Summary
In response to a cell cycle signal, the cytoskeletal protein FtsZ assembles into a ring structure that establishes the location of the division site and serves as a framework for assembly of the division machinery. A battery of factors control FtsZ assembly to ensure that the ring forms in the correct position and at the precise time. EzrA, a negative regulator of FtsZ ring formation, is important for ensuring that the ring forms only once per cell cycle and that cytokinesis is restricted to mid-cell. EzrA is distributed throughout the plasma membrane and localizes to the ring in an FtsZ-dependent manner, suggesting that it interacts directly with FtsZ to modulate assembly. We have performed a series of experiments examining the interaction between EzrA and FtsZ. As little as twofold overexpression of EzrA blocks FtsZ ring formation in a sensitized genetic background, consistent with its predicted function. A purified EzrA fusion protein interacts directly with FtsZ to block assembly in vitro. Although EzrA is able to inhibit FtsZ assembly, it is unable to disassemble preformed polymers. These data support a model in which EzrA interacts directly with FtsZ at the plasma membrane to prevent polymerization and aberrant FtsZ ring formation.
Introduction
The ability of cells to respond to both internal and external signals depends in part on the intrinsically dynamic nature of their cytoskeleton. In eukaryotes, microtubule dynamics change dramatically between interphase and mitosis when chromatin-dependent stabilization of microtubules leads to the formation of the mitotic spindle (Desai and Mitchison, 1997). Moreover, despite their seemingly static appearance, spindle microtubules are highly dynamic structures. Lowering microtubule subunit turnover through the addition of the drug paclataxel leads to hyperstabilization of microtubules and a block in mitosis and cell cycle progression (Downing, 2000). Similarly, in bacteria, the spatial and temporal regulation of cytokinesis is dependent on the precisely balanced assembly dynamics of the essential cell division protein FtsZ (Romberg and Levin, 2003).
In response to an unidentified cell cycle signal, FtsZ, a tubulin-like GTPase, assembles into a ring structure (the Z ring) at mid-cell (Romberg and Levin, 2003). Z-ring formation is the earliest event identified to date in bacterial cytokinesis. The Z ring establishes the location of the division site and is responsible for recruiting cell division proteins to the nascent septum (Daniel et al., 1998; Rothfield et al., 1999; Weiss et al., 1999). As with tubulin, GTP binding enhances FtsZ assembly (Mukherjee and Lutkenhaus, 1994). Dimerization of two FtsZ monomers leads to the formation of a GTPase active site (Löwe and Amos, 1999; Scheffers et al., 2002). It is not known whether the Z ring consists of FtsZ bound to GDP or GTP; however, in vitro, GTP hydrolysis leads to a conformational change in polymer structure and rapid disassembly (Mukherjee and Lutkenhaus, 1998a; Romberg et al., 2001). Although the ring appears to be a static structure by immunofluorescence microscopy, fluorescence recovery after photobleaching (FRAP) experiments suggest that the half-time for subunit replacement in the Escherichia coli Z ring is between 8 and 30 s (H. P. Erickson, personal communication; Stricker et al., 2002). Despite an intracellular concentration of FtsZ approximately an order of magnitude greater than the critical concentration required for assembly in vitro (Lu et al., 1998), only ≈30% of FtsZ in the cell is in the ring at a given time (Stricker et al., 2002). Factors that inhibit FtsZ polymerization are therefore critical for maintaining the pool of unassembled FtsZ and preventing aberrant Z-ring formation.
Shifting the balance in FtsZ assembly dynamics can have dramatic and, in some cases, catastrophic consequences in growing cells. Stabilizing FtsZ polymers by increasing the intracellular concentration of FtsZ two- to fivefold leads to the formation of rings at cell poles and non-productive polar septation (Ward and Lutkenhaus, 1985; Weart and Levin, 2003). Overexpressing FtsZ to higher levels disrupts Z-ring formation entirely, resulting in lethal filamentation (Ward and Lutkenhaus, 1985; R. B. Weart, unpublished data).
A number of proteins have been identified that act to modulate FtsZ assembly in vivo (Romberg and Levin, 2003). ZipA (Hale and Boer, 1997; RayChaudhuri, 1999) and FtsA (Beall and Lutkenhaus, 1989; 1992; Din et al., 1998) are essential proteins that work synergistically in E. coli to promote Z-ring formation (Pichoff and Lutkenhaus, 2002; Geissler et al., 2003). The stabilizing factor ZapA is important for maintaining the delicate balance between polymer and monomer, although it is dispensable for growth in Bacillus subtilis under normal laboratory conditions (Gueiros-Filho and Losick, 2002). Several inhibitors of FtsZ assembly have also been identified. The best characterized of these, MinC and MinD, function as a complex to prevent Z-ring formation and subsequent septation at cell poles (Rothfield et al., 1999). FtsA, ZapA, MinC and MinD are more widely conserved (Rothfield et al., 1999; Gueiros-Filho and Losick, 2002). ZipA is found only in the γ-proteobacteria.
We previously identified EzrA, a transmembrane protein involved in the spatial regulation of FtsZ in B. subtilis (Levin et al., 1999). EzrA is found throughout the low-GC Gram-positive bacteria. Null mutations in ezrA lower the critical concentration of FtsZ required for assembly and lead to the formation of polar Z rings and septa (Levin et al., 1999). Polar Z rings in ezrA mutants are most probably caused by hyperstabilized FtsZ polymers overcoming the activity of MinCD at the cell poles. Loss-of-function mutations in ezrA compensate for the destabilizing effect of 15-fold MinCD over-expression on Z-ring formation (Levin et al., 2001). EzrA is uniformly distributed throughout the plasma membrane in non-dividing cells. In dividing cells, EzrA remains distributed throughout the plasma membrane, but also concentrates at the cytokinetic ring in an FtsZ-dependent manner (Levin et al., 1999). This localization pattern suggests that EzrA is able to prevent FtsZ assembly anywhere along the inner surface of the membrane. However, upon the initiation of cell division, a positive acting factor(s) presumably overcomes EzrA activity in a position-specific manner, permitting the formation of a medial Z ring.
EzrA’s ability to co-localize with FtsZ to the cytokinetic ring suggests that it may inhibit Z-ring formation through direct interactions. In confirmation of EzrA’s function as an inhibitor of FtsZ assembly, we have determined that twofold overexpression of EzrA is sufficient to block Z-ring formation and cell division in a sensitized genetic background. In addition, we have conducted a series of in vitro experiments exploring the mechanism by which EzrA destabilizes FtsZ polymers. Our data indicate that EzrA interacts directly with FtsZ at the plasma membrane to prevent polymerization and aberrant Z-ring formation.
Results
Overexpression of EzrA inhibits FtsZ assembly in vivo
Previous work suggests that EzrA prevents aberrant Z-ring formation by inhibiting FtsZ assembly (Levin et al., 1999). This model predicts that EzrA overexpression should inhibit Z-ring formation in vivo. To test this, we put a second copy of ezrA under the control of a strong IPTG-inducible, Lac I-repressible promoter (Pspachy) (Quisel et al., 2001) at the amylase locus (see Experimental procedures) Three hours after the addition of 1 mM IPTG to cells encoding amyE∷Pspachy-ezrA (PL1031), EzrA levels were approximately twofold greater than wild type as detected by quantitative immunoblot (data not shown). This modest increase in EzrA levels did not appear to alter the phenotype or viability of otherwise wild-type cells. There was no significant change in either the frequency or the position of Z-ring formation as detected by immunofluorescence microscopy (data not shown).
