Abstract
Zygosaccharomyces bailii virus Z (ZbV-Z) is a monosegmented dsRNA virus that infects the yeast Zygosaccharomyces bailii and remains unclassified to date despite its discovery >20 years ago. The previously reported nucleotide sequence of ZbV-Z (GenBank AF224490) encompasses two nonoverlapping long ORFs: upstream ORF1 encoding the putative coat protein and downstream ORF2 encoding the RNA-dependent RNA polymerase (RdRp). The lack of overlap between these ORFs raises the question of how the downstream ORF is translated. After examining the previous sequence of ZbV-Z, we predicted that it contains at least one sequencing error to explain the nonoverlapping ORFs, and hence we redetermined the nucleotide sequence of ZbV-Z, derived from the same isolate of Z. bailii as previously studied, to address this prediction. The key finding from our new sequence, which includes several insertions, deletions, and substitutions relative to the previous one, is that ORF2 in fact overlaps ORF1 in the +1 frame. Moreover, a proposed sequence motif for +1 programmed ribosomal frameshifting, previously noted in influenza A viruses, plant amalgaviruses, and others, is also present in the newly identified ORF1–ORF2 overlap region of ZbV-Z. Phylogenetic analyses provided evidence that ZbV-Z represents a distinct taxon most closely related to plant amalgaviruses (genus Amalgavirus, family Amalgaviridae). We conclude that ZbV-Z is the prototype of a new species, Zygosaccharomyces bailii virus Z, which we propose to assign as type species of a new genus of monosegmented dsRNA mycoviruses in family Amalgaviridae. Comparisons involving other unclassified mycoviruses with RdRps apparently related to those of plant amalgaviruses, and having either mono- or bisegmented dsRNA genomes, are also discussed.
Keywords: Amalgaviridae, dsRNA, Fungal virus, Mycovirus, Ribosomal frameshifting, Totiviridae
1. Introduction
The budding yeast Zygosaccharomyces bailii (family Saccharomycetaceae, phylum Ascomycota) is a widespread cause of food spoilage. A monosegmented (i.e., nonsegmented) dsRNA virus from Z. bailii, Zygosaccharomyces bailii virus Z (ZbV-Z) (Radler et al., 1993; Schmitt and Neuhausen, 1994), has been tentatively assigned by some authors to genus Totivirus in family Totiviridae (Wickner et al., 2012) and is currently listed as such by the USA National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/taxonomy). ZbV-Z has, however, not yet been recognized and classified by the International Committee on Taxonomy of Viruses (ICTV) (http://ictvonline.org/virusTaxonomy.asp).
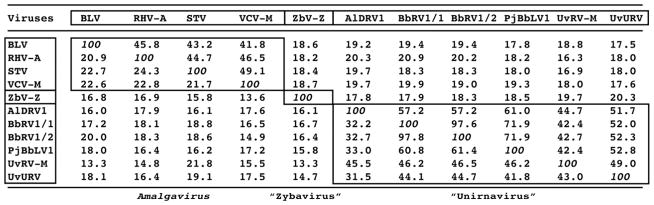
The reported genome length of ZbV-Z is 3157 bp (GenBank AF224490; no peer-reviewed journal publication), and its plus-strand sequence includes two nonoverlapping long ORFs, ORF1 encoding the 34-kDa putative coat protein (CP) (see evidence in this regard in Schmitt and Neuhausen (1994)) and ORF2 encoding the RdRp, separated by 190 nt in the same frame. Interestingly, the results of NCBI PSI-BLAST searches of the Non-redundant Protein Sequences (NR) database (e.g., Table 1) showed that the RdRp of ZbV-Z is only distantly related to those of family Totiviridae members. Instead, the RdRp of ZbV-Z is more closely related to those of plant amalgaviruses, which are monosegmented dsRNA viruses constituting the only currently approved genus, Amalgavirus, in family Amalgaviridae (originally proposed name “Amalgamaviridae”) (Liu and Chen, 2009; Martin et al., 2011; Sabanadzovic et al., 2009, 2010), as well as those of monosegmented dsRNA mycoviruses constituting the proposed genus “Unirnavirus” (Jiang et al., 2015; Koloniuk et al., 2015; Kotta-Loizou et al., 2015; Lin et al., 2015; Nerva et al., 2015; Zhu et al., 2015; name suggested by Kotta-Loizou et al.) and those of family Partitiviridae members, which are bisegmented dsRNA viruses infecting fungi, plants, or protists (Nibert et al., 2014). Other recent authors, too, have observed that the RdRp of ZbV-Z is more related to those of amalgaviruses than to those of family Totiviridae members (Liu et al., 2012a, b).
Table 1.
NCBI PSI-BLAST search results for ZbV-Z RdRp (GenBank AAF37275)
| Virus | Family:genus | GenBank no. | E values: |
|---|---|---|---|
| Blueberry latent virus | Amalgaviridae:Amalgavirus | ADO14116 | 4e–17 |
| Blueberry latent virus | Amalgaviridae:Amalgavirus | ADO14118 | 5e–17 |
| Ustilaginoidea virens unassigned | “Unirnavirus”b | AKM52549 | 1e–16 |
| RNA virus HNND-1 | |||
| Beauveria bassiana RNA virus 1 | “Unirnavirus” | CEF90232 | 1e–16 |
| Rhododendron virus A | Amalgaviridae:Amalgavirus | ADM36020 | 2e–16 |
| Beauveria bassiana RNA virus 1 | “Unirnavirus” | AKC57301 | 7e–16 |
| Fragaria chiloensis cryptic virus | Partitiviridae:Deltapartitivirus | AAZ06131 | 6e–15 |
| Ustilaginoidea virens RNA virus M | “Unirnavirus” | AIT56395 | 7e–15 |
| Rose cryptic virus 1 | Partitiviridae:Deltapartitivirus | ABY60412 | 1e–14 |
The top 10 hits after ZbV-Z from the first-iteration PSI-BLAST search are listed in order of their E values.
Name of proposed genus suggested by Kotta-Loizou et al. (2015)
One notable apparent difference between ZbV-Z and the plant amalgaviruses is that the latter have partially overlapping ORF1 (encoding a 40- to 45-kDa protein of unclear function but possible CP) and ORF2 (encoding the RdRp), with ORF2 in the +1 frame relative to ORF1 (Table 2), whereas ZbV-Z has nonoverlapping ORF1 and ORF2 as described above (Table 2, Fig. 1A). Upon examining the ZbV-Z sequence from GenBank AF224490, however, we observed that a properly positioned 1-nt insertion somewhere between positions 1088 and 1100 would extend ORF2 by 221 nt upstream, allowing it to overlap ORF1 in the +1 frame as in the plant amalgaviruses (Fig. 1B). We therefore undertook to redetermine the nucleotide sequence of ZbV-Z in order to address this prediction.
Table 2.
Properties of amalgaviruses, ZbV-Z, and unirnaviruses relating to ORF2 translation by proposed PRF
| Virus | Genus | GenBank no. | ORF1 rangea | ORF2 rangea | Overlap (nt)b | FSc |
|---|---|---|---|---|---|---|
| Blueberry latent virus | Amalgavirus | HM029246d | 131–1291 | 930–3329 | 362 | +1 |
| Rhododendron virus A | Amalgavirus | HQ128706 | 38–1306 | 696–3326 | 611 | +1 |
| Southern tomato virus | Amalgavirus | EF442780 d | 87–1268 | 976–3324 | 293 | +1 |
| Vicia cryptic virus M | Amalgavirus | EU371896 | 20–1324e | 966–3314e | 359 | +1 |
| Zygosaccharomyces bailii virus Z | “Zybavirus”f | AF224490g | 38–916 | 1091–3034 | 0 | 0 |
| KU200450g | 38–916 | 870–3083 | 47 | +1 | ||
| Alternaria longipes dsRNA virus 1 | “Unirnavirus” h | KJ817371 | 298–1500 | 1428–3308 | 73 | −1 |
| Beauveria bassiana RNA virus 1/1 | “Unirnavirus” | LN610699k | 249–1265 | 1256–3097 | 10 | −1 |
| Beauveria bassiana RNA virus 1/2 | “Unirnavirus” | KM233415k | 212–1228 | 1219–3060 | 10 | −1 |
| Penicillium janczewskii Beauveria bassiana-like virus 1 | “Unirnavirus” | KT601106 | ≤1m–951 | 915–2777 | 37 | −1 |
| Ustilaginoidea virens RNA virus M | “Unirnavirus” | KJ101567 | ≤3m–413 | 404–2236 | 10 | −1 |
| Ustilaginoidea virens unassigned | “Unirnavirus” | KR106133 | 31–975 | 1032–2828 | 0 | −1 |
| RNA virus HNND-1 | 936n–2828 | 40 | −1 |
The range of each ORF as defined here reflects the span between flanking stop codons, with no effort to predict which if any start codon(s) may be used. This is the appropriate way to define ORFs for detecting overlaps consistent with ribosomal frameshifting mechanisms. The flanking stop codons are not included in the specified range.
