Abstract
In a recent comprehensive investigation, we largely failed to identify significant genetic markers associated with P3 amplitude or to corroborate previous associations between P3 and specific single nucleotide polymorphisms (SNPs) or genes. In the present study we extended this line of investigation to examine time-frequency (TF) activity and intertrial phase coherence (ITPC) in the P3 time window, both of which are associated with P3 amplitude. Previous genome-wide research has reported associations between P3-related theta and delta activity and individual genetic variants. A large, population-based sample of 4211 subjects, comprising male and female adolescent twins and their parents, was genotyped for 527,828 single nucleotide polymorphisms (SNPs), from which over six million SNPs were accurately imputed. Heritability estimates were greater for TF energy than ITPC, whether based on biometric models or the combined influence of all measured SNPs (derived from genome-wide complex trait analysis). The magnitude of overlap in the specific SNPs associated with delta energy and ITPC and P3 amplitude was significant. A genome-wide analysis of all SNPs, accompanied by an analysis of approximately 17,600 genes, indicated a region of chromosome 2 around TEKT4 that was significantly associated with theta ITPC. Analysis of candidate SNPs and genes previously reported to be associated with P3 or related phenotypes yielded one association surviving correction for multiple tests: between theta energy and CRHR1. However, we did not obtain significant associations for SNPs implicated in previous genome-wide studies of TF measures. Identifying specific genetic variants associated with P3 amplitude remains a challenge.
Keywords: Genome-wide association study, Endophenotype, P300, Heritability, Time-frequency energy, Inter-trial phase coherence
1. Introduction
1.1. Endophenotypes and gene discovery
Since a recent article by Gottesman and Gould popularized the concept (Gottesman and Gould, 2003), endophenotypes have seen increasingly widespread use in psychiatric genetics. A number of slightly different definitions of an endophenotype have been proposed (e.g., Almasy and Blangero, 2001; Cannon and Keller, 2006; de Geus, 2010; Gottesman and Gould, 2003; Iacono and Malone, 2011; Kendler and Neale, 2010; Miller and Rockstroh, 2013), yet at the heart of all of them is the notion that endophenotypes are closer to the actual gene product than psychiatric categories and are likely genetically less complex than psychiatric disorders. This characteristic implies that endophenotypes are likely to produce larger effect sizes in relation to specific genetic markers than psychiatric categories and disorders, which has led to the hope that endophenotypes will facilitate identifying genes associated with psychopathology (e.g., Iacono and Malone, 2011).
We and our colleagues recently conducted a comprehensive set of molecular-genetic analyses of 17 psychophysiological measures that can, to different degrees, be considered candidate endophenotypes for various psychiatric disorders (Iacono, 2014; Malone et al., 2014a,b; Vaidyanathan et al., 2014a,b,c; Vrieze et al., 2014a,b). Our sample consisted of >4000 subjects, most measures were at least moderately heritable in our data, and our approach was comprehensive, including sequencing rare variants in addition to genome-wide association study (GWAS). Nevertheless, our results failed to produce compelling evidence of specific genetic variants associated with these candidate endophenotypes. Although we did obtain several novel findings across the set of studies, and these may well ultimately provide insight into the endophenotypes and the disorders with which they are commonly associated, significant findings were few in number and at least some are likely false positives. These findings collectively challenge the notion that endophenotypes will facilitate gene finding.
1.2. P3 amplitude, its time-frequency constituents, and phase resetting
Although P3 amplitude is a robust endophenotype for different forms of disinhibitory psychopathology, including substance abuse (Euser et al., 2012; Iacono and Malone, 2011; Iacono et al., 2003; Porjesz et al., 2005), we uncovered virtually no evidence of specific genetic variants that were significantly associated with it; only one gene (MYEF2) was significantly associated with P3 amplitude, and its relevance is unclear. In a paper that in some ways anticipated these findings, Miller and Rockstroh (2013) argued that the kinds of measures psychologists and psychiatrists commonly consider as candidate endophenotypes are themselves too far downstream from the action and expression of specific genes, and the causal pathway from susceptibility genetic markers to manifest psychopathology is nonlinear and complex. They suggested that endophenotypes for our endophenotypes are necessary. P3 amplitude is certainly not a unitary phenomenon, reflecting at least two different subcomponents (P3a and P3b). Moreover, waveforms recorded at the scalp are made up of sources of activity from different brain areas that combine in very particular ways at the recording electrode. It may be that measures that are more elementary than P3 or that reflect characteristics of brain activity that contribute to what we observe as the P3 wave are closer to gene action than P3 itself, providing greater sensitivity to detecting genetic effects (cf. Ford, 2014).
Current models of ERPs that attempt to describe the relationship between them and ongoing EEG activity emphasize three main phenomena. The classic view holds that ERPs reflect evoked activity: activity of neural ensembles elicited by events in the experimental task, independent of ongoing activity. More recent views, although not without their critics, hold that events influence ongoing activity in a way that gives rise to the observed ERP, either by modulating the amplitude of ongoing activity in the absence of stimulus locking or by partially resetting the phase of ongoing oscillations (Klimesch et al., 2007; Makeig and Onton, 2009; Makeig et al., 2002; Sauseng et al., 2007). Of course, these models are not mutually exclusive (Fell et al., 2004; Min et al., 2007).
At the same time, approaches that simultaneously measure brain activity with respect to its time course and underlying frequencies have become popular in recent years. Such approaches lend themselves directly to describing specific ways that events modulate ongoing activity, both in time and frequency. Our own work has suggested that such approaches may offer incremental validity or other advantages over traditional approaches that focus on voltage deflections in the time domain only (Gilmore et al., 2010a). Moreover, preliminary evidence indicates that measures of time-frequency activity have some of the properties of an endophenotype (Gilmore et al., 2010a,b; Jones et al., 2006b; Kamarajan et al., 2004, 2006; Padmanabhapillai et al., 2006a,b; Rangaswamy et al., 2007; Yoon et al., 2013).
1.3. Theta and delta activity and specific molecular-genetic markers
Indeed, the only published studies using a genome-wide search for genetic markers other than our previous GWAS of P3 amplitude focused on time-frequency features rather than P3 amplitude (Chen et al., 2009; Jones et al., 2004, 2006a; Kang et al., 2012; Zlojutro et al., 2011), examining energy in the theta and delta ranges during a time window that spans the P3 response. This focus accords with evidence that P3 amplitude is primarily composed of activity (energy) in the theta and delta ranges (Basar et al., 2001; Karakas et al., 2000; Yordanova et al., 2000), with the former more prominent at frontal electrodes and the latter more prominent at parietal ones (see Fig. 1 in Karakas et al., 2000). In a genome-wide linkage analysis in a large family sample of over 1300 individuals in 253 families with a dense history of alcoholism, followed by linkage disequilibrium analysis in a subset of Caucasian families, Jones and colleagues observed linkage between frontal theta activity and a region of chromosome 7, containing CHRM2, which codes for a muscarinic cholinergic receptor. Given the likely importance of acetylcholine to ERP generation (Frodl-Bauch et al., 1999; Kenemans and Kähkönen, 2010), follow-up association analysis was conducted for several SNPs in this gene. One showed a strong association with frontal theta, while another showed a strong association with central-parietal delta.
This initial study was followed by two additional investigations using essentially the same sample but a slightly modified time-frequency window, or time-frequency region of interest (TFROI), in order to focus especially on midline frontal theta activity. The first investigation also expanded the number of CHRM2 SNPs examined (Jones et al., 2006a), while the second examined SNPs in GRM8 (Chen et al., 2009), a gene coding for a glutamate receptor, which is in the same region as CHRM2. Jones and colleagues found that six SNPs in CHRM2, in addition to rs2359786, the SNP associated with frontal theta activity in their original report, were associated with midline frontal theta energy. Four of these, including rs2359786, were associated with midline parietal theta energy as well. Chen and colleagues reported significant associations between eight SNPs in GRM8 and frontal theta activity. Seven also showed significant associations with central and parietal theta activity.
Two GWASs were conducted subsequently. Zlojutro and colleagues conducted a two-stage analysis (Zlojutro et al., 2011). The first stage consisted of a GWAS of frontal theta energy in a case-control sample of 1064 individuals, 640 of them with alcohol dependence and the remainder controls. In the second stage, SNPs were prioritized for follow-up analysis in a non-overlapping family replication sample, based on significance in the first stage, as well as putative function and other factors. Although neither stage yielded any associations that were genome-wide significant, four SNPs produced similar effect sizes and the same direction of effect across the two stages. The second GWAS consisted of a family-based analysis of frontal theta energy in >1500 individuals from 117 Caucasian families (Kang et al., 2012). A cluster of SNPs on chromosome 21 showed particularly small p-values, with seven being genome-wide significant. Fully half of the 22 SNPs with p-values < 10−5 and all of the genome-wide significant SNPs were contained in KCNJ6. The protein encoded by this gene is an inward rectifier potassium channel, which is widely distributed in the brain and is an important element in the transmission of many neurotransmitters, including dopamine, acetylcholine, GABA, and glutamate.
1.4. Rationale for and aims of the current study
Taken collectively, these studies indicate that potentially important genetic variants are associated with theta energy in particular, especially at frontal electrode sites. We undertook the present investigation to expand on our previous GWAS of P3 amplitude, following the lead of these two GWASs. Our primary aim was to determine whether time-frequency measures might provide a window onto the relevant molecular-genetic influences on P3-related measures that P3 itself failed to do. Our secondary aim was to determine whether our results would be consistent with previous findings concerning these time-frequency measures, which would provide converging evidence concerning the influence of specific genetic variants on P3-related activity. We adopted the general analytic approach of our previous study, which comprised several aspects: In a family sample of approximately 4200 individuals, the largest sample used to conduct genome-wide analyses of time-frequency measures to date, we 1) conducted biometric analyses of twin and family data to confirm that the phenotypes examined are heritable; 2) determined the proportion of variance in each phenotype accounted for by all SNPs on our genotyping array in aggregate (“SNP heritability”), a molecular-genetic analog of biometric heritability analyses using measured genetic variants rather than phenotypic relationships; 3) carried out a GWAS of the association between each of >6 million SNPs in turn and our phenotypes; and 4) conducted a similar analysis of individual genes, rather than SNPs. In addition to genome-wide analyses of SNPs and genes with a focus on discovery, we examined SNPs and genes that might be considered candidates for association with our phenotypes because they have previously been associated with theta energy or P3 amplitude. In a similar vein, we examined 111 SNPs that have recently been determined to be associated with schizophrenia due to the robust association between schizophrenia and P3 amplitude (Ford, 1999) and time-frequency measures (Ethridge et al., 2015).
Using TFROIs corresponding to the P3 window, we examined total theta energy at electrode Fz, the primary focus of previous studies (Chen et al., 2009; Jones et al., 2006a; Kang et al., 2012; Zlojutro et al., 2011), and total delta energy at Pz. This electrode-TFROI pair reflects the predominant topographic distribution of theta and delta and these two measures are likely the primary contributors to P3 amplitude. In addition, we examined inter-trial phase coherence (ITPC) within each TFROI (theta-Fz and delta-Pz). ITPC is a measure of the consistency of the phase of the signal at each time-frequency bin across trials, and thus permits one to quantify the degree to which brain activity shows time- and frequency-dependent synchronization to stimulus onset (Tallon-Baudry and Bertrand, 1999). Although ITPC measures have not been previously examined, ITPC is reduced in externalizing disorders (Burwell et al., 2014), like P3 amplitude, and examining ITPC measures might shed light on our P3 findings.
2. Material and method
2.1. Participants
The sample for the present investigation was identical to the sample in our previous GWAS of P3 amplitude, which consisted of all twins from the Minnesota Twin Family Study (MTFS; Iacono et al., 1999; Keyes et al., 2009; McGue et al., 2007) who completed a laboratory assessment when they were approximately 17 years old and all parents who completed an identical assessment (N = 4214). The MTFS comprises three different cohorts of twin families. In two of the three, twins were approximately 11 years old during the year they were enrolled in the study, whereas in the third, twins were approximately 17 years old. The MTFS uses a longitudinal design, with assessment of twins repeated at 3- to 5-year intervals. Data for the present study are from the age-17 assessment, which was the second follow-up assessment of younger twins and the intake assessment of older twins. Because the midline frontal electrode was not part of the recording montage used at the beginning of the MTFS, fewer subjects have data from Fz (N = 3447) than Pz (N = 4167). There were 2422 adolescent subjects (1247 females, or 51%) and 1745 parents (1183 fathers, or 68%). Adolescent participants ranged in age from 16.6 to 20.0 (mean, 17.7), while parents ranged in age from 28.4 to 65.3 (mean, 44.5). (A few parents of younger-cohort twins were <30 years old at the time the family was enrolled.) Participants gave written consent or assent as appropriate, depending on their age.
The sample is broadly representative ethnically of the state of Minnesota during the relevant birth years, and it is predominantly Caucasian (96%). To avoid population stratification, which confounds molecular-genetic analyses if ethnic differences exist in allele frequencies and mean levels of the phenotype under study, we only included Caucasian subjects in these analyses, as in our previous study. Ethnicity was based on self-report, combined with principal component analysis (PCA) of genotype data (Miller et al., 2012).
2.2. Experimental task and time-frequency measures
Event-related potentials (ERPs) were elicited by means of the Begleiter rotated heads task (Begleiter et al., 1984). Subjects viewed a sequence of 240 stimuli, each appearing for 100 ms. One third were designated targets and consisted of a superior view of a stylized head with one ear. Subjects were instructed to press a button on the left arm of their chair if viewing a head with a left ear or the button on the right arm of their chair if viewing a head with a right ear. Half the target stimuli were rotated 180°. The remaining 160 stimuli consisted of plain ovals, which subjects were instructed to ignore.
2.2.1. Data collection
Data were collected over the course of >20 years using two different stimulus delivery methods and EEG recording systems. For approximately three-fourths of the sample, stimuli were presented using in-house software, while EEG data were collected using Grass Neurodata 12 systems, with a passband of 0.1to30Hz(half amplitude;6dBper octave rolloff), and digitized at 256 Hz with 12 bits resolution. Each trial consisted of 2 s, with a 500-ms prestimulus baseline. The intertrial interval varied between 1 and 2 s. Data were recorded from four electrodes (Fz, Pz, P3, and P4) with a linked-ear reference. Eyeblinks and other eye movements were recorded by means of a pair of electrodes in a transverse arrangement, with one superior to the eye and one over the outer canthus. For the remaining subjects, who were in the Enrichment Sample (ES; Keyes et al., 2009), data were collected continuously from 61 scalp electrodes using a Biosemi ActiveTwo system, a sampling rate of 1024 Hz, and 24 bits resolution. ActiveTwo amplifiers are DC-coupled. To avoid aliasing, all signals were filtered by means of a 5th-order Bessel sinc filter with a cutoff frequency of 205 Hz (3 dB attenuation), imposed in the ActiveView software. Four additional monopolar electrodes were placed superior to each eye and over each outer can-thus. A script written in E-Prime software version 1.2 (Psychology Software Tools, Pittsburgh, PA) mimicked the original Pascal program as closely as possible.
