Abstract
Stem cell based therapy is a promising intervention for ischemic heart diseases. However, the functional integrity of stem cells is impaired in an ischemic environment. Here, we report a novel finding that heat shock significantly improves Sca-1+stem cell survival in an ischemic environment by the regulation of the triangle: Heat Shock Factor 1(HSF1), HSF1/miR-34a, Heat Shock Protein 70(HSP70). Initially we prove that HSP70 is the key chaperone mediating cytoprotective effect of heat shock in Sca-1+cells and then we establish miR-34a as a direct repressor of HSP70. We found that HSP70 was down-regulated in heat shocked Sca-1+ stem cells (HSSca-1+ cells). Intriguingly, we demonstrate that the down-regulation of miR-34a is attributed to HSF1-mediated epigenetic repression through histone H3 Lys27 trimethylation (H3K27me3)on miR-34a promoter. Moreover, we show that heat shock induces exosomal transfer of HSF1 fromSca-1+ cells, which directs ischemic cardiomyocytes towards a prosurvival phenotype by epigenetic repression of miR-34a. In addition, our in vivo study demonstrates that transplantation of HSSca-1+ cells significantly reduces apoptosis, attenuates fibrosis and improves global heart functions in ischemic myocardium. Hence, our study not only provides novel insights into the effects of heat shock on stem cell survival and paracrine behavior but may have therapeutic values for stem cell therapy in ischemic heart diseases.
Keywords: Sca-1, Stem Cell Transplantation, Hypoxia, Cardiac, miR-34a, Chaperone, Histone modification, Exosome
Introduction
Ischemic heart diseases (IHD) lead to severe morbidity and mortality in humans. Stem cell therapy is a promising approach to compensate the loss of cardiac cells by ischemia in addition to the interventions such as percutaneous coronary intervention (PCI) and coronary artery bypass graft (CABG) in the treatment of IHD1. However, stem cell therapy is hampered due to massive cell death of transplanted stem cells, because only 5% of the cells are detectable in the myocardium at 2 hours and 1% detectable at 18 hours after transplantation2. Hence, there is an urgent need for formulation of interventions that can significantly improve stem cell survival in the ischemic environment. In the past, it has been reported that transgenic overexpression of anti-apoptotic proteins or growth factors can enhance stem cell survival in harsh environment3. Nevertheless, the potential risks of immune rejection and carcinogenesis in hosts limits its application in clinical settings. While it has been proposed that heat shock could promote adult cell survival4,5, its effect on stem cells survival particularly their paracrine behaviors poorly understood.
In the present study, we established a heat shock model which significantly reduced stem cell death in vitro and in vivo and showed that HSP70 is the key regulator mediating the cytoprotective effect. Although in the past two decades, much work has shown that HSP70 was prosurvival protein against cellular stress, its underlying mechanism is still not fully understood6. Currently, microRNAs are emerging as new regulators of stress responses7. Here we showed that miR-34a directly targets HSP70 in heat shocked stem cells. Furthermore, we proved that the repression of miR-34a is mediated via HSF1 remodeling of chromatin structure on its promoter. We also showed that transplantation of heat shocked stem cells significantly attenuates cell death and fibrosis in ischemic heart and improves global heart functions. Even more interestingly, our study suggests that improved cardiac outcome is potentially mediated by heat shock induced exosomal transfer of HSF1 from Sca-1+ cells, which directs ischemic cardiomyocytes towards a prosurvival phenotype by remodeling the chromatin structure on miR-34a promoter.
Methods
A detailed methods section is available in the Online Supplemental Information.
Results
Pre-Induction Of HSP70 By Heat Shock Rescues Sca-1+ Stem Cells From Death After Oxygen Glucose Deprivation
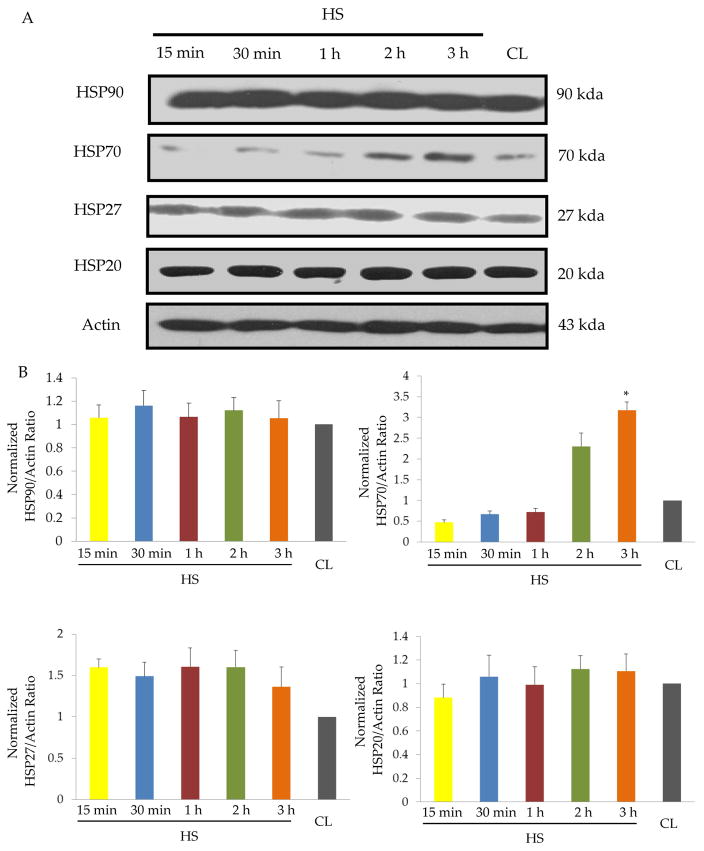
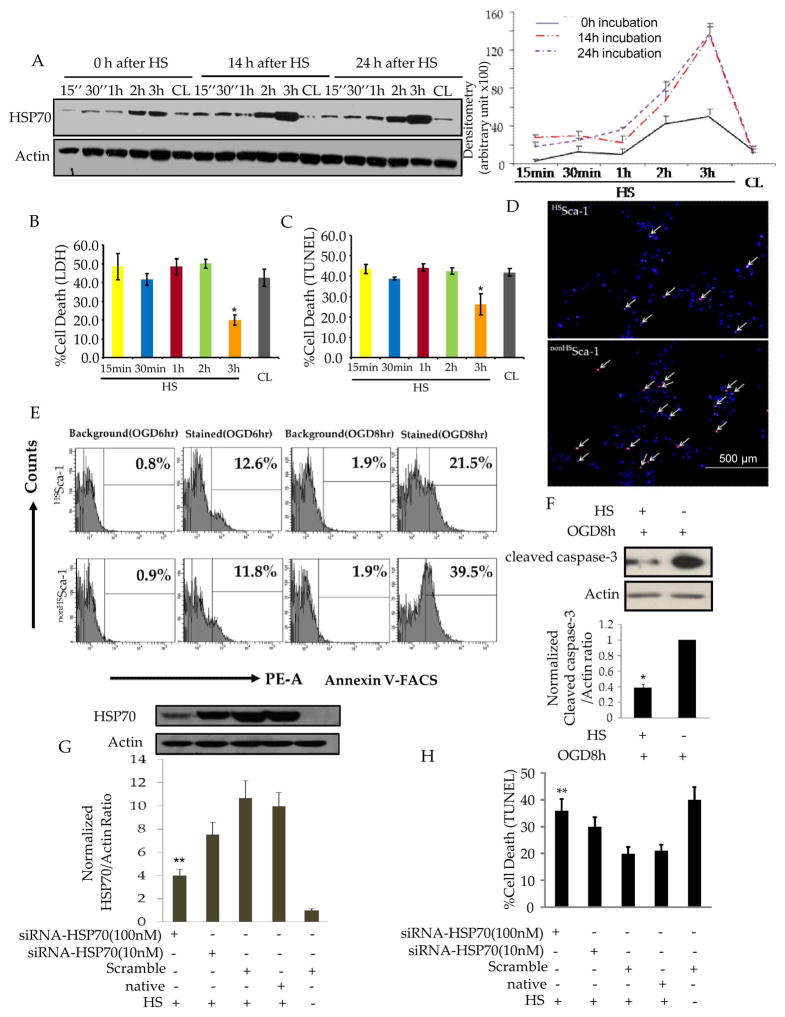