Although it did not cause a detectable difference in wild-type cells, twofold overexpression of EzrA was sufficient to impair division severely in cells encoding ftsZts, a heat-sensitive allele of ftsZ consisting of wild-type FtsZ fused to a green fluorescent protein (GFP) moiety (Levin et al., 1999), at permissive temperatures. At 37°C, ftsZts cells are near wild type for growth, although they are slightly longer and exhibit rings at quarter positions, presumably because of a delay in division at mid-cell. However, the GFP fusion renders FtsZ unable to form rings at the restrictive temperature of 45°C leading to extensive filamentation and cell death. A loss-of-function mutation in ezrA was previously shown to suppress the heat sensitivity of this conditional allele at 45°C (Levin et al., 1999). Taking advantage of the GFP tag on the C-terminus of FtsZts to visualize Z rings in live cells, we determined that MO54 (amyE∷Pspachy-ezrA ftsZts) cells were near normal for division and had a high frequency of Z rings after growth in the absence of IPTG at the permissive temperature of 37°C (Fig. 1A and C). In contrast, 3 h after the addition of IPTG, exponentially growing amyE∷Pspachy-ezrA ftsZts cells within the field of view were on average four to five times longer than those grown in the absence of inducer. This is probably an underestimate as many filaments extended well beyond the field of view making them difficult to measure (Fig. 1B). Although there were a few Z rings in the filaments, they were irregularly spaced, and the distance between any two rings was significantly greater than the length of a typical wild-type cell (Fig. 1D). Consistent with the absence of Z rings and extensive filamentation, the viability of amyE∷Pspachy-ezrA ftsZts cells decreased 100- to 1000-fold after induction of EzrA, as determined by plating efficiency in the presence and absence of IPTG. These data support our model in which EzrA prevents aberrant Z-ring formation by altering FtsZ assembly dynamics in the cell.
Fig. 1.
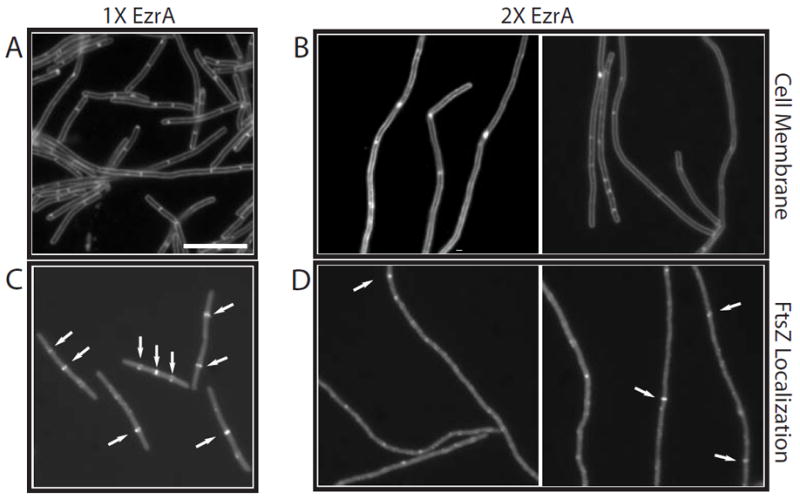
The effect of EzrA overexpression on cell division and Z-ring formation in ftsZts mutant cells.
A and B. Cells were stained with the vital membrane stain FM4-64 to visualize septa.
C and D. FtsZts-GFP.
A and C. MO54 (amyE∷Pspachy-ezrA∷ftsZts) cells grown in the absence of inducer.
B and D. MO54 cells 3 h after the addition of IPTG. EzrA levels are twofold above wild type. Arrows point to FtsZ rings. Scale bar = 5 μm.
EzrA inhibits sedimentation of purified FtsZ assembled in the presence of GTP and DEAE-dextran
Genetic and cell biological data (Levin et al., 1999; 2001) suggest that EzrA prevents aberrant Z-ring formation by modulating FtsZ assembly dynamics. To test EzrA’s ability to inhibit FtsZ assembly in vitro, we added increasing concentrations of EzrA to B. subtilis FtsZ assembly reactions and assayed its effect on FtsZ polymerization by two different methods: sedimentation and 90° angle light scattering.
For these experiments, we used an EzrA fusion protein in which we have replaced EzrA’s 26-residue membrane-spanning domain with E. coli thioredoxin to enhance solubility. We have also added a 6×His tag to the C-terminus of EzrA in this construct to facilitate purification. The 26-residue membrane-spanning domain is not required for EzrA localization to the cytokinetic ring (Fig. 2). However, both an EzrA protein missing only the membrane anchor and the Thio–EzrA fusion protein fail to complement an ezrA null mutation for the defect in FtsZ localization (data not shown). This lack of complementation is presumably due to a role for the transmembrane anchor in concentrating EzrA at the plasma membrane. To demonstrate that any effect on FtsZ assembly resulted from EzrA itself, thioredoxin-6×His was used as a control. B. subtilis FtsZ was purified using a modified version of the strategy described by Wang and Lutkenhaus (1993) (see Experimental procedures).
Fig. 2.
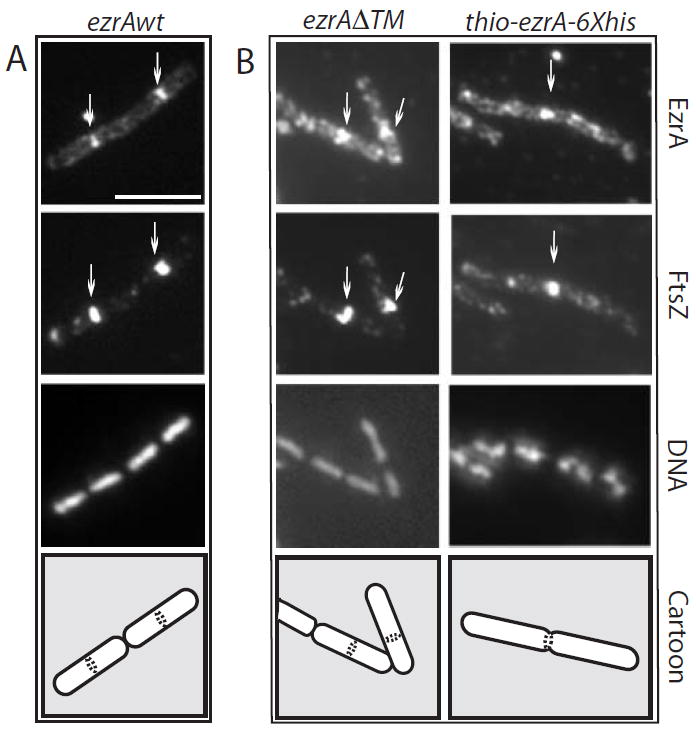
The transmembrane anchor is dispensable for EzrA localization. Cells were grown to mid-exponential phase (OD600 = ≈ 0.5) in the presence of IPTG and fixed and stained for immunofluorescence microscopy using antiserum against FtsZ (Levin and Losick, 1996) or EzrA (this work). A single field of view is shown in each column. From top to bottom: EzrA, FtsZ, DNA and a schematic showing cell boundaries and the position of the EzrA/FtsZ rings.
A. ezrA∷Pspachy-ezrA (PL923).
B. ezrA∷Pspachy-ezrAΔTM (PL925). The high background in the anti-EzrA panels in (B) results from cytoplasmic staining of EzrA in the absence of the transmembrane domain. Scale bar = 5 μM.
In the sedimentation assay, we relied on the ability of large bundles of FtsZ polymers to pellet efficiently at 250 000 g. FtsZ assembled in the presence of GTP forms single-stranded polymers (protofilaments), paired protofilaments and, occasionally, small hoop-shaped bundles of polymers (Fig. 3A–C). Efficient sedimentation of polymerized FtsZ requires the addition of bundling agents such as divalent cations or DEAE-dextran to encourage the formation of high-molecular-weight polymer bundles (Yu and Margolin, 1997; Mukherjee and Lutkenhaus, 1998b; Trusca et al., 1998). B. subtilis FtsZ assembly reactions conducted in the presence of DEAE-dextran and GTP indicate that the majority of protein is in large macromolecular bundles, each consisting of tens of single-stranded polymers in parallel arrays (Fig. 3D). Although DEAE-dextran-induced assembly does not require GTP (Trusca et al., 1998), we found that B. subtilis FtsZ sedimentation was reduced up to ≈40% when GDP was used in place of GTP in the reaction, although this was somewhat variable (data not shown). DEAE-dextran-induced sedimentation of FtsZ has been used previously to measure the effect of the division inhibitor SulA on the in vitro assembly of E. coli FtsZ (Trusca et al., 1998).
Fig. 3.

Electron micrographs (EMs) of B. subtilis FtsZ assembled under different conditions. A. ‘Deep-etch’ EM of FtsZ polymers assembled in the presence of GTP. Micrograph kindly provided by Dr John Heuser.