The overlap region as defined here is determined by the specified ORF1 and ORF2 ranges, not including any of the nucleotides within the flanking stop codons.
FS, frame shift: the frame in which ORF2 is found relative to ORF1 (either −1, 0, or +1), not attempting to specify the nature of the predicted PRF mechanism. For example, a −1 frame shift as shown in this table could be mechanistically either −1 or +2 to bring ORF2 in frame with ORF1.
Sequences of representative strains were used for these viruses.
These ORFs are found in the reverse strand sequence of that deposited in GenBank.
Newly proposed genus name (this study)
The original and newly amended sequences were used for this virus.
Name of proposed genus suggested by Kotta-Loizou et al. (2015)
The sequences of two different strains have been used for this virus.
ORF1 remains open to the plus-strand 5′ end of these viruses, suggesting that the published sequences are truncated at this end.
Including a predicted U or C substitution at nt position 1031 and thereby extending ORF2 toward the 5′ end of plus strand such that it now overlaps ORF1 in the −1 frame.
Fig. 1.
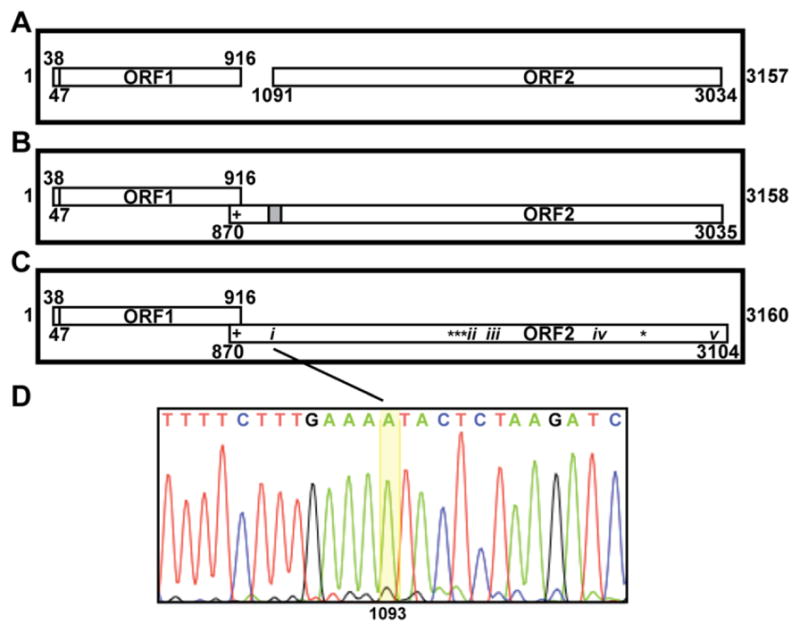
Genome diagrams and sequencing results. (A–C) The genome of ZbV-Z/412 is depicted by the thicker-lined box in each panel, with the spans of ORF1 and ORF2 indicated by the thinner-lined boxes inside. Whether ORF1 and ORF2 are in frame 1, 2, or 3 is indicated by vertical position top to bottom in each panel (e.g., in panel A, both ORF1 and ORF2 are in frame 2). The limits of each ORF as defined by flanking stop codons are numbered by nucleotide position. The first nucleotide in the putative start codon of ORF1 is also numbered. Scales in all three panels are approximate. (A) Diagram of the ZbV-Z/412 genome from GenBank AF224490. (B) Our prediction for how a single 1-nt insertion (within the gray-shaded area) would allow ORF2 to overlap ORF1 in the +1 frame (frame 3). $, Position of a proposed slippery sequence for +1 PRF, allowing translation of an ORF1/ORF2 fusion product. (C) Diagram of the ZbV-Z/412 genome from new sequencing results (GenBank KU200450). i–v, Positions of the respective indels; *, Positions of four single-nucleotide substitutions; $, same as described for panel B. (D) Sequencing electropherogram across the region of indel i in the ZbV-Z plus strand, showing the 1-nt (A) insertion at position 1093.
In fact, we made another observation that further encouraged us to redetermine the nucleotide sequence of ZbV-Z. Firth et al. (2012), Jagger et al. (2012), and Shi et al. (2012) have characterized UCC_UUU_CGU (underlines, ORF1 codon boundaries; CGN are all rare Arg codons in most eukaryotes) as a motif for mechanistically +1 programmed ribosomal frameshifting (PRF), allowing translation of the virulence-modulating PA-X protein of influenza A viruses. Additionally, Firth et al. (2012) have noted that there are similar, putative +1 slippery sequences in the ORF1–ORF2 overlap region of three of the plant amalgaviruses: UCU_UUU_CGU in blueberry latent virus, ACU_UUU_CGC in rhododendron virus A, and ACU_UUU_CGU in Vicia cryptic virus M. Notably, in the ZbV-Z sequence from GenBank AF224490, and specifically within the predicted region of ORF1–ORF2 overlap that we describe above, the related sequence CUU_UUU_CGA is found at nt positions 905–913 (Fig. 1B). Thus, there appears to be a possible +1 slippery sequence in the proper frame and position in the ZbV-Z plus strand to allow expression of an ORF1/ORF2 fusion product if, that is, a 1-nt insertion could be found further downstream (in the 1088–1100 region of GenBank AF224490 as described above) to provide a region of ORF1–ORF2 overlap in which the putative +1 PRF could productively occur.
2. Materials and methods
2.1. Yeast culture and harvest
Z. bailii 412 cells (from our lab clone DD1) were grown in liquid YPG medium (1% yeast extract, 2% peptone, 2% glucose) at 30°C with shaker agitation at 250 rpm until reaching an optical density of ~1.0 as measured at 600 nm. Cells were then harvested by centrifugation at 1500 × g for 5 min at 4 C. Resulting pellets (from 10 mL of culture each) were resuspended in 1 mL cold deionized water, flash-frozen on 100% alcohol and dry ice, and stored at −80 C until use.
2.2. RNA purifications
After thawing the yeast pellet, 250 μL of RNA buffer (500 mM NaCl, 10mM EDTA, 200 mM Tris plus HCl to pH 7.5) and 750 μL of TRIzol LS reagent (Ambion) were added, along with a volume of glass beads (diameter, 0.5 mm; BioSpec) equivalent to 100 μL. Samples were incubated for 5 min at room temperature, after which 150 μL of chloroform was added and the sample was subjected to vortex agitation for 2 min (to allow breakage of the yeast cell walls). The TRIzol extraction protocol was then completed per the manufacturer’s instructions. To enrich for dsRNA from the TRIzol extract of total RNA, treatment with microgranular cellulose (cellulose powder MN 301, Macherey-Nagel) was performed largely as described by Castillo et al. (2011). Cellulose was equilibrated with STE buffer (100 mM NaCl, 50 mM Tris plus HCl to pH 7.5, 1 mM EDTA) plus 16% ethanol, followed by pelleting and removal of the buffer, and then resuspension in 1.8 mL fresh STE buffer plus ethanol. Total RNA from a TRIzol extraction was then added in a volume of ≤200 μL, followed by incubation with vortex agitation for 1 h. Using a syringe for each step, the suspension was pushed through an empty mini-column (Promega), and the retained cellulose was washed with 2 mL fresh STE buffer plus ethanol. After placing the mini-column in an empty microtube, excess buffer was removed by microcentrigation at 13,000 × g for 1 min. After transferring the mini-column to a new empty microtube, 20 μL of STE buffer without ethanol was added, followed by microcentrigation at 13,000 × g for 1 min. The collected eluate was then reapplied to the mini-column, followed again by microcentrigation at 13,000 × g for 1 min to yield the final eluate containing dsRNA.