To evaluate the heat shock proteins (HSPs) expressed after heat shock in Sca-1+ cells, we examined HSP20, HSP27, HSP70, HSP90 by different heat shock (HS) intensity (42°C;15 min, 30min, 1hr, 2hrs, 3hrs, respectively). Western blots (Figure 1.A) and quantitative results (Figure 1.B) showed significant up-regulation of HSP70 at the end of HS for 3 hrs (HS3h)(3.5 folds vs. control, P<0.05), while no significant change was observed for HSP20, HSP27, HSP90 protein expression after HS. Western blot and quantitative results showed that the peak protein expression of HSP70 was detected at 14h after HS-3h (HS3h-incu14h)(>12 folds vs. control(CL-incu14h), and maintained up to 24h after HS (Figure 2.A). In the following description, HS3h-incu14h and CL-incu14h were designated as HSSca-1+ cells and non-HSSca-1+ cells respectively. In survival studies, HSSca-1+ cells underwent significantly reduced cell death after oxygen glucose deprivation(OGD) 8h as compared to non-HSSca-1+ cells (LDH release: 20.0%±2.7 vs. 42.5%±4.5%, p<0.01; TUNEL:15.2%±2.3% vs. 38.6%±1.8%, p<0.01; Annexin V-PE Flow Cytometry:21.5%±1.6 vs.39.5%±2.1%, p<0.01)(Figure 2.B–E). HSSca-1+ cells consistently showed marked reduction of cleaved caspase-3 in HSSca-1+ cells (Figure 2.F). The effectiveness of HS on Sca-1+ cells against cell death and the selective and marked upregulation of HSP70 led us to interrogate the role of HSP70 for enhanced Sca-1+ cell survival. We addressed this question through a loss-of-function study. Western blot confirmed the siRNAs against HSP70 could efficiently diminish the upregulation of HSP70 in HSSca-1+ cells (Figure 2.G) and consequently sensitized the cells to OGD (p<0.01, vs. Scramble, Figure 2.H). Thus, these studies proved that HSP70 is the key cytoprotective protein to rescue Sca-1+ cells from death after OGD.
Figure 1. Profiling of heat shock proteins (HSPs) after different heat shock durations in Sca-1+ cells.
A, Western blot analysis was performed to detect HSP90, HSP70, HSP27, HSP20 protein expression using total cellular proteins (20 μg/lane) obtained from Sca-1+ cells that were heat shock (HS) treated at 42°C for 15min, 30min, 1h, 2h, 3h and non-treated Sca-1+ cells as control(CL). B, Densitometry for HSP90, HSP70, HSP27, HSP20 after normalization against Actin levels. The data shown are the average of three independent experiments with error bars denoting standard deviation. (*) denotes P<0.05 for significant differences between HS3h and CL.
Figure 2. Up-regulation of HSP70 is essential for enhanced survival of HSSca-1+ cells after oxygen glucose deprivation.
A. Left panel: Western blot analysis was performed to exhibit the expression pattern of HSP70 in heat shocked Sca-1+ cells which were incubated at 37°C for different time periods (0, 14, 24 hours, designated as incu 0 h, incu 14 h, incu 24 h respectively) after heat shock. Right panel: Densitometry for HSP70 after normalization against Actin levels. B. After OGD 8 h, culture supernatants were collected and assayed for LDH (for the group-incu 14h). At that time point, cells were fixed and followed by TUNEL assay (C) and the representative pictures for TUNEL assay in HSSca-1+ and non-HS Sca-1+ cells are shown in D(Red: TUNEL; Blue: DAPI). (*) denotes P<0.05 for significant differences between HSSca-1+ and non-HS Sca-1+ cells. White arrow indicates TUNEL positive cells. E. Apoptosis was analyzed by Annexin V–FITC staining followed by FACS. The percentage of apoptotic cells was quantitated. F. upper panel: Western blot showing reduced cleaved caspase-3 expression in HSSca-1+ cells as compared with non-HSSca-1+ cells; (*) denotes P<0.05 for significant differences between HSSca-1+ and non-HSSca-1+ cells. Lower panel: Densitometry for cleaved caspase-3 after normalization against Actin levels. G. Western blot showing siRNA successfully diminished HSP70 expression in HSSca-1+ cells as compared with Scramble. (**) denotes P<0.01 for significant differences between siRNA-HSP70 (100 nM) and Scramble treated cells. H. siRNA-HSP70 significantly reversed cytoprotective effect of heat shock as determined by TUNEL assay. (**) denotes P<0.01 for significant differences between siRNA-HSP70 (100 nM) and Scramble treated cells. The data shown are the average of three independent experiments with error bars denoting standard deviation.