2.2.2. Data processing
Data were processed in Matlab (The Mathworks, Natick, MA). After data from the Biosemi system were transformed to match the format of the Grass system data (Malone et al., 2014b), the same pipeline was used for all data, based on functions in the Psychophysiology Toolbox for Matlab (http://sourceforge.net/projects/psychophys/) and custom scripts. Observations made when the data were collected guided us in identifying potentially problematic data. Trials containing artifacts such as excessively large voltage deflections and sudden transients were marked for exclusion. Eye blinks and other movements were corrected by means of the regression method of Gratton et al. (1983).
2.2.3. Time-frequency transform and dependent measures
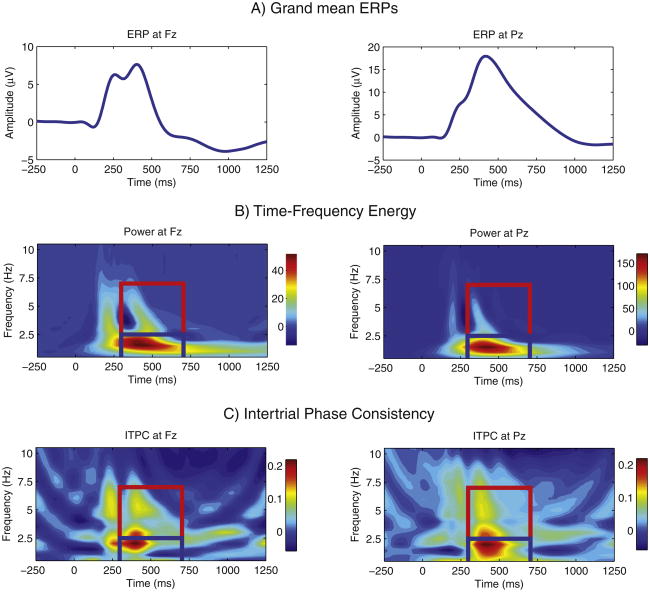
Time-frequency transforms of all target trials of valid, artifact-corrected EEG were obtained by means of the reduced interference distribution (RID; Williams, 2001). The RID is a member of Cohen's class of bilinear time-frequency transforms (TFDs) (Cohen, 1989), and it has several advantages over wavelet methods: it preserves energy while satisfying the time and frequency marginals, meaning that it faithfully reproduces the signal energy at each time-frequency bin, and its resolution is uniform in time and frequency (Bernat et al., 2005; Weis et al.,2010; Williams and Jeong, 1989). RIDs can be represented in a form that is equivalent to multiplying a signal's autocorrelation function, the inverse Fourier transform of the signal's power spectrum, by a kernel, which acts as a filter, and transforming the resulting “ambiguity function” back into the time-frequency domain. Using the RID with a binomial kernel, as in previous work (Williams, 2001), we derived the (squared) magnitude of the signal at each time-frequency bin for all artifact-free target trials, separately for Fz and Pz. To reduce computational load, each time series was resampled at 64 Hz, resulting in a time resolution (sampling interval) of 15.625 ms and a frequency resolution of 0.5 Hz. These values were then averaged over trials to produce estimates of total energy (power) to capture all activity whether stimulus-locked or not (cf. Jones et al., 2006a). This is similar to the procedure used in previous genome-wide analyses (Kang et al., 2012; Zlojutro et al., 2011), although using a different transform to represent time-frequency energy (RID versus S-transform). Power values were corrected for baseline variation by removing the mean of the 200 ms preceding stimulus onset separately for each frequency bin. The mean energy distribution for each electrode is plotted in Fig. 1B.
Fig. 1.
The grand mean ERP and mean time-frequency measures, by electrode. The grand mean ERP responses to target stimuli are plotted in panel a. The two plot abcissas are separately scaled. The mean time-frequency representation of the data for each electrode is plotted in panel b. This was derived by averaging the RID time-frequency transform of all artifact-free EEG waveforms for target trials. The heat maps for these two plots are also separately scaled. Panel c provides a plot of the mean intertrial phase coherence (ITPC), derived for the same set of EEG waveforms that went into panel b. Because we subtracted the frequency-specific mean ITPC in a 200-ms prestimulus baseline period from the vector of ITPC values in that frequency, small negative values are possible, where poststimulus ITPC was slightly less than prestimulus ITPC was at the same discrete frequency. For the figures in both panels, the theta and delta time-frequency regions of interest are outlined (in red and blue, respectively).
Although it faithfully represents time- and frequency-varying energy, the RID is a real-valued distribution, which makes it uninformative about signal phase, because phase is derived from the imaginary part of a complex-valued representation of a signal. We therefore used a parallel procedure to extract inter-trial phase coherence (ITPC) from the same data by means of the RID-Rihaczek (Aviyente et al., 2011), a recently developed application of the RID to the Rihaczek distribution, also a member of Cohen's class but one that yields a complex energy density function. Its time-frequency resolution is determined by the rate of change of instantaneous frequency, providing precise localization in time-frequency space of phase modulation (Aviyente et al., 2011). We used the RID-Rihaczek on all valid and artifact-free target trials to produce the time-varying complex spectrum representing each trial, similar to the time-varying energy spectrum produced by the RID (Tallon-Baudry et al., 1996). For each electrode, the intertrial variability of the RID-Rihaczek phase estimate for each discrete time, t, and frequency, ω, was estimated as , where j represents trial, N is the total number of target trials, i is the imaginary unit , eiϕ is the complex representation of the phase angle Φ relative to the average of all phase estimates, and the vertical bars indicate the length of the vector contained within them (Burwell et al., 2014). Phase angles are normalized to unit length; as a result, ITPC is independent of magnitude. This is analogous to the approach of Tallon-Baudry et al. (1996). ITPC ranges from 0, indicating that phase are randomly distributed across trials, to 1, indicating identical phase estimates across trials. ITPC estimates were baseline-corrected by subtracting the average ITPC for the 200 ms before stimulus onset from each post-stimulus time-frequency bin, separately for each frequency bin, as for TF power (Aviyente et al., 2011; Burwell et al., 2014; Cavanagh et al., 2010; Roach and Mathalon, 2008). The resulting values were averaged over all bins in each TFROI to produce estimates of the magnitude of theta and delta stimulus locking.
We derived average energy and ITPC in the recorded signals for two TFROIs. Both spanned 300 to 700 ms (the P3 time window). One comprised 2.5 to 7 Hz and the other, 0.5 to 2.5 Hz. Choice of ROIs was informed by visual inspection of the average ERP (Fig. 1A) and the average TF energy in our sample (Fig. 1B) as well as the roadmap provided by the two previous GWASs. Although appropriately tuned to the TF characteristics of our data, these ROIs thus conform closely to those used previously and are arguably most likely to aid in our goal of identifying genetic variants related to P3 amplitude. In particular, our first TFROI is virtually identical to the theta ROI used in Kang et al., which successfully identified genetic variants related to theta power: 3 to 7 Hz in the same time window. (The GWAS of Zlojutro et al. defined theta as spanning 4 to 7 Hz but did not produce genome-wide significant hits.). Owing to this similarity in TFROIs, and to avoid using more precise but potentially awkward and lengthy descriptors, we refer to our first ROI as theta, despite the fact that its lower frequency boundary is somewhat lower than typical, and the second as delta, despite the fact that it represents only the lower frequencies in the traditional delta band. We hope that what we gain in economy of description by using these terms more than offsets any potential confusion or misunderstanding.
Because theta is most prominent at frontal scalp locations and delta most prominent over parietal regions, we examined theta energy and ITPC at Fz and delta energy and ITPC at Pz. The mean ITPC for each electrode is displayed in Fig. 1C.
2.3. Molecular genetic data and genotyping
The pipeline for extracting and processing DNA and quality control procedures for the molecular-genetic data are described extensively elsewhere (Iacono et al., 2014a; Miller et al., 2012). Quality control filters were carefully applied to samples and markers on the Illumina array to eliminate problematic instances of either. Samples were dropped for having too many uncalled markers (at least 5000), low scores on a metric produced by the array manufacturer, extreme heterozygosity, and sample mix-ups or failure to confirm known genetic relationships with other subjects. Markers were eliminated if identified as problematic by Illumina; if the call rate was <99%; if mismatches occurred in duplicate samples; if the minor allele frequency (MAF) was <1%; if showing a significant deviation from Hardy-Weinberg genotype frequencies or Mendelian inconsistencies across families; if a significant association with participant gender or processing batch was obtained; and if there were excessive heterozygous calls for markers on the X chromosome in males or in mitochondrial DNA in the sample. PCA was subsequently conducted specifically on the genotypes of all Caucasian subjects in order to determine the major dimensions of genetic variation, and the first 10 components were used subsequently to statistically control any confounding due to residual variation in allele frequencies (Price et al., 2006).
SNPs were imputed from the 529,827 markers on the Illumina array that remained after these quality control filters had been applied using minimac (Howie et al., 2012). Genotypes were first phased in Beagle (Browning and Browning, 2009), which uses known familial structure to improve phasing accuracy, and then imputed using 1000 Genomes reference haplotypes (1000 Genomes Project Consortium, 2012). A total of 9,331,500 SNPs were imputed. We used only those SNPs with a MAF of at least 0.01 (which eliminated 1,131,558 markers) and that had been imputed accurately, with an imputation r2 of at least 0.90 (which eliminated an additional 2,164,929 markers), leaving us with a final set of 6,035,013 markers. Although we restricted our primary analyses to SNPs with imputation r2 of 0.90 or greater, we used all SNPs in our analysis of candidate SNPs drawn from the relevant literature. Whereas association analysis with measured SNPs typically uses a count of the number of minor alleles, ranging from 0 to 2,imputation instead produces an allele dosage for each variant. Each genotype (AA, Aa, and aa, represented as 0, 1, and 2, respectively) is weighted by the posterior probability of that genotype as estimated by the imputation algorithm and the three weighted allele counts summed.
2.4. Statistical analyses
Measures of delta-Pz and theta-Fz energy and ITPC were adjusted for covariates likely to affect them: gender, age cohort, chronological age, and recording system (Biosemi or Grass). We also adjusted all outcome measures for the 10 PCs from Eigenstrat reflecting the major dimensions of genetic variation in the sample, as is customary in molecular-genetic research (Price et al., 2006). The specific method of adjusting for these covariates depended on the analysis, as described below: they were included as covariates of no interest in GWAS analyses and analyses of all SNPs in aggregate, whereas the phenotypes were adjusted via linear regression prior to biometric analysis.
2.4.1. Biometric heritability
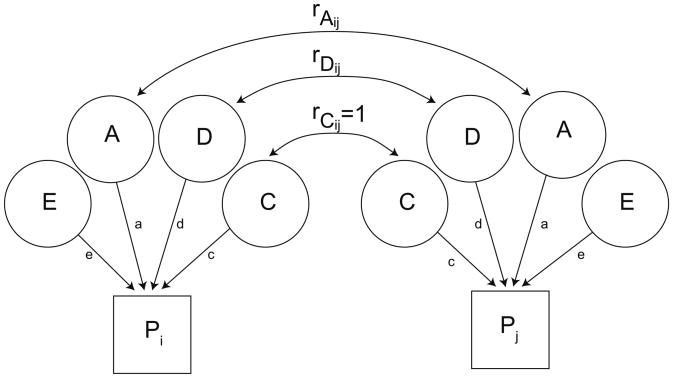
We derived estimates of the amount of heritable variance in outcome measures using standard biometric approaches to twin-family data (Neale et al., 2003) implemented in the OpenMx package for R (Boker et al., 2011). We fit models to data from only the twins as well as from the entire four-member family. Biometric models represent the (covariate-adjusted) time-frequency measures as due to (“caused by”) latent variables representing additive genetic influence (A), dominance genetic influence (D), reflecting nonadditive effects at a single allele, common, or shared, environment (C), and unique, or unshared, environment (E) (see Fig. 2). The logic of biometric model-fitting relies on the known correlations between family members with respect to the latent variables to derive the expected covariance matrix, which is compared to the observed covariance matrix. MZ twins are genetically identical, and the genetic correlation between them is therefore 1, whether for additive or dominance influences. DZ twins share half their segregating genes, on average, while parent-offspring pairs share half their genes by descent; the additive genetic correlation in these pairs is 0.5. The DZ twin correlation for dominance effects, which are based on inheritance from both parents, is 0.52, or 0.25. Parents are unrelated genetically. All family members by definition share the common environment, whereas E reflects environmental factors that are unique to each individual; it does not contribute to within-family correlations. Our models did not allow for assortative mating, the tendency for people with similar characteristics tend to marry (“like marries like”). We also did not allow for “vertical” environmentally-mediated influences between parents and twins.
Fig. 2.
A schematic of the biometric latent variable model. Two family members (individuals i and j) are included to illustrate the model. Their phenotypes are represented by the boxes labeled Pi and Pj. Each phenotype is due to the influence of the four latent variables (ADCE). The variance of each family member's phenotype and the covariance between each pair of family members are determined by the path estimates from each latent variable to the phenotype (a, d, c, and e) and the covariance between family members. The variances of each latent variable are fixed at 1. Their covariances are a function of the family relationships. The genetic covariance between pairs of individuals is depicted as rAij and rDij for additive and dominance variance, respectively. It equals 1 for MZ twins for both types of inheritance, whereas the covariance due to A is ½ for DZ twins and parent-offspring pairs. The covariance due to D is ¼ for DZ twins. C is assumed the same for all pairs and is equal to 1. E is by definition unique to each individual (uncorrelated for pairs of family members).
Fitting models to four-member families allows us to estimate the variance of all four latent variables, and we report the results of such ADCE models in addition to ACE models. Parent-offspring correlations are often smaller in magnitude than DZ twin or sibling correlations. Although this can be consistent with dominance effects, which are shared by siblings but not parents and their offspring, it can also reflect gene-environment interactions, which cause different genetic effects to be expressed during different developmental periods or in different cohorts (Eaves et al., 1978), or simply reduced environmental sharing between parents and offspring. Twin data are unconfounded in this way and may produce a truer estimate of D effects. For twin data, we fit both ACE and ADE models and report the results of each. Only when the estimate of D is statistically significant do we consider it potentially meaningful.
