Abstract
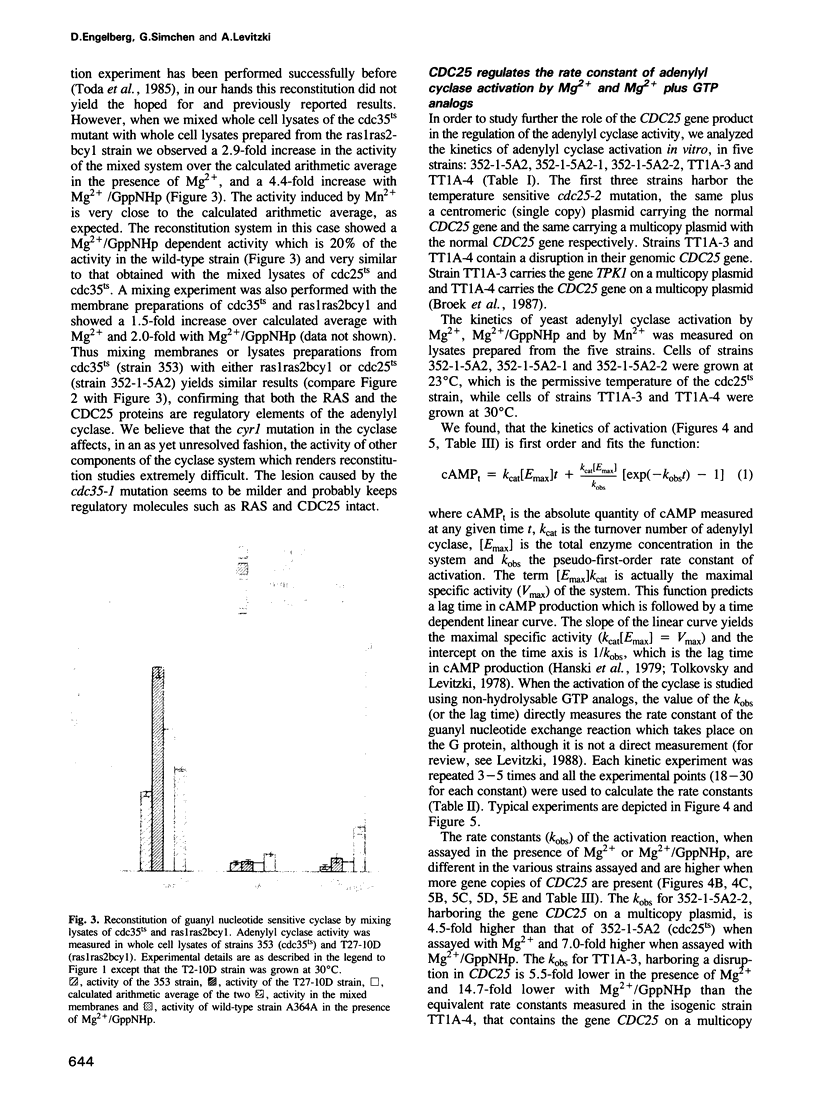
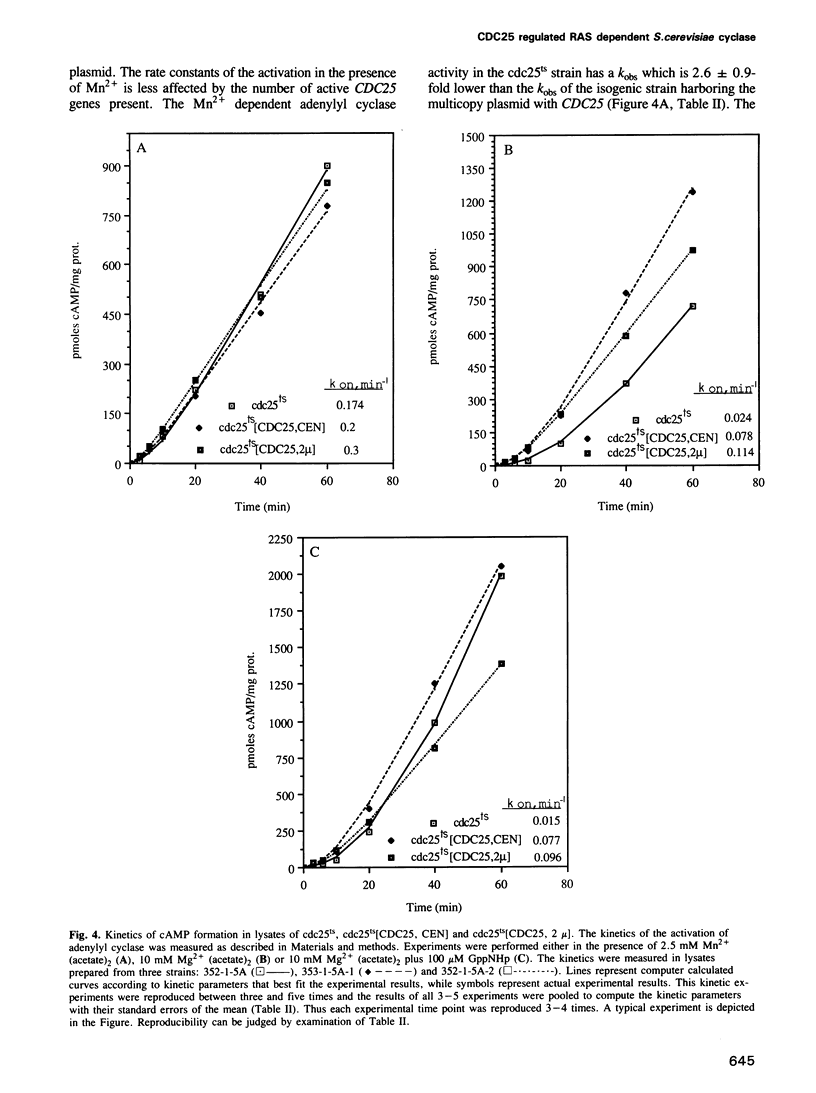
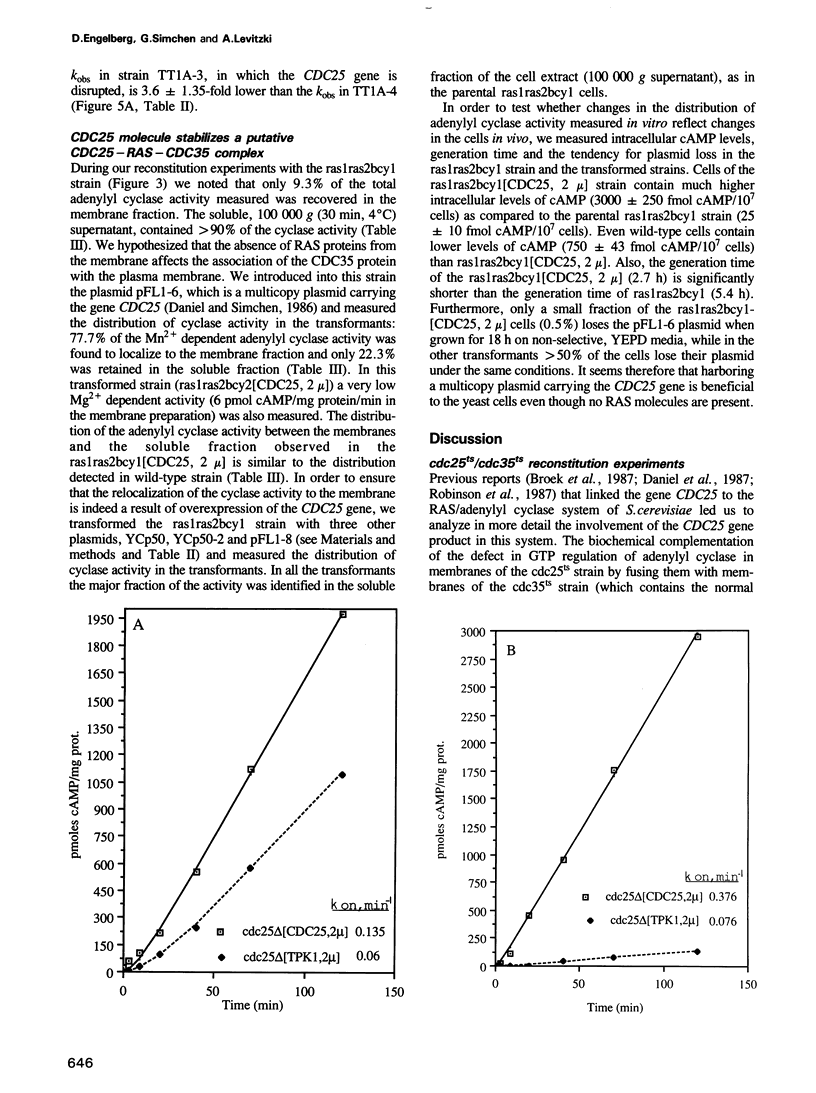
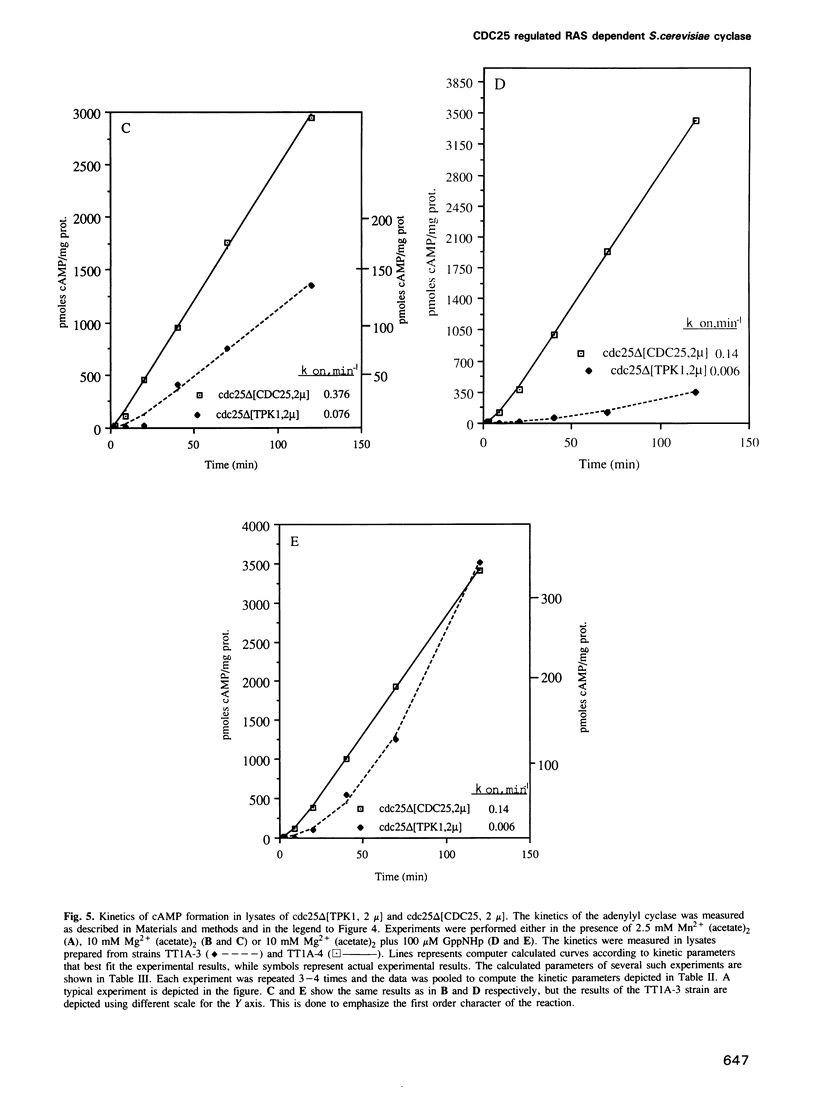
The attenuated GTP regulation adenylyl cyclase (CDC35) lysates or membranes prepared from cells of a cdc25ts strain is enhanced 2.5- to 6-fold by mixing these lysates or membranes with lysates or membranes from a cdc35ts strain harboring wild-type CDC25. The kinetics of activation of the Saccharomyces cerevisiae adenylyl cyclase in vitro is first order, as is the activation of mammalian adenylyl cyclase. The rate of enzyme activation in the presence of non-hydrolysable analogs of GTP increases with the number of CDC25 gene copies present in the cell. When GppNHp was used the rate of activation of the cyclase in a strain harboring a multicopy plasmid of CDC25 was 7.0-fold higher than the rate in an isogenic strain with the cdc25-2 mutation. The rate of adenylyl cyclase activation from a strain with a disrupted CDC25 gene is 14.7-fold lower than the rate in an isogenic strain containing the CDC25 gene on a multicopy plasmid. The reconstitution experiments described provide direct biochemical evidence for the role of the CDC25 protein in regulating the RAS dependent adenylyl cyclase in S.cerevisiae. The reconstitution experiments and the kinetic experiments may also provide a biochemical assay for the CDC25 protein and can form the basis for its characterization. In this study we also show that adenylyl cyclase activity in ras1ras2byc1 cells is found in the soluble fraction, whereas in wild-type strain it is found in the membrane fraction. Overexpression of the gene CDC25 in the ras1ras2bcy1 strain relocalizes adenylyl cyclase activity to the membrane fraction. This finding suggests a biochemical link between CDC25 and CDC35 in the absence of RAS, in addition to its role in regulating RAS dependent adenylyl cyclase.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Broek D., Samiy N., Fasano O., Fujiyama A., Tamanoi F., Northup J., Wigler M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell. 1985 Jul;41(3):763–769. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- Broek D., Toda T., Michaeli T., Levin L., Birchmeier C., Zoller M., Powers S., Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987 Mar 13;48(5):789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Camonis J. H., Jacquet M. A new RAS mutation that suppresses the CDC25 gene requirement for growth of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2980–2983. doi: 10.1128/mcb.8.7.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camonis J. H., Kalékine M., Gondré B., Garreau H., Boy-Marcotte E., Jacquet M. Characterization, cloning and sequence analysis of the CDC25 gene which controls the cyclic AMP level of Saccharomyces cerevisiae. EMBO J. 1986 Feb;5(2):375–380. doi: 10.1002/j.1460-2075.1986.tb04222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casperson G. F., Walker N., Bourne H. R. Isolation of the gene encoding adenylate cyclase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5060–5063. doi: 10.1073/pnas.82.15.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casperson G. F., Walker N., Brasier A. R., Bourne H. R. A guanine nucleotide-sensitive adenylate cyclase in the yeast Saccharomyces cerevisiae. J Biol Chem. 1983 Jul 10;258(13):7911–7914. [PubMed] [Google Scholar]
- Cerione R. A., Staniszewski C., Caron M. G., Lefkowitz R. J., Codina J., Birnbaumer L. A role for Ni in the hormonal stimulation of adenylate cyclase. Nature. 1985 Nov 21;318(6043):293–295. doi: 10.1038/318293a0. [DOI] [PubMed] [Google Scholar]
- Clark S. G., McGrath J. P., Levinson A. D. Expression of normal and activated human Ha-ras cDNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Oct;5(10):2746–2752. doi: 10.1128/mcb.5.10.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J., Becker J. M., Enari E., Levitzki A. The activation of adenylate cyclase by guanyl nucleotides in Saccharomyces cerevisiae is controlled by the CDC25 start gene product. Mol Cell Biol. 1987 Oct;7(10):3857–3861. doi: 10.1128/mcb.7.10.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J., Simchen G. Clones from two different genomic regions complement the cdc25 start mutation of Saccharomyces cerevisiae. Curr Genet. 1986;10(9):643–646. doi: 10.1007/BF00410911. [DOI] [PubMed] [Google Scholar]