B–D. Negative stain EM of (B) and (C) FtsZ assembled in the presence of GTP and (D) FtsZ assembled in the presence of GTP and 0.1 mg ml−1 DEAE-dextran. Note the single-stranded polymers, paired protofilaments and small bundles of FtsZ in (B), hoop-shaped bundle in (C) and large bundles of filaments in (D). FtsZ was at 5 μM for both the ‘deep etch’ and the negative stain EM. GTP was at 1 mM for all experiments. The scale bar is 50 nm in (A) and 20 nm in (B) to (D).
EzrA efficiently inhibits FtsZ sedimentation when it is added to the assembly reaction before GTP but before the addition of DEAE-dextran. We began to see some inhibition of FtsZ assembly at a 1:5 molar ratio of EzrA to FtsZ and to get near maximal inhibition at a 1:1 ratio of EzrA to FtsZ (Fig. 4A and C). The addition of the thioredoxin control protein did not significantly affect FtsZ assembly at any of the concentrations tested (Fig. 4A and C). We estimate that there are between 10 000 and 20 000 EzrA molecules per cell during exponential growth in Luria broth (Fig. 5). Previous estimates place the intracellular concentration of B. subtilis FtsZ at ≈5000 molecules per cell (Feucht et al., 2001). The ratio at which the EzrA fusion protein maximally inhibits FtsZ assembly in vitro is approximately equivalent to the in vivo ratio of EzrA to FtsZ.
Fig. 4.
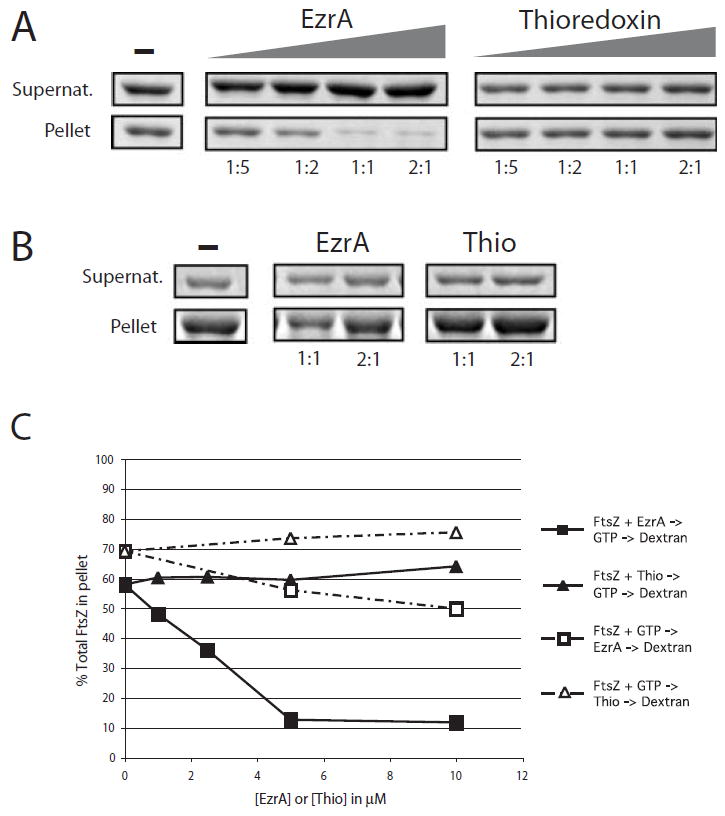
EzrA inhibits sedimentation of FtsZ assembled in vitro. FtsZ is 5 μM in all reactions. The samples were spun at 250 000 g to separate unassembled protein from protofilament bundles, and the relative concentration of protein in the supernatants (unassembled FtsZ) and pellets (assembled FtsZ) was analysed by SDS–PAGE.
A. EzrA and thioredoxin were added to final concentrations of 1, 2.5 μM, 5 μM and 10 μM before the addition of GTP and DEAE-dextran. Left. Increasing concentration of FtsZ in the supernatants and decreasing concentration of FtsZ in the pellets result from the inhibition of FtsZ assembly by purified EzrA. Right. The thioredoxin control protein does not significantly inhibit FtsZ assembly. The ratio of EzrA or thioredoxin to FtsZ is shown under each lane. B. Order of addition. EzrA and thioredoxin were added to the reaction 1 min after the addition of GTP. Ratios of EzrA and thioredoxin to FtsZ are shown below each lane.
C. Plot of percentage of total FtsZ in the pellet. EzrA, squares; thioredoxin, triangles. Closed symbols represent data from the experiment shown in (A). Open symbols represent data from the experiment shown in (B). Differences in the sedimentation efficiency of FtsZ in the absence of EzrA or thioredoxin between the experiments shown in (A) and (B) reflect normal variations between protein preparations.
Fig. 5.
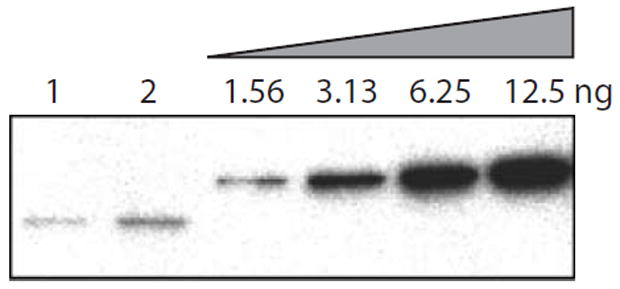
Quantification of EzrA protein in B. subtilis. Wild-type cells (JH642) grown to mid-exponential phase (OD600 = 0.5) in Luria broth (LB) were sampled for colony-forming units and lysed as described in Experimental procedures. Different amounts of lysate (6.29 × 103 cfu μl−1) were loaded in lane 1 (5 μl) and lane 2 (10 μl). EzrA was detected by immunoblotting with whole sera. EzrA was quantified by comparing the intensity of the bands in lanes 1 and 2 with that of a twofold serial dilution of purified EzrA fusion protein (four right hand lanes) taking into account the number of colony-forming units. Based on these data, we estimate that there are 10 000–20 000 copies of EzrA per cell during exponential growth in LB.
The order of addition appears to be critical to the ability of EzrA to block FtsZ assembly. We achieved the greatest inhibition of FtsZ sedimentation when FtsZ was preincubated with EzrA before GTP was added to the reaction. In contrast, when EzrA was added after FtsZ polymerization was stimulated by the addition of 1 mM GTP, the degree to which EzrA inhibited FtsZ assembly was significantly reduced (Fig. 4B and C). Based on data from 90° angle light scattering experiments detailed below, we presume that the small amount of inhibition we observed resulted from EzrA’s effect on the unassembled FtsZ still extant 1 min after the addition of GTP. If EzrA was added after DEAE-dextran, we saw little if any change in FtsZ assembly (data not shown). These data suggest that EzrA normally acts to prevent the formation of new polymers but is unable to disassemble preformed polymers or protofilament bundles.
EzrA inhibits FtsZ polymerization in a 90° angle light scattering assay
The requirement for the bundling agent DEAE-dextran may affect the results of the sedimentation assay described above. For example, the bundling agent can stabilize otherwise unstable polymers and can also induce significant sedimentation in the presence of GDP as well as in the absence of nucleotide (Trusca et al., 1998). To address these issues and to distinguish between a model in which EzrA inhibits FtsZ polymerization and one in which EzrA prevents the DEAE-dextran-facilitated formation of large macromolecular bundles of FtsZ polymers, we tested EzrA’s effect on FtsZ assembly in a 90° angle light scattering assay.
An alternative method for quantifying FtsZ assembly, 90° angle light scattering provides a means of measuring relative amounts of FtsZ polymerization in real time in the absence of a bundling agent (Mukherjee and Lutkenhaus, 1999). Light scattering relies on the fact that single-stranded FtsZ polymers and small polymer bundles diffract light better than FtsZ monomers. The addition of GTP to an FtsZ assembly reaction results in the formation of single-stranded polymers and small polymer bundles (Fig. 3B and C) and thus leads to a rapid increase in the degree of light scattering at 310 nm. In contrast to DEAE-dextran-stimulated sedimentation, light scattering is GTP-dependent rendering it more representative of physiological conditions.
