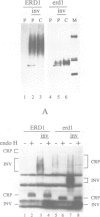
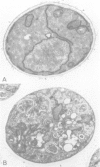
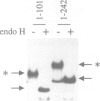
Abstract
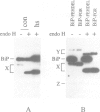
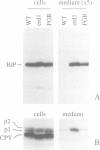
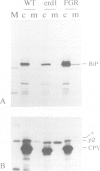
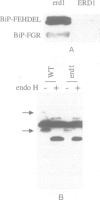
We have previously shown that the C-terminal sequence HDEL acts as a retention signal for luminal endoplasmic reticulum (ER) proteins in Saccharomyces cerevisiae, and that it is possible to isolate mutants that fail to retain an invertase fusion protein bearing this signal. Analysis of many such mutants defines two genes, ERD1 and ERD2. Cells lacking the ERD1 gene secrete the endogenous ER protein, BiP. Under normal growth conditions, the rate of secretion is equivalent to the rate at which wild-type cells secrete a modified form of BiP that lacks the HDEL signal altogether. Thus, erd1 cells show a profound disruption of the retention system. The mutant cells have no gross abnormality of their intracellular membrane system, but show defects in the Golgi-dependent modification of glycoproteins. We suggest that sorting of luminal ER proteins normally occurs in the Golgi, and that the function of ERD1 is required for the correct interaction of an HDEL receptor with its ligands. The sequence of ERD1 predicts a membrane protein with several transmembrane domains, a conclusion supported by analysis of ERD1-SUC2 fusion proteins.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banta L. M., Robinson J. S., Klionsky D. J., Emr S. D. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988 Oct;107(4):1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bole D. G., Hendershot L. M., Kearney J. F. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J Cell Biol. 1986 May;102(5):1558–1566. doi: 10.1083/jcb.102.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A., Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 1989 Sep;8(9):2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal P. K., Ballou C. E. Regulation of the protein glycosylation pathway in yeast: structural control of N-linked oligosaccharide elongation. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8824–8828. doi: 10.1073/pnas.84.24.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings B. A., Zubenko G. S., Hasilik A., Jones E. W. Mutant defective in processing of an enzyme located in the lysosome-like vacuole of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Jan;78(1):435–439. doi: 10.1073/pnas.78.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Johnson L. M., Bankaitis V. A., Emr S. D. Distinct sequence determinants direct intracellular sorting and modification of a yeast vacuolar protease. Cell. 1987 Mar 13;48(5):875–885. doi: 10.1016/0092-8674(87)90084-5. [DOI] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984 Mar;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Banta L. M., Emr S. D. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol Cell Biol. 1988 May;8(5):2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukuruzinska M. A., Bergh M. L., Jackson B. J. Protein glycosylation in yeast. Annu Rev Biochem. 1987;56:915–944. doi: 10.1146/annurev.bi.56.070187.004411. [DOI] [PubMed] [Google Scholar]
- Munro S., Pelham H. R. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987 Mar 13;48(5):899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Munro S., Pelham H. R. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986 Jul 18;46(2):291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- Normington K., Kohno K., Kozutsumi Y., Gething M. J., Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989 Jun 30;57(7):1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980 Aug;21(1):205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. Control of protein exit from the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:1–23. doi: 10.1146/annurev.cb.05.110189.000245. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. Evidence that luminal ER proteins are sorted from secreted proteins in a post-ER compartment. EMBO J. 1988 Apr;7(4):913–918. doi: 10.1002/j.1460-2075.1988.tb02896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Hardwick K. G., Lewis M. J. Sorting of soluble ER proteins in yeast. EMBO J. 1988 Jun;7(6):1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Misra L. M., Vogel J. P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989 Jun 30;57(7):1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Rothman J. H., Howald I., Stevens T. H. Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO J. 1989 Jul;8(7):2057–2065. doi: 10.1002/j.1460-2075.1989.tb03614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J. H., Hunter C. P., Valls L. A., Stevens T. H. Overproduction-induced mislocalization of a yeast vacuolar protein allows isolation of its structural gene. Proc Natl Acad Sci U S A. 1986 May;83(10):3248–3252. doi: 10.1073/pnas.83.10.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph H. K., Antebi A., Fink G. R., Buckley C. M., Dorman T. E., LeVitre J., Davidow L. S., Mao J. I., Moir D. T. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell. 1989 Jul 14;58(1):133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- Sengstag C., Hinnen A. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual aminoacid composition. Nucleic Acids Res. 1987 Jan 12;15(1):233–246. doi: 10.1093/nar/15.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982 Sep;30(2):439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]