed the lack of limb anomalies in published patients with dup 17p13.3 as probably due to non-penetrance [Klopocki et al., 2012]. However, the striking SHFLD phenotype was rare in our cohort of individuals with larger duplications (only 1 had SHFLD), even though most had duplications of the same genes. We therefore formally compared penetrance of SHFLD between the two groups. The first group includes individuals ascertained because of SHFLD or a relative with SHFLD; all have 17p13.3 duplications that include BHLHA9. We estimated penetrance of the SHFLD phenotype using data from Family 1 in the first paper [Armour et al., 2011] and all 17 families in the second [Klopocki et al., 2012]. After removing one affected individual from each family as a proband, 29/80 duplication carriers had SHFLD.
The second group consists of individuals ascertained because of a developmental disorder or relative with a developmental disorder. We included only individuals from our series and four prior reports [Bi et al., 2009; Bruno et al., 2010; Hyon et al., 2011; Roos et al., 2009] with 17p13.3 duplications that also include BHLHA9, as shown in Figure 1. We found that only one of 21 duplication carriers had SHFLD (P<0.0031, Fisher’s Exact Test), a difference that is statistically significant. We then divided both groups into males and females and repeated the analysis, finding the same statistically significant result for males but not for females, likely because of fewer females in our cohort and lower penetrance in females in the SHFLD cohorts.
While this analysis is confounded by variable ascertainment and incomplete phenotype data, the results suggest that duplication of BHLHA9 alone is not enough to cause SHFLD. That is, duplication of the gene may be necessary to cause the SHFLD phenotype, but is not sufficient. We compared the 17p13.3 duplications between the two groups and found two potentially important differences. First, the SHFLD group duplications are smaller (average 244 kb and mean 263 kb) than most developmental disorder group duplications (average 1.55 Mb and mean 1.12 Mb). Second, most SHFLD group duplications have one of the two duplication breakpoints close to BHLHA9, with 19/20 located between the start of ABR and the end (exon 7) of TUSC5 (chr17:853509-1151031). In contrast, only 4/17 duplications in our series contained BHLHA9 and had a breakpoint in this region. This includes Pt6 with SHFLD, cleft palate and additional dysmorphic and ocular features not seen in other subjects. We believe that our data imply a more complex genomic mechanism, and hypothesize that both duplication of BHLHA9 and disruption or separation from nearby regulatory elements are necessary to cause the SHFLD phenotype.
The data from SHFLD families with dup 17p13.3 showed significantly higher penetrance in males than females [Klopocki et al., 2012]. We asked whether the same phenomenon occurs in patients with larger duplications presenting with developmental phenotypes. Among our 17 probands in Groups 1 and 2, 13 were male and only 4 female. The excess of males in this still relatively small series was not statistically significant, but suggests a trend that bears further observation. Further, Groups 1 and 2 included four instances of mother to son, two instances of father to son, and interestingly no instances of either mother or father to daughter transmission (Table I and Supplementary Table I; we excluded Families 7 and 8 for reasons indicated in the Tables). The sons born to duplication carrier mothers were more severely affected than their mothers, but we have insufficient data to determine whether the sons born to carrier fathers were more severely affected than their fathers. In the entire series, we did not find greater severity among male compared to female probands.
Marfanoid habitus
Two patients in our series and two from the literature have what was termed by their physicians as a “marfanoid habitus”. The first reported patient (Patient 4 in the report) was described as tall, with large hands and long face [Bi et al., 2009]. He had aortic root dilation and mitral valve prolapse treated with Losartan. No molecular studies were performed as he was thought not to meet criteria for either Marfan or Loeys-Dietz syndrome. The second reported patient (Patient 1 in the report) also had severe hypotonia, and muscle biopsy revealed findings suggestive of congenital muscle fiber-type disproportion [Roos et al., 2009]. He was tall (height +3.5 SD) but his parents were also tall. He had long limbs and a long face. Echocardiographic findings and molecular testing were not reported.