2.3. Sequence determinations
We used the previously reported ZbV-Z sequence (GenBank AF224490) to design oligonucleotide primers for performing RT–PCR on total RNA extracted from Z. bailii 412. The primers were designed to amplify four overlapping regions of the ZbV-Z/412 genome: positions 63–1070, 570–1580, 1473–2543, and 2194–3133 (numbered according to GenBank AF224490). The RT step was performed according to manufacturer’s instructions for the SuperScript III First-Strand Synthesis System (Invitrogen), except that the reaction was allowed to incubate for 45 min at 55°C, followed by 15 min at 70°C. The PCR step was performed according to manufacturer’s instructions for Taq DNA Polymerase with Standard Taq Buffer (NEB), except that 34 cycles were performed with denaturation at 95 °C for 30 s and hybridization at 50°C for 30 s in each cycle. PCR products were separated by electrophoresis (20 min at 120 volts) on a 0.7% agarose gel and stained with ethidium bromide. The DNA bands were excised from the gel during visualization with a UV transilluminator, followed by purification according to manufacturer’s instructions for the QIAquick Gel Extraction Kit (Qiagen). DNA and the same primers as used for PCR were then sent to the Dana Farber/Harvard Cancer Center DNA Resource Core for Sanger sequencing in both directions for each amplicon.
For de novo determination of the terminal sequences of ZbV-Z/412, RLM-3′RACE was performed on cellulose-purified dsRNA from Z. bailii 412 using modifications of previously described methods (Coutts and Livieratos, 2003). The dsRNA was first denatured by incubation in 90% dimethyl sulfoxide for 15 min at 65°C. The denatured dsRNA was precipitated by addition of NaCl to a concentration of 150 mM, plus 2.5 volumes of 100% ethanol. The resulting RNA pellet was washed with 70% ethanol and then air-dried, followed by resuspension in ligation mix (1×T4 RNA Ligase Reaction Buffer (NEB), 1 mM DTT, 1mM ATP, 2 U/μL RNasin (Promega), 20% PEG 8000 (NEB), 40 pmoles DNA adapter 5′phosphate-CAATACCTTCTGACCATGCAGTGACA GTCAGCATG-3′amino modifier (IDT), and 10 U T4 RNA ligase 1 (NEB), plus DEPC-treated water treated to reach 20 μL). Ligation was allowed to proceed for 16 h at 16°C. Next, 80 μL of DEPC-treated water and 1 mL of TRIzol LS reagent were added, and the sample was incubated at room temperature for 5 min, followed by addition of 200 μL chloroform and microcentrifugation at 1200 × g for 5 min at 4°C before recovering the aqueous phase. 10 μL 3M sodium acetate pH 5.2 and 220 μL 95% ethanol were then added, and the sample incubated on dry ice for 10 min before microcentrifugation at 13,000 × g for 20 min at 4°C. The resulting pellet was washed with 70% ethanol and air dried, then resuspended in 10 μL DEPC-treated water. The RT step was performed as described above, but this time using outer anti-adapter primer CATGCTGACTGTCACTGCATGG. PCR amplifications were performed as described above, with each reaction including the outer anti-adapter primer and an internal primer based on the ZbV-Z/412 sequence beginning at nt position 283 or 2850 for the amplicon corresponding to the 5′ or 3′ end of plus strand, respectively. The amplicon corresponding to the 5′ end of plus strand was obtained and prepared for sequencing as described above. The amplicon corresponding to the 3′ end of plus strand was not obtained, however, and we therefore performed a second, nested reaction using inner anti-adapter primer CTGTCACTGCATGG TCAGAAGG and an internal primer based on the ZbV-Z/412 sequence beginning at nt position 2884. Nested PCR included the following steps: initial denaturation at 95°C for 2 min, followed by 24 cycles of denaturation at 95°C for 30 s, hybridization at 58°C for 30 s, and elongation at 72°C for 30 s, followed by a final extension at 68°C for 8 min. The amplicon was obtained in this case and prepared for sequencing as described above. Sanger sequencing was performed in both directions for both RLM-3′RACE amplicons, and the sequences were found to match 100%, in the regions of overlap, with the sequences obtained as described in the preceding paragraph.
2.4. Sequence-based analyses
Table S1 lists abbreviations and GenBank accession numbers for the nucleotide sequences of all dsRNA viruses included in this study. Searches of the NR database with protein sequence queries deduced from the nucleotide sequences were performed using NCBI PSI-BLAST (Schäffer et al., 2001) as implemented with defaults at http://blast.ncbi.nlm.nih.gov/Blast.cgi. ORFs were identified in nucleotide sequences using EMBOSS getorf as implemented with defaults at http://www.bioinformatics.nl/emboss-explorer/. Molecular weight and pI values for proteins were calculated using Compute pI/MW as implemented with defaults at http://web.expasy.org/compute_pi/. Global pairwise comparisons of protein sequences were performed using Needle or Needleall as implemented at http://www.bioinformatics.nl/emboss-explorer/, with the following parameter differing from the defaults: Apply end gap penalties, Yes. As a simple convention for comparing sequences from the different viruses in the absence of certainty about translational initiation and frameshifting sites, the ORF1 and ORF2 translation product sequences used for Fig. 4 began with the first Met codon in each of the two ORFs, except for the comparisons involving the ORF1 translation product of UvRV-M (GenBank KJ101567) as described in the Fig. 4 legend. Local pairwise comparisons of nucleotide sequences were performed with EMBOSS Matcher as implemented with defaults at http://www.ebi.ac.uk/Tools/psa/.
Fig. 4.
Pairwise identity scores. Sequences of the ORF1 and ORF2 translation products of the indicated viruses were compared pairwise using EMBOSS Needle as described in Materials and Methods. The resulting identity scores are shown in %, with ORF1:ORF1 product scores at lower left and the ORF2:ORF2 product (RdRp:RdRp) scores at upper right. Distinguishable subsets of higher scores, reflecting three different taxa (approved or proposed genera) of viruses, are boxed. Because the ORF1 sequences of UvRV-M appear to be truncated at the 5′ end, only the C-terminal 137 residues of each of the other ORF1 products has been used for comparison with the 137-residue ORF1 product of UvRV-M. Names of approved and proposed genera are indicated at bottom.
For phylogenetic analyses, multiple sequence alignments were performed with the RdRp sequences using MAFFT 7.27 (L-INS-i) (Katoh and Standley, 2013) as implemented with defaults at http://mafft.cbrc.jp/alignment/server/. The convention of using sequences beginning with the first Met residue in the RdRp-encoding ORF was applied again here, including for Fig. 3. Phylogenetic relationships were then determined using PhyML 3.0 (Guindon et al., 2010) as implemented at http://www.hiv.lanl.gov/content/sequence/PHYML/interface.html with the following parameters differing from the defaults: Sequence type/model, Amino acids/LG; Proportion of invariable sites, estimated from data; Gamma shape parameter, estimated from data; Starting tree(s) optimization, Tree topology and Branch length; Tree improvement, Best of NNI and SPR; Branch support, Approximate Likelihood Ratio Test (aLRT), SH-like supports. The results in Newick format were then submitted to TreeDyn 198.3 as implemented at http://www.phylogeny.fr/ for displaying branch support values in % and collapsing branches with support values below 50%. The output in Newick format was then opened in FigTree v1.4.0 (downloaded from http://tree.bio.ed.ac.uk/software/figtree/) for refining the phylogram for presentation. For Fig. 3A, values estimated from the data were Proportion of invariable sites, 0.018, and Gamma shape parameter, 2.445. For Fig. 3B, estimated values were Proportion of invariable sites, 0.003, and Gamma shape parameter, 1.643. Alternative use of the RtREV substitution model for PhyML 3.0 yielded results very similar to those shown in Fig. 3.