Repressed miR-34a Contributed To Upregluation of HSP70 InHSSca-1+ Cells
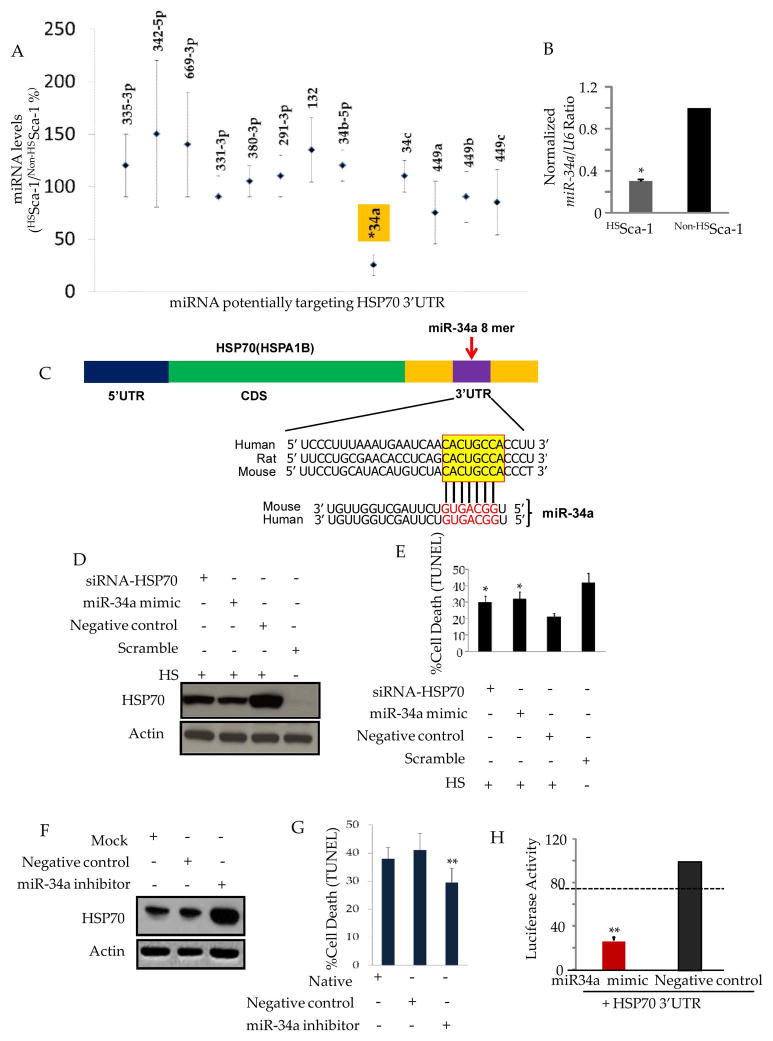
Previously, it was commonly believed that the heat shock induced upregulation of HSP70 was due to the direct transactivation of HSF1 on the promoter of HSP708. However, the role of microRNAs in HSP70 regulation is poorly understood at the present time. In order to address this question, we employed customer miRNA array (including 13 putative miRNAs targeting HSP70 3′UTR based on the prediction by Targetscan (Suppl. Information-Figure S1.) to detect specific changes in miRNAs distributed in HSSca-1+ cells. Of the 13 putative miRNAs targeting HSP70 3′UTR, only miR-34a was decreased significantly following heat shock (Figure 3.A and B). The computational analysis showed the putative binding of miR-34a and a conserved 8-mer target ‘seed’ in the 3′UTR of HSP70 (Figure 3.C). We therefore hypothesized that miR-34a targets and regulates HSP70 levels. To test this hypothesis, we first checked the protein levels of HSP70 in HSSca-1+ cells with miR-34a overexpression. Western blot showed that miR-34a mimic could significantly reduce the elevated HSP70 in HSSca-1+ cells (Figure 3.D) and sensitize the cells to OGD (Figure 3.E). We then used specific LNA inhibitor to silence miR-34a. Western blot showed marked upregulation of HSP70 after transfection of miR-34a LNA inhibitor (Figure 3.F) and subsequently increased the tolerance of HSSca-1+ to OGD (Figure 3.G). To confirm whether miR-34a can inhibit the synthesis of HSP70, we performed a luciferase reporter assay in which the luciferase reporter gene was fused to the wild-type HSP70 3′UTR. This assay resulted in approximately 70% less expression of luciferase activity compared with control cells transfected with miR-34a mimic (Figure 3.H). From this data we concluded that the miR-34a is a novel negative regulator of HSP70 and its repressionin HSSca-1+ cells contributed to the upregulation of HSP70 protein level.
Figure 3. miR-34a is novel negative regulator of HSP70.