In our series, one proband (Pt17a1, Figure 8) has a severe and distinctive connective tissue and craniofacial phenotype that appears more striking than classical Marfan syndrome, although no alternative diagnosis was apparent. His geneticist excluded MEN2B and other recognizable syndromes associated with a marfanoid habitus. He presented as a newborn with a hiatal hernia and bilateral Morgagni hernias that were repaired in infancy, and two ventricular septal defects. He also had a large umbilical hernia, asthenic habitus, kyphosis, arachnodactyly and dilatation of the aortic root, first noted at 13 years. Sequencing and deletion/duplication analysis of FBN1, TGFBR1 and TGFBR2 were normal. Another patient in our series (Pt1) was described as having a “marfanoid” appearance but no photographs or echocardiographic data were available. He was originally referred for significant hypotonia and had prominent ears, long chest, long lean body and flat feet. No information was available regarding aortic root dilation or molecular testing.
All four of these patients were male and none were described in sufficient detail to allow us to determine their similarity to one another, or the appropriateness of the designation “marfanoid habitus”. Examination of clinical photographs does not suggest a similar craniofacial phenotype for these patients and we are not convinced that they have the same cause for their “marfanoid” appearance. The critical region is large and does not contain an attractive candidate gene (Figure 11). Further reports with clinical and molecular data will be needed to determine if this represents another rare and distinct duplication 17p13.3 phenotype, a coincidence of marfanoid features in families with tall stature, or several different phenotypes with heterogeneous causes. From the data available, it seems reasonable to consider echocardiographic screening of patients with duplication 17p13.3.
Conclusions
In summary, we reviewed 34 individuals from 21 families with duplication 17p13.3 associated with heterogeneous breakpoints, and found several overlapping phenotypes. However, none were pathognomonic or likely to allow recognition of this genomic disorder on clinical grounds alone. Within families we found significant variability of phenotype with parents generally being less severely affected than their child. This suggests that the minimally affected individual with 17p13.3 duplication is unlikely to be ascertained unless a more severely affected child is born.
Our review of 51 patients with duplication 17p13.3 (34 reported here and 16 from the literature) confirms most abnormalities noted in previous reports, and adds information regarding prenatal features, dysmorphic facial features, brain malformations and several less common medical problems. Most importantly, our data demonstrate that incomplete penetrance and highly variable expressivity are characteristic of this duplication syndrome. This suggests that regulation of gene expression in this region must be complex. Further, our data suggest that prior reports linking duplication of specific genes such as LIS1 with a specific phenotype were premature.
The features associated with telomeric duplications of 17p13.3 (Group 1) include early developmental delay, mild to moderate ID or LD, an increased rate of autism, hypotonia and mildly myopathic facial features in infants and young children, mild but characteristic facial dysmorphic features at older ages, and occasional mild brain malformations involving the corpus callosum, cerebellum and posterior fossa. The features associated with larger duplications of 17p13.3 (Group 2) include the features enumerated above as well as more striking facial dysmorphic findings at older ages consisting of a long face, small mouth, prominent jaw and pointed chin, and malformations of the cerebellum and posterior fossa that are more consistent and severe than those found with smaller duplications. Several probands of varying ages had persistent hypotonia, hyperextensible joints and mild gait instability irrespective of their group. Most but not all older children and adults were overweight.
The features associated with centromeric duplications (Group 3) were less severe and less specific, consisting of frequent failure-to-thrive in infancy, developmental delay, mild to moderate ID or LD generally less severe than in Groups 1–2, seizures in one, behavioral problems especially temper tantrums and attention deficit disorder, and less distinct dysmorphic facial features. We found no association in Group 3 with autism or brain malformations.
Our analysis of dup 17p13.3-associated phenotypes proved far more challenging than we expected. In retrospect, we attribute this to the lack of any highly penetrant abnormality, the presence of several relatively common and nonspecific features such as failure-to-thrive, obesity, intellectual disability and autism, and to the complex inheritance we found for rare variant phenotypes (CLP, SHFLD and marfanoid habitus). Despite the many CNV-associated syndromes described so far, most genomic regions have not yet been associated with deletion or duplication syndromes. While some are likely to be early lethal, we expect that many more will have non-specific and variable phenotypes, and propose that duplication 17p13.3 may serve as a paradigm for this class of disorders.