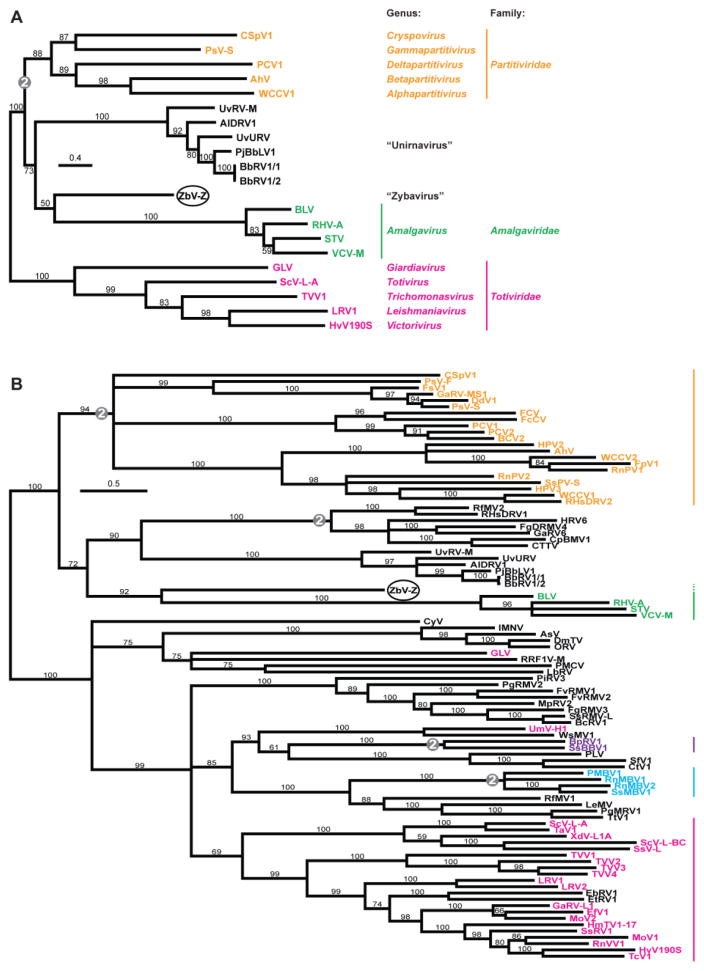
Fig. 3.
Phylogenetic analyses. As described in Materials and Methods, RdRp (ORF2 product) sequences of the indicated mono- and bisegmented dsRNA viruses were compared in multiple sequence alignments, followed by maximum-likelihood phylogenetic analyses. The trees are displayed as unrooted rectangular phylograms. Branch support values are shown in %; branches with <50% support are collapsed to the preceding node. Scale bar, average number of substitutions per alignment position. See Table S1 for a summary of abbreviations and GenBank numbers. Viruses representing approved species in families Amalgaviridae, Partitiviridae, and Totiviridae are labeled respectively in green, orange, and magenta, unclassified viruses in black. ZbV-Z is highlighted with an oval. Clades of bisegmented viruses are indicated by “2” in a gray circle. (A) Names of approved and proposed genera are indicated. (B) Family ranges are indicated by vertical lines, colored as in panel A. Additional families in this panel are Botybirnaviridae (purple) and Megabirnaviridae (cyan). Our proposal to expand family Amalgaviridae to encompass ZbV-Z (proposed genus Zybavirus) is indicated by the dotted portion of the green bar. Two currently approved members of family Totiviridae, GLV and UmV-H1, appear to fall outside the range of this family in this analysis and others.
3. Results
3.1. Redetermination of the ZbV-Z nucleotide sequence
Z. bailii 412 is the specific isolate of this yeast species from which ZbV-Z has been previously characterized (Radler et al., 1993; Schmitt and Neuhausen, 1994; GenBank AF224490). Anticipating that ZbV-Z may later be characterized from other Z. bailii isolates (e.g., see gel evidence for ZbV-Z in Z. bailii 427 in Radler et al. (1993)), we henceforth refer to the strain of this virus from Z. bailii 412 as ZbV-Z/412. A culture of Z. bailii 412 was obtained as a kind gift from Manfred J. Schmitt and Frank Breinig (Saarland University, Saarbrücken, Germany). After being grown to mid- to late-log phase in YPG media, cells were harvested, disrupted by vortex agitation with glass beads, and subjected to extraction for total RNA. After enriching this extract for dsRNA by cellulose affinity and then performing agarose gel electrophoresis, three bands were observed (Fig. 2), consistent in size with the expected L (~4.0 kbp), Z (~3.0 kbp), and M (~2.0 kbp) dsRNA segments carried by Z. bailii 412 (Radler et al., 1993; Schmitt and Neuhausen, 1994). Although only the Z segment is the focus of this report, the L segment is thought to be the genome of an uncharacterized totivirus (ZbV-L/412) and the M segment its toxin-encoding satellite (Schmitt and Neuhausen, 1994; Weiler et al., 2002). Primer pairs were next designed from GenBank AF224490 and used for amplification by RT-PCR and direct Sanger sequencing of the amplicons. In this manner, sequences were determined in full from both genomic RNA strands of ZbV-Z/412, except for limited terminal portions of each strand outside the span of these initial primers. RNA-ligase-mediated 3′ rapid amplification of cDNA ends (RLM-3′RACE) was then performed on ZbV-Z/412 dsRNA to determine these unread terminal sequences at both ends of the genome. Our final, full-length nucleotide sequence of ZbV-Z/412 has been deposited in GenBank with accession number KU200450.
Fig. 2.
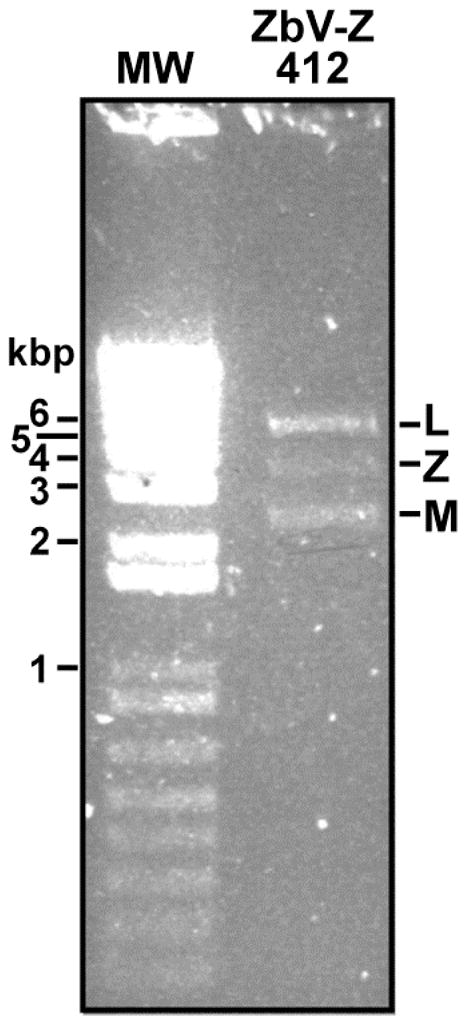
RNA gel. A lysate of Z. bailii 412 was enriched for dsRNA as described in Materials and Methods. A 10-μl aliquot of the lysate was then subjected to electrophoresis in a 1% agarose gel, followed by staining with ethidium bromide. Positions of selected DNA molecular weight (MW) markers are labeled at left in kbp.
The new sequencing results for ZbV-Z/412 included five insertions or deletions (indels) relative to GenBank AF224490 (Fig. 1C): (i) a 1-nt insertion after position 1092 (run of 4, not 3, A’s) (shown in Fig. 1D); (ii) a 2-nt deletion of positions 1811 and 1812 (run of 2, not 4, T’s); (iii) a 2-nt insertion after position 1845 (run of 6, not 4, T’s); (iv) a 3-nt insertion (TTG) after position 2331; and (v) a 1-nt deletion of position 3015 (run of 2, not 3, G’s). Indel i introduces a frame shift that extends ORF2 by 221 nt upstream such that ORF2 now overlaps ORF1 by 47 nt in the +1 frame (Fig. 1C). Notably this is precisely the type of predicted change to the previous sequence of ZbV-Z, a 1-nt insertion somewhere between positions 1088 and 1100 as described in Introduction, which led us to undertake this study. Indel ii introduces a frame shift that changes the encoded protein sequence over the downstream 10 residues, and indel iii introduces a compensatory frame shift that returns the reading frame to the same as that before indel ii. Indel iv results in insertion of a new Trp residue into the encoded protein sequence. Indel v introduces a frame shift that extends ORF2 by 45 nt downstream, changing the formerly last 7 residues of the encoded protein sequence and also extending it by 15 residues to the next downstream stop codon in the new reading frame. In addition to these five indels, our new results for ZbV-Z/412 identified four single-base substitutions relative to GenBank AF224490 at positions 1805, 1809, 1810, and 2619, yielding three further changes to the encoded protein sequence (Fig. 1C). All of these observed indels and substitutions are located within ORF2.