A. Customer microarray data of miRNA expression levels from 3 independent experiments. Relative levels of 13 miRNAs potentially targeting the HSP70 (HSPa1b) 3′UTR under heat shock are shown. miRNA levels in non-HSSca-1+ cells are considered to be equal to 100%. B. Real-time PCR showing expression of miR-34a was repressed inHSSca-1+ cells as compared to non-HSSca-1+ cells. (*) denotes P< 0.01, relative to non-HSSca-1 cells. C. Schematic layout of the HSP70 (HSPa1b) 5′UTR, mRNA coding sequence (CDS) and 3′UTR, with the relative location of the miR-34a binding site. Depiction is not to scale. Sequence of mouse and human miR-34a and predicted interaction with conserved 8-mer miR-34a seeds found within the HSP70 (HSPa1b) 3′UTRs from different species (highlighted in yellow box) are shown. D. Western blot showing overexpression of miR-34a by transfection of miR-34a mimic diminished upregulation of HSP70 in HSSca-1+ cells. E. The cytoprotective effect of heat shock was compromised by overexpression of miR-34a via transfection of miR-34a mimic, as determined by TUNEL assay. (*) denotes P< 0.01, relative to negative control-transfected HSSca-1+ cells. F. Loss of miR-34a by transfection of miR-34a LNA inhibitor caused elevation of HSP70 detected by Western blot and rescued apoptosis as determined TUNEL assay (G). (**) denotes P< 0.01, relative to the negative control-transfected Sca-1+ cells. H. Co-transfection of plasmid pEZX-MT01-HSP70-3′UTR with miR-34a mimic or negative control. Plasmid pEZX-MT01-HSP70-3′UTR (GeneCopoeia) expressing firefly luciferase (hLuc) followed by 3′UTR of mouse HSP70, and renilla luciferase (hRLuc) under constitutive control of the cytomegaly virus (CMV) promoter. Every firefly luciferase reading was normalized to that of the control Renilla luciferase. The luciferase activity of miR-34a mimic-transfected cells were presented as percentages relative to the level of luciferase in the negative control-transfected cells (this control luciferase level is considered as 100%). The values represent mean ± SEM (n= 3). The dashed line represents the threshold of luciferase activity (75%), suppression of luciferase level below which indicates positive binding. (**) denotes P< 0.01, relative to the negative control-transfection.
HSF1 Repressed miR-34a
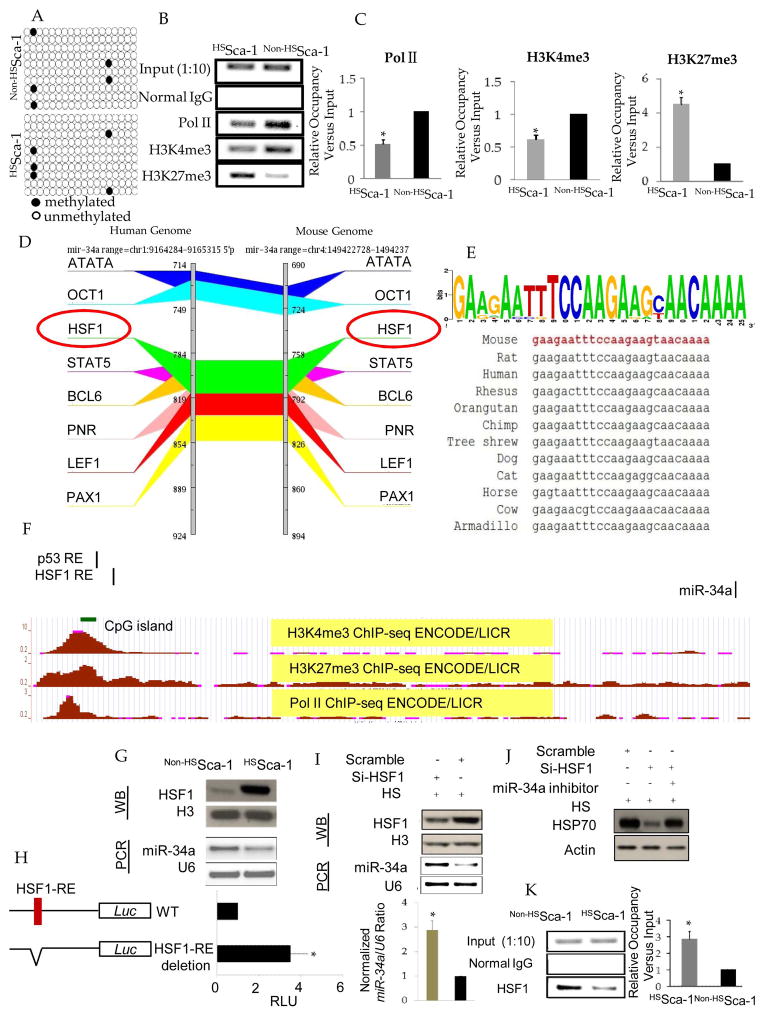
As the repression of miR-34a contributed to the upregulation of HSP70 in HSSca-1+ cells, we then investigated the underlying factors for miR-34a repression. First, we ruled out the possibility that the miR-34a genomic loci were deleted in HSSca-1+ cells, since equal amounts were detected in the same amounts of genomic DNA extracted from HSSca-1+ cells and non-HSSca-1+ cells using real-time PCR (Suppl. Information-Figure S2.). Although previous studies showed that DNA methylation on miR-34a promoter was one of the most common cause for its downregulation9, 10, we found the CpG island on miR-34a promoter was hypomethylated in both HSSca-1+ cells and non-HSSca-1+ cells through bisulfate sequencing (Figure 4.A and Suppl. Information-Figure S3.), thus indicating DNA methylation is less likely to regulate their expression in HSSca-1+ cells. While trimethylation of histone H3 at Lys4 (H3K4me3) is linked to transcriptional activation, trimethylation of H3 at Lys 27(H3K27me3) is linked to transcriptional repression, next we performed ChIP-qPCR to detect the relative occupancy of H3K4me3, H3K27me3 and Pol II (the RNA polymerase responsible for miR-34a transcription) on the genomic region extracted from ChIP-seq data from ENCODE project in UCSC genome browser. The results showed a reduction in enrichment of H3K4me3, Pol II as opposed to increased enrichment of H3K27me3 on miR-34a promoter in HSSca-1+ cells (Figure 4.B and C). These results indicate that the repression of miR-34a in HSSca-1+ cells is mediated by transcriptional repression via histone methylation instead of DNA methylation. Then we further investigated the mechanism of transcriptional repression for miR-34a. By searching the GenomeTrafac database, we found a putative HSF1 responsive element (HSF1-RE) in the mouse miR-34a promoter region (Figure 4.D). Deeper homology analysis found the putative HSF1 responsive element is highly conserved in multiple species (Figure 4.E). Furthermore, the putative element is near the previously reported p53 response element (p53-RE), which has been shown to be responsible for transactivation of miR-3411, 12. The putative HSF1-RE along with the CpG islands, pol II, H3K4me3, and H3K27me3 peaks in miR-34a promoter were displayed using the UCSC genome browser (Figure 4.F). Based on the computational analysis and the inverse relationship of HSF1 and miR-34a (Figure 4.G), we determined that whether HSF1 regulates the expression of miR-34a. To test our hypothesis, we cloned the wide type (WT) and mutated construct (HSF1-RE deletion) of promoter region of mouse miR-34a upstream of a luciferase gene in a reporter plasmid (pGL4) respectively. Interestingly, transfection of reporter plasmid with mutated construct into HSSca-1+ cells led to significant upregulation of luciferase activity as compared to wide type, indicating HSF1-RE is a repressor element for miR-34a transcription(Figure 4.H). Moreover, knock down of HSF1 by siRNA in HSSca-1+ cells led to upregulation of miR-34a(Figure 4.I) and miR-34a inhibitor rescued the loss of HSP70 caused by knock down of HSF1. Finally, ChIP-qPCR validated the binding of HSF1 on the putative HSF1-RE and found HSF1 was enriched on this cis element in HSSca-1+ cells as compared to non-HSSca-1+ cells (Figure 4.M). Taken together, we concluded that miR-34a repression by chromatin remodeling is attributed to direct binding of HSF1 on miR-34a promoter.