Supplementary Material
Figure 3.
Facial photos of eight patients from Group 2. Note the round faces and facial hypotonia in younger Patients 9, 10 and 16 (A–F), and the long faces with prominent jaw and pointed chins in older Patients 11, 12, 13 and 15 (G–L).
Figure 5.
Facial photos of five patients from Group 3. Note an inconsistent phenotype with normal appearance of Patient 19.
Acknowledgments
We wish to thank all of the children and families in this study for their help with this project over several years. The work was funded by the US National Institutes of Health under NINDS grant NS058721 (to W.B.D.).
Footnotes
URLs
DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources, http://decipher.sanger.ac.uk; UCSC Genome Browser, http://genome.ucsc.edu.
References
- Adamek N, Geeves MA, Coluccio LM. Myo1c mutations associated with hearing loss cause defects in the interaction with nucleotide and actin. Cell Mol Life Sci. 2011;68(1):139–150. doi: 10.1007/s00018-010-0448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldinger KA, Lehmann OJ, Hudgins L, Chizhikov VV, Bassuk AG, Ades LC, Krantz ID, Dobyns WB, Millen KJ. FOXC1 is required for normal cerebellar development and is a major contributor to chromosome 6p25.3 Dandy-Walker malformation. Nat Genet. 2009;41(9):1037–1042. doi: 10.1038/ng.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour CM, Bulman DE, Jarinova O, Rogers RC, Clarkson KB, DuPont BR, Dwivedi A, Bartel FO, McDonell L, Schwartz CE, Boycott KM, Everman DB, Graham GE. 17p13.3 microduplications are associated with split-hand/foot malformation and long-bone deficiency (SHFLD) Eur J Hum Genet. 2011;19(11):1144–1151. doi: 10.1038/ejhg.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Sapir T, Shchelochkov OA, Zhang F, Withers MA, Hunter JV, Levy T, Shinder V, Peiffer DA, Gunderson KL, Nezarati MM, Shotts VA, Amato SS, Savage SK, Harris DJ, Day-Salvatore DL, Horner M, Lu XY, Sahoo T, Yanagawa Y, Beaudet AL, Cheung SW, Martinez S, Lupski JR, Reiner O. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41(2):168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno DL, Anderlid BM, Lindstrand A, van Ravenswaaij-Arts C, Ganesamoorthy D, Lundin J, Martin CL, Douglas J, Nowak C, Adam MP, Kooy RF, Van der Aa N, Reyniers E, Vandeweyer G, Stolte-Dijkstra I, Dijkhuizen T, Yeung A, Delatycki M, Borgstrom B, Thelin L, Cardoso C, van Bon B, Pfundt R, de Vries BB, Wallin A, Amor DJ, James PA, Slater HR, Schoumans J. Further molecular and clinical delineation of co-locating 17p13.3 microdeletions and microduplications that show distinctive phenotypes. J Med Genet. 2010;47(5):299–311. doi: 10.1136/jmg.2009.069906. [DOI] [PubMed] [Google Scholar]
- Cardoso C, Leventer RJ, Ward HL, Toyo-Oka K, Chung J, Gross A, Martin CL, Allanson J, Pilz DT, Olney AH, Mutchinick OM, Hirotsune S, Wynshaw-Boris A, Dobyns WB, Ledbetter DH. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am J Hum Genet. 2003;72(4):918–930. doi: 10.1086/374320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MG, Johns MA, Zeng X, Zhou L, Zink MC, Mankowski JL, Donovan DM, Baylin SB. Mice deficient in the candidate tumor suppressor gene Hic1 exhibit developmental defects of structures affected in the Miller-Dieker syndrome. Hum Mol Genet. 2000;9(3):413–9. doi: 10.1093/hmg/9.3.413. [DOI] [PubMed] [Google Scholar]
