Abstract
FtsZ is a prokaryotic tubulin homolog that assembles into a ring at the future site of cell division. The resulting “Z ring” forms the framework for the division apparatus, and its assembly is regulated throughout the bacterial cell cycle. A highly dynamic structure, the Z ring exhibits continual subunit turnover and the ability to rapidly assemble, disassemble, and, under certain circumstances, relocalize. These in vivo properties are ultimately due to FtsZ’s capacity for guanosine triphosphate (GTP)-dependent, reversible polymerization. FtsZ polymer stability appears to be fine-tuned such that subtle changes in its assembly kinetics result in large changes in the Z ring structure. Thus, regulatory proteins that modulate FtsZ’s assembly dynamics can cause the ring to rapidly remodel in response to developmental and environmental cues.
Keywords: cytokinesis, cytoskeleton, polymerization, cell cycle, tubulin
INTRODUCTION
Seemingly static cellular structures can in reality be remarkably dynamic, undergoing continual regeneration of constituent parts. Subunit turnover in the eukaryotic cytoskeleton allows cells to rapidly reorganize in response to environmental and developmental signals. Populations of microtubules and actin filaments are maintained through continual and carefully balanced assembly and disassembly (24, 107). Bacteria have cytoskeletal elements that are related to actin and tubulin (137), and accumulating evidence shows that the prokaryotic cytoskeleton is also a dynamic system in which multiple inputs allow for precisely tuned and rapid responses to a changing environment.
The dynamic assembly of FtsZ, the prokaryotic homolog of tubulin, is fundamental to the regulation of bacterial cell division. In this review we focus on the molecular basis of FtsZ assembly and its modulation during cell growth and development. FtsZ is conserved throughout bacteria and archaea; it also exists in chloroplasts and in the mitochondria of certain eukaryotes (37, 82, 101, 120). In bacteria, FtsZ forms a macromolecular structure called the Z ring that provides the framework for the division apparatus and determines the site of cytokinesis. In vitro, the stability of FtsZ polymers, like that of microtubules, is precisely tuned. FtsZ polymerizes in the presence of GTP but tends to depolymerize again once the bound GTP has been hydrolyzed. In vivo, FtsZ assembly is also transient. Subunits in the Z ring are continually exchanged with those in the cytoplasm. The constant balance between assembly and disassembly means that small changes in FtsZ polymerization dynamics are able to trigger large and rapid changes in the location or morphology of FtsZ structures in the cell. Proteins that induce such changes dictate the timing and location of cytokinesis.
In this review we discuss (a) Z ring assembly and dynamics in vivo, (b) FtsZ polymer assembly in vitro, and (c) the counterbalancing effects of proteins known to regulate FtsZ assembly. Available data suggest that the mechanisms governing Z ring formation are fundamentally conserved. We have highlighted any differences in FtsZ assembly between species where they exist.
PART I: Z RING DYNAMICS THROUGHOUT THE CELL CYCLE
During different stages of the cell cycle FtsZ exhibits a series of different behaviors: assembly to form the Z ring at midcell, maintenance of the ring through continual subunit turnover, and constriction and disassembly of the ring. The transitions between Z ring growth, maintenance, and constriction are likely to be regulated by factors that influence FtsZ’s assembly dynamics. However, the precise nature of the signals driving these transitions remains unclear. In eukaryotes DNA replication, chromosome segregation, and cell division occur in strict succession, whereas in rapidly growing prokaryotes these processes often overlap (26). Nonetheless, in all organisms cytokinesis must be tightly coupled to the cell cycle so that each daughter cell receives a complete copy of the genome.
A strong correlation exists between Z ring behavior and cell cycle progression (Figure 1). Z ring formation begins after the initiation of DNA replication and the separation of replication origins, and it roughly coincides with nucleoid segregation (23, 47, 67, 110, 117). Data suggest that FtsZ assembly may start at a single point on the inner surface of the cell membrane and then rapidly extend bidirectionally to form an arc and then a closed circle (3). This ring serves as the framework for assembly of other cell division proteins (82, 120). Although the ring maintains its shape and size for a large portion of the cell cycle (23, 146), individual FtsZ molecules in the ring are rapidly exchanged with cytoplasmic FtsZ (130). When an unknown signal triggers cytokinesis, new cell wall is deposited and the Z ring contracts along with the cytoplasmic membrane (1, 10, 62). By the end of the septation process the Z ring has disassembled (130). Below we discuss the dynamics of Z ring assembly and its coordination with events in the cell cycle.
Figure 1.
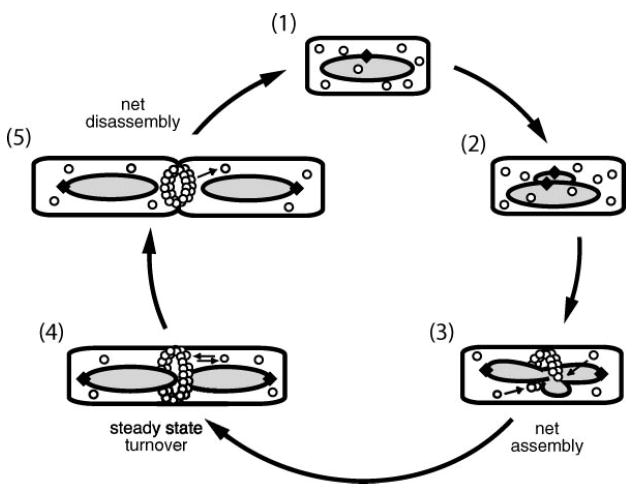
The bacterial division cycle. FtsZ assembly is coordinated with DNA replication and segregation. Step 1: In newborn cells FtsZ (○) is unassembled. A circular chromosome with a single origin of replication (◆) is located at midcell. Steps 2–4: After chromosome replication initiates, the polymerase machinery remains at midcell, but origins of replication separate and move to opposite poles of the cell. Once replication is complete, the condensed chromosomes separate, leaving a nucleoid-free space. Step 3: Z ring formation coincides with chromosome segregation. Assembly starts from a single point at midcell and extends bidirectionally. Step 5: During cytokinesis the Z ring constricts at the leading edge of the invaginating septum. Small arrows indicate the assembly and disassembly of FtsZ subunits.
Factors Affecting Z Ring Initiation and Localization
Z ring formation establishes the location of the future division site and is an integral part of the temporal regulation of cytokinesis. The initiation of FtsZ assembly must be tightly controlled both temporally and spatially to prevent aberrant septation. Z ring formation depends on at least three factors: FtsZ concentration, the initiation of DNA replication, and the presence or absence of the nucleoid mass.
A MINIMUM FtsZ CONCENTRATION IS NECESSARY BUT NOT SUFFICIENT FOR Z RING FORMATION
FtsZ assembles in vitro in a concentration-dependent manner (see Part II: Biochemical Properties of FtsZ Protein, below), which suggests that FtsZ levels in a cell might control the timing of Z ring formation. In this model FtsZ concentration in newborn cells would be too low to support assembly but would gradually increase during the course of the cell cycle and reach a critical concentration immediately prior to Z ring formation. A minimum FtsZ concentration is in fact necessary for division in Escherichia coli and for Z ring assembly in Bacillus subtilis (17, 61), but under most circumstances fluctuations in FtsZ concentration are not sufficient to explain the timing of ring formation.
Both E. coli and B. subtilis can modulate transcription of the ftsZ operon (4, 35, 55, 140), but FtsZ protein levels remain constant throughout the cell cycle of both organisms during steady state growth (146). Altering FtsZ levels is not sufficient to change the timing of medial ring formation. Lowering ftsZ expression below wild-type levels does not change the timing of cytokinesis in E. coli, though cell size increases slightly (102). Increasing FtsZ levels twofold in B. subtilis does not alter the frequency or timing of medial Z ring formation (146), though it does lead to an increase in the formation of polar Z rings. Similarly, two- to sevenfold increases in FtsZ levels in E. coli lead to an increase in the frequency of polar septation (145). These data suggest that although high FtsZ expression levels do not change medial Z ring formation, they can overcome a division inhibitor that normally prevents assembly at the cell poles. (In E. coli and B. subtilis this inhibitor is most likely the MinCD complex; see Part III: Proteins that Modulate FtsZ Assembly.) In support of this model, during sporulation in B. subtilis, increased FtsZ expression is required to initiate Z ring formation at previously suppressed locations adjacent to the cell poles (7, 36, 38).
Unlike B. subtilis and E. coli, Caulobacter crescentus modulates FtsZ levels in a cell cycle–dependent manner. In this organism transcriptional regulation and proteolysis ensure that FtsZ is undetectable in swarmer cells, which do not initiate DNA replication or division (57, 111), but is highly expressed prior to cytokinesis in the reproductively competent stalk cells (57, 109, 121). Following cytokinesis FtsZ is cleared from stalk cells (109). Nonetheless, ectopic expression of ftsZ in swarmer cells is not sufficient to drive medial Z ring formation (111). Ectopic FtsZ expression can only stimulate ring formation after cell differentiation and the subsequent initiation of DNA replication (110). Thus, it is ultimately cell cycle regulation that dictates the initiation of Z ring formation even when elaborate regulation of FtsZ levels exists.
REPLICATION INITIATION IS NECESSARY FOR POSITIONING THE Z RING AT MIDCELL
Z ring formation initiates after the onset of DNA synthesis and the separation of newly replicated origin DNA, which suggests that assembly might be triggered by events at the start of chromosome replication (23, 47, 110, 117). However, blocking the initiation of DNA replication does not inhibit Z ring formation in either E. coli or B. subtilis (47, 131). The terminal phenotype of B. subtilis DNA replication initiation mutants is a cell in which the Z ring is located off-center, adjacent to the medially positioned, unreplicated nucleoid (47). Similarly, a Z ring forms adjacent to the nucleoid in E. coli cells in which either DNA replication or nucleoid segregation are defective (131, 132).
While DNA replication is not essential for FtsZ assembly in E. coli and B. subtilis, correct positioning of the Z ring at midcell does require replication initiation, but not elongation of the replication fork. Permitting DNA replication in germinating B. subtilis spores, but preventing elongation with a thymine auxotroph, leads to degradation of the origin region and the correct positioning of a medial Z ring over the unreplicated nucleoid (47, 117). Because the FtsZ ring forms proximal to the previous location of the origin of replication, and because replication initiation is required for proper ring localization, it is tempting to speculate that the same machinery that recruits the origin of replication and the polymerase machinery to midcell also helps to unmask a preferred FtsZ nucleation site at midcell. In support of this idea, the polar placement of the division septum during sporulation in B. subtilis is partially dependent on the anchoring of the origin region at the cell pole (41).