Figure 4. miR-34a repression in HSSca-1+ cells is attributed to HSF1 mediated chromatin remodeling.
A. Bisulfite sequencing showing no hypermethylation on miR-34a promoter in HSSca-1+ cells. Genomic DNA derived from HSSca-1+ and non-HSSca-1+ cells was subjected to bisulfite sequencing and examined at the region on CpG island of miR-34a, as indicated in the map. Methylated CpG are presented by black circles and unmethylated sites by open circles. B. ChIPqRT-PCR analysis of RNA polymerase II (Pol II), H3K4me3, and H3K27me3 on the promoter region of miR-34a in HSSca-1+ and non-HSSca-1+ cells. Data are normalized to inputs from three experiments (mean ± SEM). (*) denotes P<0.05 for significant differences related to non-HSSca-1+ cells (C). D. Mouse-human conserved HSF1 responsive element (red circle) in miR-34a promoter region (downloaded from the GenomeTrafac database). E. Sequence logo showing high conservation of the putative HSF1-RE. The logo was constructed from 12 mammal species. The height of each letter is proportional to the frequency of the base, and the height of the letter stack shows the conservation at that position. F. AUCSC genome browser screenshot showing the location of validated p53 response element (p53-RE), the putative HSF1 response element (HSF1-RE), CpG island, Pol II, H3K4me3, and H3K27me3 tracks on miR-34a promoter in mouse genome. G. Western blot (upper panel) and PCR (lower panel) showing inverse correlation between HSF1 and miR-34a expression. Nuclear protein and total RNA was extracted from the same batch of cells. H. Luciferase (Luc) reporter constructs contain the miR-34a promoter with the potential HSF1-RE upstream of a luciferase gene (WT), or the miR-34a promoter with the deletion of potential HSF1-RE (HSF1-RE deletion). Red indicates the potential HSF1-RE (chr4:149,423,492-149,423,516). Sca-1+ cells were transfected with WT or HSF1-RE deletion plasmids for 24 h, followed by heat shock treatment. Luciferase activity was measured 14 h after heat shock. Luciferase activity is expressed as the ratio of luciferase activity of HSF1-RE deletion to WT luciferase activity and Renilla luciferase as a normalizing control. RLU, relative light units. Data represent mean ± SEM (n= 3).(*) denotes P< 0.05, relative to WT. I. Knock down of HSF1 by siRNA as shown by western blot significantly up-regulated miR-34a expression detected by real-time PCR. Data represent mean ± SEM (n= 3).(*) denotes P< 0.05, relative to Scramble-transfected HSSca-1+ cells. J. Western blot showing knock down of HSF1 by siRNA down regulated HSP70 expression which was rescued by miR-34a inhibitors. K. ChIP-qPCR verified the binding of HSF1 on the putative HSF1-RE, showing HSF1 was significantly enriched on the promoter region of miR-34a in HSSca-1+ cells as compared to non-HSSca-1+ cells. Data represent mean ± SEM (n= 3).(*) denotes P< 0.05, relative tonon-HSSca-1+ cells.