The addition of 1 mM GTP to a reaction containing 2.5 μM FtsZ and 5 μM thioredoxin control protein led to a rapid increase in light scattering (Fig. 6A). In contrast, when the same reaction was conducted in the presence of a 2:1 molar ratio of EzrA to FtsZ, the increase in light scattering was approximately one-tenth that observed with thioredoxin (Fig. 6A and B). GDP does not stimulate FtsZ assembly under these conditions (Fig. 6A and B). A titration of EzrA indicated that, as in the sedimentation assay, maximal inhibition occurred at a 2:1 molar ratio of EzrA to FtsZ (Fig. 6B). These data corroborate the results of the sedimentation assay (Fig. 4) and suggest that EzrA is able to inhibit the formation of single-stranded FtsZ polymers and/or protofilament bundles.
Fig. 6.

EzrA inhibits FtsZ polymerization as measured by 90° angle light scattering. Light scattering is in arbitrary units (AU). FtsZ is at 2.5 μM in all reactions.
A. EzrA or thioredoxin was added to the reaction before the addition of GTP to a final concentration of 5 μM. Arrow indicates the time of addition of 1 mM GTP or 1 mM GDP. The addition of GDP reduces FtsZ light scattering below baseline, presumably because it inhibits GTP-independent association of FtsZ monomers.
B. Concentration-dependent inhibition of FtsZ assembly by EzrA. The ratio of EzrA to FtsZ is shown underneath each bar. The ratio of thioredoxin to FtsZ is 2:1. Margins of error were calculated using data from three independent experiments.
C. EzrA or GDP were added to an FtsZ assembly reaction 1 min after the addition of GTP. The first arrow indicates the addition of GTP and the second the addition of EzrA or GDP. EzrA was at a final concentration of 5 μM in this experiment. GDP was used at a final concentration of 1 mM.
As with the sedimentation experiments, the order of addition was critical to the ability of EzrA to inhibit FtsZ assembly in the light scattering assay. We detected efficient inhibition of FtsZ assembly only when the EzrA fusion protein and FtsZ were combined before the addition of GTP. If EzrA was added to the reaction 1 min subsequent to FtsZ and GTP, after the initiation of FtsZ assembly, we did not see a reduction in light scattering, although we did observe an inhibition of further assembly (Fig. 6C). The addition of 1 mM GDP resulted in a decrease in assembly, indicating that the polymers were dynamic under these experimental conditions. This observation reinforces the idea that EzrA normally functions to prevent FtsZ assembly but is unable to disassemble preformed polymers at the ratios that we tested in vitro.
Interestingly, in both the sedimentation and the light scattering assays, we did not see complete inhibition of FtsZ assembly at approximately physiological ratios of EzrA to FtsZ. We have confirmed this incomplete inhibition by electron microscopy of negatively stained FtsZ assembly reactions conducted in the presence and absence of EzrA (data not shown). We believe that the contrast between the incomplete inhibition of FtsZ assembly that we observed in vitro and EzrA’s ability to prevent aberrant Z-ring formation in vivo results from two key differences. First, as discussed in detail below, in vivo, the effective ratio of EzrA to FtsZ at the plasma membrane is likely to be an order of magnitude higher than we can assay the activity of our soluble EzrA fusion protein in vitro. Secondly, EzrA does not act alone inside the cell. Experiments demonstrating that a loss-of-function mutation in ezrA overcomes the block in Z-ring formation associated with 15-fold overexpression of MinCD (Levin et al., 2001) suggest that EzrA acts in concert with the Min proteins and potentially other as yet unidentified inhibitors of FtsZ assembly. Finally, preventing FtsZ-ring formation in vivo may not require complete inhibition of FtsZ assembly. Simply reducing either polymer length or stability may render FtsZ unable to support the assembly of the division apparatus.
EzrA interacts directly with FtsZ
EzrA localizes to the cytokinetic ring in an FtsZ-dependent manner (Levin et al., 1999). This observation, coupled with the ability of a purified EzrA fusion protein to inhibit FtsZ assembly in both the sedimentation and 90° angle light scattering assays (Figs 4 and 6), suggests that EzrA modulates FtsZ assembly dynamics through direct interactions. To investigate this possibility, we examined the capacity of EzrA to interact directly with FtsZ using a cross-linking agent in conjunction with affinity purification.
When the EzrA fusion protein and native FtsZ were combined in an equimolar ratio in the presence of the cross-linker DSP (linker arm = 12 Å), we observed the formation of a high-molecular-weight complex that presumably contains both FtsZ and EzrA (Fig. 7A, lane 3). We did not see this high-molecular-weight FtsZ complex in reactions containing FtsZ alone or in the presence of thioredoxin or a His-tagged version of Spo0J, a 32 kDa cytoplasmic protein involved in chromosome segregation in B. subtilis (Lin et al., 1997) (Fig. 7A, lanes 1, 4 and 5). Notably, EzrA forms high-molecular-weight complexes regardless of the presence of FtsZ, suggesting that it is able to interact with itself to some degree (Fig. 7B, lanes 2 and 3).
Fig. 7.
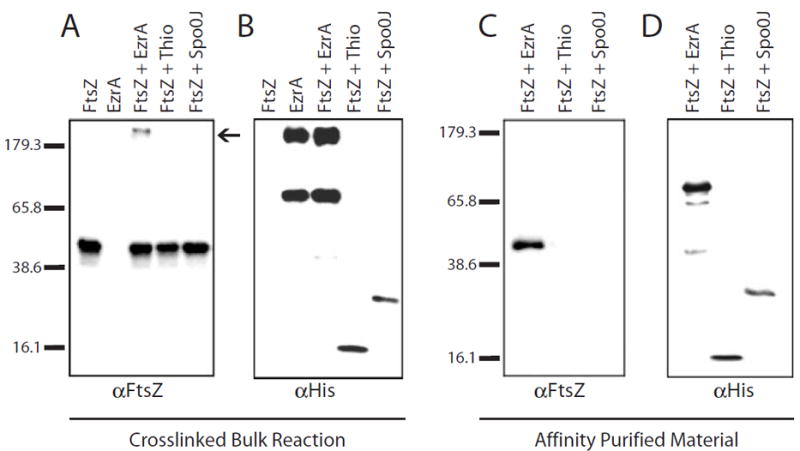
EzrA binds to FtsZ.
A and B. Immunoblots of cross-linking reaction products.
A. Anti-FtsZ.
B. Anti-6×His.
FtsZ alone (lane 1); EzrA alone (lane 2); FtsZ plus EzrA-6×His (lane 3); FtsZ plus Thio-6×His (lane 4); FtsZ plus Spo0J-6×His (lane 5).
C and D. Co2+ affinity purification of 6×His-tagged cross-linked products. Eluants were separated by SDS–PAGE and subjected to immunoblotting with antisera against FtsZ (C) or 6×His (D). FtsZ plus EzrA-6×His (lane 1); Thio-6×His (lane 2); or Spo0J-6×His (lane 3). Breakdown products of EzrA-6×His can be seen in (D), lane 1. Molecular weight markers in kDa are shown to the right of blots (A) and (C). Variations in band intensity reflect differences in transfer efficiency between high- and low-molecular-weight material.
To confirm that the high-molecular-weight FtsZ complexes were the product of an EzrA–FtsZ interaction, we took advantage of metal affinity resin charged with Co2+ to purify His-tagged products of the cross-linking reactions. FtsZ co-purified with His-tagged EzrA (Fig. 7C, lane 1), but not with either thioredoxin or the Spo0J-6×His control protein (Fig. 7C, lanes 2 and 3). To simplify analysis, the cross-link was broken before SDS–PAGE in Fig. 7C and D (see Experimental procedures).
These experiments indicate that the interaction between EzrA and FtsZ was specific. Based on quantitative analysis using imagegauge software (Fuji), we estimate that ≈ 19% of the total FtsZ is bound to EzrA after cross-linking. We were unable consistently to co-purify FtsZ with the EzrA fusion protein in the absence of cross-linker, suggesting that their interaction is probably transient. The addition of GTP or GDP had no effect on the ability of EzrA to bind FtsZ in these assays, indicating that EzrA is capable of interacting with a range of multimeric forms of FtsZ (data not shown).
The effect of EzrA on FtsZ’s intrinsic GTPase activity
FtsZ’s ability to hydrolyse GTP is dependent on the dimerization of two FtsZ monomers to form a GTPase active site (Scheffers et al., 2002). To determine how EzrA modulates polymer dynamics upon binding to FtsZ, we assessed the effect of EzrA on the intrinsic GTPase activity of FtsZ.