LOCAL INHIBITION OF Z RING FORMATION BY THE UNSEGREGATED NUCLEOID
The correlation between Z ring formation and the separation of the bacterial nucleoid has led to the long-standing hypothesis that the bacterial nucleoid is a determinant in the timing and placement of the division septum [the nucleoid occlusion model (97, 149)]. According to this model, cell division can take place at any point along the length of a cell, but early in the cell cycle the presence of the unsegregated nucleoid prevents septation at midcell. Later, DNA segregation reveals a small, nucleoid-free zone at midcell that is able to support assembly of the division apparatus. Z ring formation in the nucleoid-free space adjacent to the cell poles is prevented by the presence of the MinCD division inhibitors (see Part III: Proteins that Modulate FtsZ Assembly, below) (155).
The nucleoid appears to play a role in preventing aberrant Z ring formation, but in both E. coli and B. subtilis its presence alone is not sufficient to dictate the precise spatiotemporal regulation of FtsZ assembly. Z ring formation is clearly influenced by the location of the nucleoid under some circumstances. When DNA replication or partitioning has been blocked, the chromosome remains at midcell, and nonmedial Z rings form adjacent to the nucleoid (47, 131, 132). In addition, when chromosome partioning is blocked in the absence of the Min proteins, the Z ring in E. coli is free to form at any position along the nucleoid-free space (155). Under certain circumstances, however, the position of the nucleoid coincides with that of the Z ring and of cytokinesis. During sporulation in B. subtilis the nucleoid is bisected by the asymmetrically positioned septum and the DNA must subsequently be pumped across the septum into the forespore (128, 150). Moreover, Z rings can form directly over the nucleoid in cells with mutations in chromosome condensation genes (smc in B. subtilis and mukB in E. coli) (14, 40, 156). One potential explanation is that Z ring formation and septation can occur in regions of the nucleoid that are relatively less dense. If this is indeed the case, this would explain why medial FtsZ ring formation occurs only after degradation of the DNA adjacent to the origin of replication in germinating B. subtilis spores blocked for DNA replication (117).
Maintenance of the Z Ring as a Dynamic Framework
Once formed, the Z ring is present for a significant portion of the cell cycle [e.g., 85% of the mass doubling period in B. subtilis cells growing in Luria broth medium] (23, 146). In B. subtilis the proportion of cells with Z rings increases with increasing growth rate, indicating that the Z period (the time the Z ring is present in a given cell) remains relatively constant even when doubling time decreases (67, 146). The prolonged duration of the Z period suggests there may be a minimum time required for proper assembly of the entire division apparatus.
During the Z period, although no gross changes in the Z ring can be detected by immunofluorescence, the ring is extremely dynamic, with subunit turnover on par with that of the eukaryotic cytoskeleton (103). For example, the Z ring can disappear and reassemble within minutes after shifting cells expressing a heat-sensitive ftsZ allele (ftsZ84) into and out of the nonpermissive temperature (2). More recently, the halftime for replacement of FtsZ subunits in the E. coli cytokinetic ring has been found to be between 8 and 30 sec, as indicated by fluorescence recovery after photobleaching (FRAP) experiments on cells expressing FtsZ-GFP (green fluorescent protein) [(130); H.P. Erickson, personal communication]. Similar fluorescence recovery rates are observed for GFP fused to ZipA, a protein associated with the Z ring in E. coli (130), which hints that the entire division machinery might also be continually turning over. The fluorescence intensity of the ring before and after FRAP shows that only ~30% of the FtsZ in a cell is in the ring at any given time, suggesting that a large pool of soluble FtsZ is available for exchange. In eukaryotic cells the detection of large pools of unassembled actin and tubulin prompted successful searches for proteins that regulate polymerization.
This rapid subunit turnover requires energy but does not appear to be critical for normal cytokinesis. Turnover rates of FtsZ in the ring appear to correspond to FtsZ’s in vitro ability to hydrolyze GTP, but lowering the hydrolysis rate by an order of magnitude is not lethal to the cell. For example, FtsZ84 protein hydrolyzes GTP in vitro at rates 10 times slower than wild-type FtsZ (20, 115), and FRAP experiments conducted at the permissive temperature indicate that recovery rates for FtsZ in ftsZ84 cells are also approximately 10 times slower than in wild type. Nonetheless, cell division still occurs with no apparent defect at these permissive temperatures. Many other FtsZ mutations also dramatically lower GTP hydrolysis rates without preventing cytokinesis (130). Thus, cells appear able to tolerate considerable variation in the subunit turnover rates of the Z ring.
Remodeling the Z Ring
If the rapid turnover of FtsZ subunits within the Z ring is not essential for cytokinesis under normal growth conditions, why does it occur? One function may be to allow cells to disassemble or remodel the division apparatus in response to DNA damage, starvation, or other environmental stresses. We review two examples below.
DNA damage induces the SOS response in bacteria, during which cell division is prevented until the DNA has been repaired (68). In E. coli, DNA damage results in the expression of SulA, which causes the Z ring to be dismantled (11, 39). Once the DNA lesions are repaired, SulA levels rapidly decline (88, 89), permitting FtsZ to once again assemble and division to resume.
Z ring remodeling also plays an important role during development in B. subtilis. In response to cell crowding and nutritional deprivation, B. subtilis produces environmentally resistant endospores [for reviews, see (15)]. A key step in sporulation is the switch from binary fission to polar septation (32, 63). Polar septation is preceded by the relocalization of FtsZ from midcell to positions adjacent to both cell poles (62). The shift in Z localization involves the transformation of the medial ring into a spiral that extends the length of the cell and then coalesces into rings at both cell poles (7). [The choice of which ring to use is largely stochastic (127).] Formation of the spiral depends both on an increase in FtsZ expression levels and on the expression of SpoIIE, a protein that colocalizes with FtsZ during sporulation and may help stabilize the polymer (5, 58, 64, 77, 153). Intriguingly, if sporulating cells that have not yet divided are given fresh nutrients, FtsZ can relocalize again from the cell poles to midcell (7), indicating that the medial site is still available.
Z Ring Constriction
When a cell divides, the Z ring constricts at the leading edge of the invaginating septum (10). There is limited evidence as to FtsZ’s precise function during this process. At minimum, FtsZ is needed to maintain the medial localization of the other division proteins. To do this, the FtsZ framework must be able to retain its coherence despite an ever-shrinking structure. It has also been suggested that FtsZ might generate the force that directs cytokinesis, similar to the actomyosin ring in eukaryotes (31).
Cytokinesis occurs in two discrete stages: the formation of an initial, small invagination at the site of the septum, followed by the completion of cell division. FtsZ is necessary throughout both stages. If at any time during cytokinesis conditional alleles of ftsZ are shifted to nonpermissive temperatures, the Z ring disappears, all other proteins in the division apparatus delocalize, and the septum ceases to invaginate (2, 134). If invagination has not yet begun, cells are left with smooth walls, indicating that septum formation cannot begin without FtsZ. In contrast, when conditional alleles in later-acting genes such as ftsI (which encodes PBP3) are used to block division, cytokinesis ceases only after the initial invagination step (134). Qualitative changes appear to occur after constriction has begun. When heat-sensitive ftsZ84 cells are raised to the nonpermissive temperature, Z rings rapidly disassemble. After returning to the permissive temperature, FtsZ can reoccupy some sites but not those where invagination of the cell wall is visible (2).
The molecular mechanism of Z ring constriction during septum formation remains unclear. One model suggests that as the diameter of the ring grows smaller, FtsZ subunits are released from the ring. Alternatively, filaments might be sliding against each other without disassembling, as occurs in the actomyosin ring in eukaryotes (13). Regardless of the mechanism of constriction, once cytokinesis is complete, the vast majority of FtsZ in E. coli and B. subtilis appears to be cytoplasmic, although FtsZ foci are occasionally observed at cell poles (62). These polar foci of FtsZ presumably reflect the remaining presence of potential FtsZ binding sites at these locations.
PART II: BIOCHEMICAL PROPERTIES OF FtsZ PROTEIN
Z ring initiation, maintenance, and constriction are regulated at least in part by the modulation of FtsZ assembly dynamics. The investigation of tubulin polymerization in vitro has provided invaluable insights into microtubule assembly dynamics and function in vivo. Similarly, understanding FtsZ behavior in vivo will require the detailed analysis of FtsZ polymer assembly and structure in vitro. Because FtsZ dynamics have been investigated and interpreted in the context of the eukaryotic cytoskeleton, we first review the mechanisms behind microtubule and actin assembly and disassembly. We next describe the structure of FtsZ subunits and polymers. Finally, we discuss the dynamic behavior that FtsZ exhibits in vitro and the possible molecular mechanism behind those dynamics.
Eukaryotic Cytoskeletal Polymers: Paradigms for Nucleotide-Dependent, Reversible Assembly
In eukaryotic cells, microtubules continually grow and shrink, probing the cytoplasm to locate disperse targets and to build and maintain the mitotic spindle. Tubulin’s GTP hydrolysis provides the energy for microtubule polymer dynamics; it is the continual input of energy that ultimately allows the cell to rapidly rearrange its cytoskeleton [reviewed in (24)].
The reversibility of microtubule polymerization is built into the tubulin subunits’ inherent assembly and hydrolysis properties. Tubulin is a stable heterodimer consisting of highly homologous α and β subunits, both of which bind GTP (Figure 2b). The GTP on α-tubulin is sandwiched between the subunits of the dimer and does not hydrolyze or exchange with nucleotide in solution. The nucleotide bound to the β subunit can both exchange and hydrolyze. For the remainder of the paper, when referring to tubulin, only the β-tubulin nucleotide binding site is discussed. When β-tubulin is bound to GTP, the protein assembles into a microtubule comprised of 13 straight protofilaments. After assembly the GTP is hydrolyzed and the resulting guanosine diphosphate (GDP)-containing microtubule is destabilized. An analogous situation occurs for actin polymerization with adenosine triphosphate (ATP) [reviewed in (108)].
Figure 2.