HSSca-1+ Cells Transplantation Reduced Cardiomyocyte Apoptosis Post Myocardial Infarction
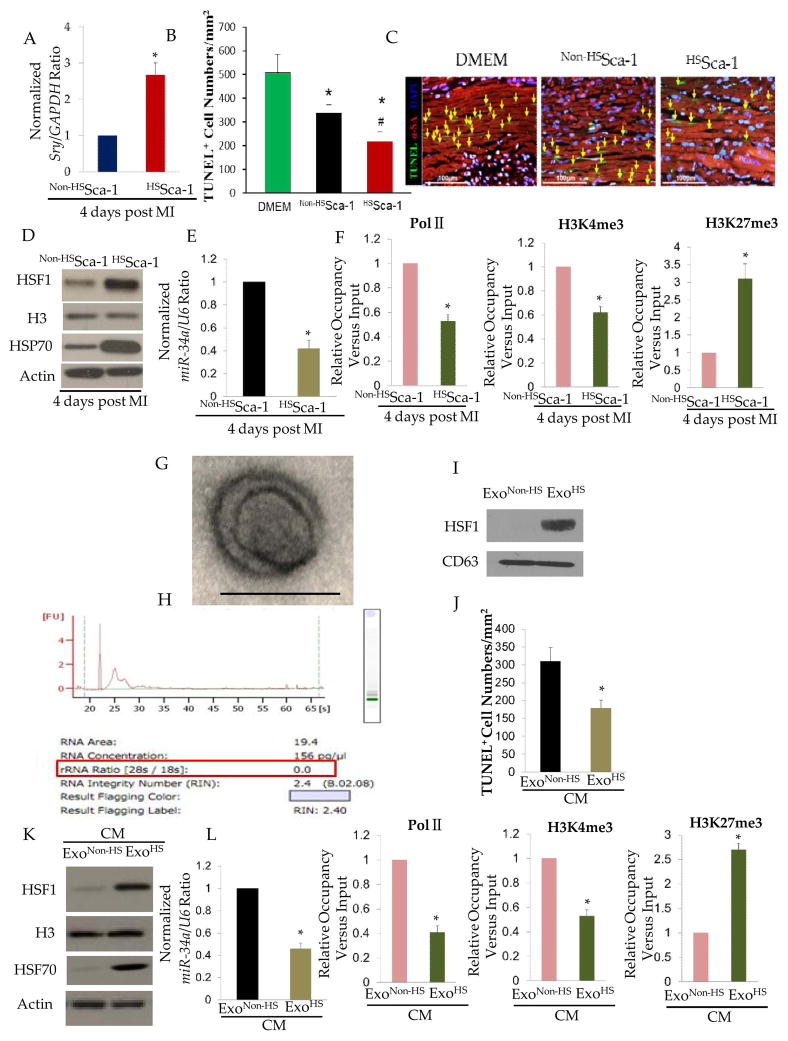
To assess the functional benefits of HSSca-1+ cells in infarcted hearts compared with non-HS Sca-1+ cells, we performed a survival study by detecting the Sry gene expression in female hearts with male Sca-1+ cells transplantation. Real-time PCR showed that Sry gene expression was significantly higher in HSSca-1+ cells transplanted hearts 4 days post transplantation (approximately 2.5 folds vs. control)(n= 4/group)(Figure 5.A). Moreover, by genetic lineage tracing, we found HSSca-1+ cells had a better survival as compared to non-HS Sca-1+ in a chronic setting (7 days, 14 days, 28 days post transplantation)(Suppl. Information-Figure S4). Since body temperature might influence the survival and functional integrity of Sca-1+ stem cells, we examined the body temperatures of animals post MI. However, we did not find a significant change in body temperatures of animals used in different groups (DMEM, non-HS Sca-1+ and HS Sca-1+)(data not shown). Hence, we excluded the possibility that stem cell survival is as the result of the disparity of body temperatures of animals used for non-HS Sca-1+ and HS Sca-1+ cells. Moreover, HSSca-1+ cells transplantation significantly reduced cardiomyocytes apoptosis post myocardial infarction (Figure 5.B and C). Furthermore, molecular analysis revealed thatHSF1 and HSP70 expression was significantly increased and miR-34a was decreased in left ventricle (LV) 4 days post transplanted with HSSca-1+ cells(Figure 5.D and E). ChIP-qPCR further demonstrated the repression of miR-34a in myocardium was associated with the chromatin remodeling on miR-34a promoter (Figure 5.F) as similarly as we showed in HSSca-1+ cells. Therefore, we hypothesized whether HSSca-1+ cells could influence the chromatin landscape of ischemic myocardium via paracrine behavior. Since exosomes have been recently reported as critical carriers for paracrine effects, we isolated exosomes from the culture medium of HSSca-1+ cells, which was characterized by a typical ultrastructure (Figure 5.G) with minimal ribosomal RNA indicating cellular contamination was minimal (Figure 5.H) and expression of typical exosome marker CD63 (Figure 5.I). Interestingly we found HSF1 was significantly enriched in exosomes isolated from culture medium of HSSca-1+ cells (ExoHS) as compared to exosomes isolated from culture medium of non-HSSca-1+ cells (Exonon-HS)(Figure 5.I). To test whether exosomes could rescue cardiomyocytes apoptosis, we administered ExoHS and Exonon-HS into the culture medium of neonatal cardiomyocytes respectively and incubated for 24 h then subjected the neonatal cardiomyocytes to OGD for 4 hrs. The cardiomyocytes incubated with ExoHS showed significantly reduced apoptosis as compared to the ones incubated with Exonon-HS (Figure 5.J). Molecular analysis revealed that ExoHS increased HSF1 and HSP70 expression (Figure 5.K) and decreased miR-34a expression (Figure 5.L) in cardiomyocytes subjected to OGD, which was possible by influencing the chromatin structure of miR-34a as shown by ChIP-qPCR (Figure 5.M). In order to verify HSF1-loaded exosomes from Sca-1+ cells could be uptaken by neonatal cardiomyocytes, we transduced the lentivirus pCDH-CMV-HSF1-GFP (overexpressing fusion protein HSF1-GFP and pCT-CD63-RFP (overexpressing fusion proteinCD63-RFP) (System Biosciences, SBI) into Sca-1+ cells. After transduction for 24 hrs, the Sca-1+ cells were co-cultured with neonatal cardiomyocytes in a non-contact manner. Interestingly, confocal imaging showed that neonatal cardiomyocytes uptook the HSF1-loaded exosomes from Sca-1+ cells under OGD)(Suppl.Information-FigureS5), implying this scenario might take place in vivo.
Figure 5. HSSca-1+ cells transplantation reduced cardiomyocyte apoptosis.