- De Vendittis E., Vitelli A., Zahn R., Fasano O. Suppression of defective RAS1 and RAS2 functions in yeast by an adenylate cyclase activated by a single amino acid change. EMBO J. 1986 Dec 20;5(13):3657–3663. doi: 10.1002/j.1460-2075.1986.tb04696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder D., Im M. J., Klein H. W., Hekman M., Holzhöfer A., Dees C., Levitzki A., Helmreich E. J., Pfeuffer T. Reconstitution of beta 1-adrenoceptor-dependent adenylate cyclase from purified components. EMBO J. 1986 Jul;5(7):1509–1514. doi: 10.1002/j.1460-2075.1986.tb04390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988 May;8(5):2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman A. G. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Hanski E., Rimon G., Levitzki A. Adenylate cyclase activation by the beta-adrenergic receptors as a diffusion-controlled process. Biochemistry. 1979 Mar 6;18(5):846–853. doi: 10.1021/bi00572a017. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973 Jun;74(2):267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heideman W., Casperson G. F., Bourne H. R. Adenylyl cyclase in yeast. Hydrodynamic properties and activation by trypsin. J Biol Chem. 1987 May 25;262(15):7087–7091. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Broek D., Wigler M. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell. 1985 Dec;43(2 Pt 1):493–505. doi: 10.1016/0092-8674(85)90179-5. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Powers S., Cameron S., Fasano O., Goldfarb M., Broach J., Wigler M. Functional homology of mammalian and yeast RAS genes. Cell. 1985 Jan;40(1):19–26. doi: 10.1016/0092-8674(85)90304-6. [DOI] [PubMed] [Google Scholar]