Although systematic variation in mean levels of the time-frequency measures was accommodated through adjusting them for relevant co-variates, we accommodated gender and, especially, age-cohort differences in phenotypic variances by means of scalar moderators of the model path coefficients (a, d, c, and e in Fig. 2). Although estimates of variance moderation were not significant in all analyses, this parameter was retained in all models for the sake of uniformity in model-fitting approach and ease of presentation.
We extended our biometric model to the bivariate case to estimate the magnitude of additive genetic variance shared by each phenotype and P3 amplitude. In addition, we estimated the genetic variance shared by the two energy measures, the two ITPC measures, and by energy and ITPC within a frequency band. These analyses provide an indication of how much genetic overlap likely exists between pairs of time-frequency phenotypes as well as between each such phenotype and P3 amplitude.
2.4.2. Genome-wide complex trait analysis: SNP heritability
Genome-wide complex trait analysis (GCTA) (Yang et al., 2011), also known as genomic restricted maximum likelihood (GREML), assesses the additive effect of all SNPs in aggregate. SNPs are characterized by high levels of linkage disequilibrium, or LD, due to the fact that certain combinations of SNPs are more likely to be inherited together than others. LD creates a correlation among SNPs, which aids in imputing markers. However, it also inflates GCTA estimates (Speed et al., 2012), and the behavior of GCTA with imputed markers is not yet well understood. We therefore opted to use only the genotyped SNPs in our GCTA analyses, which also facilitates comparisons with our previous results for P3 amplitude. GCTA treats each SNP as a random effect in a linear mixed model (LMM). Fixed effects consisted of the covariates described above. A matrix of genotypes comprises the matrix of random effects in LMMs, which can be parameterized as a matrix of pairwise genetic relationships among all participants (the genetic relatedness matrix, or GRM). Restricted maximum likelihood is used to estimate the random effect variance, which is the total variance in the phenotype accounted for by all SNPs. (Strictly speaking, it is the phenotypic variance accounted for by variants on the genotyping array or in LD with them.) GCTA thus estimates the degree to which phenotypic similarity is due to genotypic similarity, whereas biometric models estimate genetic influence on the basis of phenotypic covariances.
In samples made up of related individuals, estimates of the additive genetic variance in a phenotype are driven by the phenotypic correlations among individuals, which are therefore biased because they can be inflated by common environmental effects or nonadditive genetic effects. We used two approaches to address this. First, GCTA filters the sample using a user-chosen threshold of relatedness to produce a sub-sample of individuals who are only very distantly related genetically in such a way as to maximize sample size. We used a threshold of relatedness of 0.05 in order to be consistent with the second approach, described next.
The goal of GCTA is to determine whether genetic similarity among individuals unrelated by pedigree is associated with phenotypic similarity, and filtering a family-based sample such as ours produces a sample of genetically largely unrelated individuals. However, this results in imprecise estimates (large SEs). We therefore also adopted an alternate method that has been recently proposed for estimating SNP heritability in samples of related individuals (Zaitlen et al., 2013), based on the fact that heritability can be estimated from identity-by-descent (IBD) co-variance matrices. The IBD covariance matrix in turn can be approximated from the identity-by-state (IBS) matrix representing related individuals. Following Zaitlen et al., we used a relatedness threshold of 0.05 to identify related individuals, setting to 0 off-diagonal elements of the GRM for distantly related subjects (those below this threshold, the same subjects used to derive our first estimate of SNP heritability). Using GCTA with the full GRM and the IBS-based approximation to the IBD matrix simultaneously yields estimates of two variance components. One is an estimate of SNP heritability similar to the estimate derived from unrelated individuals, whereas the other reflects the portion of heritability captured by phenotypic relationships not due to genotyped SNPs. Zaitlen and colleagues labeled this “ unexplianed heritability” analogous to the notion of “missing heritability” (Maher, 2008). Total narrow-sense heritability is the sum of these two variance components. Because this method uses the whole sample while taking into account both closely related and unrelated, or distantly related, pairs, it produces more precise estimates than those based on a subsample of unrelated pairs. The estimate of narrow-sense heritability it produces can be compared to corresponding estimates from our biometric models.
GCTA can be extended to bivariate analyses, which produce estimates of the genetic correlation between phenotypes and its standard error, an indication of the degree to which the same SNPs contribute to two different phenotypes. We conducted bivariate analyses for the same pairs of variables as in our biometric analyses. As in the univariate case, whereas shared genetic influence between phenotypes is estimated in biometric models based on phenotypic covariances (correlations), in bivariate GCTA analyses it is estimated from measured SNPs. In order to obtain estimates paralleling our univariate SNP heritability estimates, we adopted a genetic relatedness threshold of 0.05 for these analyses as well.
Finally, GCTA has recently been extended to estimate dominance genetic influences from measured SNPs (Zhu et al., 2015).As with additive genetic influence, dominance is estimated from genetic similarity among unrelated individuals. We used the same threshold of 0.05 for this purpose as all other GCTA analyses.
2.4.3. Genome-wide association study (GWAS)
Our genome-wide analysis of individual SNPs consisted of regression analyses of the effects on our time-frequency measures of each of SNP. The nested structure of our sample, which induces a correlation among family members, is problematic for standard linear regression analyses. To account for the lack of independence in family data, we used Rapid Feasible Generalized Least Squares (RFGLS; Li et al., 2011). RFGLS is a computationally efficient form of generalized least squares (GLS), which can be appropriate when residuals are correlated (or heteroskedastic). The unknown residual covariance structure within higher-order units is estimated based on observed variance-covariance matrix. In the present case, data were clustered in families comprising one to four members, with three family types: MZ and DZ families and step-parents. RFGLS estimates the residual covariance matrix separately for each type.
Rather than estimating the residual covariance structure (conditional on model covariates and a given SNP) for each SNP, which can result in several million different regression analyses, RFGLS estimates the residual covariance matrix once, conditional only on model covariates, based on the assumption that SNP effects on the residual covariances will be negligible. This produces significant savings in computational time and minimal bias or loss of power (Li et al., 2011). Constraints are imposed on elements of the residual covariance matrix in order to reduce the number of parameters to be estimated, thereby avoiding problems with algorithm convergence. The mother-offspring and father-offspring correlations are constrained equal, as are variances for the two members of a twin pair. In all, four correlations (MZ or DZ twin pair, mother-offspring, father-offspring, mother-father) and four variances (twin, mother, father, step-parent) were estimated for each time-frequency measure. The “working correlation matrix” of within-family correlations produced by RFGLS is reported in Table 2 for each phenotype. The independent variable in each analysis consisted of the allele dosage, and an additive model of association tested for each SNP association (with 1 df). We adopted the customary significance threshold of 5 × 10−8.
Table 3a.
Biometric heritability estimated from family data.
| Measure | Model | A | D | C | E | |
|---|---|---|---|---|---|---|
| Energy | Theta-Fz | ACE | 0.593 (0.540–0.641) | – | 0.000 (0.000–0.017) | 0.407 (0.359–0.460) |
| ADCE | 0.376 (0.239–0.474) | 0.268 (0.165–0.378) | 0.000 (0.000–0.057) | 0.355 (0.314–0.403) | ||
| Delta-Pz | ACE | 0.564 (0.516–0.607) | – | 0.000 (0.000–0.017) | 0.436 (0.393–0.484) | |
| ADCE | 0.352 (0.158–0.439) | 0.262 (0.170–0.385) | 0.000 (0.000–0.092) | 0.386 (0.347–0.429) | ||
| ITPC | Theta-Fz | ACE | 0.329 (0.265–0.392) | – | 0.000 (0.000–0.020) | 0.671 (0.608–0.735) |
| ADCE | 0.139 (0.000–0.238) | 0.283 (0.169–0.417) | 0.000 (0.000–0.000) | 0.578 (0.516–0.644) | ||
| Delta-Pz | ACE | 0.374 (0.318–0.429) | – | 0.000 (0.000–0.015) | 0.626 (0.571–0.682) | |
| ADCE | 0.140 (0.000–0.231) | 0.334 (0.231–0.461) | 0.000 (0.000–0.000) | 0.527 (0.476–0.582) |
In addition to a genome-wide scan aimed at discovering significant SNPs, we examined two subsets of SNPs. The first consisted of 183 SNPs that have been reported to be associated with P3 amplitude or theta or delta energy (see Malone et al., 2014b). Fewer tests are involved in analyzing these SNPs than in a genome-wide analysis, and significance was therefore assessed against a Bonferroni criterion of 2.73 × 10−4 (0.05/183). The second subset was derived from a recent report of a multi-stage case-control GWAS of schizophrenia, which identified 128 genome-wide significant associations from 108 independent loci (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Because schizophrenics characteristically show P3 amplitude reductions (Ford, 1999) and reduced low-frequency ERP activity (Ethridge et al., 2015), we examined associations between these SNPs and our phenotypes, both individually and together, in the form of a genetic risk score.
2.4.4. Versatile gene-based association study (VEGAS)
We tested associations between individual genes, rather than SNPs, and each time-frequency measure using VEGAS (Liu et al., 2010), which aggregates the results for all SNPs in a gene into a single score. This approach can be particularly powerful when several SNPs located in a gene are causally related to the phenotype, in which case the p-value associated with any one of them may not be small enough to be statistically distinguishable from noise. VEGAS assigns SNPs to a gene by reference to the UCSC Genome Browser assembly, including all SNPs within 50 kb of the 3′ and 5′ untranslated region (UTR) of a given gene in order to capture regulatory SNPs and SNPs in LD with those in the gene itself. Individual p-values for each SNP are converted into chi-squared statistics with 1 df and summed. Because the p-values were produced by RFGLS, they accurately reflect the nested structure of our data. VEGAS thus easily accommodates the clustered nature of our sample. LD causes SNPs and their p-values to be correlated, which requires determining the null distribution of the gene score in the presence of LD. VEGAS uses Monte Carlo methods and the LD structure of a reference sample from the International HapMap Project (International HapMap Consortium, 2005). We selected the CEPH sample of Utah residents of European ancestry in HapMap (CEU) for this purpose.
We used VEGAS to conduct gene-based tests of association in a manner parallel to our analyses of individual SNPs. We tested the association between each of approximately 17,600 autosomal genes and our time-frequency measures in a genome-wide scan comparable to our GWAS of SNPs. A threshold of p < 2.84 × 10−6 was used for determining statistical significance, which corrects for the number of different genes. In addition, we evaluated a subset of 19 candidate genes that have been reported to be associated with theta or delta energy or P3 amplitude, using a significance threshold of 2.78 × 10−3. This list included MYEF2, which we found was associated with P3 amplitude in a genome-wide analysis (Malone et al., 2014b).
3. Results
3.1. Descriptive statistics
Fig. 1 illustrates sample averages for the grand mean ERPs, time-frequency energy, and ITPC. Amplitude of the grand mean was less overall at Fz than at Pz, and plots of the ERPs and of TF energy are separately scaled for the two electrodes in order to reveal detail in each. Whereas the Pz ERP is dominated by the P3 and slow activity, a P2-N2-P3 sequence is clear in the Fz grand mean. These well-resolved peaks suggest theta activity, which, although small in magnitude, is captured in the plot of TF energy in Fig. 1B. The plots of ITPC in panel C indicate two regions of increased theta phase consistency that correspond to the positive deflections in the ERP (P2 and P3), especially at Fz, although ITPC is strongest in the delta TFROI as well as in the low end of the theta range, both corresponding well in time to the peak of the P3. Table 1 presents descriptive statistics. Energy and ITPC values were greater for delta-Pz than theta-Fz. Energy and ITPC values were greater for female adolescents than males in all cases. Energy values were also greater for females than males among the parents, whereas the reverse was true for ITPC. These differences justify including these gender and age cohort as co-variates in all subsequent analyses.
Table 1.
Descriptive statistics for the sample.
| Measure | N | Female twins | Male twins | Mothers | Fathers | Step-parents | |
|---|---|---|---|---|---|---|---|
| Energy | Theta Fz | 3429 | 4.01 (0.46) | 3.76 (0.46) | 3.38 (0.53) | 3.24 (0.49) | 3.24 (0.44) |
| Delta Pz | 4150 | 5.14 (0.50) | 5.00 (0.51) | 4.51 (0.59) | 4.24 (0.55) | 4.32 (0.51) | |
| ITPC | Theta Fz | 3448 | 0.054 (0.044) | 0.045 (0.039) | 0.052 (0.048) | 0.060 (0.052) | 0.066 (0.052) |
| Delta Pz | 4178 | 0.180 (0.093) | 0.172 (0.092) | 0.149 (0.102) | 0.166 (0.106) | 0.180 (0.107) |
Note: Entries in the table are means, with SDs in parentheses. Energy measures (squared magnitude, or μV2) are log-transformed. ITPC values range from 0 (random phase distribution) to 1 (complete phase locking). The vast majority of step-parents were fathers.
Table 2 presents correlations for the different pairs of family members: MZ twins, DZ twins, mothers and their adolescent children, and fathers and their adolescent children. These were derived from RFGLS and represent the “working correlation matrix” used to accommodate within-family dependency in our data. The pattern of within-family correlations is informative about the likelihood and relative magnitude of genetic and environmental influences. MZ twin correlations were substantial, especially for the two energy measures. They were also at least twice the magnitude of the DZ twin correlations, indicating that genetic influences are clearly important. DZ twin and parent-offspring correlations were generally equivalent in magnitude, although there were exceptions. Overall, these correlations suggest that additive genetic effects, and maybe dominance effects as well, are important. There is little evidence for shared environmental influences.
Table 2.
Within-family correlations.
| Measure | MZ twins | DZ twins | Mother-offspring | Father-offspring | Mother-father | |
|---|---|---|---|---|---|---|
| Energy | Theta Fz | 0.646 | 0.173 | 0.167 | 0.204 | −0.030 |
| Delta Pz | 0.611 | 0.339 | 0.176 | 0.158 | −0.028 | |
| ITPC | Theta Fz | 0.409 | 0.156 | 0.073 | 0.046 | −0.065 |
| Delta Pz | 0.465 | 0.199 | 0.119 | 0.038 | −0.033 |
Note: All correlations were produced by RFGLS, after adjusting measures for effects of age, gender, age cohort, recording system and the 10 Eigenstrat PCs.