A. Real-time PCR for mice Sry gene in female mouse hearts (n= 4/group) with myocardial infarction (MI) on day-4 post-transplantation of male HSSca-1+ or non-HSSca-1+ cells. B. TUNEL assay and representative pictures (C) showing HSSca-1+ cells transplantation reduced apoptosis of cardiomyocytes on day-4 post myocardial infarction. (*) denotes P< 0.01. vs. DMEM; (#) denotes P< 0.01. vs. non-HSSca-1+ cells. D. Representative western blot image showing elevated HSF1 and HSP70 expression in hearts(n=4/group) on day-4 post-transplantation of HSSca-1+ cells. E. Real-time PCR showing miR-34a was significantly downregulated in hearts (n=4/group) on day-4 post-transplantation of HSSca-1+ cells. (*) denotes P< 0.01. vs.non-HSSca-1+ cells treated hearts. F. ChIP qRT-PCR analysis of RNA polymerase II (Pol II), H3K4me3, and H3K27me3 on the promoter region of miR-34a in hearts (n=4/group) on day-4 post-transplantation of HSSca-1+ cells. Data are normalized to inputs. (*) denotes P< 0.01. vs.non-HSSca-1+ cells treated hearts. G. A representative electron microscopy image of an isolated exosome from culture medium of HSSca-1+ cells. Scale bar, 100 nm. H. Bioanalyzer analysis showing total RNA isolated from exosomes contained no or little ribosomal RNA (18S- and 28S- rRNA). I. Western blot showing isolated exosomes express typical exosomes marker CD63 and HSF1 was enriched in exosomes from HSSca-1+ cells. J. TUNEL assay showing administration of exosomes isolated from culture medium of HSSca-1+ cells (ExoHS) rescued apoptosis of neonatal cardiomyocytes under OGD. Data represent mean ± SEM (n= 3).(*) denotes P< 0.05, relative to cardiomyocytes administered with isolated exosomes from culture medium of non-HSSca-1+ cells (Exonon-HS). K. Western blot showing ExoHS administration increased HSF1 and HSP70 protein levels in neonatal cardiomyocytes under OGD as compared to Exonon-HS. L. Real-time PCR showing ExoHS administration decreased miR-34a expression in neonatal cardiomyocytes under OGD. Data represent mean ± SEM (n= 3).(*) denotes P< 0.05, relative to cardiomyocytes with Exonon-HS administration. M. ChIP qRT-PCR analysis of RNA polymerase II (Pol II), H3K4me3, and H3K27me3 on miR-34a promoter in neonatal cardiomyocytes under OGD with ExoHS and Exonon-HS administration. Data represent mean ± SEM (n= 3).(*) denotes P< 0.05, relative to cardiomyocytes with Exonon-HS administration.
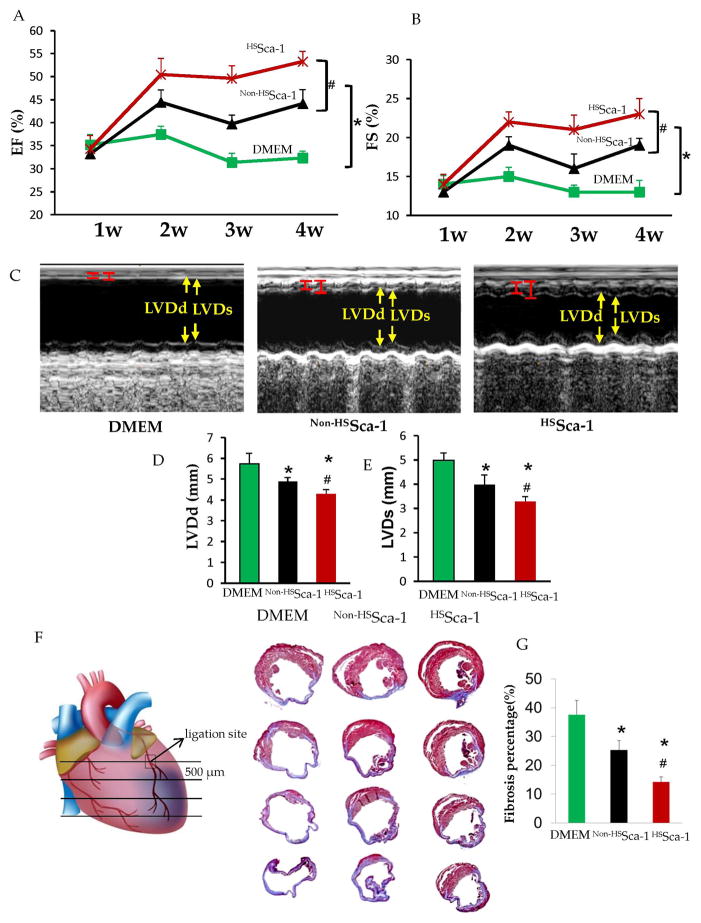
HSSca-1+ Cells Transplantation Improved Global Heart Function
Echocardiography analysis revealed ejection fraction (EF) and fractional shortening (FS) were progressively improved from 1 week to 4 weeks in the hearts post transplantation with HSSca-1+ cells as compared to the ones treated with non-HSSca-1+ cells and DMEM (Figure 6.A and B). LV diastolic dimension (LVDd) and LV systolic dimension (LVDs) were significantly decreased in the hearts with HSSca-1+ cells transplantation 4 weeks post MI (Figure 6.C, D and E). Moreover, Masson’s trichrome staining and subsequent calculation of fibrosis area revealed that the hearts with HSSca-1+ cells transplantation showed a decreased fibrosis as compared to the ones treated with non-HSSca-1+ cells and DMEM 4 weeks post myocardial infarction (Figure 6.F and G). These results suggest that transplantation of HSSca-1+ cells played an important role in attenuating LV remodeling after MI.
Figure 6. HSSca-1+cells transplantation improved global heart function and ameliorated cardiac fibrosis after myocardial infarction.
Serial LV performance parameters (left ventricle ejection fraction (LVEF) and left ventricle fractional shortening (LVFS)) post MI (1w, 2w, 3w, 4w) were measured as shown in A and B (n=8/group). (*) denotes P< 0.01. vs. DMEM; (#) denotes P< 0.01. vs. non-HSSca-1+ cells. C. Representative M-mode echocardiography 4 weeks post MI. Left ventricle diastolic dimension (LVDd) and left ventricle systolic dimension (LVDs) were measured as shown in D and E respectively. (*) denotes P< 0.01. vs. DMEM; (#) denotes P< 0.01. vs. non-HSSca-1+ cells. F. Representative Masson’s trichrome-stained pictures from 4 continuous sections (skip 500 μM) for fibrosis measurement in hearts with DMEM injection, non-HSSca-1+ and HSSca-1+ cells transplantation 4 weeks post MI (n=8/group). Scar tissue and viable myocardium are identified in blue and red, respectively. G. Quantitative analysis of fibrosis. (*) denotes P< 0.01. vs. DMEM; (#) denotes P< 0.01. vs. non-HSSca-1+ cells.