- Kataoka T., Powers S., McGill C., Fasano O., Strathern J., Broach J., Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984 Jun;37(2):437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Levitzki A. Beta-adrenergic receptors and their mode of coupling to adenylate cyclase. Physiol Rev. 1986 Jul;66(3):819–854. doi: 10.1152/physrev.1986.66.3.819. [DOI] [PubMed] [Google Scholar]
- Levitzki A. From epinephrine to cyclic AMP. Science. 1988 Aug 12;241(4867):800–806. doi: 10.1126/science.2841758. [DOI] [PubMed] [Google Scholar]
- Lowy D. R., Willumsen B. M. The ras gene family. Cancer Surv. 1986;5(2):275–289. [PubMed] [Google Scholar]
- Martegani E., Baroni M., Wanoni M. Interaction of cAMP with the CDC25-mediated step in the cell cycle of budding yeast. Exp Cell Res. 1986 Feb;162(2):544–548. doi: 10.1016/0014-4827(86)90358-7. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Oshima Y., Ishikawa T. Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2355–2359. doi: 10.1073/pnas.79.7.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May D. C., Ross E. M., Gilman A. G., Smigel M. D. Reconstitution of catecholamine-stimulated adenylate cyclase activity using three purified proteins. J Biol Chem. 1985 Dec 15;260(29):15829–15833. [PubMed] [Google Scholar]
- Mbonyi K., Beullens M., Detremerie K., Geerts L., Thevelein J. M. Requirement of one functional RAS gene and inability of an oncogenic ras variant to mediate the glucose-induced cyclic AMP signal in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1988 Aug;8(8):3051–3057. doi: 10.1128/mcb.8.8.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F. ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989 Jan 13;56(1):5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- Munder T., Küntzel H. Glucose-induced cAMP signaling in Saccharomyces cerevisiae is mediated by the CDC25 protein. FEBS Lett. 1989 Jan 2;242(2):341–345. doi: 10.1016/0014-5793(89)80498-3. [DOI] [PubMed] [Google Scholar]
- Nikawa J., Cameron S., Toda T., Ferguson K. M., Wigler M. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1987 Nov;1(9):931–937. doi: 10.1101/gad.1.9.931. [DOI] [PubMed] [Google Scholar]