We measured FtsZ’s GTP hydrolysis rate in the presence and absence of EzrA using a spectrophotometric method for quantifying free phosphate (Molecular Probes). At a concentration of 5 μM, B. subtilis FtsZ was able to hydrolyse GTP at a rate of ≈0.65 ± 0.07 GTP/FtsZ min−1. The addition of either the EzrA fusion protein or the thioredoxin control protein did not have a strong effect on GTP hydrolysis. The average rate of GTP hydrolysis by FtsZ in the presence of 10 μM EzrA, a 2:1 molar ratio of EzrA to FtsZ, was 0.88 ± 0.11 GTP/FtsZ min−1. Similar results were obtained at a 2:1 ratio of thioredoxin to FtsZ (0.70 ± 0.26 GTP/FtsZ min−1). Taking into account the margin of error, these values are essentially equivalent to the rate of GTP hydrolysis by FtsZ alone. The order of addition did not have an appreciable effect on GTPase activity in this assay (data not shown). The inability of EzrA to alter significantly FtsZ’s GTPase activity at a 2:1 molar ratio suggests that EzrA does not substantially disrupt the interaction between FtsZ monomers despite its ability to inhibit FtsZ assembly at these concentrations in both the sedimentation and the 90° angle light scattering assays (Figs 4 and 6).
Discussion
Genetic and cell biological data suggest that EzrA interacts directly with FtsZ to prevent the formation of aberrant Z rings and polar septa (Levin et al., 1999; 2001) (Fig. 1). The results presented here indicate that EzrA interacts physically with FtsZ to inhibit the assembly of polymers and/or small bundles of protofilaments. Moreover, we find that, although EzrA prevents FtsZ assembly, it is not able to disassemble preformed polymers nor does it have a significant effect on FtsZ’s intrinsic GTPase activity. Taken together, these data support a model in which EzrA contributes to the spatial regulation of Z-ring formation in vivo by blocking FtsZ assembly at aberrant locations along the cell membrane (Fig. 8).
Fig. 8.

Model for EzrA inhibition of FtsZ assembly at the cell membrane. Left. EzrA functions at the membrane to inhibit FtsZ assembly and prevent aberrant Z-ring formation. At mid-cell, a positive acting factor (arrows) promotes FtsZ assembly and overcomes EzrA activity allowing for medial ring formation. Right. In the absence of EzrA, FtsZ is free to assemble anywhere along the inner surface of the membrane leading to aberrant FtsZ assembly at cell poles. Despite the formation of polar Z rings, a positive acting factor (arrows) still promotes FtsZ assembly at mid-cell and leads to a situation in which this position is favoured for cytokinesis.
EzrA exhibits activity similar to the cell division inhibitor MinC. In B. subtilis and E. coli, MinC acts in conjunction with MinD to prevent polar Z-ring formation and polar septation. MinD keeps MinC sequestered at the cell poles where it appears to interact directly with FtsZ to inhibit assembly. Twenty-five- to 50-fold overexpression of E. coli MinC inhibits division in vivo (de Boer et al., 1992), and a MalE–MinC fusion protein blocks sedimentation of FtsZ polymers in vitro when both proteins are at approximately equimolar ratios (Hu et al., 1999). Like EzrA, MinC does not alter FtsZ’s GTP hydrolysis activity even when MinC is in molar excess (Hu et al., 1999).
In contrast to MinC, however, fluorescence microscopy indicates that EzrA is distributed throughout the plasma membrane and concentrates at mid-cell in an FtsZ-dependent manner (Levin et al., 1999). This localization pattern presents something of a conundrum; how can a protein that co-localizes with FtsZ at mid-cell prevent polar septation but not block medial ring formation? The order of addition experiments may provide a partial explanation. Both sedimentation and 90° angle light scattering experiments indicate that, although EzrA blocks polymerization of FtsZ monomers, it is unable to disassemble preformed polymers. This supports a situation in vivo in which EzrA acts throughout the plasma membrane to prevent FtsZ assembly at inappropriate locations. However, at mid-cell, a temporally regulated positive acting factor, perhaps a component of FtsZ’s putative nucleation site (Addinall and Lutkenhaus, 1996), overcomes EzrA activity, allowing for FtsZ polymerization and Z-ring formation. Once formed, FtsZ polymers are resistant to EzrA; although EzrA still interacts with FtsZ in the ring, it is unable to disassemble the polymers that constitute the Z ring. The stability of polymers in the medial Z ring is further enhanced through the activity of stabilizing proteins such as FtsA and ZapA (Gueiros-Filho and Losick, 2002; Pichoff and Lutkenhaus, 2002).
Localization to the plasma membrane appears to be important for both EzrA and MinC activity. Deleting EzrA’s membrane-spanning domain does not block localization to the Z ring (Fig. 2B), nor does it prevent EzrA from inhibiting FtsZ assembly in vitro (Figs 4 and 6). The loss of its membrane anchor, however, severely impairs EzrA’s ability to inhibit FtsZ assembly at cell poles (data not shown). Similarly, MinC requires interaction with the membrane-associated protein MinD to prevent polar Z-ring formation and septation (Rothfield et al., 1999). One explanation for this phenomenon may be a simple difference in effective concentration. Localizing two proteins to the surface of a membrane enhances their interaction by several orders of magnitude (Gelb, 1997). Thus, in wild-type cells, the effective ratio of EzrA or MinC to FtsZ at the cell membrane is likely to be orders of magnitude higher than if both proteins were cytoplasmic.
The proteins that regulate tubulin and actin polymerization provide a range of models for how EzrA might act to inhibit FtsZ assembly. For example, EzrA could function in a manner similar to the actin-associated protein thymosin beta-4 by binding to monomers of FtsZ, restricting the pool of free subunits available for polymerization (Safer et al., 1997; Huff et al., 2001). EzrA might also sever FtsZ polymers such as the microtubule-interacting protein katanin (McNally et al., 1996). Alternatively, EzrA could ‘cap’ polymerizing FtsZ, binding to one end of a growing polymer and preventing the addition of new subunits.
Our results argue against EzrA functioning as either a monomer-binding protein or a severing factor. Nucleotide hydrolysis requires the dimerization of two FtsZ monomers (Scheffers et al., 2002). At sufficiently high concentrations, a protein that binds to and sequesters free FtsZ monomers should significantly decrease GTP hydrolysis. SulA, an SOS-inducible protein that inhibits cell division in E. coli after DNA damage, blocks both FtsZ assembly and GTP hydrolysis when both proteins are equimolar in solution (Mukherjee et al., 1998; Trusca et al., 1998). The recently solved crystal structure of SulA indicates that it binds to FtsZ monomers, thereby preventing the formation of the GTPase active site and polymerization (Cordell et al., 2003). In contrast to SulA, EzrA does not significantly alter FtsZ’s ability to hydrolyse GTP at a 2:1 ratio of EzrA to FtsZ. This suggests that EzrA does not normally disrupt the interaction between FtsZ monomers and thus does not support a role for EzrA as a monomer-sequestering protein. Similarly, factors that sever FtsZ polymers should disrupt assembly regardless of when they are added to a polymerization reaction. Data from the sedimentation and light scattering assays indicate that, although EzrA prevents FtsZ polymerization, it is unable to disassemble preformed polymers (Figs 4B and C and 6C). Thus, it is unlikely that EzrA severs or otherwise alters the stability of FtsZ polymers.
Although the methods used here do not allow us to distinguish between models, our data are consistent with EzrA acting by either preventing the formation of stabilizing lateral interactions between FtsZ protofilaments or binding to the ends of growing polymers and preventing the addition of new subunits, a function analogous to actin modulatory proteins such as CapZ and capping protein (Pollard et al., 2000). However, these are just two of many possible models for EzrA activity. Extensive analysis will be required to understand the precise mechanism by which EzrA modulates FtsZ assembly in vivo.