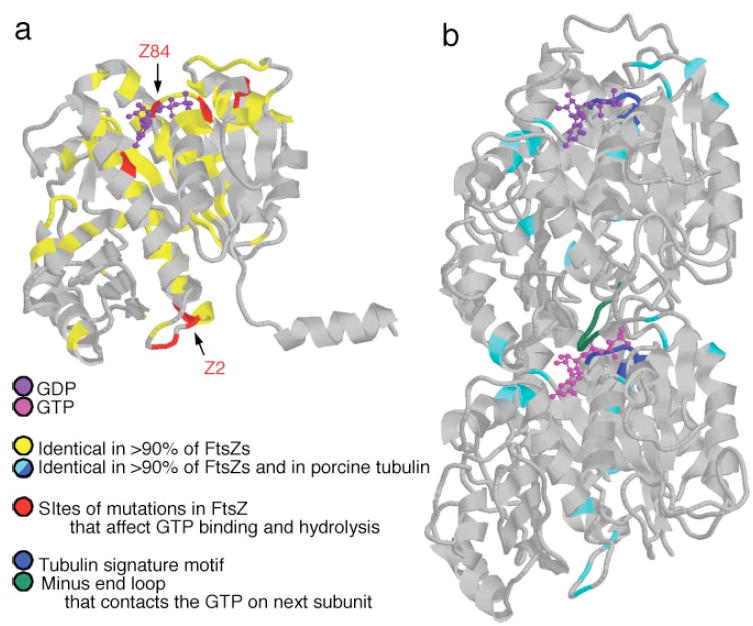
FtsZ and tubulin atomic structures. FtsZ and tubulin share minimal sequence homology but similar protein folds. (a) Methanococcus jannaschii FtsZ (70). Highly conserved residues (yellow) concentrate around the GTP binding site. Mutations that affect GTP binding and hydrolysis (red, e.g., Z84, Z2) are located at the plus end of the protein, where GTP binds, and at the minus end of the protein. (b) Porcine α/β tubulin dimer (98). A small number of residues in pig tubulin are also highly conserved in FtsZ (cyan). They include the tubulin signature motif (blue), which is important for GTP binding in both proteins, and a loop at the minus end of the subunits (green), which contacts the GTP in the neighboring subunit after assembly.
The coupling of nucleotide hydrolysis to polymer assembly also allows for complicated, nonequilibrium behaviors such as dynamic instability and treadmilling. The term dynamic instability is often misused and does not mean that polymers containing GTP are more stable than those containing GDP. Instead, it refers to the fact that at steady state, individual microtubules undergo stochastic and reversible transitions between growth and rapid shrinkage [reviewed in (24)]. This occurs because microtubules are only metastable structures: The GDP-bound protofilaments would prefer to curl outwards, but they are held together by the lateral interaction in the microtubule wall. When this balance of forces is disturbed, protofilaments separate and rapidly peel away.
Treadmilling is another nonequilibrium phenomenon. One end of a polymer steadily grows while the other end of the same polymer steadily shrinks. Treadmilling occurs in microtubules, but it has been best characterized in actin [reviewed in (108)]. In actin filaments, an asymmetric distribution of ATP, adenosine diphosphate (ADP)-Pi, and ADP-bound subunits develops. At steady state, ATP subunits associate at one end of the filament while ADP subunits dissociate from the other.
Currently there is no evidence whether FtsZ polymers exhibit either dynamic instability or treadmilling. Nonetheless, FtsZ and tubulin share many properties. Like tubulin, FtsZ polymerizes in the presence of GTP to form straight polymers that can curl and become more labile after hydrolysis to GDP. Much is still being learned about FtsZ polymerization. How does GTP hydrolysis affect polymer structure and dynamics? What higher-order structures does FtsZ form, and how do their dynamics differ from those of single-stranded protofilaments? How is a new polymer initiated? Do polymers assemble and disassemble as a population, or do they undergo dynamic instability or treadmilling? The most common techniques used to study these FtsZ assembly dynamics in vitro, along with the limitations of each, are described in Table 1. Unless otherwise noted, FtsZ proteins from all species behave similarly when tested.
TABLE 1.
Assays for FtsZ’s biochemical activities
| Method | Purpose | Advantages | Disadvantages |
|---|---|---|---|
| Blot overlays or UV cross-linkinga | Detecting bound nucleotide | Easy |
|
| Isolation of FtsZ and bound nucleotide by sedimentation or filter bindingb | Detecting bound nucleotide | Quantitative |
|
| Conventional sedimentationc | Quantifying assembly | Easy |
|
| Analytical ultracentrifugatione | Quantifying assembly | Quantitative |
|
| 90° light scatteringc | Quantifying assembly | Detection in real time |
|
| Electron microscopyf | Visualizing polymers | High resolution |
|
| Fluorescence microscopyg | Visualizing polymers | Visualization in real time |
|
FtsZ Structures
To understand the dynamic behavior of FtsZ, it is first necessary to review the data on its structure. Below we describe the structure of (a) the FtsZ subunit, (b) the GTP binding site, (c) the protofilaments assembled in GTP and GDP, and (d) the higher-order FtsZ structures. These static pictures of FtsZ subunits and polymers will form the basis for discussing dynamic FtsZ polymer populations in the next section.
THE SUBUNIT
Despite sharing minimal sequence homology (~10%) (94), FtsZ and tubulin have strikingly similar protein folds and GTP binding interactions (98) (Figure 2). FtsZ is a ~40-kDa polypeptide whose atomic structure was determined using FtsZ from the thermophillic archaeon Methanococcus jannaschii (70). The structure contains a major β sheet surrounded by α helices; perpendicular to this is a smaller subdomain, which also consists of a short β sheet surrounded by α helices (Figure 2a). The FtsZ and tubulin structures fall into a subclass different from that of small G proteins, dynamin, and all other GTP binding proteins possessing a P-loop (59, 98). Instead, they appear most closely related to that of the NADH binding protein, glyceraldehyde-3-phosphate dehydrogenase (98).
Not visible in the atomic structure is a flexible, highly charged C-terminal region. It is extremely short in M. jannaschii but is longer and essential for cytokinesis in E. coli and C. crescentus (25, 80, 81, 143). A ~15-amino-acid conserved sequence near the very C terminus is necessary for binding ZipA and FtsA, two essential Z ring proteins in E. coli (46, 72, 81, 90, 139, 143) and for binding FtsA in C. crescentus (25) and S. aureus (151). This sequence is also present in species that lack ZipA and FtsA (30), but otherwise, there is little conservation between the C termini of divergent organisms (6, 83). Tubulin’s C terminus, like that of FtsZ, is unstructured, charged, and the binding site for many microtubule-associated proteins, but the sequences are unrelated.
THE GTP BINDING SITE
The GTP binding residues are the amino acids that are best conserved between FtsZ and tubulin (70, 98) (Figure 2b), reflecting similarities in the way the two proteins interact with nucleotide during polymer assembly and disassembly. The nucleotide binds on the surface that will form the interaction site between subunits in a protofilament. In analogy with the microtubule structure, we call the protein bonds within an FtsZ protofilament longitudinal bonds and the GTP binding surface of the monomer the plus end of the protein. Unlike tubulin, unassembled FtsZ exists as a monomer rather than as a heterodimer, and all subunits can exchange nucleotide (93, 129). Once a longitudinal bond has formed, the nucleotide is likely to be almost completely buried in the interface between FtsZ subunits (98). Conserved sequences at both the plus and minus ends of a subunit are essential to FtsZ’s ability to bind and hydrolyze GTP.
Monomeric FtsZ uses conserved residues at the plus end of the protein to bind GTP tightly [~5 μM Kd (93)]. Seven of these amino acids constitute the tubulin signature motif, a sequence conserved in all tubulins and originally used to identify FtsZ as a distant tubulin homolog (20, 93). ftsZ84, a mutation in a conserved glycine in this motif (G105S in E. coli) (Figure 2a), is the best-characterized mutation at the plus end of FtsZ. This mutation disrupts nucleotide binding, thereby decreasing both polymer assembly and GTP hydrolysis in vitro (13, 20, 115). ftsZ84 renders E. coli cells heat-sensitive for division (79), but the FtsZ84 protein does not exhibit temperature-sensitive defects in vitro (13, 20, 116). Thus it is unclear how the defects in nucleotide binding and hydrolysis relate to FtsZ84 function in vivo.
Residues at the minus end of the subunit participate in both protofilament bond formation and GTP hydrolysis (29). Thus, FtsZ assembly is necessary for GTP hydrolysis (20, 129, 144). Several of the minus end residues are highly conserved in FtsZ and tubulin (98) (Figure 2). Mutations in many of these residues (Figure 2a) do not eliminate the in vitro ability of FtsZ to bind GTP or assemble, but they do reduce the protein’s nucleotide hydrolysis rate (19, 75, 78, 123, 124, 136, 143).
PROTOFILAMENTS
The conserved GTP binding residues reflect similarities in the structures that FtsZ and tubulin form with GTP and GDP. When GTP is present, protofilaments form (94) in which subunits are stacked linearly without any net twist or curvature (31, 71) (Figure 3a). FtsZ protofilaments can exist in isolation, whereas tubulin protofilaments do not form except in the context of a multi-stranded polymer. In the presence of GDP, the interaction between FtsZ subunits is substantially weakened (13, 95, 118, 129). In addition, GDP-bound protofilaments can curve sharply to form rings or spirals with a diameter of 15–25 nm (24, 31, 76, 114, 136) (Figure 3b). Because FtsZ protofilaments readily assemble under a wide variety of conditions and are structurally almost indistinguishable from those of tubulin, it is likely that FtsZ protofilaments are a genuine component of the Z ring in vivo.
Figure 3.
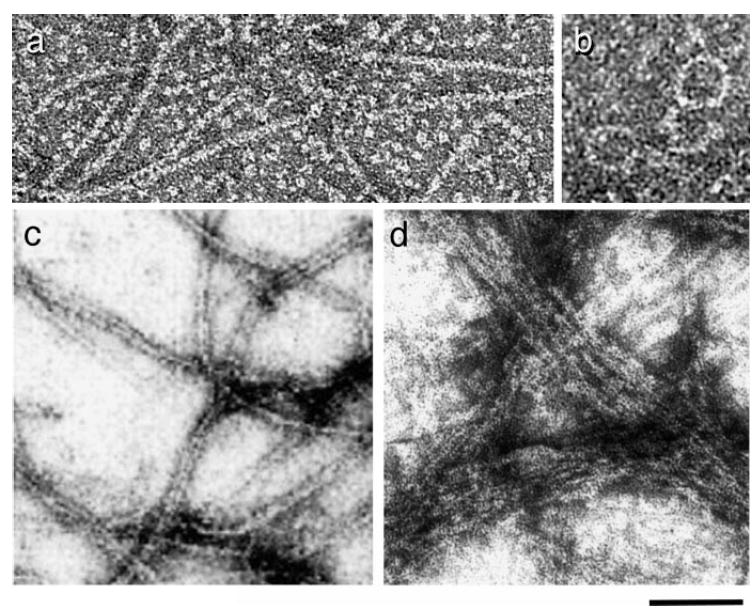
FtsZ polymers. (a) FtsZ protofilaments assembled with GTP. Monomeric FtsZ is also visible in the background (micrograph by L. Romberg). (b) FtsZ rings assembled with GDP (119). (c) Protofilament pairs and bundles assembled in reactions with GTP (96). (d) Large FtsZ bundles formed in the presence of ZipA and GTP (114). Bar is 50 nm in (a) and (b), 100 nm in (c) and (d). Images reprinted with permission.