3.2. Biometric heritability
Parameter estimates from fitting biometric models appear in Tables 3a and 3b. The first are based on the whole four-member family, whereas the second are based only on MZ and DZ twin pairs. Results are consistent in indicating the importance of genetic factors, more so for energy measures than ITPC measures. The ADCE model fits the family data better than the ACE model in every case, although, as noted earlier (Section 2.4.1), twin models are arguably more useful for assessing the significance of D. For three of the four twin models, D could be constrained equal to 0 without significant loss of fit; chi-squared statistics with 1 df were all <1. The sole exception was for theta-Fz energy, Δχ2 = 8.5 with 1 df, p < 0.002, indicating that D effects might be important for this phenotype. Estimates of A from ACE models may provide an upper bound on the likely degree of additive genetic influence, whereas estimates of A from ADE or ADCE models may provide a lower bound.
Table 3b.
Biometric heritability estimated from twin data
| Measure | Model | A | D | C | E | |
|---|---|---|---|---|---|---|
| Energy | Theta-Fz | ACE | 0.633 (0.563–0.677) | – | 0.000 (0.000–0.056) | 0.367 (0.323–0.416) |
| ADE | 0.046 (0.000–0.453) | 0.599 (0.187–0.685) | – | 0.356 (0.315–0.403) | ||
| Delta-Pz | ACE | 0.516 (0.335–0.647) | – | 0.093 (0.000–0.262) | 0.390 (0.350–0.435) | |
| ADE | 0.613 (0.373–0.652) | 0.000 (0.000–0.241) | – | 0.387 (0.348–0.430) | ||
| ITPC | Theta-Fz | ACE | 0.400 (0.227–0.462) | – | 0.000 (0.000–0.151) | 0.600 (0.538–0.665) |
| ADE | 0.232 (0.157–0.457) | 0.174 (0.000–0.463) | – | 0.593 (0.531–0.660) | ||
| Delta-Pz | ACE | 0.460 (0.315–0.511) | – | 0.000 (0.000–0.128) | 0.540 (0.489–0.595) | |
| ADE | 0.309 (0.000–0.507) | 0.156 (0.000–0.502) | – | 0.535 (0.483–0.591) |
Note: Proportions of the variance in each phenotype due to additive genetic influence (A), dominance influence (D), common or shared environment (C), and unshared or unique environment (E). Ninety-five percent confidence intervals are in parentheses. Estimates from family data use four-member families, whereas estimates from twin data are based only on the twins. Results of family models are for ACE and ADCE. Twin data can only be used to estimate ADE or ACE models; C and D cannot be estimated simultaneously in the same model.
3.3. SNP heritability
Results from GCTA are presented in Table 4. They parallel the biometric heritability results in that estimates of additive variance (SNP heritability) were greater for the two energy measures than for the ITPC measures. The SNP heritability estimate for delta-Pz was significant; the likelihood-ratio test statistic was 3.27 with 1 df, p < 0.035. (The test statistic distribution is treated as a mixture of chi-square distributions with 0 and 1 df and p = 0.50, and the test is one sided.) The test statistic for theta-Fz was 2.07, p < 0.075. None of the estimates of dominance effects was significant, with the largest point estimate being 0.086 (SE = 0.239).
Table 4.
GCTA SNP heritability results.
| Unrelated | Whole sample | ||||
|---|---|---|---|---|---|
|
|
|||||
| Domain | Measure | h2SNP | h2SNP | h2unexplained | h2total |
| Energy | Theta-Fz | 0.304 (0.213) | 0.273 (0.129) | 0.350 (0.130) | 0.623 |
| Delta-Pz | 0.417 (0.178)* | 0.413 (0.109) | 0.174 (0.110) | 0.587 | |
| ITPC | Theta-Fz | 0.176 (0.204) | 0.102 (0.121) | 0.242 (0.125) | 0.344 |
| Delta-Pz | 0.264 (0.175) | 0.180 (0.102) | 0.205 (0.105) | 0.385 | |
h2SNP is the heritability due to additive influences estimated from genotyped SNPs. “Unrelated” refers to a subsample based on filtering the genetic relatedness matrix (GRM) with a threshold of 0.05 to select unrelated pairs of subjects. Ns are 1678 for theta measures and 2040 for delta measures. h2SNP in the whole sample is based on the method of Zaitlen et al. (2013) to estimate the SNP heritability in samples of related individuals. h2unexplained reflects the portion of heritability captured by phenotypic relationships that is not due to genotyped SNPs, analogous to ‘missing heritability.’ The sum of the two (h2total) provides an estimate of narrow-sense heritability.
p < 0.05 by likelihood-ratio test.
GCTA results estimated separately for different allele frequency categories appear in Table 5. The energy measures tend to be accounted for primarily by relatively more common SNPs, particular those with MAFs that are somewhat intermediate (neither low nor high). Several of the estimates for delta-Pz energy were significant by LRT, with values of the test statistic ranging from 2.83 to 5.80 (on 1 df), all p-values <0.05. Estimates for theta-Fz were not, which was in part likely due to the larger SEs accompanying them, but followed the same general pattern as estimates for delta-Pz. Results for the two ITPC measures are less consistent and it is difficult to discern any pattern, particularly in light of the large SEs, although SNPs with MAFs between 0.30 and 0.40 were significantly associated with delta-Pz ITPC, Δχ2 = 3.14 on 1 df, p = 0.038. Owing to the correlation between markers induced by LD, these estimates are not independent; they cannot be summed to obtain an overall SNP heritability estimate.
Table 5.
SNP heritability by minor allele frequency bins.
| Domain | Measure | 0.01 to 0.10 (90,956) | 0.10 to 0.20 (122,662) | 0.20 to 0.30 (104,898) | 0.30 to 0.40 (96,430) | 0.40 to 0.50 (92,592) |
|---|---|---|---|---|---|---|
| Energy | Theta-Fz | 0.000 (0.158) | 0.207 (0.156) | 0.167 (0.142) | 0.145 (0.135) | 0.144 (0.124) |
| Delta-Pz | 0.051 (0.124) | 0.185 (0.130) | 0.287 (0.117)a | 0.209 (0.113)a | 0.192 (0.103)a | |
| ITPC | Theta-Fz | 0.220 (0.152) | 0.081 (0.154) | 0.090 (0.147) | 0.076 (0.130) | 0.000 (0.119) |
| Delta-Pz | 0.151 (0.126) | 0.099 (0.127) | 0.089 (0.114) | 0.209 (0.112)a | 0.000 (0.099) |
Note: Estimates are for the proportion of variance accounted for by SNPs with MAFs in the specified range (with SEs in parentheses). A threshold of 0.05 was used to select unrelated individuals. Numbers in parentheses below the limits for each bin are the numbers of SNPs in the bin.
Significant by one-tailed likelihood-ratio test.
3.4. Bivariate biometric and SNP heritability
The biometric model used to estimate phenotypic (univariate) heritability is readily extended to accommodate two phenotypes, which permits estimating the magnitude of shared genetic variance (covariance) between two phenotypes. Shared genetic variance, typically expressed as a genetic correlation, indicates the degree to which genetic influences are common to the two phenotypes. We examined the genetic correlation between each time-frequency measure and P3 amplitude as well as between pairs of time-frequency measures. The model also permits estimating the bivariate phenotypic correlation between these pairs of measures. Like the genetic correlation, this is a model-implied correlation based on data from our four-member families, which makes the two correlations directly comparable. Point estimates for these and 95% confidence intervals around them are presented in the columns of Table 6 labeled “Biometric Phenotypic r” and “Biometric Genetic r.”
Table 6.
Bivariate biometric and GCTA analyses.
| Measure 1 | Measure 2 | Biometric | GCTA Genetic r (SE) | |
|---|---|---|---|---|
|
| ||||
| Phenotypic r (95% CI) | Genetic r (95% CI) | |||
| Delta-Pz energy | P3 | 0.799 (0.787–811) | 0.864 (0.835–0.890) | 0.962 (0.122)a |
| Delta-Pz ITPC | P3 | 0.428 (0.400–0.455) | 0.593 (0.522–0.661) | 1.000 (0.362)a |
| Theta-Fz energy | P3 | 0.303 (0.268–0.337) | 0.414 (0.339–0.487) | 0.490 (0.377) |
| Theta-Fz ITPC | P3 | 0.165 (0.129–0.200) | 0.190 (0.081–0.298) | 0.285 (0.617) |
| Delta-Pz energy | Theta-Fz energy | 0.413 (0.381–0.444) | 0.469 (0.396–0.538) | 0.852 (0.274)a |
| Delta-Pz energy | Delta-Pz ITPC | 0.348 (0.319–0.377) | 0.432 (0.346–0.515) | 0.289 (0.340) |
| Theta-Fz energy | Theta-Fz ITPC | 0.201 (0.165–0.235) | 0.172 (0.059–0.282) | 1.000 (0.917) |
| Delta-Pz ITPC | Theta-Fz ITPC | 0.241 (0.207–0.274) | 0.412 (0.280–0.539) | 1.000 (0.812) |
“Phenotypic r” is the correlation based on the model-expected family covariance matrix. “Biometric Genetic r” is the correlation for the latent variable A representing additive genetic influences, based on the same model as the phenotypic correlation. The GCTA “Genetic r” is the correlation between each pair of traits based on the SNPs on the Illumina array. A threshold of 0.05 was used on the GRM to select unrelated individuals.
Significant by likelihood ratio test.
We used GCTA to conduct parallel bivariate analyses, which also produces estimates of the genetic correlation between pairs of phenotypes. However, such estimates are based on measured SNPs rather than inferred from phenotypic covariances among family members. They can therefore provide a more direct estimate of pleiotropy – the degree to which individual genetic variants affect two traits. To maintain parallelism with our univariate GCTA analyses, we used a relatedness threshold of 0.05 to produce subsamples of unrelated individuals.
For biometric and GCTA estimates, correlations between each time-frequency measure and P3 amplitude appear in the top of the table, while correlations between pairs of time-frequency measures appear in the bottom (with the exception of cross-frequency, cross-domain correlations, which were not estimated). Genetic correlations, whether derived from biometric model or GCTA, were larger for the association between delta-Pz measures (energy and ITPC) and P3 amplitude, and these were all significant; confidence intervals around the biometric genetic correlation estimates did not include 0 and likelihood-ratio tests of the GCTA genetic correlations were significant (χ2 = 4.71 and 2.91 with 1 df, p-values <0.015 and 0.044 for delta-Pz energy and delta-Pz ITPC, respectively, by GCTA's one-tailed test). Theta-Fz energy was modestly associated with P3 amplitude as well as delta-Pz energy, the latter correlation also being significant for both biometric and GCTA estimates (χ2 = 5.88, p < 0.008 for the GCTA correlation). Biometric genetic correlations indicated that energy and ITPC were more strongly related for delta-Pz than theta-Fz; the two sets of confidence intervals did not overlap. A similar comparison for genetic correlations based on measured SNPs cannot be made, because correlations involving theta-Fz ITPC could not be estimated with any precision; all SEs were quite large. The point estimates therefore cannot be considered trustworthy and the large SEs make a comparison between biometric and GCTA genetic correlations meaningless.
3.5. SNP effects: Genome-wide analysis
GWAS results are presented in two forms for each measure: a Q-Q plot and a Manhattan plot. The former (Figs. 3-6) plots the observed distribution of p-values against the expected distribution under the null hypothesis, although a — log10 transformation is used in order to emphasize the most significant results (those with the smallest p-values). Because the vast majority of SNPs are expected not to have an effect on a given phenotype, the points produced should adhere closely to a line at a 45-degree angle, indicating that observed and expected values agree closely. Only small p-values, which might reflect true effects, are expected to deviate from this line. Excessive deviation from the line can indicate that the test results are biased, due to population stratification or other factors. The overall degree to which the observed points conform to expected values is quantified by genomic control (GC) statistics, which are printed in the upper left corner of each Q-Q plot. Both mean and median GC values were close to 1, where 1 indicates perfect agreement, indicating that there was no meaningful inflation (or deflation) in these data. For all measures but delta-Pz ITPC, the points corresponding to the strongest associations deviate from the expected line, although most do not exceed the significance threshold of 5 × 10−8 (discussed in the next paragraph), suggesting that there may be meaningful associations that are not quite strong enough to be statistically significant.
Fig. 3.
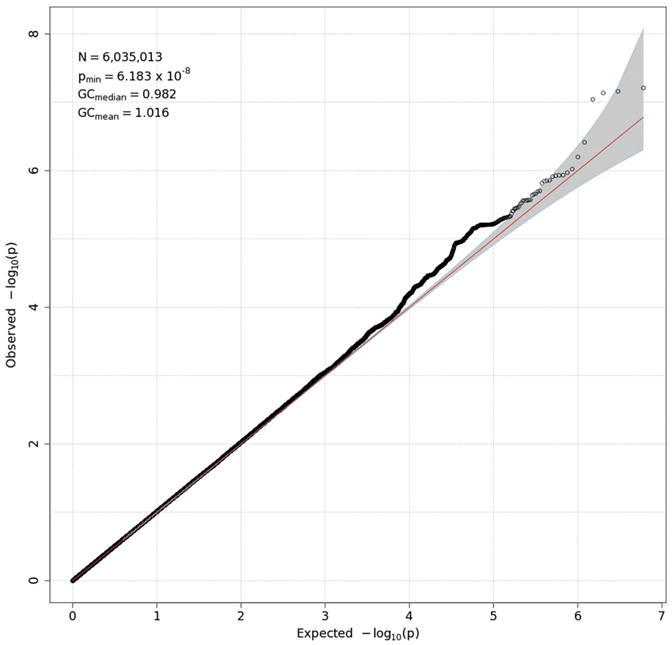
Q-Q plot for theta-Fz energy. Observed p-values are plotted against expected p-values under the null distribution. The vast majority of associations are expected to be nonsignificant, and their distribution therefore should conform closely to the null distribution, indicated by the red 45° line. A significant deviation from this line can indicate inflated power resulting from residual population stratification in allele frequencies. None is evident here. A value of 1 for genomic control (GC) statistics (mean and median), which are provided in the upper left corner of the plot, indicates complete absence of inflation. N in the upper-left box is the number of SNPs in the analysis, while pmin is the smallest p-value across all SNPs. To emphasize the smallest p-values, which may reflect true associations, they are typically represented as −log10(p-value). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
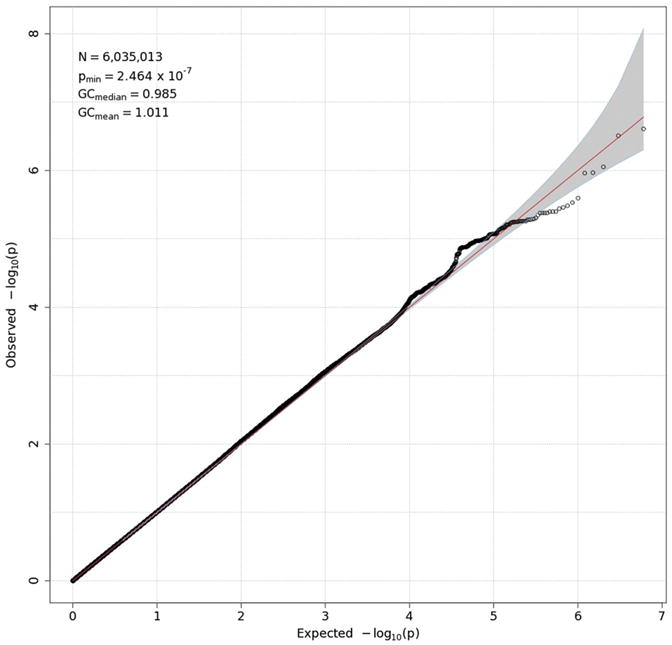
Q-Q plot for delta-Pz ITPC. See the caption for Fig. 3 for details.