In wild-type cells, FtsZ exists in a precise balance between soluble monomers, small multimers and the membrane-associated polymers that constitute the Z ring (Justice et al., 2000). Maintaining this balance requires a network of regulatory proteins to modulate FtsZ assembly. At the cell poles, EzrA, MinC and, potentially, the bacterial nucleoid (Margolin, 2001) act together to prevent aberrant Z-ring formation. However, during cytokinesis at mid-cell, instead of working in conjunction with MinC, EzrA has to counteract the effects of positive regulators of FtsZ assembly such as ZapA, FtsA and possibly components of the putative FtsZ nucleation site (Addinall and Lutkenhaus, 1996; Regamey et al., 2000). The balance at mid-cell is thus shifted in favour of FtsZ assembly leading to medial Z-ring formation. FtsZ levels in E. coli and B. subtilis are well above the critical concentration required for polymerization (Lu et al., 1998; Feucht et al., 2001). FtsZ assembly must therefore be tightly controlled both spatially and temporally to ensure that the Z ring forms only once per cell cycle and is appropriately positioned. Regardless of the mechanism by which EzrA inhibits FtsZ assembly, factors such as EzrA are essential for preventing ectopic FtsZ polymerization and maintaining the dynamic nature of the Z ring. Our data highlight the importance of precisely balanced polymer stability in the spatial and temporal regulation of Z-ring formation and reinforce the need to identify other members of the regulatory network responsible for modulating FtsZ assembly dynamics during the cell cycle.
Experimental procedures
General methods
Cloning and genetic manipulation were performed using standard techniques (Sambrook et al., 1989; Harwood and Cutting, 1990). KlentaqLA was used for polymerase chain reactions (PCRs). Unless otherwise noted, all strains were cultured at 37°C in Luria broth (LB). Chloramphenicol was used at a final concentration of 5 μg ml−1, spectinomycin at 100 μg ml−1 and ampicillin at 100 μg ml−1. FtsZ and Spo0J concentrations were determined using the bicinchoninic acid (BCA) protein assay and the Coomassie plus assay respectively (Pierce). EzrA and thioredoxin concentration were determined using a combination of OD280 and the BCA assay.
Bacterial strains
All B. subtilis strains are derivatives of the strain JH642 (Perego et al., 1988). PL1031 (amyE∷Pspachy-ezrA) was engineered to express ezrA from the strong IPTG-inducible promoter Pspachy (Quisel et al., 2001). ezrA was amplified from chromosomal DNA and ligated into the amylase Pspachy vector pPL82 (Levin et al., 2001). This vector integrates by homologous recombination into the amylase locus. Primer sequences are available on request. In the course of our investigation, we determined that even limited EzrA expression is lethal to exponentially growing E. coli cells because of a block in FtsZ assembly and division (P. A. Levin, unpublished data). Although this observation supports our model in which EzrA inhibits FtsZ assembly, it has made it difficult to clone ezrA through E. coli, even under the control of a B. subtilis inducible promoter, on account of unregulated ezrA expression. To circumvent this issue, the ligation mix was transformed directly into JH642 selecting for resistance to chloramphenicol, and transformants were screened for loss of amylase activity (Harwood and Cutting, 1990). MO54 is a derivative of PL1031 that encodes both amyE∷Pspachy-ezrA and the heat-sensitive allele of ftsZ, ftsZts (Levin et al., 1999).
The ezrA∷Pspachy-ezrA construct was made by cloning a 170 bp fragment encoding ezrA’s native ribosome binding site to basepair 37 of the ezrA open reading frame (ORF) behind the IPTG-inducible Pspachy promoter (Quisel et al., 2001) to create plasmid pPL78. ezrA∷Pspachy-ezrAΔTM was made by cloning the ribosome binding site, start codon and basepairs 73–233 of the ezrA ORF behind the Pspachy promoter to create plasmid pPL79. Both plasmids were integrated into the B. subtilis chromosome by single cross-over selecting for chloramphenicol resistance. Cells were screened by PCR and immunoblot to ensure that the integration resulted in an intact copy of ezrA or ezrAΔTM that was expressed in response to IPTG. The addition of 1 mM IPTG to these cells results in approximately wild-type levels of EzrA expression.
The His-tagged EzrA fusion protein was cloned under the control of the Pbad promoter in E. coli Top10 cells (Invitrogen) and then transferred to the slyD null strain BB101 [X90 = lac pro, F′ (lacIq) ara, lacpro, nalA argEam] (Chivers and Sauer, 1999) for expression. Pbad (Guzman et al., 1995) allows for tightly regulated expression of ezrA in E. coli. B. subtilis FtsZ was expressed in PL1184, a derivative of E. coli strain ER2566 (F− lambda− fhuA2 [lon] ompT lacZ∷T7 gene1 gal sulA11 D(mcrC-mrr)114∷IS10 R(mcr-73∷miniTn10-TetS)2 R(zgb-210∷Tn10) (TetS) endA1 [dcm]) (NEB) encoding the plasmid pBS58. pBS58 overexpresses E. coli ftsQAZ and allows for the cloning of B. subtilis ftsZ in E. coli (Wang and Lutkenhaus, 1993). For expression in B. subtilis, the Thio-EzrA-6×His fusion protein was amplified from the pBAD expression vector and placed behind the Pspachy promoter at the amylase locus for expression in B. subtilis.
Determination of EzrA concentration in cells
Quantitative immunoblotting was used to determine the amount of EzrA in exponentially growing B. subtilis cells. Wild-type cells (JH642) were grown to mid-exponential phase (OD600 = 0.5) in Luria broth, sampled for colony-forming units (cfu) and lysed with lysozyme and detergent. Colony-forming units were determined in duplicate. Five and 10 μl of cell lysate (6.29 × 103 cfu μl−1) were separated by 12% SDS–PAGE. Immunoblotting was performed using sera raised against the EzrA fusion protein (above) and goat anti-rabbit antibodies conjugated to horseradish peroxidase (Jackson Immunoresearch) (Harlow and Lane, 1988). Blots also included a twofold serial dilution of purified EzrA fusion protein from 12.5 ng to 1.56 ng. Immunoblots were developed using the ChemiDetect kit (Bio-Rad) and visualized and quantified with the luminescent image analyser LAS-1000plus in conjunction with IMAGEGAUGE software, version 3.41 (Fuji Film).
Microscopy
Cells were prepared for fluorescence microscopy as described previously (Levin et al., 2001). Images were captured using an Olympus BX51 microscope with an OrcaERG camera (Hamamatsu) in conjunction with the openlabs, version 3.0.9 software (Improvision). Images were processed with Adobe photoshop, version 6.0.1. ‘Deep-etch’ electron microscopy of FtsZ (Fig. 2A) protofilaments was done as described previously (Heuser, 1983; 1989) and visualized using a JEOL transmission electron microscope operating at 100 kV. Negative stain electron microscopy of FtsZ assembled in the presence of GTP (Fig. 2B and C) or GTP and DEAE-dextran (Fig. 2D) was done according to the method described by Romberg et al. (2001). Filaments were visualized using a Hitachi transmission electron microscope.
Expression plasmids
Codons 26–562 of the ezrA ORF were amplified by PCR from chromosomal DNA and cloned into the pBAD/Thio-TOPO vector (Invitrogen). The resulting plasmid, pRS3, encodes a 79 kDa protein that consists of EzrA fused in frame to an N-terminal thioredoxin moiety with a C-terminal 6×His tag to assist purification. Primer sequences are available on request. The 6×His-tagged thioredoxin fusion control protein was expressed from the pBAD/Thio vector (Invitrogen). B. subtilis FtsZ was expressed under the control of an IPTG-inducible promoter from pCXZ (Wang and Lutkenhaus, 1993). Spo0J-6×His was expressed from pDL3, a kind gift from Dr Alan Grossman (Lin et al., 1997).
EzrA, thioredoxin and Spo0J protein purification
Overnight cultures of MB101 cells containing pRS3 or pBAD/Thio were diluted 1:100 in LB medium containing 100 μg ml−1 ampicillin. At an OD600 of ≈0.4, protein expression was induced by the addition of 0.2% l-(+)-arabinose, and cells were allowed to grow for 4 more hours. Cultures were centrifuged at 2000 g for 10 min at 4°C, and the pellets were washed once and resuspended in ice-cold buffer (50 mM NaH2PO4, pH 8.0;, 300 mM NaCl) to a final volume of 10 ml. Cell pellets were stored at −80°C.