The role of nucleotide hydrolysis in microtubule assembly has been studied using GMPCPP, a nonhydrolyzable analog of GTP. Equivalent studies of FtsZ have been hindered by the lack of GTP analogs that interact similarly with FtsZ. GMPCPP allows assembly of FtsZ polymers but is slowly hydrolyzed (76, 119). Chelating magnesium prevents GTP hydrolysis (20, 93, 115) and allows protofilament assembly (76, 96), but this assembly is not efficient at low protein concentrations (118, 129). GMPPNP does not bind FtsZ well (125), and GTPγ S, although it binds FtsZ, does not support assembly (13, 125, 154). Interestingly, GTPγ S has been reported to stabilize preformed FtsZ polymers (125), which suggests that subunits bound to this analog may cap the ends of filaments.
MULTISTRANDED POLYMERS
The Z ring is thought to contain 6 to 7 protofilaments (130), but how these protofilaments are associated with each other is unknown. The filaments could interact directly with each other through their lateral surfaces or indirectly through cross-linking by other cell division proteins. The amino acids on the lateral surfaces of FtsZ and tubulin are not homologous; thus, microtubules do not provide a good model for the higher-order association of FtsZ protofilaments (98). For the moment the assembly of multistranded FtsZ structures in vitro provides the best information on the higher-order structures that might form in vivo.
Many different surfaces of the FtsZ protofilament can be used to form a wide variety of multistranded structures, including protofilament pairs and bundles (Figure 3c,d), sheets, spirals, hoops, and tubes (13, 31, 71, 74, 76, 95, 96, 143, 148). [FtsZ tubes are not analogous to microtubules: Protofilaments are paired and tightly spiraled (76), not straight as in the microtubule wall.] Lateral associations are affected by protein concentration, pH (31, 95), and the presence of multivalent cations including DEAE dextran (13, 76), calcium (76, 96, 154), and magnesium (96, 148). FtsZ’s tendency to bundle also varies between species. For example, FtsZ from Mycobacterium tuberculosis appears to bundle more readily than that of E. coli (148).
Tubulin, in addition to forming microtubules, can also assemble in vitro into a wide array of other structures that are not observed in vivo (27). Thus, many of the lateral interactions described above for FtsZ may not be relevant in vivo. It will be important to determine what structures assemble in the context of the Z ring, with or without the help of other division proteins, because these structures will affect the pathway by which the Z ring assembles and turns over.
FtsZ Polymer Dynamics
During the cell cycle the Z ring goes through phases of net assembly, steady state turnover, and net disassembly. To understand these processes, we first need to understand the behavior of pure FtsZ polymers in vitro. At steady state, which nucleotide intermediates (GTP, GDP-Pi, or GDP) do the polymers contain, and how does this affect the assembly dynamics? Might FtsZ exhibit dynamic instability or treadmilling? Is the initial assembly of FtsZ polymers cooperative? Below we discuss what is known about FtsZ’s in vitro steady state assembly dynamics and the initiation of FtsZ polymers.
THE NUCLEOTIDE CONTENT OF FtsZ POLYMERS AT STEADY STATE
Turnover of the Z ring appears to be powered by FtsZ’s GTP hydrolysis activity (130). Knowing the nucleotide intermediate that predominates in polymers at steady state may suggest how the energy from hydrolysis is used. For example, the net twist in an actin filament changes after phosphate release and alters the filament’s affinity for proteins that sever or nucleate actin (107, 147). In addition, hydrolysis and phosphate release cause changes in the polymerization properties of actin and tubulin. These changes are fundamental to the mechanisms of dynamic instability and treadmilling and thus to the regulation of polymer turnover in eukaryotic cells.
For FtsZ each hydrolysis event requires many steps (GTP binding, hydrolysis, phosphate release, GDP release) (Figure 4), and the slowest step in the cycle determines the intermediate that predominates. If hydrolysis is rapid and product release is slow, GDP or GDP-Pi will build up in the polymer. If product release is relatively fast, the polymers will contain GTP. Further complicating matters, during each hydrolysis cycle an individual subunit may go through a cycle of assembly and disassembly. Because the GTP is sandwiched between subunits in FtsZ polymers, the assembly state of a subunit may affect the rate at which the GDP and phosphate can be released into solution.
Figure 4.
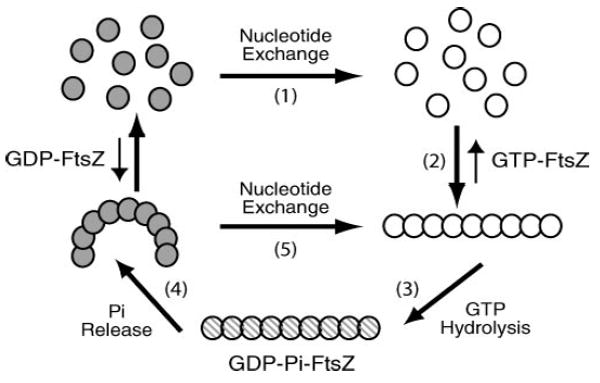
Model for FtsZ’s GTP hydrolysis cycle. FtsZ’s hydrolysis cycle is connected to polymer assembly and disassembly. 1: Monomers rapidly exchange nucleotide with solution. In vivo, more GTP than GDP exists in the cytoplasm. 2: When bound to GTP, FtsZ assembly is enhanced. 3: Once assembled, FtsZ can hydrolyze the GTP. 4: Phosphate release follows. GDP-containing polymers are more likely to be curved or to disassemble than GTP-containing polymers. 5: Nucleotide exchange might occur without complete disassembly of the polymer.
FtsZ polymers may not contain the same nucleotide as microtubules. Micro-tubules contain GDP-bound subunits because after tubulin assembly and GTP hydrolysis, the GDP is trapped between subunits in the microtubule wall. Since it is rare for longitudinal bonds in the middle of a microtubule to break, nucleotide exchange occurs only when subunits dissociate from the ends of a microtubule. Indirect evidence suggests that FtsZ polymers behave differently. FtsZ polymers have a much higher steady state GTP turnover rate than microtubules do: ~5/FtsZ/min (95, 96, 129, 144) versus 0.06/tubulin/min (98a). This suggests either that FtsZ subunits release from the polymer at a high rate or that even without releasing, subunits in the center of the polymer can exchange nucleotide. If such reactions were more rapid than hydrolysis itself, steady state FtsZ polymers would contain GTP.
Two groups have addressed the steady state nucleotide content of FtsZ polymers (87, 126). Both groups found one equivalent of 32P bound to FtsZ polymerized with γ 32P-GTP, but it remains unclear whether hydrolysis or phosphate release is the rate-limiting step in the cycle. One group proposed that hydrolysis is rate limiting and polymers contain GTP. They found the exchange of nucleotide between polymer and solution to be faster than the steady state hydrolysis rate (87). The other group suggested that hydrolysis is rapid and phosphate release is slow (126). They determined that the vast majority of 32P bound to FtsZ was inorganic phosphate rather than the terminal phosphate on a GTP. However, in these latter experiments it is unclear whether the hydrolysis occurred in solution or during experimental handling. Clearly, more experiments need to be done to resolve this tricky issue.
In vivo, the nucleotide content of FtsZ polymers may be affected by Z ring proteins that induce protofilament bundling. The kinetics of GTP hydrolysis changes when protofilament bundles are stabilized by calcium or DEAE dextran. Depending on reaction conditions, these cations can either increase or decrease the steady state hydrolysis rate (74, 96, 154). Calcium bundles exchange nucleotide only slowly (87) and may contain GDP-bound subunits (although as above, hydrolysis might have occurred during the time the polymers were sedimented) (125). Interestingly, the degree of curvature in a protofilament may also affect the ability of nucleotide to be exchanged. When curved protofilaments were assembled in DEAE dextran/GDP/Mg++ and then excess GTP/EDTA was added, the curved protofilaments immediately straightened, whereas straight protofilaments could not rapidly exchange nucleotide and curve (76). In tightly associated, straight protofilaments multiple hydrolysis events may be needed in order for complete breakage of the bundle and release of the GDP (87).
Implications for dynamic instability and treadmilling
The distribution of nucleotide intermediates in FtsZ polymers will help determine whether FtsZ turnover occurs through dynamic instability or treadmilling. In vivo, dynamic instability might trigger sudden depolymerization or force generation during ring contraction. Polymer treadmilling could participate in relocalization of the Z ring during B. subtilis sporulation. However, because individual FtsZ polymers are too small to be observed growing and shrinking under the light microscope (Table 1), no direct data currently exist. Neither phenomenon is likely if FtsZ polymers predominantly contain GTP. For dynamic instability to occur in microtubules, the energy from GTP hydrolysis must be stored as strain in the GDP-containing microtubule wall. It is unlikely that individual FtsZ protofilaments could exhibit dynamic instability by the same mechanism. They lack the lateral bonds that could prevent GDP-bound interfaces from breaking or relaxing to a lower-energy, curved conformation. In the various multistranded FtsZ polymers the ability to store the energy from GTP hydrolysis will be influenced by the relative rates of hydrolysis, nucleotide exchange, and protofilament disassembly. Likewise, if FtsZ polymers do not build up a region of GDP at one end of the filament, they could not treadmill with the same mechanism as actin filaments.
NUCLEATION AND COOPERATIVE ASSEMBLY
Many biological polymers assemble cooperatively, meaning that it is more difficult for a new polymer to be established than for polymerization to continue from pre-established nuclei or polymer ends (100). Eukaryotic cells take advantage of nucleated assembly to control polymer localization [reviewed in (24, 147)]. The creation of new microtubules is largely limited to the microtubule organizing center, where γ-tubulin, a specialized tubulin isoform, is thought to form the nucleation sites. This localized nucleation allows interphase eukaryotic cells to form a single, radial array of microtubules that organizes the layout of the cell as a whole.
Similarly, cooperative assembly of FtsZ might help limit the number of Z rings in a cell by making polymer addition to a ring that already exists more favorable than initiation of a new ring elsewhere. Although a threshold level of FtsZ must be expressed before Z rings will form (61), potential nucleating factors in the bacterial membrane have remained elusive. In addition, it is unknown whether protofilaments preassemble in the cytoplasm and are only subsequently added to the Z ring or whether assembly of the Z ring occurs solely through direct monomer addition.
To understand the initiation of FtsZ assembly in vivo, it is necessary to determine the role of cooperativity in the assembly of both single and multistranded polymers in vitro. Below we describe the theoretical mechanisms behind cooperative assembly and the methods that can be used to detect it. We then review the evidence for cooperativity in (a) single-stranded polymers assembled in the absence of hydrolysis, (b) single-stranded polymers assembled with Mg++ and GTP, and (c) multistranded FtsZ polymers.