Manhattan plots (Figs. 7–10) also provide — log10(p-values) for each SNP, but SNPs are ordered by their position in the genome (and thus by chromosome), rather than by p-value, which permits one to determine the location of any significant or interesting associations. A horizontal line indicates the threshold for genome-wide significance, which is 7.30 on the — log10 scale (p-value of 5 × 10−8). A second horizontal line is drawn at 5, which corresponds to a p-value of 10−5 and which is sometimes used to indicate suggestive significance. For theta-Fz energy (Fig. 7), four SNPs on chromosome 13 have p-values <10−7 and lie near the genome-wide significant threshold. p-Values this small often subsequently replicate, leading some to suggest that this an appropriate significance threshold (Panagiotou et al., 2012). These four SNPs thus might reflect a true association. However, they are located in an intergenic region. The Manhattan plot for delta-Pz energy (Fig. 8) indicates several regions with small p-values, especially on chromosomes 2, 3, 16 and 18, but none were significant. The Manhattan plot for delta-Pz ITPC (Fig. 10) is unremarkable.
Fig. 7.
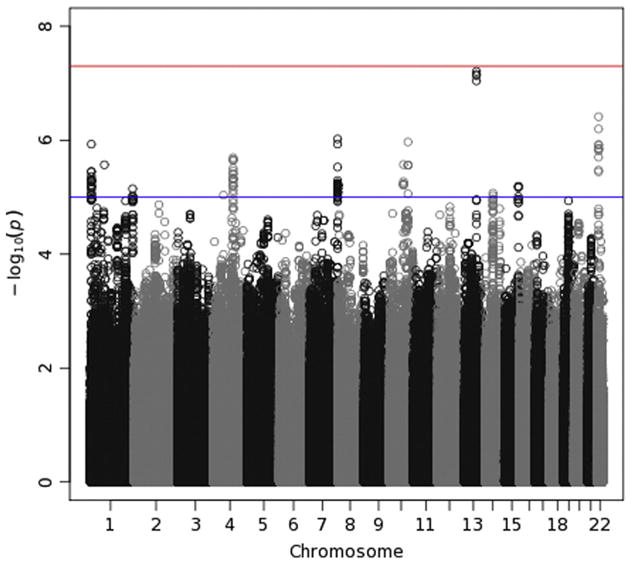
Manhattan plot for theta-Fz energy. This plots −log10(p-values) against their location, grouped by chromosome, to permit one to locate regions of significant and subthreshold associations in the genome. Vertical streams of points indicate a region harboring a likely association; LD among nearby SNPs creates a neighborhood of large −log10(p-values). A horizontal red line at 7.3 reflects the genome-wide significance level (5 × 10−8). A second, blue line is drawn at 5 (p-value of 10−5), which is sometimes used to indicate suggestive significance. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 10.
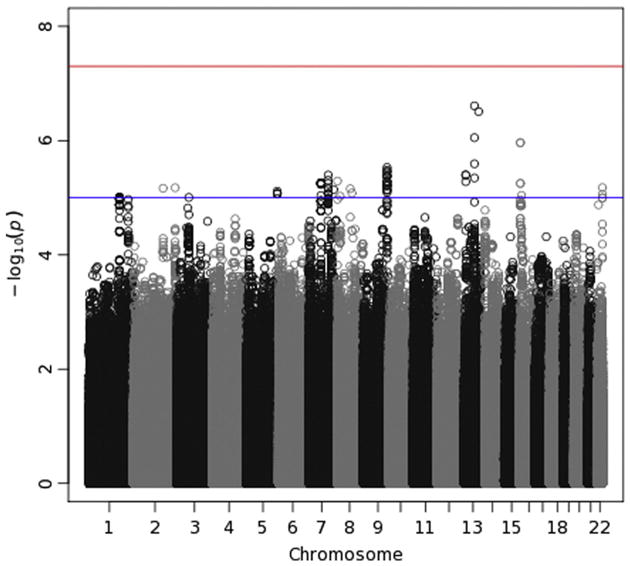
Manhattan plot for delta-Pz ITPC. See the caption for Fig. 7 for details.
Fig. 8.
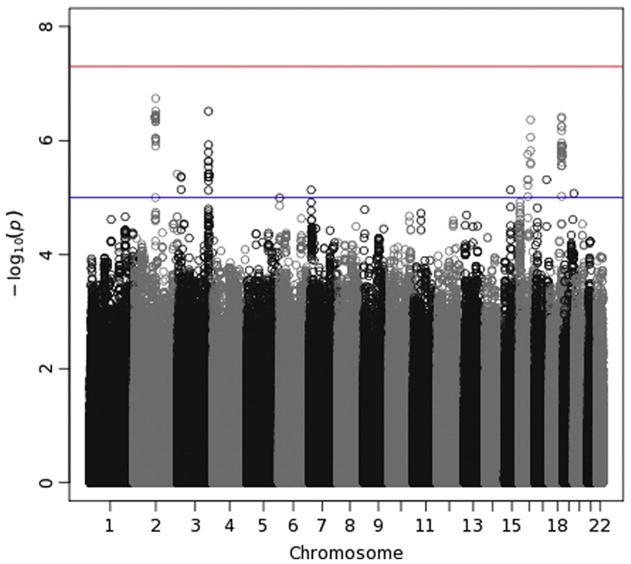
Manhattan plot for delta-Pz energy. See the caption for Fig. 7 for details.
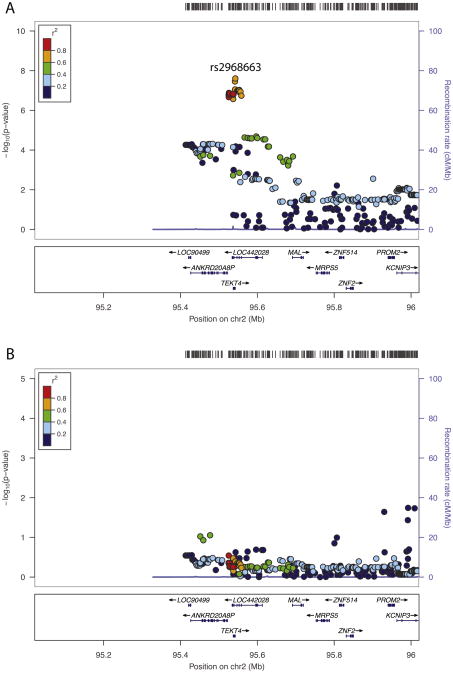
On the other hand, the Manhattan plot for theta-Fz ITPC (Fig. 9) clearly shows an area of chromosome 2 with elevated — log-transformed p-values. Two exceed the genome-wide significance threshold and 25 others in the same region have p-values < 10−6. Given that neighboring SNPs tend to be correlated, one would expect this type of cluster of small p-values if there is a clear association with the phenotype. Fig. 11 was produced by the locuszoom software tool (http://csg.sph.umich.edu/locuszoom/) to “zoom in” on this region, providing greater detail than is permitted by the Manhattan plot. In the top panel (labeled A), the — log-transformed p-values for all SNPs within 500 kb of rs2968663, the SNP producing the smallest p-value, are plotted. Points are color-coded by the average LD between the SNP represented by that point and the others in the plot. Most of the SNPs associated with the smallest p-values are characterized by high average LD, although not all are. Fig. 11 also displays known genes in the region by their location, and the most significant SNPs, those with — log10(p-values) greater than approximately 6, are contained in or flank two genes in particular: ANKRD20A8P and TEKT4. LOC442028, which also contains some of these SNPs, is a pseudogene – a nonfunctional gene that is likely defunct. A smaller region of chromosome 7 produced SNPs with p-values that were almost significant, with one approximately 10−7. Thus, these SNPs may truly be associated with theta ITPC. They are located within 15 to 30 kb of forkhead box P2 (FOXP2), which codes for a transcription factor expressed in brain and required for the proper development of speech and language regions during embryogenesis. Mutations involving different SNPs in FOXP2 are associated with schizophrenia (Sanjuán et al., 2006) and autism (Li et al., 2005). However, its relevance to theta-Fz ITPC is questionable, and these SNPs are nowhere near CHRM2 or GRM8, genes on chromosome 7 that have previously been reported to be associated with theta energy (Chen et al., 2009; Jones et al., 2006a).
Fig. 9.
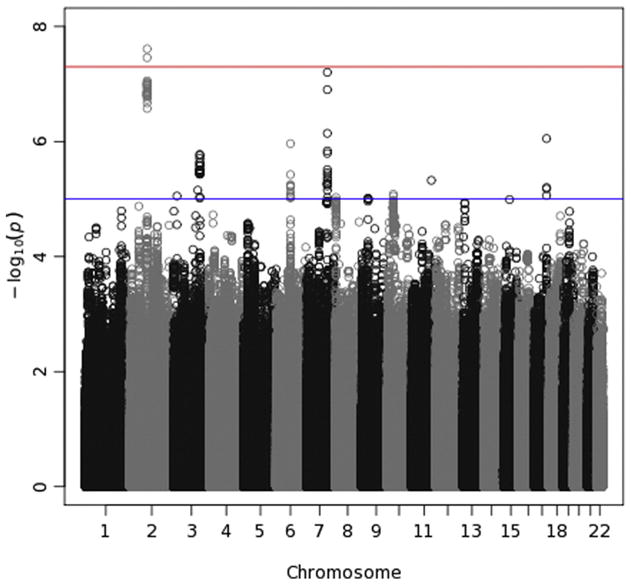
Manhattan plot for theta-Fz ITPC. See the caption for Fig. 7 for details.
Fig. 11.
Detailed view of the region of chromosome 3 containing significant associations with theta-Fz ITPC. Minus log-transformed significance levels for this region are plotted using the locuszoom software tool to providea detailed viewof this region. A rug plot at the top indicates the distribution of SNP locations. The color of each point in the plot indicates the average LD r2 of that SNP with other SNPs in the region. Genes in the region are displayed at the bottom of the plot. Panel A plots association results for the genome-wide scan. The SNP with the smallest p-value (rs2968663) is labeled. Panel B plots the results of a subsequent analysis that included each subject's value for this SNP as an additional covariate to determine whether there was more than one signal in this region, which would be indicated if strong associations remained after controlling for rs2968663.
To determine whether the signal on chromosome 2 is due to more than one source, we repeated our analysis using the dosage for rs2968663 as an additional covariate. Results are presented in the bottom panel of Fig. 11 (labeled B). All associations are clearly nonsignificant, suggesting that the SNPs with small p-values in the top panel (large — log10[p-values]) do not represent an association with theta-Fz ITPC that is independent of the association with rs2968663. There thus appears to be only one signal related to theta-Fz ITPC from this region of chromosome 2.
3.6. SNP effects: Candidate SNPs
We examined a set of 183 candidate SNPs reported to be associated with P3-related time-frequency measures or P3 amplitude (the same candidate SNPs assessed in Malone et al., 2014b). Table 7 provides information about the 15 associations (for 13 different SNPs) that met a nominal significance level of p < 0.05. The majority is in GRM8 and several others are in CRHR1. None of the SNPs exceeded a significance threshold of 2.7 × 10−4, necessary to correct for all 183 tests (per time-frequency phenotype), which itself ignores the fact that four phenotypes were tested against these 183 SNPs.
Table 7.
GWAS results for all candidate SNPs producing nominally significant associations (p < 0.05).
| SNP | Chr | Position | Gene | Alleles | MAF | r2 | Beta | SE | t-Statistic | df | p-Value | Measure | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs13247338 | 7 | 126,090,509 | GRM8 | C | T | 0.143 | 0.998 | 0.336 | 0.169 | 1.99 | 3428 | 0.047 | Theta-ITPC |
| rs2299456 | 7 | 126,168,766 | GRM8 | A | G | 0.160 | – | 0.040 | 0.018 | 2.23 | 4131 | 0.026 | Delta |
| rs2402816 | 7 | 126,191,176 | GRM8 | A | G | 0.382 | – | −0.675 | 0.240 | −2.81 | 4158 | 0.005 | Delta-ITPC |
| rs2299459 | 7 | 126,195,368 | GRM8 | A | C | 0.216 | 1.000 | −0.050 | 0.016 | −3.15 | 4131 | 0.002 | Delta |
| rs1158720 | 7 | 126,208,306 | GRM8 | C | T | 0.223 | 0.991 | −0.045 | 0.016 | −2.87 | 4131 | 0.004 | Delta |
| rs7797602 | 7 | 126,287,827 | GRM8 | C | T | 0.220 | 0.997 | −0.049 | 0.016 | −3.10 | 4131 | 0.002 | Delta |
| rs7797602 | 7 | 126,287,827 | GRM8 | C | T | 0.220 | 0.997 | −0.568 | 0.283 | −2.01 | 4158 | 0.045 | Delta-ITPC |
| rs1074728 | 7 | 126,312,844 | GRM8 | G | A | 0.459 | – | −0.026 | 0.013 | −2.01 | 4131 | 0.045 | Delta |
| rs1074728 | 7 | 126,312,844 | GRM8 | G | A | 0.459 | – | −0.628 | 0.232 | −2.71 | 4158 | 0.007 | Delta-ITPC |
| rs4731323 | 7 | 126,324,030 | GRM8 | A | G | 0.220 | – | 0.035 | 0.015 | 2.30 | 4131 | 0.021 | Delta |
| rs1424558 | 7 | 136,531,671 | CHRM2 | C | G | 0.340 | 0.997 | −0.260 | 0.125 | −2.07 | 3428 | 0.038 | Theta-ITPC |
| rs1396862 | 17 | 43,902,997 | CRHR1 | G | A | 0.204 | 0.998 | 0.035 | 0.016 | 2.22 | 3410 | 0.026 | Theta |
| rs878886 | 17 | 43,912,490 | CRHR1 | C | G | 0.204 | 0.998 | 0.035 | 0.016 | 2.22 | 3410 | 0.026 | Theta |
| rs878887 | 17 | 43,912,582 | CRHR1 | C | T | 0.204 | 0.998 | 0.035 | 0.016 | 2.22 | 3410 | 0.026 | Theta |
Note: Chr is the chromosome on which each SNP is located, while Position is its base pair position (GRCh37/hg19). Gene gives the nearest gene, while Alleles give the major and minor alleles, respectively, and MAF the frequency of the minor allele. r2 is the imputation r2, a measure of imputation quality. If it is not provided, then the SNP was called. Beta is the estimated coefficient from RFGLS, with SE its standard error, t-statistic the test statistic. Measure indicates which time-frequency measure was associated with a given SNP in our data. All SNPs are located in genes, except rs1424558, which flanks CHRM2. The Bonferroni-adjusted p-value threshold was 2.73 × 10−4.