3.2. Initial analysis of the amended ZbV-Z nucleotide sequence
The new results show the overall length of the ZbV-Z/412 genomic plus strand to be 3160 nt, from 5′-GUAAAAGAAC to UAUGCCUUGG-3′. ORF1 spans positions 38–916, between stop codons at positions 35–37 and 917–919 (Fig. 1C). Its 5′-most AUG codon is at positions 47–49 and is in a strong sequence context for translation initiation (AGUAUGG). The protein-coding region of ORF1 is therefore predicted to span positions 47–916 and to encode a 290-aa, 34-kDa putative CP (pI, 6.2). These details for ORF1 and its predicted translation product are the same as those from GenBank AF224490. ORF2 spans positions 870–3104, between stop codons at positions 867–869 and 3105–3107 and thereby overlaps ORF1 by 47 nt (positions 870–916) (Fig. 1C). Within this 47-nt overlap is found the sequence CUU_UUU_CGA (underlines, ORF1 codon boundaries) at positions 905–913 (Fig. 1C), representing a putative +1 PRF motif per Firth et al. (2012), as described in Introduction. If +1 PRF indeed occurs near the 3′ end of this motif, then the resulting ORF1/ORF2 fusion is expected to span positions 47–910:912–3104 and to encode a 1012-aa, 119-kDa putative CP/RdRp (pI, 9.2). Based on these analyses, the nontranslated regions at the ends of the ZbV-Z/412 genomic plus strand are fairly short: 46 nt at the 5′ end and 77 nt at the 3′ end including the ORF2 stop codon (Fig. 1C).
3.3. Sequence comparisons and phylogenetic analyses
To begin to address the phylogeny of ZbV-Z/412, we compared its ORF2-encoded amino acid (RdRp) sequences with those from several other mono- and bisegmented dsRNA viruses. Given the original, tentative assignment of ZbV-Z to family Totiviridae, we included sequences representing the type species of the five approved genera in that family (Saccharomyces cerevisiae virus L-A from genus Totivirus, Helminthosporium victoriae virus 190 from genus Victorivirus, Giardia lamblia virus from genus Giardiavirus, Leishmania RNA virus 1 from genus Leishmaniavirus, and Trichomonas vaginalis virus 1 from genus Trichomonasvirus). Also, given the findings in Table 1 described above, we included sequences representing the four approved species of plant viruses in genus Amalgavirus, family Amalgaviridae (Blueberry latent virus, Rhododendron virus A, Southern tomato virus, and Vicia cryptic virus M); sequences from six strains of monosegmented dsRNA mycoviruses that constitute proposed genus Unirnavirus (Alternaria longipes dsRNA virus 1 from A. longipes isolate HN28 (AlDRV1), Beauveria bassiana RNA virus 1 from B. bassiana isolates EABb-92/11-Dm (BbRV1/1) and A24 (BbRV1/2), Penicillium janczewskii Beauveria bassiana-like virus 1 from P. janczewskii isolate MUT4359 (PjBbLV1), Ustilaginoidea virens RNA virus M from U. virens isolate GX-1 (UvRV-M), and Ustilaginoidea virens unassigned RNA virus from U. virens isolate HNND-1 (UvURV)) (Jiang et al., 2015; Koloniuk et al., 2015; Kotta-Loizou et al., 2015; Lin et al., 2015; Nerva et al., 2015; Zhu et al., 2015); and sequences representing the type species of the five approved genera in family Partitiviridae (White clover cryptic virus 1 from genus Alphapartitivirus, Atkinsonella hypoxylon virus from genus Betapartitivirus, Penicillium stoloniferum virus S from genus Gammapartitivirus, Pepper cryptic virus 1 from genus Deltapartitivirus, and Cryptosporidium parvum virus 1 from genus Cryspovirus). The results from maximum-likelihood phylogenetic analyses provided evidence that ZbV-Z/412 represents a distinct taxon relative to the other analyzed viruses, more closely related to plant amalgaviruses and unirnaviruses than to Totiviridae and Partitiviridae members (Fig. 3A). Sequence identity scores from pairwise comparisons of the ORF1 and ORF2 product sequences of ZbV-Z/412 with those of plant amalgaviruses and unirnaviruses were consistent with the phylogenetic results and provided further evidence that ZbV-Z/412 represents a distinct taxon (Fig. 4). We conclude that ZbV-Z/412 is the prototype strain of a new species, Zygosaccharomyces bailii virus Z, which we further propose to assign as type species of a new genus of monosegmented dsRNA mycoviruses outside family Totiviridae. We suggest the provisional name “Zybavirus” for discussing this genus (Fig. 3A), as derived from the name of the prototype host Zygosaccharomyces bailii.
4. Discussion
4.1. Additional considerations regarding taxonomic classification of ZbV-Z
A relevant question is whether ZbV-Z/412 and its proposed genus Zybavirus should be assigned to family Amalgaviridae. Based on the RdRp-based phylogenetic tree in Fig. 3A, one might conclude (i) that proposed genera Zybavirus and Unirnavirus should both be assigned to family Amalgaviridae, (ii) that genus Zybavirus should be assigned to family Amalgaviridae but genus Unirnavirus should not, or (iii) that neither genus Zybavirus nor genus Unirnavirus should be assigned to family Amalgaviridae. In an effort to resolve this ambiguity, we undertook several additional analyses.
Terminal sequences of the genomic RNA strands are often conserved among related dsRNA viruses, reflecting important roles in RNA transcription and/or replication. Among the plant amalgaviruses, although their plus-strand 3′ sequences appear more variable, their plus-strand 5′ sequences are more conserved, with the consensus being 5′-GWWWWWWWWW (W = A or U). ZbV-Z/412 fits this consensus in part, with its plus-strand 5′ sequence being 5′-GUAAAAGAAC. The consensus sequence for the plant amalgaviruses and ZbV-Z/412 combined is thus 5′-GWWWWWNWW (N = potentially any base). The plus-strand 5′ sequences reported to date for unirnaviruses are also more variable, perhaps because several of them are incomplete, such that they seem to add little to this analysis.
Table 3 highlights other notable similarities—in genome, ORF1 product, and predicted ORF1/ORF2 product lengths—among plant amalgaviruses, ZbV-Z/412, and unirnaviruses. The genome lengths of them all fall within a range of 2890 to 3437 bp, their ORF1 product lengths in a range of 290 to 404 aa (excluding UvRV-M/GX-1 since its plus-strand sequence appears to be substantially truncated at its 5′ end; see Table 2), and their predicted ORF1/ORF2 product lengths in a range of 926 to 1077 aa (again excluding UvRV-M/GX-1). Table 3 further shows that even the uppermost values in these ranges are substantially smaller than the lowermost values of family Totiviridae members. Their distinctively similar genome and protein lengths are thus discrete characteristics that might be used to support the assignment of all three taxa—genus Amalgavirus and proposed genera Zybavirus and Unirnavirus—to family Amalgaviridae. On the other hand, three differences of note in Table 3 are that unirnaviruses seem to have longer 5′ nontranslated regions than do the other viruses; amalgaviruses have much longer regions of ORF1–ORF2 overlap than do the other viruses; and unirnaviruses have ORF2 in the −1 frame relative to ORF1, instead of the +1 frame as do the other viruses. This last characteristic is discussed additionally below.
Table 3.