Cooperative assembly in multistranded polymers emerges by necessity (100). During polymer initiation, when two monomers associate, only a single new bond is formed, whereas when a subunit adds to the end of a multistranded polymer, both longitudinal and lateral bonds can form simultaneously. As a result, subunit addition to the end of a multistranded structure is more favorable than de novo formation of a polymer. For microtubules the problem of nucleation is complicated by GTP hydrolysis. Microtubule nuclei may consist of two protofilaments aligned laterally (141). If the protofilaments hydrolyze GTP, they may curl and separate (16). In single-stranded polymers other mechanisms are needed to produce nucleated assembly. A nucleus is defined as the smallest polymer that is more likely to extend than to fall apart again (100). Cooperative assembly of individual protofilaments might therefore emerge if, following the association of a minimal number of subunits, a conformational change increased the affinity for binding of additional subunits.
Cooperative polymer assembly can be detected in a number of ways. The Kd for the formation of dimers should be dramatically higher than that for subunit addition to polymer ends. There is a sharp critical concentration for assembly, below which no polymer forms and above which all additional protein is converted into polymer. Dimers and other complexes smaller than the size of the minimal nucleus are extremely rare. Finally, at protein concentrations just above the critical concentration there are prominent, concentration-dependent lags in assembly because nuclei need to slowly accumulate before polymer extension can rapidly proceed.
Assembly of small polymers in the absence of nucleotide hydrolysis
In the absence of nucleotide hydrolysis, FtsZ assembly does not appear to be highly cooperative. Two groups used analytical ultracentrifugation to show that FtsZ dimers are nearly as stable as small FtsZ polymers. Sossong et al. (129) studied assembly in both GDP and magnesium-free GTP and found that FtsZ dimers, trimers, and tetramers were prevalent. In addition, the Kd for association of FtsZ-GDP dimers was identical to that of trimers (31 μM). Rivas et al. (118) determined the dissociation constants for assembly of up to 12 GDP-FtsZ subunits. As polymer length increased, the Kd for binding of additional subunits decreased only by a factor of two. In contrast, there may be up to seven orders of magnitude difference between the Kd for association of actin dimers and that for the addition of actin subunits to filament ends (147).
Assembly with Mg++ and GTP
Polymerization with Mg++ and GTP is more representative of in vivo assembly but is technically more challenging to study. Currently, data exist in support of both noncooperative and nucleated assembly models. In contrast to the hours required for low concentrations of tubulin to polymerize (106, 141), FtsZ assembly can reach steady state in 1 to 60 sec (13, 96, 119). Lags in initial FtsZ assembly have been irreproducibly observed, but they are not likely to be caused by nucleation reactions requiring multiple subunits to come together. Instead, they may be due to aggregated FtsZ needing to disperse before polymerizing or to subunits needing to bind nucleotide (20, 93, 119, 143, 144). Nonetheless, FtsZ polymerization and GTP hydrolysis often initiate rather abruptly at a critical concentration of ~1 μM [(74, 95, 96, 118, 129, 144, 148) but see also (119)]. Perhaps polymerization of FtsZ requires a model for cooperative assembly different from those previously developed for microtubules and actin.
Assembly of multistranded polymers
Data on the cooperativity of multistranded FtsZ polymers are also somewhat ambiguous. DEAE dextran and calcium appear to enhance FtsZ assembly (73, 94, 154). However, in one report the critical concentration did not appear to be lowered by calcium (~1 μM) (154). While no lags have been detected in reactions with calcium and DEAE dextran (124, 125), M. tuberculosis FtsZ protofilaments bundle in the absence of these cations and assemble slowly, with brief lags in hydrolysis rates (148).
PART III: PROTEINS THAT MODULATE FtsZ ASSEMBLY
The polymerization properties described above are regulated in vivo by proteins that interact directly with FtsZ. Changes in FtsZ polymer structure or GTPase activity can alter the balance between assembly and disassembly. Positive-acting factors promote Z ring formation and stabilize the ring during assembly of the division apparatus. Negative-acting factors prevent FtsZ assembly at inappropriate locations and ensure that the ring is dynamic enough to respond to the signals governing cytokinesis. Together they form a regulatory network that dictates the spatial and temporal control of cytokinesis (Figure 5a).
Figure 5.
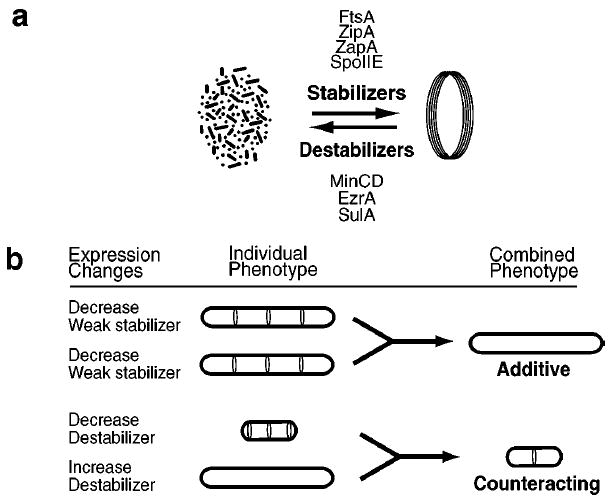
Synergistic interactions. (a) In a cell some FtsZ is in the cytoplasm while other FtsZ is assembled into a membrane-associated ring. The balance between these states is maintained by the combined actions of stabilizing proteins and destabilizing proteins. (b) Synergistic effects are seen when the expression levels of two regulatory proteins are changed simultaneously. (Bottom) A counteracting effect. Loss of a destabilizing factor leads to the formation of extra FtsZ rings at cell poles. Increasing expression of a destabilizing protein inhibits Z ring formation and leads to cell filamentation. Together these two changes can restore normal Z ring formation and cell viability. (Top) An additive effect. Removal of a single, weakly stabilizing protein allows FtsZ assembly but prevents division. Simultaneous removal of two weakly stabilizing proteins can block Z ring formation altogether.
Below we discuss the proteins known to affect FtsZ assembly. Several of these proteins are not essential for cytokinesis, and none are required for Z ring formation. Their role in regulating Z ring assembly is often confirmed by genetic experiments in which the expression of two proteins is simultaneously altered. Moreover, despite the dramatic evolutionary conservation of FtsZ itself, only two of these proteins, FtsA and ZapA, are themselves broadly conserved. Thus, the proteins that control FtsZ polymerization can have subtle effects that are tailored to the needs of individual species.
Stabilizing Proteins
FtsZ assembly is the first known event in bacterial cytokinesis and is required to localize all other components of the division machinery to the nascent septal site (82, 120). However, the Z ring is not sufficiently stable to support cytokinesis without the assistance of accessory division proteins. To date, the proteins FtsA, ZipA, and ZapA are known to interact directly with FtsZ to stabilize the Z ring.
FtsA
FtsA is a widely conserved protein (120) that binds directly to FtsZ (25, 45, 46) and recruits other cell division proteins to the Z ring (82, 120). Although FtsA is not required for Z ring formation (143), the stability and function of the ring are affected by FtsA levels. FtsA is essential in E. coli and the ratio of FtsA to FtsZ must be maintained within a narrow range for cells to divide (18); this ratio is achieved by cotranscription of the two genes. FtsA is required for Z ring formation in situations where the Z ring is artificially destabilized (105).
The ability of FtsA to modulate FtsZ polymer stability in vitro has not yet been directly examined. Purified FtsA is phosphorylated and can bind and hydrolyze ATP (34, 122). The phosphorylation state of FtsA appears to correlate with ATP binding and membrane association, but a mutation that prevents phosphorylation and in vitro ATP binding still rescues conditional ftsA alleles (122). FtsA is part of the actin/hexokinase/HSP70 superfamily (139) and has been shown to dimerize (34, 152), but to date there is no evidence that it polymerizes. MreB, a protein required for maintaining cell shape, is a better candidate for a functional prokaryotic homolog of actin (54, 138).
ZipA
Like FtsA, ZipA is a Z ring protein that is dispensable for ring formation but is required for recruitment of other proteins to the septal site (44, 69, 82, 120). While ZipA is essential in E. coli, it is only conserved in the γ subdivision of gram-negative bacteria. Genetic experiments suggest that ZipA stabilizes FtsZ polymers in the cytokinetic ring. A twofold overexpression of ZipA suppresses the heat sensitivity of ftsZ84, a mutation that results in unstable Z rings at high temperatures (114). ZipA may be enhancing the association of FtsZ with the cell membrane, or it may be stabilizing the interaction between FtsZ protofilaments. The ZipA protein consists of a hydrophobic membrane anchor that is tethered to a cytoplasmic FtsZ binding domain via a flexible peptide (43, 46, 90, 91, 99). In vitro, the cytoplasmic domain of ZipA enhances the bundling of FtsZ protofilaments (45, 114) (Figure 3d).
ZapA
ZapA is a Z ring protein that is conserved in a wide range of bacteria (42), but in contrast to both FtsA and ZipA, it appears to be dispensable for division under normal growth conditions. B. subtilis zapA null mutants are phenotypically unchanged, with wild-type frequency and positioning of Z rings (42). ZapA becomes essential for efficient division in the absence of EzrA or DivIVA, two proteins that normally modulate Z ring assembly, or in cells in which FtsZ concentrations have been artificially lowered (42). In vitro, ZapA interacts directly with FtsZ and induces protofilaments to associate into large bundles (42). The genetic experiments that originally identified ZapA (42) suggest that in vivo this bundling activity may help stabilize Z rings.
THE ADDITIVE EFFECTS OF STABILIZING FACTORS
Although individual cell division proteins may be dispensible for Z ring formation, the simultaneous removal of two weakly stabilizing factors can disrupt FtsZ assembly (Figure 5b, top). Such synergistic interactions confirm that ZipA and FtsA promote Z ring stability. For example, whereas the loss of ZipA or FtsA alone does not prevent FtsZ assembly, depletion of both FtsA and ZipA in exponentially growing E. coli cells blocks the formation of new Z rings and renders previously formed ones unstable (105). Similarly, a mutation in FtsZ that renders it unable to interact with either ZipA or FtsA inhibits its ability to form rings (105).
Destabilizing Proteins
Despite FtsZ concentrations in E. coli cells that are well above the critical concentration for in vitro polymer assembly (74), much of the protein is cytoplasmic (130). To prevent aberrant assembly and maintain subunit turnover in the Z ring, destabilizing factors must counter the actions of the polymer-stabilizing proteins discussed above. Below we describe three sets of destabilizing factors: the MinCD complex, EzrA, and SulA.