A total of 111 out of the 128 markers identified in the recent large study of schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) were available in our imputed data. Twenty-three SNPs were associated with one or more phenotypes at an uncorrected significance level of p < 0.05 (27 associations in all), with delta-Pz energy showing the most such nominal associations (10). However, none of the associations exceeded a per-phenotype Bonferroni-corrected significance threshold of 4.50 × 10−4 (0.05/111).
To construct a genetic risk score, we multiplied the allele dosage for each marker by the (log-odds) regression coefficient from the PGC report, after alleles had been aligned between datasets, and computed the sum of weighted dosages. RFGLS was used to assess the association between this risk score and each time-frequency phenotype, with the same covariate set as in our other analyses (age, age cohort, sex, recording system, and the first 10 Eigenstrat PCs). Parameter estimates were in the expected direction in all four analyses, with a higher risk score associated with reduced energy or ITPC. The strongest effect was for theta-Fz ITPC, t = 1.85, p = 0.064; the zero-order correlation between it and the risk score was − 0.037.
3.7. Gene effects: Genome-wide analysis
In an analysis that paralleled our GWAS, we conducted a genome-wide examination of all autosomal genes, with a significance criterion of 2.8 × 10−6. One association was significant at this level, that between theta-Fz ITPC and TEKT4 on chromosome 2. As Fig. 9 indicates, and as discussed above, TEKT4 (tektin 4) is a small gene located near the SNPs with the smallest p-values; of the 25 SNPs on chromosome 2 with p-values < 10−6, nine are located in TEKT4 or flank it. This association thus likely reflects the same signal as is evident in Fig. 9. TEKT4 is a protein-coding gene but its relevance to theta ITPC is unclear. It appears to be a structural component of microtubules (cilia and flagella), and it has been linked to breast and colorectal cancer.
3.8. Gene effects: Candidate genes
Table 8 presents results from examining associations with 18 genes that might be candidates for association with P3-related time-frequency measures or P3 amplitude itself. Although the set of candidate genes is a subset of all genes examined in the previous genome-wide analysis, we adopted a significance threshold in these analyses that adjusts only for the number of candidate genes, rather than all genes in the genome, resulting in a threshold of 2.8 × 10−3. An association between CRHR1 and theta-Fz energy was observed with a p-value exceeding this threshold (p = 1.47 × 10−3). As indicated previously, several candidate SNPs in CRHR1 were associated with theta-Fz energy at a threshold of p < 0.05. In addition, GRM8 was nominally associated (p < 0.05) with both delta measures (energy and ITPC). Although we previously obtained a genome-wide significant association with P3 amplitude and MYEF2, none of the associations between time-frequency phenotypes and MYEF2 were significant here.
Table 8.
VEGAS gene-based test associations for candidate genes.
| Gene | Chr | N SNPs | Start position | End position | Theta-Fz energy | Theta-Fz ITPC | Delta-Pz energy | Delta-Pz ITPC | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| Test-statistic | p-Value | Test-statistic | p-Value | Test-statistic | p-Value | Test-statistic | p-Value | |||||
| DISC1 | 1 | 543 | 229,829,183 | 230,243,641 | 383.31 | 0.895 | 859.68 | 0.092 | 636.08 | 0.333 | 785.77 | 0.517 |
| ARID5A | 2 | 46 | 96,566,190 | 96,582,098 | 88.07 | 0.147 | 118.18 | 0.036 | 10.09 | 0.980 | 29.57 | 0.605 |
| DRD3 | 3 | 145 | 115,330,246 | 115,380,589 | 228.56 | 0.148 | 212.56 | 0.190 | 244.06 | 0.123 | 126.91 | 0.507 |
| GSK3B | 3 | 214 | 121,028,235 | 121,295,203 | 138.55 | 0.700 | 222.08 | 0.408 | 663.98 | 0.014 | 109.84 | 0.835 |
| MME | 3 | 131 | 156,280,129 | 156,384,212 | 101.98 | 0.650 | 67.87 | 0.910 | 76.65 | 0.884 | 140.82 | 0.378 |
| FABP2 | 4 | 124 | 120,457,852 | 120,462,764 | 40.52 | 0.859 | 100.18 | 0.457 | 48.84 | 0.802 | 9.72 | 1.000 |
| CNR1 | 6 | 137 | 88,906,303 | 88,911,775 | 99.68 | 0.719 | 71.53 | 0.892 | 84.87 | 0.821 | 156.15 | 0.345 |
| ABCB1 | 7 | 228 | 86,970,883 | 87,180,500 | 444.58 | 0.058 | 137.49 | 0.791 | 229.71 | 0.401 | 105.44 | 0.898 |
| CHRM2 | 7 | 293 | 136,203,938 | 136,352,311 | 97.41 | 0.999 | 186.34 | 0.817 | 252.84 | 0.586 | 485.29 | 0.116 |
| GRM8 | 7 | 1338 | 125,865,887 | 126,679,664 | 1077.12 | 0.702 | 667.54 | 0.962 | 2544.71 | 0.042 | 2927.19 | 0.020 |
| ANXA13 | 8 | 209 | 124,762,214 | 124,818,828 | 228.41 | 0.408 | 214.90 | 0.472 | 278.91 | 0.211 | 328.66 | 0.113 |
| HTR7 | 10 | 233 | 92,490,555 | 92,607,651 | 235.46 | 0.435 | 386.14 | 0.182 | 344.19 | 0.175 | 84.82 | 0.911 |
| BDNF | 11 | 96 | 27,633,017 | 27,699,872 | 193.04 | 0.078 | 115.37 | 0.306 | 33.08 | 0.933 | 52.04 | 0.787 |
| DRD2 | 11 | 202 | 112,785,526 | 112,851,211 | 153.85 | 0.617 | 117.38 | 0.815 | 176.80 | 0.525 | 201.69 | 0.435 |
| DRD4 | 11 | 60 | 627,304 | 630,703 | 105.67 | 0.132 | 45.88 | 0.625 | 29.95 | 0.830 | 68.39 | 0.338 |
| MYEF2 | 15 | 12 | 46,218,920 | 46,257,850 | 4.75 | 0.465 | 27.23 | 0.099 | 7.46 | 0.429 | 12.46 | 0.289 |
| CRHR1 | 17 | 47 | 41,217,448 | 41,268,973 | 272.32 | 0.001 | 46.54 | 0.384 | 11.79 | 0.911 | 53.22 | 0.311 |
| KCNJ6 | 21 | 495 | 37,918,656 | 38,210,566 | 298.26 | 0.940 | 289.99 | 0.939 | 357.49 | 0.819 | 399.33 | 0.709 |
| COMT | 22 | 146 | 18,309,308 | 18,336,530 | 259.82 | 0.078 | 316.40 | 0.032 | 89.53 | 0.812 | 61.00 | 0.954 |
Note: Chr is the chromosome on which each gene is located, NSNPs is the total number of SNPs included by VEGAS within the gene boundary. Its start and end position (using build hg37) are provided as well. The test-statistic for each phenotype is a chi-square statistic. Nominally significant associations (p < 0.05) are italicized. Only one association (bold face and italicized) surpassed the Bonferroni threshold of 2.64 × 10−3: CRHR1 and theta-Fz energy.
4. Discussion
Biometric analyses confirmed the importance of genetic influences, primarily additive influences, for the four P3-related TF measures considered here. GCTA results supported this inference and suggest that these phenotypes are highly polygenic. For both theta-Fz and delta-Pz, TF energy was more heritable than ITPC, both in biometric models and GCTA analyses. Biometric heritability estimates were only slightly lower in magnitude than those reported previously by our group for a TF measure based on the ERP, rather than trial-level data as in the present investigation (Gilmore et al., 2010b). Estimates of h2SNP from GCTA analyses were significant or nearly significant for both energy measures in GCTA analyses with unrelated individuals. Although the method for assessing h2SNP in family data does not produce significance tests of the two variance components estimated, h2SNP estimates were at least twice the SEs, suggesting that a significant proportion of variance is accounted for by all SNPs in aggregate.
SNPs with relatively large MAFs appeared to be most responsible for theta and delta energy, despite the fact that the number of SNPs is relatively uniform across the different MAF bins. Although estimates of h2SNP were significant only for delta-Pz energy, the pattern of estimates was similar for the two phenotypes; SNPs with MAFs between 0.10 and 0.30 yielded the largest h2SNP estimates. SNPs with the smallest MAFs appeared to have a negligible influence on both. It is difficult to discern much of a pattern to the ITPC results. If anything, SNPs with the smallest MAFs seemed to be particularly important, in contrast to the case with energy measures. The pattern of h2SNP estimates for the two energy measures is consistent with stabilizing selection pressures. However, the large SEs around these estimates and the arbitrariness of any approach to creating MAF bins requires more than a grain of salt to accompany any inferences about specific patterns of influence.
Bivariate analyses indicated that both delta-Pz energy and ITPC were highly related genetically to P3 amplitude, although the analogous bivariate GCTA analysis result was not quite significant. Shared SNPs also accounted for a significant amount of the energy in the two bands, but the overlap was not due to SNPs with the largest effects. For instance, the number of SNPs with p-values < 10−5 varied from 77 to 137 for the four phenotypes, but there was no overlap among the four sets. The genetic influence common to these phenotypes is likely due to many SNPs with small effects.
Analyses of candidate SNPs and genes failed to produce compelling results, including analyses of a risk score based on markers robustly associated with schizophrenia. We obtained nominally significant associations between GRM8 and both delta-Pz measures (p < 0.05). However, GRM8 has previously been reported to be associated with theta activity, not delta activity (Chen et al., 2009). The RID used in the present investigation is particularly effective in resolving very low frequency activity, which may account for the fact that we obtained an association between GRM8 and delta activity, whereas Chen and colleagues obtained one for theta activity. However, the two associations we obtained for GRM8 did not meet our threshold for correcting for multiple tests. Only one association – that between theta-Fz energy and CRHR1 – survived such correction. However, the significance of this is uncertain. CRHR1 is located on chromosome 17, and there is no evidence of small p-values on chromosome 17 in previous GWASs of TF activity (Figs.4 and 1 in Zlojutro et al., 2011 and Kang et al., 2012, respectively). Our study may have been the first that was adequately powered to detect this association. CRHR1 was on our list of candidate genes because it was reported to be associated with P3 amplitude (Chen et al., 2010). In our previous GWAS, we obtained a nominally significant association between CRHR1 and a P3 genetic factor score (p ≤ 0.047) but not with measured P3 itself (p ≤ 0.475), with the magnitude of the p-values themselves as well as the discrepancy between them suggesting that there was no true effect (see online supplement at http://onlinelibrary.wiley.com/doi/10.1111/psyp.12345/suppinfo). Thus, it seems quite possible that this association represents a false positive.
4.1. Variation in findings across tasks and samples
The primary goal of the present investigation was to determine whether TF measures might provide insight into molecular genetic influences on P3 amplitude. Our approach was directly informed by two previous GWASs of TF activity elicited by a simple oddball task in the hope that using a similar approach might lead to significant findings. However, we did not obtain significant associations with any SNPs previously found to be associated with TF activity (Kang et al., 2012; Zlojutro et al., 2011). This may reflect differences in tasks and in samples. The rotated heads task used in the present study to elicit ERPs is somewhat more challenging than the simple oddball tasks used in those previous studies, requiring subjects to determine whether the stylized head displayed on screen for a particular trial includes a left or right ear, rather than simply that it has an ear (for example). That we did not obtain findings consistent with those previously reported may indicate that the variants associated with theta-Fz energy in previous studies are specific to the cognitive processes involved in simpler oddball tasks, such as attention and simple target detection. These variants may not be as relevant to the processes involved in identifying the type of stimulus (head with left or right ear), although both tasks presumably tap attentional processes to varying degrees. The TF activity elicited by this task has a clear posterior maximum, reflected in the lesser magnitude of TF power in the signal at Fz relative to the signal at Pz. Most power is in the low frequencies comprising our delta TFROI even at Fz. However, GCTA analyses indicated the presence at a gross level at least of a detectable genetic signal in theta-Fz energy, particularly by the method of Zaitlen and colleagues adapted for family data that improves the precision of SNP heritability estimates (Zaitlen et al., 2013).
Our sample included a large number of adolescents, and the distribution of TF energy may be subtly different between adolescents and adults. However, the ratio of theta energy to total energy in our data was virtually identical for the four age-gender groups, ranging from 0.428 to 0.438 (see Table 1). More important is the fact that the sample for the present investigation was population based whereas previous GWASs used case-control samples, in which cases were drawn from families with a dense history of alcoholism. Enriching one's sample in this way likely increases variation at the lower end of the distributions of psychopathology and associated phenotypes. Yet the present sample, being representative of the broader population of Minnesota, is certainly not lacking for psychopathology (see discussion in Iacono et al., 2014b). Moreover, conditioning subject selection on a dense family history of alcoholism also creates the potential for spurious associations. If a specific genetic marker and a putative endophenotype are both associated with the characteristic on which subject selection is conditioned, such as alcoholism, this can produce an apparent association between the two even in the absence of a causal influence of the marker on the putative endophenotype; in this case the association is simply due to the fact that both are associated with alcoholism. Thus, it may be that the associations between specific genetic markers and TF phenotypes observed in previous GWASs are spurious, which is why we did not obtain similar results despite having a larger sample.