The EzrA and thioredoxin fusion proteins were both purified via the same protocol. Briefly, cells were thawed on ice, and 4-(2-aminoethyl)benzenesulphonyl fluoride (AEBSF), a protease inhibitor, and lysozyme were added to final concentrations of 1 mM and 0.4 mg ml−1 respectively. Cells were incubated for 30 min on ice and lysed by sonication. Lysates were cleared by centrifugation at 120 000 g for 45 min at 4°C, and imidazole was added to the resulting supernatant to a final concentration of 5 mM. The supernatant was then subjected to affinity chromatography using a Hi-Trap chelating HP column (Amersham) charged with Co2+ on an ÄKTAprime low-pressure purification system (Amersham). The column was washed extensively with a buffer containing 50 mM Hepes, pH 7.5, 350 mM NaCl and 5 mM imidazole, and the fusion proteins were eluted in a step gradient of imidazole concentrations between 25 mM and 200 mM (25 mM, 75 mM, 100 mM, 150 mM and 200 mM). Both fusion proteins eluted in high concentration at 150 mM imidazole. The fractions containing the fusion protein were pooled, applied to PD-10 columns (Amersham) to exchange them into buffer A (50 mM Hepes, pH 7.5, 175 mM NaCl) and concentrated using Centricon filters (Millipore) with the appropriate molecular weight cut-offs (YM-50 for the EzrA fusion protein and YM-10 for the thioredoxin fusion proteins). Glycerol was added to 10% final concentration, and the aliquots were frozen in liquid nitrogen and stored at −80°C. Purified proteins were estimated to be ≥95% pure by SDS–PAGE. The purified EzrA fusion protein described above was used to generate antibodies against EzrA in rabbits (Covance). Spo0J was purified as described previously (Lin and Grossman, 1998); however, PD-10 columns were used for buffer exchange in place of dialysis.
FtsZ purification
Overnight cultures of PL1184 cells (ER2566 pCXZ pBS58) were diluted 1:100 in LB medium containing 100 μg ml−1 ampicillin. Cells were grown to OD600 of ≈0.4 and induced with 1 mM IPTG for 4 h at 37°C. Cultures were pelleted at 2000 g for 10 min at 4°C, washed once and resuspended in ice-cold buffer (50 mM Tris, pH 8.8, 100 mM NaCl, 1 mM EDTA), and stored at −80°C.
FtsZ was expressed and purified essentially as described previously (Wang and Lutkenhaus, 1993) with the exception that a 30–35% ammonium sulphate cut was used in place of the 25–35% cut. We determined that the gel filtration step did not significantly enhance the purity or activity of the final product (P. A. Levin, unpublished data). Precipitated proteins from the 35% ammonium sulphate cut were washed and resuspended in 5 ml of polymerization buffer (50 mM MES, pH 6.5, 2.5 mM MgAc, 1 mM EGTA). Glycerol was added to 10% and GDP to 50 μM final concentration. Aliquots were frozen in liquid nitrogen and stored at −80°C. Purified protein was estimated to be ≥95% pure by SDS–PAGE.
Analysis of EzrA overexpression phenotype
To assess the effect of twofold EzrA overexpression on cell viability, we determined the plating efficiency of wild-type and ftsZts mutant cells after induction of an IPTG-inducible allele of ezrA. Briefly, overnight cultures of ftsZ+ or ftsZts mutant cells encoding amyE∷Pspachy-ezrA (PL1031 and MO54 respectively) were diluted twice into fresh media without IPTG and cultured at 37°C to an OD600 of ≈0.600. Cells were then serially diluted on LB plates in the presence and absence of 1 mM IPTG. Plating efficiencies were calculated based on the number of cfu per ml of culture.
For fluorescence microscopy and quantitative immunoblotting, overnight cultures of PL1031 and MO54 were diluted 10-fold into fresh LB and cultured at 37°C to an OD600 of ≈0.600. Cells were then diluted a second time in the presence and absence of 1 mM IPTG. Samples were removed and prepared for fluorescence microscopy and/or quantitative immunoblotting (Weart and Levin, 2003). Average cell lengths were determined after 3 h of growth in the presence and absence of IPTG. Samples were stained with the vital membrane stain FM4-64 (Molecular Probes) (Pogliano et al., 1999), and cell lengths were measured as the distance between two septa. Immunofluorescence microscopy in conjunction with affinity-purified antiserum against FtsZ (Levin and Losick, 1996) was used to visualize FtsZ rings in PL1031. A Chroma GFP filter set (no. 41012) was used to visualize FtsZts–GFP rings in MO54. Quantitative immunoblotting to determine relative levels of EzrA was performed as described previously (Weart and Levin, 2003) using polyclonal antiserum raised in rabbits against EzrA (this work).
Sedimentation assay
Sedimentation assays were similar to those described by Trusca et al. (1998). Before sedimentation, purified FtsZ was precleared by spinning at 250 000 g for 15 min at 4°C to remove any aggregates or pre-existing polymers. FtsZ was then diluted in polymerization buffer at room temperature to 5 μM final concentration. GTP was added to a final concentration of 1 mM to stimulate polymerization, and bundle formation was initiated by the addition of 0.1 mg ml−1 diethylaminoethyl-dextran hydrochloride (DEAE-dextran). Depending on the order of addition, varying concentrations of EzrA or the thioredoxin control protein in a constant volume of buffer A were added to the reaction before or after GTP but before the addition of DEAE-dextran. The final volume for all reactions was 100 μl. Reactions were spun at 250 000 g for 10 min at 20°C. To avoid touching the pellet, only 80 μl of supernatant was withdrawn for analysis. The remaining supernatant was then removed, and the pellets were gently resuspended in 100 μl (the same volume as the reaction) of polymerization buffer. Samples from pellets and supernatants were subjected to 12% SDS–PAGE to determine relative FtsZ concentrations. Gels were stained with Coomassie brilliant blue and documented and analysed using a Kodak DC290 camera in conjunction with Kodak ID 3.5.3 software.
90° angle light scattering assay
Light scattering assays were conducted essentially as described in by Romberg et al. (2001). Precleared FtsZ was diluted to a final concentration of 2.5 μM in polymerization buffer and placed in a quartz cuvette (Starna Cells) in a FluoroMax-2 fluorimeter. The excitation and emission wavelengths were set at 310 nm, and the slit width was 1 mm. Readings were taken once per second at room temperature, and a baseline was gathered for 1 min before the addition of 1 mM GTP or GDP to the cuvette. For the experiment shown in Fig. 6A and B, EzrA or the thioredoxin control protein was added before establishing a baseline and before the addition of GTP. For the experiment in Fig. 6C, EzrA and GDP were added 1 min after the addition of GTP. The final concentration of EzrA in this experiment was 5 μM and of GDP was at 1 μM. Data were normalized to the baseline reading in the presence of EzrA or thioredoxin alone and plotted using Microsoft excel.
Co-affinity purification of EzrA and FtsZ
Purified proteins were diluted to 5 μM final concentrations in buffer A and 5 μM BSA. The cross-linker dithiobis(succinimidylpropionate) (DSP; Pierce), which interacts with primary amines, was added to the diluted proteins to a final concentration of 250 μM, and the solution was incubated at room temperature for 30 min. To quench the cross-linking reaction, Tris (pH 7.4) was added to a final concentration of 20 mM at room temperature for 15 min. This solution was bound to Co2+-charged affinity resin (Clontech) overnight at 4°C, and the resin was washed five times with 33 bed volumes of buffer A. The fusion proteins and any cross-linked FtsZ were eluted with buffer A plus 250 mM imidazole. The disulphide bond in DSP was cleaved by heating eluants in SDS sample buffer with β-mercaptoethanol at 100°C for 5 min before separation by SDS–PAGE. Immunoblot analysis was performed as described previously (Weart and Levin, 2003) using anti6×His sera (Santa Cruz Biotechnology) and affinity-purified sera against FtsZ (Levin and Losick, 1996). Immunoblots were analysed using an LAS-1000plus luminescent image analyser in conjunction with imagegauge software, version 3.41 (Fuji Film).