THE MinCD COMPLEX
In E. coli and B. subtilis the best-characterized proteins affecting FtsZ assembly are the Min proteins. These proteins were originally identified by E. coli mutations that caused aberrant septation at cell poles, which led to “minicell” formation (21). MinC and MinD form a division inhibitor (the MinCD complex) that is concentrated at the cell poles (127). Null mutations in either minC or minD result in the formation of Z rings at polar positions as well as at medial positions (11, 66). Simultaneous overexpression of MinC and MinD prevents the formation of Z rings and cytokinesis (65, 104).
MinC interacts directly with FtsZ to inhibit Z ring formation (51). It may do so by altering FtsZ polymer assembly, although current data are ambiguous. When FtsZ was incubated with MinC in vitro, significantly fewer FtsZ polymers could be detected by electron microscopy (51). However, MinC had no effect on FtsZ’s GTP hydrolysis rate (51), suggesting that MinC does not change the stability of protofilaments or their nucleotide exchange rates. In addition, overexpression of MinCD does not change the gel filtration profile of FtsZ polymers in E. coli lysates (56). It should be noted that these approaches are not equally sensitive to different aspects of polymer formation. These data would be consistent if MinC reduced the length or bundling of FtsZ polymers without completely inhibiting FtsZ self-association.
MinD is required for concentrating MinC at the membrane of cell poles (50, 84). It associates with the membrane via an N-terminal amphipathic helix (133) and has an ATPase activity that may play a role regulating this membrane localization (48, 52). The polar localization of MinD and hence MinC is dependent on a third protein that is not well conserved across evolutionary boundaries. Mutations in minE in E. coli or divIVA in B. subtilis result in the mislocalization of MinCD and extensive cell filamentation (21, 28, 85, 112, 113). MinE and DivIVA share no similarity in either their protein sequence or their mechanism of MinCD localization [reviewed in (127)]. MinE is concentrated at midcell and prevents MinCD from localizing there; instead the entire MinCD complex oscillates rapidly from pole to pole (112, 113). In contrast, DivIVA binds to MinD and statically tethers the MinCD complex to the cell poles (84, 85).
Although the Min proteins are important for preventing aberrant Z ring formation at cell poles, they are not required for establishing the nascent division site at midcell in either E. coli or B. subtilis. Septation occurs at midcell rather than at the cell poles ~90% of the time in B. subtilis minCD null mutants (66) and at a high frequency in E. coli minCDE null mutants as well (21). It is thus not surprising that the Min proteins are not conserved throughout the bacteria (120). The lack of conservation suggests either that cell poles are not competent for FtsZ assembly in certain species or that aberrant septation events are prevented through other means.
EzrA
EzrA is a nonessential protein that is conserved throughout the low GC gram-positive bacteria (61). An ezrA null mutation results in the formation of extra Z rings, primarily at cell poles, a phenotype strikingly similar to that of a minCD null mutation (61). EzrA has an N-terminal transmembrane anchor and a large cytoplasmic domain that includes four coiled-coil motifs. An EzrA-GFP fusion distributes diffusely around the periphery of the cell but also concentrates at the cytokinetic ring in an FtsZ-dependent manner (61). The extra rings in EzrA null mutants suggest that EzrA acts throughout the plasma membrane to destabilize newly formed FtsZ polymers and prevent Z ring formation at inappropriate sites. EzrA activity would need to be overcome at midcell by a factor promoting FtsZ assembly. The biochemical nature of the EzrA-FtsZ interaction has yet to be determined.
SulA
SulA is a soluble E. coli protein that is induced as part of the SOS response. SulA binds directly to FtsZ and inhibits assembly of the Z ring (49, 53, 56, 92, 135, 136). In vitro, 1:4 ratios of SulA to FtsZ can inhibit polymer assembly without affecting FtsZ’s GTP hydrolysis activity (136). At a 1:1 ratio of SulA to FtsZ, both assembly and GTP hydrolysis are inhibited (136), which suggests a model in which SulA binds to one end of FtsZ and prevents the addition of new subunits. At lower concentrations SulA would cap polymers, limiting their length, whereas at higher concentrations SulA would completely prevent FtsZ dimerization and formation of the active site. SulA’s ability to prevent assembly at relatively low concentrations suggests that in vivo even limited induction of SulA may be sufficient to rapidly block cytokinesis.
EVIDENCE FOR OTHER PROTEINS
To date, only two other proteins have been suggested to affect Z ring stability. The first, SpoIIE, colocalizes with FtsZ in polar rings during sporulation in B. subtilis (64). SpoIIE is a phosphatase that helps activate cell type–specific gene expression, but it may also play a structural role by directly stabilizing polar Z rings (7, 33, 58). The second, FtsW, is an essential cell division protein. Loss of FtsW was originally suggested to render Z rings less stable (12). However, more recent data indicate that FtsW is recruited to the Z ring late in the assembly process and functions primarily to localize its cognate transpeptidase FtsI (PBP3) (86).
THE COUNTERBALANCING EFFECTS OF STABILIZING AND DESTABILIZING FACTORS
The destabilization of FtsZ polymers by proteins that inhibit Z ring formation can be counteracted by a simultaneous stabilization of FtsZ assembly via different means (Figure 5b, bottom). Mutations that stabilize FtsZ polymers, such as ftsZ2, render FtsZ resistant to the destabilizing effects of both MinC and SulA (8, 9, 53, 136). Similarly, in B. subtilis a null mutation in the gene encoding EzrA, a destabilizing factor, suppresses the inhibition of Z ring formation and cell division associated with 15-fold overexpression of MinCD (65). Moreover, increasing MinCD levels can compensate for the loss of EzrA to some extent, as polar rings do not form in ezrA null mutant cells in which MinCD has been overexpressed (65). Synergistic effects such as these are useful for confirming the activities of nonessential genes.
PERSPECTIVES AND OUTLOOK
The forces governing Z ring assembly during bacterial cytokinesis are strikingly similar to those controlling the microtubule and actin cytoskeletons in eukaryotic cells. FtsZ polymer dynamics are modulated during the cell cycle by a precisely balanced network of positive- and negative-acting factors. Assembly is exquisitely sensitive to these factors because of the intrinsic reversibility of FtsZ polymerization. Thus, developmental and environmental signals are integrated during Z ring assembly and maintenance to ensure the proper spatiotemporal control of cytokinesis.
A comprehensive understanding of FtsZ dynamics during cell division will likely depend on the extension of the experimental techniques currently being used. For example, polymerization assays with improved time resolution and linearity will help illuminate difficult enzymatic issues in FtsZ polymer dynamics. Furthermore, the relatively recent identification of ZapA and EzrA highlights the importance of nonessential proteins in cell division. The discovery of additional proteins that exert subtle effects on FtsZ assembly may require sophisticated biochemical or genetic approaches. Ultimately, the analysis of these proteins and their effects on FtsZ polymerization, both in vivo and in vitro, will pave the way for more precise models of FtsZ dynamics within the Z ring.
Acknowledgments
We thank Debabrata RayChaudhuri for micrographs of FtsZ bundled in the presence of ZipA, Eva Nogales for FtsZ alignments with porcine tubulin, and Harold Erickson for communicating data prior to publication. We are also grateful to members of the Mitchison and Levin labs, and to Rikki Eggert, Harold Erickson, Rich Losick, Ellen Quardokus, Mimi Shirazu, Jim Skeath, and Sigal Ben-Yehuda for helpful discussions and critical reading of the manuscript.
LITERATURE CITED
- 1.Addinall SG, Bi EF, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–84. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall SG, Cao C, Lutkenhaus J. Temperature shift experiments with ftsZ84 (Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J Bacteriol. 1997;179:4277–84. doi: 10.1128/jb.179.13.4277-4284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Addinall SG, Lutkenhaus J. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol. 1996;22:231–37. doi: 10.1046/j.1365-2958.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- 4.Aldea M, Garrido T, Pla J, Vicente M. Division genes in Escherichia coli are expressed coordinately to cell septum requirements by gearbox promoters. EMBO J. 1990;9:3787–94. doi: 10.1002/j.1460-2075.1990.tb07592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arigoni F, Pogliano K, Webb CD, Stragier P, Losick R. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science. 1995;270:637–40. doi: 10.1126/science.270.5236.637. [DOI] [PubMed] [Google Scholar]
- 6.Beall B, Lowe M, Lutkenhaus J. Cloning and characterization of Bacillus subtilis homologs of Escherichia coli cell division genes ftsA and ftsZ. J Bacteriol. 1988;170:4855–64. doi: 10.1128/jb.170.10.4855-4864.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell. 2002;109:257–66. doi: 10.1016/s0092-8674(02)00698-0. [DOI] [PubMed] [Google Scholar]
- 8.Bi E, Lutkenhaus J. Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA) J Bacte-riol. 1990;172:5602–9. doi: 10.1128/jb.172.10.5602-5609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi E, Lutkenhaus J. Interaction between the min locus and ftsZ. J Bacteriol. 1990;172:5610–16. doi: 10.1128/jb.172.10.5610-5616.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escheri-chia coli. Nature. 1991;354:161–64. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 11.Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175:1118–25. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyle DS, Khattar MM, Addinall SG, Lutkenhaus J, Donachie WD. ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol. 1997;24:1263–73. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 13.Bramhill D, Thompson CM. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc Natl Acad Sci USA. 1994;91:5813–17. doi: 10.1073/pnas.91.13.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–59. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brun YV, Shimkets LJ. Prokaryotic Development. Washington, DC: ASM; 2000. p. 477. [Google Scholar]
- 16.Carlier MF, Didry D, Pantaloni D. Hydrolysis of GTP associated with the formation of tubulin oligomers is involved in microtubule nucleation. Biophys J. 1997;73:418–27. doi: 10.1016/S0006-3495(97)78081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai K, Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991;173:3500–6. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–51. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai K, Mukherjee A, Xu Y, Lutkenhaus J. Mutations in ftsZ that confer resistance to SulA affect the interaction of FtsZ with GTP. J Bacteriol. 1994;176:130–36. doi: 10.1128/jb.176.1.130-136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Boer P, Crossley R, Rothfield L. The essential bacterial cell division protein FtsZ is a GTPase. Nature. 1992;359:254–56. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 21.de Boer PA, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E coli. Cell. 1989;56:641–49. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 22.Deleted in proof