In fact, findings from previous genome-wide studies of TF activity are at variance with one another. The two GWASs failed to corroborate findings from previous studies of TF activity, which consisted of genome-wide linkage followed by association analysis. It is unclear whether any SNPs significantly associated with theta power in follow-up association analyses in these studies were on the genotyping chip used in subsequent GWASs, but there were no SNPs on chromosome 7 in either GWAS with p-values < 10−5 (see Fig. 4 in Zlojutro et al., 2011 and Fig. 1 in Kang et al., 2012). In Supplementary Table S2 associated with their paper, Kang et al. reported that six SNPs in GRM8 and two in CHRM2 were associated with theta energy at a nominal significance level of p < 0.05. However, these are different SNPs than those that were significant in the original reports, and this cannot therefore be strictly considered a replication; it requires more stringent control of Type 1 error than a significance criterion of 0.05. In addition, the significant findings of Kang et al. (2012) were different from the findings of Zlojutro et al. (2011). This may well be due to differences in samples and design. The sample in Kang et al. was primarily Caucasian, whereas a substantial number of African-Americans were included in Zlojutro et al. The latter study used a case-control sample in their first stage and a smaller family sample in the second, whereas Kang et al. used a (larger) family sample, which may in some situations provide greater power for detecting genetic variants (Sham and Purcell, 2014).
On balance, however, the lack of consistency across studies is unsettling, differences in approach and sample notwithstanding. One interpretation is that each finding is particular to a unique combination of sample, task, SNPs examined, and method. A more parsimonious interpretation is that many or all of these findings are false positives. This would be in keeping with the conclusions of a recent paper on genetic research with endophenotypes (Iacono et al., 2016). Based on a comprehensive review of a variety of biological phenotypes, including psychophysiological ones, we argued that such phenotypes, including putative endophenotypes, are themselves complex and highly polygenic. The effect sizes of genetic variants influencing them are typically quite small, accounting for a fraction of a percent of phenotypic variance. This in turn requires samples an order of magnitude or two larger than have been used in endophenotype research, including the sample used here, which itself is larger than the samples used in previous GWASs of TF activity. “Small” samples such as these provide adequate power only for detecting large effects. However, in light of what we have recently learned, true effect sizes of this magnitude are unlikely.
We ourselves failed to corroborate our own prior report of genome wide significance for an association between MYEF2 and P3 amplitude despite using TF components derived from the same data and sample. This included a null finding for delta-Pz energy despite the fact that its association with P3 amplitude, whether its phenotypic correlation (r = 0.80), its biometric genetic correlation (r = 0.86), or its GCTA genetic correlation (r = 0.96), was substantial. None of the TF measure associations with MYEF2 produced a p-value < 00.42. These disappointing findings call into question the likely robustness of theMYEF2–P3 association we previously reported. It is potentially noteworthy that only 12 SNPs were contained within this gene in VEGAS, which would tend to make estimates of statistical association less reliable than for genes defined by more SNPs.
On the other hand, we obtained one potential new discovery: an association between theta-Fz ITPC and SNPs on chromosome 2 in and around TEKT4, although its functional significance is unclear. Four SNPs on chromosome 3 were associated with theta-Fz energy, although with p-values just below the threshold for genome-wide significance. Because many associations with p-values of 10−7 or less replicate in subsequent GWASs (Panagiotou et al., 2012), this may reflect a true finding. Its relevance is uncertain, however, especially because these SNPs are located in an intergenic region of the chromosome, and we are skeptical of the validity of these findings.
5. Conclusions
Our findings assist attempts to understand molecular-genetic influences on P3 amplitude by further confirming that the effects are small and polygenic, and by showing that in this case, having an arguably simpler endophenotype comprising elements of the brain electrical activity underlying P3 amplitude did not make the problem of identifying genetic variants more tractable. Our one discovery was for theta-Fz ITPC. The significance of this association may indicate that investigations of phase resetting of theta activity will be potentially useful for understanding P3 amplitude reduction as an endophenotype, it is striking that theta-Fz ITPC was the most weakly correlated with (parietal) P3 of all time-frequency phenotypes. Despite finding that delta-Pz energy and ITPC share significant additive genetic influence with P3 amplitude, we failed to obtain any significant SNP associations with these two measures.
We obtained one genome-wide significant GWAS signal and one subthreshold signal. In addition, clusters of SNPs with small p-values (large −log10[p-values]) are evident in our Manhattan plots like skyscrapers beginning to rise above a crowded city. These plots resemble the skyline of the borough of Manhattan more than the comparable plot for P3 amplitude does, which suggests the presence of a stronger genetic signal. In addition, GCTA yielded significant results in some instances. Time-frequency measures thus appear to be slightly more sensitive to genetic signal than measured P3. However, the effect sizes were not enough larger to yield many hits. Candidate time-frequency endophenotypes appear to be highly polygenic and genetically complex, and the common SNP influence on these measures also appears to be due to many variants with small effects. We previously failed to identify variants associated with P3 amplitude (Malone et al., 2014b), arguably the most robust endophenotype for clinical phenotypes investigated to date (Miller and Rockstroh, 2013). The present results do little to advance our understanding of the nature of the specific molecular genetic influences on P3 amplitude. Nor do they suggest that endophenotypes will turn out to be useful for gene discovery, despite the hope shared by many that their greater proximity to gene action relative to DSM diagnoses or other complex phenotypes makes them particularly attractive for identifying genetic variants associated with psychopathology. Because endophenotypes are themselves complex, to the extent they tap into biological processes, their utility may best be realized by providing insight into neural mechanisms once disorder relevant genetic variants are identified and found to be associated with the endophenotype (Iacono et al., 2014b; 2016).
Fig. 4.
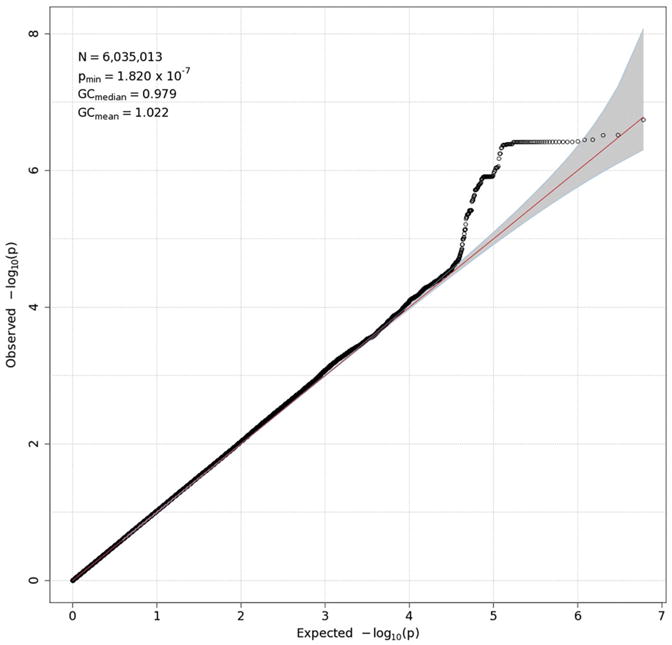
Q-Q plot for delta-Pz energy. See the caption for Fig. 3 for details.
Fig. 5.
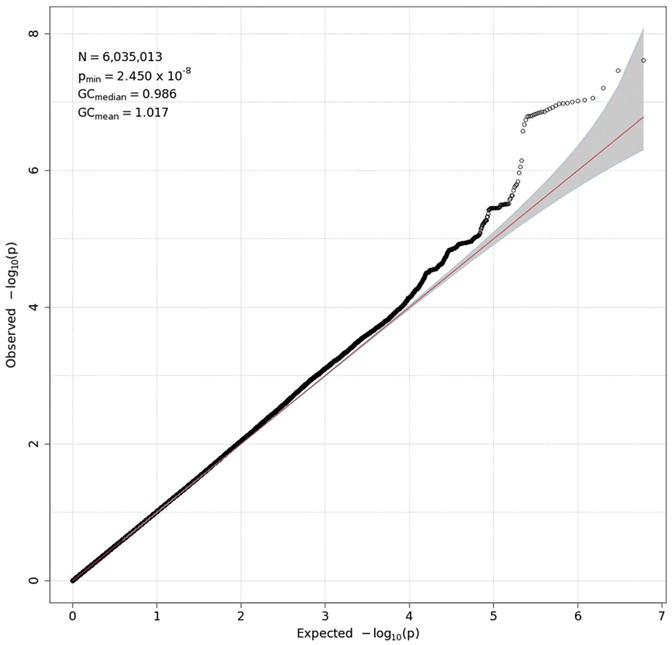
Q-Q plot for theta-Fz ITPC. See the caption for Fig. 3 for details.
Acknowledgments
We are grateful to Micah Hammer and Scott Burwell for their assistance with the EEG data, to Rachel Grazioplene for sharing her work allowing us to compute a risk score for schizophrenia, and to Mike Miller, who is without peer as curator of the molecular-genetic data. The research reported here and preparation of the manuscript were supported by grants R37 DA05147, R01 AA09367, R01 DA13240, R01 DA024417, and R01 DA036216.
References
- Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Endophenotypes as quantitative risk factors for psychiatric disease: rationale and study design. Am J Med Genet. 2001;105:42–44. [PubMed] [Google Scholar]
- Aviyente S, Bernat EM, Evans WS, Sponheim SR. A phase synchrony measure for quantifying dynamic functional integration in the brain. Hum Brain Mapp. 2011;32:80–93. doi: 10.1002/hbm.21000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basar E, Schurmann M, Sakowitz O. The selectively distributed theta system: functions. Int J Psychophysiol. 2001;39:197–212. doi: 10.1016/s0167-8760(00)00141-0. [DOI] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B, Bihari B, Kissin B. Event-related brain potentials in boys at risk for alcoholism. Science. 1984;225:1493–1496. doi: 10.1126/science.6474187. [DOI] [PubMed] [Google Scholar]
- Bernat EM, Williams WJ, Gehring WJ. Decomposing ERP time-frequency energy using PCA. Clin Neurophysiol. 2005;116:1314–1334. doi: 10.1016/j.clinph.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Boker S, Neale M, Maes H, Wilde M, Spiegel M, Brick T, Spies J, Estabrook R, Kenny S, Bates T, Mehta P, Fox J. OpenMx: An open source extended structural equation modeling framework. Psychometrika. 2011;76:306–317. doi: 10.1007/s11336-010-9200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwell SJ, Malone SM, Bernat EM, Iacono WG. Does electroencephalogram phase variability account for reduced P3 brain potential in externalizing disorders? Clin Neurophysiol. 2014;125:2007–2015. doi: 10.1016/j.clinph.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Keller MC. Endophenotypes in the genetic analyses of mental disorders. Annu Rev Clin Psychol. 2006;2:267–290. doi: 10.1146/annurev.clinpsy.2.022305.095232. [DOI] [PubMed] [Google Scholar]
- Cavanagh JF, Frank MJ, Klein TJ, Allen JJ. Frontal theta links prediction errors to behavioral adaptation in reinforcement learning. NeuroImage. 2010;49:3198–3209. doi: 10.1016/j.neuroimage.2009.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC, Tang Y, Rangaswamy M, Wang JC, Almasy L, Foroud T, Edenberg HJ, Hesselbrock V, Nurnberger J, Jr, Kuperman S, O'Connor SJ, Schuckit MA, Bauer LO, Tischfield J, Rice JP, Bierut L, Goate A, Porjesz B. Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:359–368. doi: 10.1002/ajmg.b.30818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC, Manz N, Tang Y, Rangaswamy M, Almasy L, Kuperman S, Nurnberger J, O'Connor SJ, Edenberg HJ, Schuckit MA. Single-nucleotide polymorphisms in corticotropin releasing hormone receptor 1 gene (CRHR1) are associated with quantitative trait of event-related potential and alcohol cependence. Alcohol Clin Exp Res. 2010;34:988–996. doi: 10.1111/j.1530-0277.2010.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L. Time-frequency distributions-a review. Proc IEEE. 1989;77:941–981. [Google Scholar]
- de Geus EJ. From genotype to EEG endophenotype: a route for post-genomic understanding of complex psychiatric disease? Genome Med. 2010;2:1–4. doi: 10.1186/gm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves LJ, Last KA, Young PA, Martin NG. Model-fitting approaches to the analysis of human behaviour. Heredity (Edinb) 1978;41:249–320. doi: 10.1038/hdy.1978.101. [DOI] [PubMed] [Google Scholar]
- Ethridge LE, Hamm JP, Pearlson GD, Tamminga CA, Sweeney JA, Keshavan MS, Clementz BA. Event-related potential and time-frequency endophenotypes for schizophrenia and psychotic bipolar disorder. Biol Psychiatry. 2015;77:127–136. doi: 10.1016/j.biopsych.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euser AS, Arends LR, Evans BE, Greaves-Lord K, Huizink AC, Franken IH. The P300 event-related brain potential as a neurobiological endophenotype for substance use disorders: a meta-analytic investigation. Neurosci Biobehav Rev. 2012;36:572–603. doi: 10.1016/j.neubiorev.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Fell J, Dietl T, Grunwald T, Kurthen M, Klaver P, Trautner P, Schaller C, Elger CE, Fernandez G. Neural bases of cognitive ERPs: more than phase reset. J Cogn Neurosci. 2004;16:1595–1604. doi: 10.1162/0898929042568514. [DOI] [PubMed] [Google Scholar]
- Ford JM. Schizophrenia: the broken P300 and beyond. Psychophysiology. 1999;36:667–682. [PubMed] [Google Scholar]
- Ford JM. Decomposing 300 to identify its genetic basis. Psychophysiology. 2014 doi: 10.1111/psyp.12353. [DOI] [PubMed] [Google Scholar]
- Frodl-Bauch T, Bottlender R, Hegerl U. Neurochemical substrates and neuroanatomical generators of the event-related P300. Neuropsychobiology. 1999;40:86–94. doi: 10.1159/000026603. [DOI] [PubMed] [Google Scholar]