Other properties of amalgaviruses, ZbV-Z, and unirnaviruses compared with Totiviridae family members
| Virus length (aa): | Genus | GenBank no. | Genome length (bp) | Protein ORF1a | ORF1/ORF2b |
|---|---|---|---|---|---|
| Blueberry latent virus | Amalgavirus | HM029246 | 3431 | 375 | 1054 |
| Rhododendron virus A | Amalgavirus | HQ128706 | 3427 | 404 | 1077 |
| Southern tomato virus | Amalgavirus | EF442780 | 3437 | 377 | 1062 |
| Vicia cryptic virus M | Amalgavirus | EU371896 | 3434 | 394 | 1057 |
| Zygosaccharomyces bailii virus Z | “Zybavirus”c | KU200450 | 3160 | 290 | 1012 |
| Alternaria longipes dsRNA virus 1 | “Unirnavirus”d | KJ817371 | 3415 | 394 | 997 |
| Beauveria bassiana RNA virus 1/1 | “Unirnavirus” | LN610699 | 3218 | 315 | 926 |
| Beauveria bassiana RNA virus 1/2 | “Unirnavirus” | KM233415 | 3173 | 315 | 926 |
| Penicillium janczewskii Beauveria bassiana-like virus 1 | “Unirnavirus” | KT601106 | 2890e | 317 | 926 |
| Ustilaginoidea virens RNA virus M | “Unirnavirus” | KJ101567 | (2714)e | (137)e | (745)e |
| Ustilaginoidea virens unassigned | “Unirnavirus” | KR106133 | 2903 | 314 | 932 |
| RNA virus HNND-1 | |||||
| Saccharomyces cerevisiae virus L-A | Totivirus | J04692 | 4579 | 680 | 1505 |
| Trichomonas vaginalis virus 1 | Trichomonasvirus | HQ607513 | 4684 | 678 | 1429 |
| Helminthosporium victoriae virus 190S | Victorivirus | U41345 | 5179 | 772 | 1607b |
| Leishmania RNA virus 1 | Leishmaniavirus | M92355 | 5284 | 741 | 1595 |
| Giardia lamblia virus | Giardiavirus | L13218 | 6277 | 886 | 1870 |
Encoded protein starting with the first Met codon in ORF1. Ustilaginoidea virens RNA virus M (UvRV-M) is an exception as described below.
Encoded protein starting with the first Met codon in ORF1 and incorporating the +1 or −1 PRF predicted for each virus. UvRV-M is an exception as described below. For Helminthosporium victoriae virus 190S, in which the RdRp is translated as a separate product via a stop/restart mechanism (Li et al., 2015), the shown CP/RdRp length is the sum of the CP and RdRp lengths.
Newly proposed genus name (this study)
Name of proposed genus suggested by Kotta-Loizou et al. (2015)
The reported genome sequence of these viruses appears to be truncated at the 5′ end of plus strand, and their listed lengths may thus be artifactually low. In addition, ORF1 extends to the 5′ end, and the listed lengths reflect that.
As a further examination of whether these three taxa might warrant assignment to the same family, we expanded the phylogenetic analyses to include RdRp sequences from other mono- and bisegmented dsRNA viruses. A number of unclassified mono- and bisegmented dsRNA viruses from a variety of different hosts have been described in recent years (e.g., penaeid shrimp infectious myonecrosis virus (Poulos et al., 2006)), many of which probably do not warrant assignment to existing taxonomic families, and we added their RdRps to the current analyses. We also added RdRp sequences representing other approved species in families Totiviridae, Partitiviridae, Megabirnaviridae, and Botybirnaviridae. The results provided new evidence that the RdRp of ZbV-Z/412 is most closely related to those of the plant amalgaviruses (Fig. 3B) and, combined with other findings such as shown in Table 3, that assignment of proposed genus Zybavirus to family Amalgaviridae appears reasonable. The results also provided new evidence that the RdRps of unirnaviruses, though related to those of plant amalgaviruses, are less related to them than is that of ZbV-Z/412 (Fig. 3B), making assignment of proposed genus Unirnavirus to family Amalgaviridae appear less well supported at the current stage of characterizing these different viruses.
To extend the phylogenetic analyses, we attempted to compare the ORF1 product sequences of plant amalgaviruses, ZbV-Z, and unirnaviruses with those of other mono- and bisegmented dsRNA viruses. The ORF1 product sequences, however, are more divergent than the RdRp sequences, limiting their simple utility in this regard. For example, when used as query in NCBI PSI-BLAST searches, the putative CP of ZbV-Z/412 did not identify either plant amalgavirus or unirnavirus ORF1 products as homologs. Similarly, when used as query in NCBI PSI-BLAST searches, any of the unirnavirus ORF1 products identified the other unirnavirus ORF1 products as homologs but not the ZbV-Z/412 or plant amalgavirus ORF1 products, and any of the plant amalgavirus ORF1 products identified the other plant amalgavirus ORF1 products as homologs but not the ZbV-Z/412 or unirnavirus ORF1 products. On the other hand, sequence identity scores from pairwise comparisons of the ORF1 products of these viruses are consistent with their assignment to three distinct genera (Fig. 4).
Recent studies have identified yet another distinct taxon of unclassified dsRNA mycoviruses with RdRps that are phylogenetically related to those of the plant amalgaviruses, but in this case viruses with two genome segments. These bisegmented viruses include Cryphonectria parasitica bipartite mycovirus 1 from C. parasitica isolate 09269, Curvularia thermal tolerance virus from C. protuberata (CTTV, the prototype of this taxon), Fusarium graminearum dsRNA mycovirus 4 from F. graminearum isolate DK3, Rhizoctonia fumigata mycovirus from R. fumigata isolate C-314, and Rhizoctonia solani dsRNA virus 1 from R. solani isolate AG-1-IA-B275 (Marquez et al., 2007; Yu et al., 2009; Zheng et al., 2013), and possibly also Gremmeniella abietina RNA virus 6 from G. abietina isolate P3-7 and Heterobasidion RNA virus 6 from H. parviporum isolate 195-12, for which only single genome segments have been identified to date (Botella et al., 2015; Vainio et al., 2012). These viruses were also included in the analysis for Fig. 3B, which provides further evidence that they constitute a distinct taxon and that their RdRps are most closely related to those of unirnaviruses and next most closely related to those of ZbV-Z and plant amalgaviruses. Koloniuk et al. (2015) in particular have suggested that these bisegmented mycoviruses might also warrant assignment to family Amalgaviridae. Based on the RdRp-based phylogenetic tree in Fig. 3B, however, assignment of this taxon to family Amalgaviridae does not appear to be well supported at present, similarly to the case for unirnaviruses.
4.2. Putative slippery sequences for +1 PRF in ZbV-Z and the plant amalgaviruses
ORF2 overlaps ORF1 in the +1 frame in ZbV-Z (this study) as well as in the four plant amalgaviruses described to date (Liu and Chen, 2009; Martin et al., 2011; Sabanadzovic et al., 2009, 2010) (Table 2). Moreover, sequences similar to the motif for +1 PRF that was first characterized in the PA-X-encoding RNA segment of influenza A viruses (Firth et al., 2012; Jagger et al., 2012; Shi et al., 2012) are found also in the ORF1–ORF2 overlap regions of ZbV-Z/412 (this study) and three of the plant amalgaviruses (Firth et al., 2012) (Fig. 5A). Between ZbV-Z/412 and these three plant amalgaviruses, we can propose to define their consensus motif for +1 PRF as NYU_UUU_CGN (Y = pyrimidine; N = potentially any nucleotide; underlines, ORF1 codon boundaries;), where the component sequence CGN is a rare Arg codon. This proposed consensus thus corresponds well with the motif originally defined for influenza A viruses (Fig. 5A), including the presence of a rare Arg codon at the demonstrated or proposed site of ribosomal slippage and the capacity for the P-site tRNA (anticodon 3′-AAA on codon UUU) to remain engaged after +1 slippage (moves forward to codon UUC) (Firth et al., 2012; Jagger et al., 2012).
Fig. 5.
Putative PRF motifs. Sequences encompassing the putative motifs for PRF are shown for two different sets of viruses: plant amalgaviruses and ZbV-Z/412, which are predicted to undergo +1 PRF (forward arrows) for ORF1/ORF2 fusion product translation (A), and unirnaviruses, which are predicted to undergo −1 PRF (reverse arrows) for ORF1/ORF2 fusion product translation (B). The codon boundaries shown as open spaces are those for ORF1. Green text, sequences between the upstream flanking stop codon of ORF2 and the downstream flanking stop codon of ORF1; these stop codons are outside the range of sequences shown in some cases. Magenta boxes, stop codons flanking the 3′ end of ORF1; cyan boxes, stop codons flanking the 5′ end of ORF2. An underline indicates the proposed +1 or −1 slippery sequence motif in each sequence. See text for virus abbreviations and GenBank numbers. Abbreviations used here do not include strain designations. (A) The sequences are aligned without gaps relative to the putative +1 slippery sequence motif. Rare Arg codons (CGN) are highlighted with gray shading. The region encompassing the motif for +1 PRF from influenza A virus segment 3 (Firth et al., 2012; Jagger et al., 2012; Shi et al., 2012; GenBank CY157652) is shown at the top of this panel. A possible +1 slippery sequence in STV previously suggested by Sabanadzovic et al. (2009) is overlined. (B) The sequences are aligned without gaps relative to the ORF1 stop codon. Both possible −1 slippery sequence motifs for UvURV are underlined.