- 23.Den Blaauwen T, Buddelmeijer N, Aarsman ME, Hameete CM, Nanninga N. Timing of FtsZ Assembly in Es-cherichia coli. J Bacteriol. 1999;181:5167–75. doi: 10.1128/jb.181.17.5167-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desai A, Mitchison TJ. Microtubule polymerization. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 25.Din N, Quardokus EM, Sackett MJ, Brun YV. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol Microbiol. 1998;27:1051–63. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- 26.Donachie WD. Co-ordinate regulation of the Escherichia coli cell cycle or The cloud of unknowing. Mol Microbiol. 2001;40:779–85. doi: 10.1046/j.1365-2958.2001.02439.x. [DOI] [PubMed] [Google Scholar]
- 27.Dustin P. Microtubules. Berlin: Springer-Verlag; 1984. [Google Scholar]
- 28.Edwards DH, Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol. 1997;24:905–15. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- 29.Erickson HP. Atomic structures of tubulin and FtsZ. Trends Cell Biol. 1998;8:133–37. doi: 10.1016/s0962-8924(98)01237-9. [DOI] [PubMed] [Google Scholar]
- 30.Erickson HP. The FtsZ protofila-ment and attachment of ZipA—structural constraints on the FtsZ power stroke. Curr Opin Cell Biol. 2001;13:55–60. doi: 10.1016/s0955-0674(00)00174-5. [DOI] [PubMed] [Google Scholar]
- 31.Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–23. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feucht A, Abbotts L, Errington J. The cell differentiation protein SpoIIE contains a regulatory site that controls its phosphatase activity in response to asymmetric septation. Mol Microbiol. 2002;45:1119–30. doi: 10.1046/j.1365-2958.2002.03082.x. [DOI] [PubMed] [Google Scholar]
- 34.Feucht A, Lucet I, Yudkin MD, Errington J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol Microbiol. 2001;40:115–25. doi: 10.1046/j.1365-2958.2001.02356.x. [DOI] [PubMed] [Google Scholar]
- 35.Fukuchi K, Kasahara Y, Asai K, Kobayashi K, Moriya S, Ogasawara N. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology. 2000;146:1573–83. doi: 10.1099/00221287-146-7-1573. [DOI] [PubMed] [Google Scholar]
- 36.Gholamhoseinian A, Shen Z, Piggot P. Regulation of the cell division gene ftsA during sporulation of Bacillus sub-tilis. J Bacteriol. 1992;174:4647–56. doi: 10.1128/jb.174.14.4647-4656.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilson PR, Beech PL. Cell division protein FtsZ: running rings around bacteria, chloroplasts and mitochondria. Res Microbiol. 2001;152:3–10. doi: 10.1016/s0923-2508(00)01162-1. [DOI] [PubMed] [Google Scholar]
- 38.Gonzy-Tréboul G, Karamzyn-Campelli C, Stragier P. Developmental regulation of transcription of the Bacillus sub-tilis ftsAZ operon. J Mol Biol. 1992;224:967–79. doi: 10.1016/0022-2836(92)90463-t. [DOI] [PubMed] [Google Scholar]
- 39.Gottesman S, Halpern E, Trisler P. Role of sulA and sulB in filamentation by lon mutants of Escherichia coli K-12. J Bacteriol. 1981;148:265–73. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graumann PL. SMC proteins in bacteria: condensation motors for chromosome segregation? Biochimie. 2001;83:53–59. doi: 10.1016/s0300-9084(00)01218-9. [DOI] [PubMed] [Google Scholar]
- 41.Graumann PL, Losick R. Coupling of asymmetric division to polar placement of replication origin regions in Bacillus subtilis. J Bacteriol. 2001;183:4052–60. doi: 10.1128/JB.183.13.4052-4060.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–56. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–85. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 44.Hale CA, de Boer PA. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181:167–76. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hale CA, Rhee AC, de Boer PA. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J Bacteriol. 2000;182:5153–66. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haney SA, Glasfeld E, Hale C, Keeney D, He Z, de Boer P. Genetic analysis of the Escherichia coli FtsZ-ZipA interaction in the yeast two-hybrid system. J Biol Chem. 2001;276:11980–87. doi: 10.1074/jbc.M009810200. [DOI] [PubMed] [Google Scholar]
- 47.Harry EJ, Rodwell J, Wake RG. Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol Microbiol. 1999;33:33–40. doi: 10.1046/j.1365-2958.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 48.Hayashi I, Oyama T, Morikawa K. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 2001;20:1819–28. doi: 10.1093/emboj/20.8.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higashitani A, Ishii Y, Kato Y, Horiuchi K. Functional dissection of a cell-division inhibitor, SulA, of Escherichia coli and its negative regulation by Lon. Mol Gen Genet. 1997;254:351–57. doi: 10.1007/s004380050426. [DOI] [PubMed] [Google Scholar]
- 50.Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Micro-biol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 51.Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Es-cherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci USA. 1999;96:14819–24. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu ZL, Gogol EP, Lutkenhaus J. Dynamic assembly of MinD on phospho-lipid vesicles regulated by ATP and MinE. Proc Natl Acad Sci USA. 2002;99:6761–66. doi: 10.1073/pnas.102059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J, Cao C, Lutkenhaus J. Interaction between FtsZ and inhibitors of cell division. J Bacteriol. 1996;178:5080–85. doi: 10.1128/jb.178.17.5080-5085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria. Helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–22. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 55.Joseleau-Petit D, Vinella D, D’Ari R. Metabolic alarms and cell division in Escherichia coli. J Bacteriol. 1999;181:9–14. doi: 10.1128/jb.181.1.9-14.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Justice SS, Garcia-Lara J, Rothfield LI. Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Es-cherichia coli division machinery. Mol Microbiol. 2000;37:410–23. doi: 10.1046/j.1365-2958.2000.02007.x. [DOI] [PubMed] [Google Scholar]
- 57.Kelly AJ, Sackett MJ, Din N, Quardokus E, Brun YV. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–93. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khvorova A, Zhang L, Higgins ML, Piggot PJ. The spoIIE locus is involved in the Spo0A-dependent switch in the location of FtsZ rings in Bacillus subtilis. J Bacteriol. 1998;180:1256–60. doi: 10.1128/jb.180.5.1256-1260.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kull FJ, Fletterick RJ. Is the tubulin/ FtsZ fold related to the G-protein fold? Trends Cell Biol. 1998;8:306–7. doi: 10.1016/s0962-8924(98)01319-1. [DOI] [PubMed] [Google Scholar]
- 60.Deleted in proof
- 61.Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci USA. 1999;96:9642–47. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–88. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 63.Levin PA, Losick R. Asymmetric cell division in Bacillus subtilis. See Ref 15. 2000:167–89. [Google Scholar]
- 64.Levin PA, Losick R, Stragier P, Arigoni F. Localization of the sporulation protein SpoIIE in Bacillus subtilis is dependent upon the cell division protein FtsZ. Mol Microbiol. 1997;25:839–46. doi: 10.1111/j.1365-2958.1997.mmi505.x. [DOI] [PubMed] [Google Scholar]
- 65.Levin PA, Schwartz RL, Grossman AD. Polymer stability plays an important role in the positional regulation of FtsZ. J Bacteriol. 2001;183:5449–52. doi: 10.1128/JB.183.18.5449-5452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levin PA, Shim JJ, Grossman AD. Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis. J Bacteriol. 1998;180:6048–51. doi: 10.1128/jb.180.22.6048-6051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin DC-H, Levin PA, Grossman AD. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc Natl Acad Sci USA. 1997;94:4721–26. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 69.Liu Z, Mukherjee A, Lutkenhaus J. Recruitment of ZipA to the division site by interaction with FtsZ. Mol Microbiol. 1999;31:1853–61. doi: 10.1046/j.1365-2958.1999.01322.x. [DOI] [PubMed] [Google Scholar]
- 70.Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–6. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 71.Löwe J, Amos LA. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 1999;18:2364–71. doi: 10.1093/emboj/18.9.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Löwe J, van den Ent F. Conserved sequence motif at the C-terminus of the bacterial cell-division protein FtsA. Biochimie. 2001;83:117–20. doi: 10.1016/s0300-9084(00)01210-4. [DOI] [PubMed] [Google Scholar]
- 73.Lu C, Erickson HP. Purification and assembly of FtsZ. Methods Enzymol. 1998;298:305–13. doi: 10.1016/s0076-6879(98)98027-2. [DOI] [PubMed] [Google Scholar]
- 74.Lu C, Stricker J, Erickson HP. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima—quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskelet. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 75.Lu C, Stricker J, Erickson HP. Site-specific mutations of FtsZ—effects on GTPase and in vitro assembly. BMC Microbiol. 2001;1(1):7. doi: 10.1186/1471-2180-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu CL, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–70. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lucet I, Feucht A, Yudkin MD, Errington J. Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE. EMBO J. 2000;19:1467–75. doi: 10.1093/emboj/19.7.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lutkenhaus J, Addinall SG. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 79.Lutkenhaus J, Wolf-Watz H, Donachie WD. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ) J Bacteriol. 1980;142:615–20. doi: 10.1128/jb.142.2.615-620.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–3003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma X, Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli ftsZ. J Bacteriol. 1999;181:7531–44. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Margolin W. Themes and variations in prokaryotic cell division. FEMS Micro-biol Rev. 2000;24:531–48. doi: 10.1111/j.1574-6976.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 83.Margolin W, Corbo JC, Long SR. Cloning and characterization of a Rhizo-bium meliloti homolog of the Escherichia coli cell division gene ftsZ. J Bacteriol. 1991;173:5822–30. doi: 10.1128/jb.173.18.5822-5830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marston AL, Errington J. Selection of the midcell division site in Bacillus sub-tilis through MinD-dependent polar localization and activation of MinC. Mol Microbiol. 1999;33:84–96. doi: 10.1046/j.1365-2958.1999.01450.x. [DOI] [PubMed] [Google Scholar]
- 85.Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–30. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mercer KLN, Weiss DS. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J Bacteriol. 2002;184:904–12. doi: 10.1128/jb.184.4.904-912.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mingorance J, Rueda S, Gomez-Puertas P, Valencia A, Vicente M. Escherichia coli FtsZ polymers contain mostly GTP and have a high nucleotide turnover. Mol Microbiol. 2001;41:83–91. doi: 10.1046/j.1365-2958.2001.02498.x. [DOI] [PubMed] [Google Scholar]
- 88.Mizusawa S, Court D, Gottesman S. Transcription of the sulA gene and repression by LexA. J Mol Biol. 1983;171:337–43. doi: 10.1016/0022-2836(83)90097-9. [DOI] [PubMed] [Google Scholar]
- 89.Mizusawa S, Gottesman S. Protein degradation in Escherichia coli: The lon gene controls the stability of SulA protein. Proc Natl Acad Sci USA. 1983;80:358–62. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, et al. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19:3179–91. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moy FJ, Glasfeld E, Mosyak L, Powers R. Solution structure of ZipA, a crucial component of Escherichia coli cell division. Biochemistry. 2000;39:9146–56. doi: 10.1021/bi0009690. [DOI] [PubMed] [Google Scholar]