GTPase assay
The GTPase activity of FtsZ was measured using the Enzcheck Free Phosphate assay kit (Molecular Probes). This assay takes advantage of the spectrophotometric shift from a maximum absorbance of 330 nm to 360 nm that occurs when the substrate 2-amino-6-mercapto-7-methylpurine riboside (MESG) is converted to ribose 1-phosphate and 2-amino-6-mercapto-7-methylpurine through the purine nucleoside phosphorylase (PNP)-dependent addition of Pi to MESG. FtsZ was at 5 μM, and EzrA and thioredoxin were at 10 μM for these experiments. GTP (1 mM) was incubated in the presence of kit reagents, the EzrA fusion protein or the thioredoxin control and buffer (50 mM Hepes, pH 7.5, 175 mM NaCl, 2.5 mM MgCl2, 200 mM KCl) for 10 min at room temperature to neutralize any free phosphate. FtsZ was then added to a final concentration of 5 μM, and the optical density at 360 nm was measured every 10 s in a SPECTRA-max® spectrophotometer (Molecular Devices) using soft-max® PRO, version 3.1.2 (Molecular Devices). The average GTPase rate was calculated using a best fit function and Pi standards. Intriguingly, we found that using 200 mM KAc in place of KCl increased GTP hydrolysis five- to sixfold (data not shown). This enhancement may be due to the tendency of acetate to promote hydrophobic interactions (Arakawa and Timasheff, 1984).
Acknowledgments
The authors would like to thank members of the Levin laboratory for useful discussions during the course of this research. We are also grateful to M. Chesnik, P. Chivers, H. Erickson, L. Romberg and M. Vieth for technical assistance, R. Weart for help with statistical analysis and figures, and the Pakrasi laboratory for use of the FluoroMax-2 fluorimeter. The ‘deep-etch’ electron micrograph was kindly provided by Dr John Heuser and Robin Roth of the Washington University School of Medicine. The Spo0J expression vector, pDL3, was a generous gift from Dr Alan Grossman. Finally, we are indebted to S. Ben-Yehuda, P. Chivers, F. Gueiros-Filho, R. Kranz, R. Losick and L. Romberg for insightful comments on the manuscript. This work was supported, in part, by an institutional research grant from the American Cancer Society IRG-58-010-44 and Public Health Services grant GM64671 from the NIH.
References
- Addinall SG, Lutkenhaus J. FtsZ-spirals and –arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol. 1996;22:231–237. doi: 10.1046/j.1365-2958.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- Arakawa T, Timasheff SN. Mechanism of protein salting in and salting out by divalent cation salts: balance between hydration and salt binding. Biochemistry. 1984;23:5912–5923. doi: 10.1021/bi00320a004. [DOI] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. Nucleotide sequence and insertional inactivation of a Bacillus subtilis gene that affects cell division, sporulation, and temperature sensitivity. J Bacteriol. 1989;171:6821–6834. doi: 10.1128/jb.171.12.6821-6834.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J Bacteriol. 1992;174:2398–2403. doi: 10.1128/jb.174.7.2398-2403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PAJ, Crossley RE, Rothfield LI. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers PT, Sauer RT. NikR is a ribbon-helix-helix DNA-binding protein. Protein Sci. 1999;8:2494–2500. doi: 10.1110/ps.8.11.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell SC, Robinson EJ, Lowe J. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc Natl Acad Sci USA. 2003;100:7889–7894. doi: 10.1073/pnas.1330742100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Harry EJ, Katis VL, Wake RG, Errington J. Characterization of the essential cell division gene ftsL (ylld) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol Microbiol. 1998;29:593–604. doi: 10.1046/j.1365-2958.1998.00954.x. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Din N, Quardokus EM, Sackett MJ, Brun YV. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol Microbiol. 1998;27:1051–1063. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- Downing KH. Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu Rev Cell Dev Biol. 2000;16:89–111. doi: 10.1146/annurev.cellbio.16.1.89. [DOI] [PubMed] [Google Scholar]
- Feucht A, Lucet I, Yudkin MD, Errington J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol Microbiol. 2001;40:115–125. doi: 10.1046/j.1365-2958.2001.02356.x. [DOI] [PubMed] [Google Scholar]
- Geissler B, Elraheb D, Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci USA. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb MH. Protein prenylation, et cetera: signal transduction in two dimensions. Science. 1997;275:1750–1751. doi: 10.1126/science.275.5307.1750. [DOI] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PAJ. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. Chichester: John Wiley and Sons; 1990. [Google Scholar]
- Heuser JE. Procedure for freeze-drying molecules adsorbed to mica flakes. J Mol Biol. 1983;169:155–195. doi: 10.1016/s0022-2836(83)80179-x. [DOI] [PubMed] [Google Scholar]
- Heuser J. Protocol for 3-D visualization of molecules on mica via the quick-freeze, deep-etch technique. J Electron Microsc Tech. 1989;13:244–263. doi: 10.1002/jemt.1060130310. [DOI] [PubMed] [Google Scholar]
- Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci USA. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff T, Muller CS, Otto AM, Netzker R, Hannappel E. beta-Thymosins, small acidic peptides with multiple functions. Int J Biochem Cell Biol. 2001;33:205–220. doi: 10.1016/s1357-2725(00)00087-x. [DOI] [PubMed] [Google Scholar]
- Justice SS, Garcia-Lara J, Rothfield LI. Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol Microbiol. 2000;37:410–423. doi: 10.1046/j.1365-2958.2000.02007.x. [DOI] [PubMed] [Google Scholar]
- Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci USA. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Schwartz RL, Grossman AD. Polymer stability plays an important role in the positional regulation of FtsZ. J Bacteriol. 2001;183:5449–5452. doi: 10.1128/JB.183.18.5449-5452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Lin DC-H, Levin PA, Grossman AD. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc Natl Acad Sci USA. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J, Amos LA. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 1999;18:2364–2371. doi: 10.1093/emboj/18.9.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Stricker J, Erickson HP. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima – quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskeleton. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- McNally FJ, Okawa K, Iwamatsu A, Vale RD. Katanin, the microtubule-severing ATPase, is concentrated at centrosomes. J Cell Sci. 1996;109:561–567. doi: 10.1242/jcs.109.3.561. [DOI] [PubMed] [Google Scholar]
- Margolin W. Spatial regulation of cytokinesis in bacteria. Curr Opin Microbiol. 2001;4:647–652. doi: 10.1016/s1369-5274(01)00264-8. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998a;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Purification, assembly, and localization of FtsZ. Methods Enzymol. 1998b;298:296–305. doi: 10.1016/s0076-6879(98)98026-0. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J Bacteriol. 1999;181:823–832. doi: 10.1128/jb.181.3.823-832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Cao C, Lutkenhaus J. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2885–2890. doi: 10.1073/pnas.95.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M, Spiegelman GB, Hoch JA. Structure of the gene for the transition state regulator abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun YL, Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Quisel JD, Burkholder WF, Grossman AD. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J Bacteriol. 2001;183:6573–6578. doi: 10.1128/JB.183.22.6573-6578.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regamey A, Harry EJ, Wake RG. Mid-cell Z ring assembly in the absence of entry into the elongation phase of the round of replication in bacteria: co-ordinating chromosome replication with cell division. Mol Microbiol. 2000;38:423–434. doi: 10.1046/j.1365-2958.2000.02130.x. [DOI] [PubMed] [Google Scholar]
- Romberg L, Levin PA. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu Rev Microbiol. 2003;57:125–154. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg L, Simon M, Erickson HP. Polymerization of FtsZ, a bacterial homolog of tubulin: Is assembly cooperative? J Biol Chem. 2001;4:11743–11753. doi: 10.1074/jbc.M009033200. [DOI] [PubMed] [Google Scholar]
- Rothfield L, Justice S, Garcia-Lara J. Bacterial cell division. Annu Rev Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- Safer D, Sosnick TR, Elzinga M. Thymosin beta 4 binds actin in an extended conformation and contacts both the barbed and pointed ends. Biochemistry. 1997;36:5806–5816. doi: 10.1021/bi970185v. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scheffers DJ, de Wit JG, den Blaauwen T, Driessen AJ. GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry. 2002;41:521–529. doi: 10.1021/bi011370i. [DOI] [PubMed] [Google Scholar]
- Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci U S A. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J Bacteriol. 1998;180:3946–3953. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lutkenhaus J. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol Microbiol. 1993;9:435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
- Ward JE, Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- Weart RB, Levin PA. Growth rate-dependent regulation of medial FtsZ ring formation. J Bacteriol. 2003;185:2826–2834. doi: 10.1128/JB.185.9.2826-2834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]