Southern tomato virus (STV) is notably the only characterized plant amalgavirus that has not been discussed in either the preceding paragraphs or the analysis by Firth et al. (2012). The ORF1–ORF2 overlap region of multiple strains of STV (also see GenBank EU413670, KT438549, KT634055, and KT852573) lacks a sequence that strictly matches the consensus motif for +1 PRF per Firth et al. (2012). It does, however, include a rare Arg codon (CGU) that is present in a somewhat similar sequence context as in the other plant amalgaviruses and ZbV-Z/412 (Fig. 5A). Of course, the major difference in STV is that the central codon in the motif is AGG, not UUU. Interestingly, however, this STV sequence might similarly allow the P-site tRNA (anticodon 3′UCC on codon AGG) to remain engaged after +1 slippage (moves forward to codon GGC, with G:U pairing in the first position). Other RNA or protein sequences that may be essential for or otherwise modulate the proposed +1 PRF activity in these viruses remain to be identified.
4.3. Putative slippery sequences for −1 PRF in unirnaviruses
The mechanism for translating RdRp from ORF2 of the viruses in proposed genus Unirnavirus has also remained unclear. By examining their genome sequences, however, we found that in five of the six unirnavirus strains described to date, ORF2 overlaps ORF1 by 10–73 nt in the −1 frame (Table 2, Fig. 5B). This finding seems to have been overlooked in some previous reports, possibly due to defining the upstream end of ORF2 by its first Met codon, rather than by its upstream flanking stop codon as is more informative when considering possible PRF mechanisms for translating a downstream ORF. Moreover, in the remaining one of these viruses described to date, UvURV, a single nucleotide substitution within the stop codon that currently defines the upstream end of ORF2 would bring this virus in line with the others, allowing ORF2 to overlap ORF1 by 40 nt in the −1 frame (Table 2, Fig. 5B). We therefore predict that the reported UvURV sequence (GenBank KR106133) contains at least this one error, or one or more other error with the same consequence. This ORF1–ORF2 overlap possibly common to all unirnaviruses then indicates the substantive possibility of −1 PRF as the mechanism for translating the RdRp as part of an ORF1/ORF2-encoded fusion protein.
The classical slippery sequence for −1 PRF is X_XXY_YYZ, where XXX is any three of the same nucleotide, although several deviations such as GGA are tolerated; YYY is AAA or UUU (do not confuse with use of Y for pyrimidine in other cases); Z is A, C, or U; and underlines indicate codon boundaries for the upstream ORF (Firth et al., 2012). Notably, in all six unirnavirus strains described to date, a matching sequence for this −1 PRF slippery motif is found immediately or soon before the ORF1 stop codon: G_GAU_UUU in AlDRV1, BbRV1/1, BbRV1/2, and UvRV-M; G_GAU_UUC in PjBbLV1; and U_UUA_AAC or G_GAU_UUA in UvURV (Fig. 5B). Moreover, other matching sequences are not identifiable in the region of ORF1–ORF2 overlap in any of these viruses, and in the case of BbRV1/1, BbRV1/2, and UvRV-M, the region of ORF1–ORF2 overlap is so short (only 10 nt) that other possible −1 PRF slippery motifs are almost out of the question. The finding of such putative −1 slippery sequences properly positioned within the ORF1–ORF2 overlap region thus strongly supports the argument for −1 PRF as the mechanism for translating the unirnavirus RdRp. Different members of family Totiviridae are known to use different PRF or other mechanisms for RdRp expression from ORF2 (e.g., see Li et al., 2015; Parent et al., 2013), such that the different PRF mechanism proposed for unirnaviruses (−1) relative to plant amalgaviruses and ZbV-Z/412 (+1) should not automatically consign these viruses to two different families, though it does represent evidence for their divergence.
4.4. Ongoing studies of ZbV-Z
One important remaining question about the approved and proposed members of family Amalgaviridae is whether any form bona fide virions. Such particles have not been visualized to date for plant amalgaviruses, but sometimes such cryptic viruses of plants accumulate particles in small numbers that can be difficult to detect (e.g., see Tzanetakis et al., 2008). Interestingly, Krupovic et al. (2015) have recently reported that the ORF1 translation product (possible CP) of plant amalgavirus STV is homologous to the nucleocapsid proteins of certain negative-strand RNA viruses. This finding raises the possibility that the plant amalgaviruses might form filamentous nucleocapsids instead of icosahedral virus-like particles (Krupovic et al., 2015). Virions have also failed to be visualized to date for the members of proposed genus Unirnavirus. Notably, Schmitt and Neuhausen (1994) have reported the presence of virus-like particles enriched for the ~3-kbp genome and the ~35-kDa putative CP of ZbV-Z/412, following fractionation on sucrose gradients. These authors have moreover visualized virus-like particles from Z. bailii 412 that appear isometric. It is important to note, however, that Z. bailii 412 is also infected with a putative totivirus (ZbV-L/412) and its toxin-expressing M-satellite RNA (Radler et al., 1993; Schmitt and Neuhausen, 1994; Weiler et al., 2002) (see Fig. 2). We therefore consider it possible that the isometric particles shown by Schmitt and Neuhausen (1994) might represent this totivirus, and not ZbV-Z/412. Further efforts to purify and characterize ZbV-Z/412 virions are in progress in our laboratory. CTTV, one of the bisegmented dsRNA viruses with an RdRp phylogenetically related to those of amalgaviruses (Koloniuk et al., 2015; Yu et al., 2009) has also been reported to form isometric virions (Marquez et al., 2007).
Confirmation that translation of the predicted ORF1/ORF2-encoded fusion protein (putative CP/RdRp) of ZbV-Z/412 involves +1 PRF at a particular sequence is another focus of current efforts in our laboratory. We hope that by purifying ZbV-Z/412 virions, we will be able to visualize this protein by gel and then subject it to tandem mass spectrometry to identify one or more of its constituent peptides that crosses the putative CP/RdRp junction. Indeed, we have recently used this same approach to confirm the site of −2 PRF during translation of the ORF1/ORF2-encoded CP/RdRp of Trichomonas vaginalis virus 1, a family Totiviridae member (Parent et al., 2013).
Supplementary Material
Highlights.
The nucleotide sequence of ZbV-Z has been redetermined and amended.
ORF2 overlaps ORF1 in the +1 frame in the amended sequence.
The ORF1–ORF2 overlap region includes a putative +1 slippery sequence.
The encoded RdRp is phylogenetically related to those of plant amalgaviruses.
We propose ZbV-Z as type species of new genus Zybavirus in family Amalgaviridae.