- Gilmore CS, Malone SM, Bernat EM, Iacono WG. Relationship between the P3 event-related potential, its associated time-frequency components, and externalizing psychopathology. Psychophysiology. 2010a;47:123–132. doi: 10.1111/j.1469-8986.2009.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore CS, Malone SM, Iacono WG. Brain electrophysiological endophenotypes for externalizing psychopathology: a multivariate approach. Behav Genet. 2010b;40:186–200. doi: 10.1007/s10519-010-9343-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Gratton G, Coles MG, Donchin E. A new method for off-line removal of ocular artifact. Electroencephalogr Clin Neurophysiol. 1983;55:468–484. doi: 10.1016/0013-4694(83)90135-9. [DOI] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG. Genome-wide scans of genetic variants for psychophysiological endophenotypes: introduction to this special issue of Psychophysiology. Psychophysiology. 2014;51:1201–1202. doi: 10.1111/psyp.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM. Developmental endophenotypes: indexing genetic risk for substance abuse with the P300 brain event-related potential. Child Dev Perspect. 2011;5:239–247. doi: 10.1111/j.1750-8606.2011.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Carlson SR, Taylor J, Elkins IJ, McGue M. Behavioral disinhibition and the development of substance use disorders: findings from the Minnesota Twin Family Study. Dev Psychopathol. 1999;11:869–900. doi: 10.1017/s0954579499002369. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, McGue M. Substance use disorders, externalizing psychopathology, and P300 event-related potential amplitude. Int J Psychophysiol. 2003;48:147–178. doi: 10.1016/s0167-8760(03)00052-7. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, Vaidyanathan U, Vrieze SI. Genome-wide scans of genetic variants for psychophysiological endophenotypes: a methodological overview. Psychophysiology. 2014a;51:1207–1224. doi: 10.1111/psyp.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Vaidyanathan U, Vrieze SI, Malone SM. Knowns and unknowns for psychophysiological endophenotypes: integration and response to commentaries. Psychophysiology. 2014b;51:1339–1347. doi: 10.1111/psyp.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, Vrieze SI. Endophenotype best practices. Int J Psychophysiol. 2016 doi: 10.1016/j.ijpsycho.2016.07.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Porjesz B, Almasy L, Bierut L, Goate A, Wang JC, Dick DM, Hinrichs A, Kwon J, Rice JP, Rohrbaugh J, Stock H, Wu W, Bauer LO, Chorlian DB, Crowe RR, Edenberg HJ, Foroud T, Hesselbrock V, Kuperman S, Nurnberger J, Jr, O'Connor SJ, Schuckit MA, Stimus AT, Tischfield JA, Reich T, Begleiter H. Linkage and linkage disequilibrium of evoked EEG oscillations with CHRM2 receptor gene polymorphisms: implications for human brain dynamics and cognition. Int J Psychophysiol. 2004;53:75–90. doi: 10.1016/j.ijpsycho.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Jones KA, Porjesz B, Almasy L, Bierut L, Dick D, Goate A, Hinrichs A, Rice JP, Wang JC, Bauer LO, Crowe R, Foroud T, Hesselbrock V, Kuperman S, Nurnberger J, Jr, O'Connor SJ, Rohrbaugh J, Schuckit MA, Tischfield J, Edenberg HJ, Begleiter H. A cholinergic receptor gene (CHRM2) affects event-related oscillations. Behav Genet. 2006a;36:627–639. doi: 10.1007/s10519-006-9075-6. [DOI] [PubMed] [Google Scholar]
- Jones KA, Porjesz B, Chorlian D, Rangaswamy M, Kamarajan C, Padmanabhapillai A, Stimus A, Begleiter H. S-transform time-frequency analysis of P300 reveals deficits in individuals diagnosed with alcoholism. Clin Neurophysiol. 2006b;117:2128–2143. doi: 10.1016/j.clinph.2006.02.028. [DOI] [PubMed] [Google Scholar]
- Kamarajan C, Porjesz B, Jones KA, Choi K, Chorlian DB, Padmanabhapillai A, Rangaswamy M, Stimus AT, Begleiter H. The role of brain oscillations as functional correlates of cognitive systems: a study of frontal inhibitory control in alcoholism. Int J Psychophysiol. 2004;51:155–180. doi: 10.1016/j.ijpsycho.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamarajan C, Porjesz B, Jones K, Chorlian D, Padmanabhapillai A, Rangaswamy M, Stimus A, Begleiter H. Event-related oscillations in offspring of alcoholics: neurocognitive disinhibition as a risk for alcoholism. Biol Psychiatry. 2006;59:625–634. doi: 10.1016/j.biopsych.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Rangaswamy M, Manz N, Wang JC, Wetherill L, Hinrichs T, Almasy L, Brooks A, Chorlian DB, Dick D. Family-based genome-wide association study of frontal theta oscillations identifies potassium channel gene KCNJ6. Genes Brain Behav. 2012;11:712–719. doi: 10.1111/j.1601-183X.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas S, Erzengin OU, Basar E. The genesis of human event-related responses explained through the theory of oscillatory neural assemblies. Neurosci Lett. 2000;285:45–48. doi: 10.1016/s0304-3940(00)01022-3. [DOI] [PubMed] [Google Scholar]
- Kendler K, Neale M. Endophenotype: a conceptual analysis. Mol Psychiatry. 2010;15:789–797. doi: 10.1038/mp.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenemans JL, Kähkönen S. How human electrophysiology informs psychopharmacology: from bottom-up driven processing to top-down control. Neuropsychopharmacology. 2010;36:26–51. doi: 10.1038/npp.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes MA, Malone SM, Elkins IJ, Legrand LN, McGue M, Iacono WG. The enrichment study of the Minnesota twin family study: increasing the yield of twin families at high risk for externalizing psychopathology. Twin Res Hum Genet. 2009;12:489–501. doi: 10.1375/twin.12.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimesch W, Sauseng P, Hanslmayr S, Gruber W, Freunberger R. Event-related phase reorganization may explain evoked neural dynamics. Neurosci Biobehav Rev. 2007;31:1003–1016. doi: 10.1016/j.neubiorev.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Li H, Yamagata T, Mori M, Momoi MY. Absence of causative mutations and presence of autism-related allele in FOXP2 in Japanese autistic patients. Brain and Development. 2005;27:207–210. doi: 10.1016/j.braindev.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Li X, Basu S, Miller MB, Iacono WG, McGue M. A rapid generalized least squares model for a genome-wide quantitative trait association analysis in families. Hum Hered. 2011;71:67–82. doi: 10.1159/000324839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Mcrae AF, Nyholt DR, Medland SE, Wray NR, Brown KM. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Makeig S, Onton J. ERP features and EEG dynamics: an ICA perspective. In: Luck S, Kappenman E, editors. Oxford Handbook of Event-Related Potential Components. Oxford University Press; New York: 2009. [Google Scholar]
- Makeig S, Westerfield M, Jung TP, Enghoff S, Townsend J, Courchesne E, Sejnowski TJ. Dynamic brain sources of visual evoked responses. Science. 2002;295:690–694. doi: 10.1126/science.1066168. [DOI] [PubMed] [Google Scholar]
- Malone SM, Burwell SJ, Vaidyanathan U, Miller MB, McGue M, Iacono WG. Heritability and molecular-genetic basis of resting EEG activity: a genome-wide association study. Psychophysiology. 2014a;51:1225–1245. doi: 10.1111/psyp.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone SM, Vaidyanathan U, Basu S, Miller MB, McGue M, Iacono WG. Heritability and molecular-genetic basis of the P3 event-related brain potential: a genome-wide association study. Psychophysiology. 2014b;51:1246–1258. doi: 10.1111/psyp.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Keyes M, Sharma A, Elkins I, Legrand L, Johnson W, Iacono WG. The environments of adopted and non-adopted youth: evidence on range restriction from the Sibling Interaction and Behavior Study (SIBS) Behav Genet. 2007;37:449–462. doi: 10.1007/s10519-007-9142-7. [DOI] [PubMed] [Google Scholar]
- Miller GA, Rockstroh B. Endophenotypes in psychopathology research: where do we stand? Annu Rev Clin Psychol. 2013;9:177–213. doi: 10.1146/annurev-clinpsy-050212-185540. [DOI] [PubMed] [Google Scholar]
- Miller MB, Basu S, Cunningham J, Eskin E, Malone SM, Oetting WS, Schork NJ, Sul JH, Iacono WG, McGue M. The Minnesota Center for Twin and Family Research genome-wide association study. Twin Res Hum Genet. 2012;15:767–774. doi: 10.1017/thg.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min BK, Busch NA, Debener S, Kranczioch C, Hanslmayr S, Engel AK, Herrmann CS. The best of both worlds: phase-reset of human EEG alpha activity and additive power contribute to ERP generation. Int J Psychophysiol. 2007;65:58–68. doi: 10.1016/j.ijpsycho.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Neale MC, Boker SM, Xie G, Maes HH. Mx: Statistical Modeling. sixth. Department of Psychiatry, Virginia Commonwealth University; Richmond, VA: 2003. [Google Scholar]
- Padmanabhapillai A, Porjesz B, Ranganathan M, Jones KA, Chorlian DB, Tang Y, Kamarajan C, Rangaswamy M, Stimus A, Begleiter H. Suppression of early evoked gamma band response in male alcoholics during a visual oddball task. Int J Psychophysiol. 2006a;60:15–26. doi: 10.1016/j.ijpsycho.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Padmanabhapillai A, Tang Y, Ranganathan M, Rangaswamy M, Jones KA, Chorlian DB, Kamarajan C, Stimus A, Kuperman S, Rohrbaugh J, O'Connor SJ, Bauer LO, Schuckit MA, Begleiter H, Porjesz B. Evoked gamma band response in male adolescent subjects at high risk for alcoholism during a visual oddball task. Int J Psychophysiol. 2006b;62:262–271. doi: 10.1016/j.ijpsycho.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Panagiotou OA, Ioannidis JP. Genome-Wide Significance Project. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int J Epidemiol. 2012;41:273–286. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- Porjesz B, Rangaswamy M, Kamarajan C, Jones KA, Padmanabhapillai A, Begleiter H. The utility of neurophysiological markers in the study of alcoholism. Clin Neurophysiol. 2005;116:993–1018. doi: 10.1016/j.clinph.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Rangaswamy M, Jones KA, Porjesz B, Chorlian DB, Padmanabhapillai A, Kamarajan C, Kuperman S, Rohrbaugh J, O'Connor SJ, Bauer LO, Schuckit MA, Begleiter H. Delta and theta oscillations as risk markers in adolescent offspring of alcoholics. Int J Psychophysiol. 2007;63:3–15. doi: 10.1016/j.ijpsycho.2006.10.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach BJ, Mathalon DH. Event-related EEG time-frequency analysis: an overview of measures and an analysis of early gamma band phase locking in schizophrenia. Schizophr Bull. 2008;34:907–926. doi: 10.1093/schbul/sbn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán J, Tolosa A, González JC, Aguilar EJ, Pérez-Tur J, Nájera C, Moltó MD, de Frutos R. Association between FOXP2 polymorphisms and schizophrenia with auditory hallucinations. Psychiatr Genet. 2006;16:67–72. doi: 10.1097/01.ypg.0000185029.35558.bb. [DOI] [PubMed] [Google Scholar]
- Sauseng P, Klimesch W, Gruber WR, Hanslmayr S, Freunberger R, Doppelmayr M. Are event-related potential components generated by phase resetting of brain oscillations? A critical discussion Neuroscience. 2007;146:1435–1444. doi: 10.1016/j.neuroscience.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014n;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Purcell SM. Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet. 2014;15:335–346. doi: 10.1038/nrg3706. [DOI] [PubMed] [Google Scholar]
- Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. Am J Hum Genet. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallon-Baudry C, Bertrand O. Oscillatory gamma activity in humans and its role in object representation. Trends Cogn Sci. 1999;3:151–162. doi: 10.1016/s1364-6613(99)01299-1. [DOI] [PubMed] [Google Scholar]
- Tallon-Baudry C, Bertrand O, Delpuech C, Pernier J. Stimulus specificity of phase-locked and non-phase-locked 40 Hz visual responses in human. J Neurosci. 1996;16:4240–4249. doi: 10.1523/JNEUROSCI.16-13-04240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Isen JD, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of electrodermal activity: a genome-wide association study. Psychophysiology. 2014a;51:1259–1271. doi: 10.1111/psyp.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Donnelly JM, Hammer MA, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of antisaccade eye tracking error rate: a genome-wide association study. Psychophysiology. 2014b;51:1272–1284. doi: 10.1111/psyp.12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of acoustic startle eye blink and affectively modulated startle response: a genome-wide association study. Psychophysiology. 2014c;51:1285–1299. doi: 10.1111/psyp.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Malone SM, Pankratz N, Vaidyanathan U, Miller MB, Kang HM, McGue M, Abecasis G, Iacono WG. Genetic associations of nonsynonymous exonic variants with psychophysiological endophenotypes. Psychophysiology. 2014a;51:1300–1308. doi: 10.1111/psyp.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Malone SM, Vaidyanathan U, Kwong A, Kang HM, Zhan X, Flickinger M, Irons D, Jun G, Locke AE, Pistis G, Porcu E, Levy S, Myers RM, Oetting W, McGue M, Abecasis G, Iacono WG. In search of rare variants: preliminary results from whole genome sequencing of 1,325 individuals with psychophysiological endophenotypes. Psychophysiology. 2014b;51:1309–1320. doi: 10.1111/psyp.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis M, Jannek D, Roemer F, Guenther T, Haardt M, Husar P. Multi-dimensional PARAFAC2 component analysis of multi-channel EEG data including temporal tracking. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:5375–5378. doi: 10.1109/IEMBS.2010.5626484. [DOI] [PubMed] [Google Scholar]
- Williams WJ. Reduced interference time-frequency distributions: scaled decompositions and interpretations. In: Debnath L, editor. Wavelet Transforms and Time-frequency Signal Analysis. Birkhauser; Bostn: 2001. pp. 381–417. [Google Scholar]
- Williams WJ, Jeong J. New time-frequency distributions: theory and applications, circuits and systems, 1989. IEEE International Symposium on IEEE. 1989:1243–1247. [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HH, Malone SM, Burwell SJ, Bernat EM, Iacono WG. Association between P3 event-related potential amplitude and externalizing disorders: a time-domain and time-frequency investigation of 29-year-old adults. Psychophysiology. 2013;50:595–609. doi: 10.1111/psyp.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordanova J, Devrim M, Kolev V, Ademoglu A, Demiralp T. Multiple time-frequency components account for the complex functional reactivity of P300. Neuroreport. 2000;11:1097–1103. doi: 10.1097/00001756-200004070-00038. [DOI] [PubMed] [Google Scholar]
- Zaitlen N, Kraft P, Patterson N, Pasaniuc B, Bhatia G, Pollack S, Price AL. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 2013;9:e1003520. doi: 10.1371/journal.pgen.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Bakshi A, Vinkhuyzen AA, Hemani G, Lee SH, Nolte IM, van Vliet-Ostaptchouk JV, Snieder H, LifeLines Cohort S, Esko T, Milani L, Magi R, Metspalu A, Hill WG, Weir BS, Goddard ME, Visscher PM, Yang J. Dominance genetic variation contributes little to the missing heritability for human complex traits. Am J Hum Genet. 2015;96:377–385. doi: 10.1016/j.ajhg.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, Bierut LJ, Goate A, Hesselbrock V, Kuperman S. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2011;156:44–58. doi: 10.1002/ajmg.b.31136. [DOI] [PMC free article] [PubMed] [Google Scholar]