- Dobyns WB, Curry CJR, Hoyme HE, Turlington L, Ledbetter DH. Clinical and molecular diagnosis of Miller-Dieker syndrome. Am J Hum Genet. 1991;48:584–594. [PMC free article] [PubMed] [Google Scholar]
- Dobyns WB, Stratton RF, Parke JT, Greenberg F, Nussbaum RL, Ledbetter DH. Miller-Dieker syndrome and monosomy 17p. J Pediatr. 1983;102:552–558. doi: 10.1016/s0022-3476(83)80183-8. [DOI] [PubMed] [Google Scholar]
- Grimm C, Sporle R, Schmid TE, Adler ID, Adamski J, Schughart K, Graw J. Isolation and embryonic expression of the novel mouse gene hic1, the homologue of HIC1, a candidate gene for the miller-dieker syndrome [In Process Citation] Hum Mol Genet. 1999;8(4):697–710. doi: 10.1093/hmg/8.4.697. [DOI] [PubMed] [Google Scholar]
- Hannes FD, Sharp AJ, Mefford HC, de Ravel T, Ruivenkamp CA, Breuning MH, Fryns JP, Devriendt K, Van Buggenhout G, Vogels A, Stewart H, Hennekam RC, Cooper GM, Regan R, Knight SJ, Eichler EE, Vermeesch JR. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46(4):223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol Biol Cell. 2006;17(11):4856–4865. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppman-Chaney N, Wain K, Seger P, Superneau D, Hodge J. Identification of single gene deletions at 15q13.3: further evidence that CHRNA7 causes the 15q13.3 microdeletion syndrome phenotype. Clin Genet. 2012 doi: 10.1111/j.1399-0004.2012.01925.x. [DOI] [PubMed] [Google Scholar]
- Hyon C, Marlin S, Chantot-Bastaraud S, Mabboux P, Beaujard MP, Al Ageeli E, Vazquez MP, Picard A, Siffroi JP, Portnoi MF. A new 17p13.3 microduplication including the PAFAH1B1 and YWHAE genes resulting from an unbalanced X;17 translocation. Eur J Med Genet. 2011;54(3):287–291. doi: 10.1016/j.ejmg.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Ingason A, Rujescu D, Cichon S, Sigurdsson E, Sigmundsson T, Pietilainen OP, Buizer-Voskamp JE, Strengman E, Francks C, Muglia P, Gylfason A, Gustafsson O, Olason PI, Steinberg S, Hansen T, Jakobsen KD, Rasmussen HB, Giegling I, Moller HJ, Hartmann A, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Bramon E, Kiemeney LA, Franke B, Murray R, Vassos E, Toulopoulou T, Muhleisen TW, Tosato S, Ruggeri M, Djurovic S, Andreassen OA, Zhang Z, Werge T, Ophoff RA, Rietschel M, Nothen MM, Petursson H, Stefansson H, Peltonen L, Collier D, Stefansson K, St Clair DM. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2011;16(1):17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaartinen V, Gonzalez-Gomez I, Voncken JW, Haataja L, Faure E, Nagy A, Groffen J, Heisterkamp N. Abnormal function of astroglia lacking Abr and Bcr RacGAPs. Development. 2001;128(21):4217–4227. doi: 10.1242/dev.128.21.4217. [DOI] [PubMed] [Google Scholar]
- Klopocki E, Lohan S, Doelken SC, Stricker S, Ockeloen CW, Soares Thiele de Aguiar R, Lezirovitz K, Mingroni Netto RC, Jamsheer A, Shah H, Kurth I, Habenicht R, Warman M, Devriendt K, Kordass U, Hempel M, Rajab A, Makitie O, Naveed M, Radhakrishna U, Antonarakis SE, Horn D, Mundlos S. Duplications of BHLHA9 are associated with ectrodactyly and tibia hemimelia inherited in non-Mendelian fashion. J Med Genet. 2012;49(2):119–125. doi: 10.1136/jmedgenet-2011-100409. [DOI] [PubMed] [Google Scholar]