Acknowledgments
We thank Daniela Silva-Ayala for helpful comments on the manuscript, and we again thank Drs. Manfred J. Schmitt and Frank Breinig (Saarland University, Saarbrücken, Germany) for providing us an initial culture of Z. bailii 412. We also thank the anonymous reviewers of our original manuscript for their very helpful suggestions about both taxonomic classification and frameshifting signals. D.D. completed her work on this project as part of her M.S. training program in Biochemistry and Molecular and Cellular Biology, at the University of Namur, Namur, Belgium; she was supported in part by a “Bourse de mobilité internationale” from the University of Namur and a “Fond d’Aide à la Mobilité Etudiante” from the French community in Belgium. M.V. completed his work on this project as part of his Ph.D. training program in Molecules, Cells, and Organisms at Harvard University, Cambridge, MA, USA; he was supported in part by NIH training grant T32-GM007598 and by support from the Department of Molecular Cellular and Biology at Harvard University. M.L.N. was supported in part for this project by a subcontract from NIH grant R01 GM033050.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Botella L, Vainio EJ, Hantula J, Diez JJ, Jankovsky L. Description and prevalence of a putative novel mycovirus within the conifer pathogen Gremmeniella abietina. Arch Virol. 2015;160:1967–1975. doi: 10.1007/s00705-015-2456-5. [DOI] [PubMed] [Google Scholar]
- Castillo A, Cottet L, Castro M, Sepúlveda F. Rapid isolation of mycoviral double-stranded RNA from Botrytis cinerea and Saccharomyces cerevisiae. Virol J. 2011;8:38. doi: 10.1186/1743-422X-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts RHA, Livieratos IC. A rapid method for sequencing the 50- and 30-termini of dsRNA viral templates using RLM-RACE. J Phytopathol. 2003;151:525–527. [Google Scholar]
- Firth AE, Jagger BW, Wise HM, Nelson CC, Parsawar K, Wills NM, Napthine S, Taubenberger JK, Digard P, Atkins JF. Ribosomal frameshifting used in influenza A virus expression occurs within the sequence UCC_UUU_CGU and is in the +1 direction. Open Biol. 2012;2:120109. doi: 10.1098/rsob.120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Zhang T, Luo C, Jiang D, Li G, Li Q, Hsiang T, Huang J. Prevalence and diversity of mycoviruses infecting the plant pathogen Ustilaginoidea virens. Virus Res. 2015;195:47–56. doi: 10.1016/j.virusres.2014.08.022. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koloniuk I, Hrabáková L, Petrzik K. Molecular characterization of a novel amalgavirus from the entomopathogenic fungus Beauveria bassiana. Arch Virol. 2015;160:1585–1588. doi: 10.1007/s00705-015-2416-0. [DOI] [PubMed] [Google Scholar]
- Kotta-Loizou I, Sipkova J, Coutts RHA. Identification and sequence determination of a novel double-stranded RNA mycovirus from the entomopathogenic fungus Beauveria bassiana. Arch Virol. 2015;160:873–875. doi: 10.1007/s00705-014-2332-8. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Dolja VV, Koonin EV. Plant viruses of the Amalgaviridae family evolved via recombination between viruses with double-stranded and negative-strand RNA genomes. Biol Direct. 2015;10:12. doi: 10.1186/s13062-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Havens WM, Nibert ML, Ghabrial SA. An RNA cassette from Helminthosporium victoriae virus 190S necessary and sufficient for stop/restart translation. Virology. 2015;474:131–143. doi: 10.1016/j.virol.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Lin Y, Zhang H, Zhao C, Liu S, Guo L. The complete genome sequence of a novel mycovirus from Alternaria longipes strain HN28. Arch Virol. 2015;160:577–580. doi: 10.1007/s00705-014-2218-9. [DOI] [PubMed] [Google Scholar]
- Liu W, Chen J. A double-stranded RNA as the genome of a potential virus infecting Vicia faba. Virus Genes. 2009;39:126–131. doi: 10.1007/s11262-009-0362-1. [DOI] [PubMed] [Google Scholar]
- Liu H, Fu Y, Xie J, Cheng J, Ghabrial SA, Li G, Peng Y, Yi X, Jiang D. Evolutionary genomics of mycovirus-related dsRNA viruses reveals cross-family horizontal gene transfer and evolution of diverse viral lineages. BMC Evol Biol. 2012a;12:91. doi: 10.1186/1471-2148-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Fu Y, Xie J, Cheng J, Ghabrial SA, Li G, Yi X, Jiang D. Discovery of novel dsRNA viral sequences by in silico cloning and implications for viral diversity, host range and evolution. PLoS One. 2012b;7:e42147. doi: 10.1371/journal.pone.0042147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez LM, Redman RS, Rodriguez RJ, Roossinck MJ. A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science. 2007;315:513–515. doi: 10.1126/science.1136237. [DOI] [PubMed] [Google Scholar]
- Martin RR, Zhou J, Tzanetakis IE. Blueberry latent virus: an amalgam of the Partitiviridae and Totiviridae. Virus Res. 2011;155:175–180. doi: 10.1016/j.virusres.2010.09.020. [DOI] [PubMed] [Google Scholar]
- Nerva L, Ciuffo M, Vallino M, Margaria P, Varese GC, Gnavi G, Turina M. Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res. 2015 doi: 10.1016/j.virusres.2015.10.028. in press. [DOI] [PubMed] [Google Scholar]
- Nibert ML, Ghabrial SA, Maiss E, Lesker T, Vainio EJ, Jiang D, Suzuki N. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. 2014;188:128–141. doi: 10.1016/j.virusres.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Parent KN, Takagi Y, Cardone G, Olson NH, Ericsson M, Yang M, Lee Y, Asara JM, Fichorova RN, Baker TS, Nibert ML. Structure of a protozoan virus from the human genitourinary parasite Trichomonas vaginalis. mBio. 2013;4:e00056–13. doi: 10.1128/mBio.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos BT, Tang KF, Pantoja CR, Bonami JR, Lightner DV. Purification and characterization of infectious myonecrosis virus of penaeid shrimp. J Gen Virol. 2006;87:987–996. doi: 10.1099/vir.0.81127-0. [DOI] [PubMed] [Google Scholar]
- Radler F, Herzberger S, Schönig I, Schwarz P. Investigation of a killer strain of Zygosaccharomyces bailii. J Gen Microbiol. 1993;139:495–500. doi: 10.1099/00221287-139-3-495. [DOI] [PubMed] [Google Scholar]
- Sabanadzovic S, Abou Ghanem-Sabanadzovic N, Valverde RA. A novel monopartite dsRNA virus from rhododendron. Arch Virol. 2010;155:1859–1863. doi: 10.1007/s00705-010-0770-5. [DOI] [PubMed] [Google Scholar]
- Sabanadzovic S, Valverde RA, Brown JK, Martin RR, Tzanetakis IE. Southern tomato virus: The link between the families Totiviridae and Partitiviridae. Virus Res. 2009;140:130–137. doi: 10.1016/j.virusres.2008.11.018. [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MJ, Neuhausen F. Killer toxin-secreting double-stranded RNA mycoviruses in the yeasts Hanseniaspora uvarum and Zygosaccharomyces bailii. J Virol. 1994;68:1765–1772. doi: 10.1128/jvi.68.3.1765-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J Virol. 2012;86:12411–12413. doi: 10.1128/JVI.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzanetakis IE, Price R, Martin RR. Nucleotide sequence of the tripartite Fragaria chiloensis cryptic virus and presence of the virus in the Americas. Virus Genes. 2008;36:267–272. doi: 10.1007/s11262-007-0186-9. [DOI] [PubMed] [Google Scholar]
- Vainio EJ, Hyder R, Aday G, Hansen E, Piri T, Doğmuş-Lehtijärvi T, Lehtijärvi A, Korhonen K, Hantula J. Population structure of a novel putative mycovirus infecting the conifer root-rot fungus Heterobasidion annosum sensu lato. Virology. 2012;422:366–376. doi: 10.1016/j.virol.2011.10.032. [DOI] [PubMed] [Google Scholar]
- Weiler F, Rehfeldt K, Bautz F, Schmitt MJ. The Zygosaccharomyces bailii antifungal virus toxin zygocin: cloning and expression in a heterologous fungal host. Mol Microbiol. 2002;46:1095–1105. doi: 10.1046/j.1365-2958.2002.03225.x. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Ghabrial SA, Nibert ML, Patterson JL, Wang CC. Totiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2012. pp. 639–650. [Google Scholar]
- Yu J, Kwon SJ, Lee KM, Son M, Kim KH. Complete nucleotide sequence of double-stranded RNA viruses from Fusarium graminearum strain DK3. Arch Virol. 2009;154:1855–1858. doi: 10.1007/s00705-009-0507-5. [DOI] [PubMed] [Google Scholar]
- Zheng L, Liu H, Zhang M, Cao X, Zhou E. The complete genomic sequence of a novel mycovirus from Rhizoctonia solani AG-1 IA strain B275. Arch Virol. 2013;158:1609–1612. doi: 10.1007/s00705-013-1637-3. [DOI] [PubMed] [Google Scholar]
- Zhu HJ, Chen D, Zhong J, Zhang SY, Gao BD. A novel mycovirus identified from the rice false smut fungus Ustilaginoidea virens. Virus Genes. 2015;51:159–162. doi: 10.1007/s11262-015-1212-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.