- 92.Mukherjee A, Cao C, Lutkenhaus J. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2885–90. doi: 10.1073/pnas.95.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–57. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–58. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–69. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mukherjee A, Lutkenhaus J. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J Bacteriol. 1999;181:823–32. doi: 10.1128/jb.181.3.823-832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mulder E, Woldringh CL. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989;171:4303–14. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nogales E, Downing KH, Amos LA, Lowe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–58. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]; a O’Brien ET, Voter WA, Erickson HP. GTP hydrolysis during microtubule assembly. Biochemistry. 1987;26:4148–56. doi: 10.1021/bi00387a061. [DOI] [PubMed] [Google Scholar]
- 99.Ohashi T, Hale CA, de Boer PAJ, Erickson HP. Structural evidence that the P/Q domain of ZipA is an unstructured, flexi-ble tether between the membrane and the C-terminal FtsZ-binding domain. J Bacteriol. 2002;184:4313–15. doi: 10.1128/JB.184.15.4313-4315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Oosawa F, Askura S. Thermodynamics of the Polymerization of Protein. New York: Academic; 1975. p. 204. [Google Scholar]
- 101.Osteryoung KW. Organelle fission in eukaryotes. Curr Opin Microbiol. 2001;4:639–46. doi: 10.1016/s1369-5274(01)00263-6. [DOI] [PubMed] [Google Scholar]
- 102.Palacios P, Vicente M, Sanchez M. Dependency of Escherichia coli cell-division size, and independency of nu-cleoid segregation on the mode and level of ftsZ expression. Mol Microbiol. 1996;20:1093–98. doi: 10.1111/j.1365-2958.1996.tb02549.x. [DOI] [PubMed] [Google Scholar]
- 103.Pelham RJ, Chang F. Actin dynamics in the contractile ring during cytokine-sis in fission yeast. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- 104.Pichoff S, Lutkenhaus J. Escherichia coli division inhibitor MinCD blocks septation by preventing Z ring formation. J Bacteriol. 2001;183:6630–35. doi: 10.1128/JB.183.22.6630-6635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–93. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pollard TD. Measurement of rate constants for actin filament elongation in solution. Anal Biochem. 1983;134:406–12. doi: 10.1016/0003-2697(83)90316-0. [DOI] [PubMed] [Google Scholar]
- 107.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–76. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 108.Pollard TD, Cooper JA. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu Rev Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- 109.Quardokus E, Din N, Brun YV. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc Natl Acad Sci USA. 1996;93:6314–19. doi: 10.1073/pnas.93.13.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Quardokus EM, Brun YV. DNA replication initiation is required for mid-cell positioning of FtsZ rings in Caulobacter crescentus. Mol Microbiol. 2002;45:605–16. doi: 10.1046/j.1365-2958.2002.03040.x. [DOI] [PubMed] [Google Scholar]
- 111.Quardokus EM, Din N, Brun YV. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol Microbiol. 2001;39:949–59. doi: 10.1046/j.1365-2958.2001.02287.x. [DOI] [PubMed] [Google Scholar]
- 112.Raskin D, de Boer P. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–76. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raskin DM, de Boer PA. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999;181:6419–24. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.RayChaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–83. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.RayChaudhuri D, Park JT. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–54. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 116.RayChaudhuri D, Park JT. A point mutation converts Escherichia coli FtsZ septation GTPase to an ATPase. J Biol Chem. 1994;269:22941–44. [PubMed] [Google Scholar]
- 117.Regamey A, Harry EJ, Wake RG. Mid-cell Z ring assembly in the absence of entry into the elongation phase of the round of replication in bacteria: coordinating chromosome replication with cell division. Mol Microbiol. 2000;38:423–34. doi: 10.1046/j.1365-2958.2000.02130.x. [DOI] [PubMed] [Google Scholar]
- 118.Rivas G, Lopez A, Mingorance J, Ferrandiz MJ, Zorrilla S, et al. Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer—the primary steps for FtsZ assembly. J Biol Chem. 2000;275:11740–49. doi: 10.1074/jbc.275.16.11740. [DOI] [PubMed] [Google Scholar]
- 119.Romberg L, Simon M, Erickson HP. Polymerization of FtsZ, a bacterial homolog of tubulin: Is assembly cooperative? J Biol Chem. 2001;276:11743–53. doi: 10.1074/jbc.M009033200. [DOI] [PubMed] [Google Scholar]
- 120.Rothfield L, Justice S, Garcia-Lara J. Bacterial cell division. Annu Rev Genet. 1999;33:423–48. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- 121.Sackett MJ, Kelly AJ, Brun YV. Ordered expression of FtsQA and FtsZ during the Caulobacter crescentus cell cycle. Mol Microbiol. 1998;28:421–34. doi: 10.1046/j.1365-2958.1998.00753.x. [DOI] [PubMed] [Google Scholar]
- 122.Sanchez M, Valencia A, Ferrandiz M-J, Sander C, Vicente M. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 1994;13:4919–25. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Scheffers DJ, de Wit JG, den Blaauwen T, Driessen AJM. Substitution of a conserved aspartate allows cation-induced polymerization of FtsZ. FEBS Lett. 2001;494:34–37. doi: 10.1016/s0014-5793(01)02310-9. [DOI] [PubMed] [Google Scholar]
- 124.Scheffers DJ, de Wit JG, den Blaauwen T, Driessen AJM. GTP hydrolysis of cell division protein FtsZ: Evidence that the active site is formed by the association of monomers. Biochemistry. 2002;41:521–29. doi: 10.1021/bi011370i. [DOI] [PubMed] [Google Scholar]
- 125.Scheffers DJ, den Blaauwen T, Driessen AJ. Non-hydrolysable GTP-gamma-S stabilizes the FtsZ polymer in a GDP-bound state. Mol Microbiol. 2000;35:1211–19. doi: 10.1046/j.1365-2958.2000.01791.x. [DOI] [PubMed] [Google Scholar]
- 126.Scheffers DJ, Driessen AJM. Immediate GTP hydrolysis upon FtsZ poly-merization. Mol Microbiol. 2002;43:1517–21. doi: 10.1046/j.1365-2958.2002.02828.x. [DOI] [PubMed] [Google Scholar]
- 127.Shapiro L, Losick R. Dynamic spatial regulation in the bacterial cell. Cell. 2000;100:89–98. doi: 10.1016/s0092-8674(00)81686-4. [DOI] [PubMed] [Google Scholar]
- 128.Sharp MD, Pogliano K. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science. 2002;295:137–39. doi: 10.1126/science.1066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sossong TM, Brigham-Burke MR, Hensley P, Pearce KH. Self-activation of guanosine triphosphatase activity by oligomerization of the bacterial cell division protein FtsZ. Biochemistry. 1999;38(1):4843–50. doi: 10.1021/bi990917e. [DOI] [PubMed] [Google Scholar]
- 130.Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci USA. 2002;99:3171–75. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sun Q, Margolin W. Influence of the nucleoid on placement of FtsZ and MinE rings in Escherichia coli. J Bacteriol. 2001;183:1413–22. doi: 10.1128/JB.183.4.1413-1422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sun Q, Yu XC, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–503. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 133.Szeto TH, Rowland SL, Rothfield LI, King GF. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci USA. 2002;99:15693–98. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Taschner PE, Huls PG, Pas E, Woldringh CL. Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Es-cherichia coli during temperature shift experiments. J Bacteriol. 1988;170:1533–40. doi: 10.1128/jb.170.4.1533-1540.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Trusca D, Bramhill D. Fluorescent assay for polymerization of purified bacterial FtsZ cell-division protein. Anal Biochem. 2002;307:322–29. doi: 10.1016/s0003-2697(02)00036-2. [DOI] [PubMed] [Google Scholar]
- 136.Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of pu-rified FtsZ cell division protein. J Bacte-riol. 1998;180:3946–53. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.van den Ent F, Amos L, Lowe J. Bacterial ancestry of actin and tubulin. Curr Opin Microbiol. 2001;4:634–38. doi: 10.1016/s1369-5274(01)00262-4. [DOI] [PubMed] [Google Scholar]
- 138.van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 139.van den Ent F, Lowe J. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 2000;19:5300–7. doi: 10.1093/emboj/19.20.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vicente M, Kushner SR, Garrido T, Aldea M. The role of the ‘gearbox’ in the transcription of essential genes. Mol Microbiol. 1991;5:2085–91. doi: 10.1111/j.1365-2958.1991.tb02137.x. [DOI] [PubMed] [Google Scholar]
- 141.Voter WA, Erickson HP. The kinetics of microtubule assembly. Evidence for a two-stage nucleation mechanism. J Biol Chem. 1984;259:10430–38. [PubMed] [Google Scholar]
- 142.Deleted in proof
- 143.Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–59. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang X, Lutkenhaus J. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol Microbiol. 1993;9:435–42. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
- 145.Ward JE, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985;42:941–49. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- 146.Weart RB, Levin PA. Growth rate dependent regulation of medial FtsZ ring formation in Bacillus subtilis. J Bacteriol. 2003;185:2826–34. doi: 10.1128/JB.185.9.2826-2834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Welch MD, Mullins RD. Cellular control of actin nucleation. Annu Rev Cell Dev Biol. 2002;18:247–88. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
- 148.White EL, Ross LJ, Reynolds RC, Seitz LE, Moore GD, Borhani DW. Slow polymerization of Mycobacterium tuberculosis FtsZ. J Bacteriol. 2000;182:4028–34. doi: 10.1128/jb.182.14.4028-4034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Woldringh CL, Mulder E, Valkenburg JA, Wientjes FB, Zaritsky A, Nanninga N. Role of the nucleoid in the toporegu-lation of division. Res Microbiol. 1990;141:39–49. doi: 10.1016/0923-2508(90)90096-9. [DOI] [PubMed] [Google Scholar]
- 150.Wu L, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–75. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 151.Yan K, Pearce KH, Payne DJ. A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus au-reus. Biochem Biophys Res Commun. 2000;270:387–92. doi: 10.1006/bbrc.2000.2439. [DOI] [PubMed] [Google Scholar]
- 152.Yim L, Vandenbussche G, Mingorance J, Rueda S, Casanova M, et al. Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J Bacteriol. 2000;182:6366–73. doi: 10.1128/jb.182.22.6366-6373.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.York K, Kenney TJ, Satola S, Moran CP, Jr, Poth H, Youngman P. Spo0A controls the sA dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J Bacteriol. 1992;174:2648–58. doi: 10.1128/jb.174.8.2648-2658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yu XC, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–63. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yu XC, Margolin W. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol. 1999;32:315–26. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]
- 156.Yu XC, Sun Q, Margolin W. FtsZ rings in mukB mutants with or without the Min system. Biochimie. 2001;83:125–29. doi: 10.1016/s0300-9084(00)01222-0. [DOI] [PubMed] [Google Scholar]