- Mignon-Ravix C, Cacciagli P, El-Waly B, Moncla A, Milh M, Girard N, Chabrol B, Philip N, Villard L. Deletion of YWHAE in a patient with periventricular heterotopias and pronounced corpus callosum hypoplasia. J Med Genet. 2010;47(2):132–136. doi: 10.1136/jmg.2009.069112. [DOI] [PubMed] [Google Scholar]
- Nagamani SC, Zhang F, Shchelochkov OA, Bi W, Ou Z, Scaglia F, Probst FJ, Shinawi M, Eng C, Hunter JV, Sparagana S, Lagoe E, Fong CT, Pearson M, Doco-Fenzy M, Landais E, Mozelle M, Chinault AC, Patel A, Bacino CA, Sahoo T, Kang SH, Cheung SW, Lupski JR, Stankiewicz P. Microdeletions including YWHAE in the Miller-Dieker syndrome region on chromosome 17p13.3 result in facial dysmorphisms, growth restriction, and cognitive impairment. J Med Genet. 2009;46(12):825–833. doi: 10.1136/jmg.2009.067637. [DOI] [PubMed] [Google Scholar]
- Ostergaard JR, Graakjaer J, Brandt C, Birkebaek NH. Further delineation of 17p13.3 microdeletion involving CRK. The effect of growth hormone treatment. Eur J Med Genet. 2012;55(1):22–26. doi: 10.1016/j.ejmg.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Pernot E, Terryn S, Cheong SC, Markadieu N, Janas S, Blockmans M, Jacoby M, Pouillon V, Gayral S, Rossier BC, Beauwens R, Erneux C, Devuyst O, Schurmans S. The inositol Inpp5k 5-phosphatase affects osmoregulation through the vasopressin-aquaporin 2 pathway in the collecting system. Pflugers Arch. 2011;462(6):871–883. doi: 10.1007/s00424-011-1028-0. [DOI] [PubMed] [Google Scholar]
- Ramalingam A, Zhou XG, Fiedler SD, Brawner SJ, Joyce JM, Liu HY, Yu S. 16p13.11 duplication is a risk factor for a wide spectrum of neuropsychiatric disorders. J Hum Genet. 2011;56(7):541–544. doi: 10.1038/jhg.2011.42. [DOI] [PubMed] [Google Scholar]
- Roos L, Jonch AE, Kjaergaard S, Taudorf K, Simonsen H, Hamborg-Petersen B, Brondum-Nielsen K, Kirchhoff M. A new microduplication syndrome encompassing the region of the Miller-Dieker (17p13 deletion) syndrome. J Med Genet. 2009;46(10):703–710. doi: 10.1136/jmg.2008.065094. [DOI] [PubMed] [Google Scholar]
- Shchelochkov OA, Cheung SW, Lupski JR. Genomic and clinical characteristics of microduplications in chromosome 17. Am J Med Genet A. 2010;152A(5):1101–1110. doi: 10.1002/ajmg.a.33248. [DOI] [PubMed] [Google Scholar]
- Shimojima K, Sugiura C, Takahashi H, Ikegami M, Takahashi Y, Ohno K, Matsuo M, Saito K, Yamamoto T. Genomic copy number variations at 17p13.3 and epileptogenesis. Epilepsy Res. 2010;89(2–3):303–309. doi: 10.1016/j.eplepsyres.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Balikova I, Schrander-Stumpel C, Fryns JP, Devriendt K. The causality of de novo copy number variants is overestimated. Eur J Hum Genet. 2011;19(11):1112–1113. doi: 10.1038/ejhg.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickenden JA, Watson CJ. Key signalling nodes in mammary gland development and cancer. Signalling downstream of PI3 kinase in mammary epithelium: a play in 3 Akts. Breast Cancer Res. 2010;12(2):202. doi: 10.1186/bcr2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.