- Powers S., O'Neill K., Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwin C., Leidig F., Holzer H. Cyclic AMP-dependent phosphorylation of fructose-1,6-bisphosphatase in yeast. Biochem Biophys Res Commun. 1982 Aug 31;107(4):1482–1489. doi: 10.1016/s0006-291x(82)80166-6. [DOI] [PubMed] [Google Scholar]
- Purwin C., Nicolay K., Scheffers W. A., Holzer H. Mechanism of control of adenylate cyclase activity in yeast by fermentable sugars and carbonyl cyanide m-chlorophenylhydrazone. J Biol Chem. 1986 Jul 5;261(19):8744–8749. [PubMed] [Google Scholar]
- Robinson L. C., Gibbs J. B., Marshall M. S., Sigal I. S., Tatchell K. CDC25: a component of the RAS-adenylate cyclase pathway in Saccharomyces cerevisiae. Science. 1987 Mar 6;235(4793):1218–1221. doi: 10.1126/science.3547648. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Salomon Y., Londos C., Rodbell M. A highly sensitive adenylate cyclase assay. Anal Biochem. 1974 Apr;58(2):541–548. doi: 10.1016/0003-2697(74)90222-x. [DOI] [PubMed] [Google Scholar]
- Shilo V., Simchen G., Shilo B. Initiation of meiosis in cell cycle initiation mutants of Saccharomyces cerevisiae. Exp Cell Res. 1978 Mar 15;112(2):241–248. doi: 10.1016/0014-4827(78)90206-9. [DOI] [PubMed] [Google Scholar]
- Sy J., Tamai Y. An altered adenylate cyclase in cdc35-1 cell division cycle mutant of yeast. Biochem Biophys Res Commun. 1986 Oct 30;140(2):723–727. doi: 10.1016/0006-291x(86)90791-6. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Matsumoto K., Toh-E A. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):757–768. doi: 10.1128/mcb.9.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Tolkovsky A. M., Levitzki A. Coupling of a single adenylate cyclase to two receptors: adenosine and catecholamine. Biochemistry. 1978 Sep 5;17(18):3811–3817. doi: 10.1021/bi00611a021. [DOI] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987 Oct 23;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]