Discussion
Our study demonstrated that pre-induction of HSP70 by heat shock can promote Sca-1+ stem cells survival, which is in line with a previous study showing lentiviral overexpression of HSP70 enhanced mesenchymal stem cells survival after oxygen deprivation13. However, lentiviral delivery may incur carcinogenesis and thus heat shock, as a non-transgenic approach, may be a safer and more appealing option for clinical application.
Over the last several decades, it is well documented that HSF1 transactivates HSP70 transcription by direct binding on the promoter of HSP70. While appreciating the importance of this classical mechanism, our data revealed that the relation between HSF1and HSP70 is more complex than we previously thought. Surprisingly, we discovered miR-34a, a previously well-known proapoptotic miRNA is a bridge between HSF1 and HSP70 and heat shock can induce epigenetic repression of miR-34a in stem cells with the participation of HSF1, which causes a derepression of miR-34a target-HSP70. Thus, our study established a new triangle of these three old molecules and provided novel insights for their roles in stem cell survival.
Another novel finding of our study is the paracrine behavior of heat shocked stem cells. In recent years, a major breakthrough in the field of paracrine research is the discovery of the bioactive vectors-exosomes, which are small vesicles secreted by many cell types and enable a potent mode of intercellular communication14. Recent studies indicated the transfer of mRNA, miRNA and protein by exosomes might alter the function of recipient cells15–18. An intriguing finding of our study is that the secretion of HSF1-enriched exosomes induced by heat shock in Sca-1+ cells could direct cardiomyocytes toward a prosurvival phenotype via remodeling the chromatin structure of miR-34a. This is the first time stem cells have been proven to influence the chromatin landscape of adult cells by transfer of exosomes. Thus, our study highlighted the importance for future studies to decipher the roles of the transportation of transcription factors or epigenetic modifiers on the epigenome in recipient cells. Although our in vitro study has shown that HSF1-loaded exosomes could reduce cardiomyocytes apoptosis and remodel the chromatin structure of miR-34a in ischemic cardiomyocytes, our study is limited due to lack of in vivo evidence to verify this event. Therefore, direct testing of isolated exosomes derived from heat shocked Sca-1+ cells in infarcted hearts would be beneficial to prove this concept. Moreover, our in vivo study showed that transplantation of HSSca-1+ cells significantly attenuated cardiac fibrosis. Although the contribution of Sca-1+ cells towards myogenesisis one of the possible reasons19, the role of HSF1-enriched exosomes on slowing the progression of fibrosis cannot be excluded. A late-breaking report has indicated that upregulated miR-34a levels is associated with age/MI induced cardiac cell death and functional decline. Thus, the results of our studies are parallel to the idea that inhibition of miR-34a dramatically reduces cardiac cell death and fibrosis following MI and aging20. Furthermore, as we discovered HSF1 is a direct repressor for miR-34a and is enriched in ExoHS, it would be worth demonstrating in future study whether administration HSF1-enriched exosomes isolated from the culture medium of heat shocked stem cells or directly generated by artificial exosomes21 could become a novel and safe therapy (exosome is natural and non-viral vector) in ageing/fibrosis associated diseases.
Besides the role in apoptosis and fibrosis, miR-34a is also a critical barrier for reprogramming and its deficiency significantly increased reprogramming efficiency and kinetics22. Thus, it would be interesting to show whether heat shock could enhance reprogramming efficiency with a role of HSF1/miR-34a/HSF70 triangle.
Finally, the signaling pathways we deciphered might also have wide-ranging implications for cancer research since HSF1 is a critical modifier for carcinogenesis23, 24 and miR-34a is inactivated in cancer stem cells25. Whether HSF1 contributes to inactivation of miR-34a in cancer stem cells and how HSF1/miR-34a/HSF70 triangle participates in carcinogenesis should be investigated in the future.
Taken together, our study not only has significant therapeutic value in translational stem cell based therapy for cardiovascular diseases but may impact many fields such as cell reprogramming and cancer biology.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R37-HL-074272, HL-095375, and HL-087246 (to M. Ashraf) and National Natural Science Foundation of China (No. 81120108003, No. 81330007) and National Key Basic Research Program of China (No. 2012CB526602) ( to X.Y. Yu).
Footnotes
Conflict of Interest Disclosures: None
Author contributions: Y.F.: conception and design, collection of data and analysis and interpretation, manuscript writing; W.H.: cell transplantation, echocardiography and collection of data and analysis; A.G.J.: bioinformatics analysis; W.M., Y.W., H.W.K., Z.P., Z.W., R.M.M.: supply of study materials; X.Y, M.A.: conception and design, data analysis and interpretation, manuscript writing, financial support and final approval of manuscript
References
- 1.Ptaszek LM, Mansour M, Ruskin JN, et al. Towards regenerative therapy for cardiac disease. Lancet. 2012;379:933–942. doi: 10.1016/S0140-6736(12)60075-0. [DOI] [PubMed] [Google Scholar]
- 2.Wu KH, Mo XM, Han ZC, et al. Stem cell engraftment and survival in the ischemic heart. Ann Thorac Surg. 2011;92:1917–1925. doi: 10.1016/j.athoracsur.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Penn MS, Mangi AA. Genetic enhancement of stem cell engraftment, survival, and efficacy. Circ Res. 2008;102:1471–1482. doi: 10.1161/CIRCRESAHA.108.175174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yellon DM, Pasini E, Cargnoni A, et al. The protective role of heat stress in the ischaemic and reperfused rabbit myocardium. J Mol Cell Cardiol. 1992;24:895–907. doi: 10.1016/0022-2828(92)91102-b. [DOI] [PubMed] [Google Scholar]
- 5.Rordorf G, Koroshetz WJ, Bonventre JV. Heat shock protects cultured neurons from glutamate toxicity. Neuron. 1991;7:1043–1051. doi: 10.1016/0896-6273(91)90348-4. [DOI] [PubMed] [Google Scholar]
- 6.Silver JT, Noble EG. Regulation of survival gene hsp70. Cell Stress Chaperones. 2012;17:1–9. doi: 10.1007/s12192-011-0290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 9.Lodygin D, Tarasov V, Epanchintsev A, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 10.Chim CS, Wong KY, Qi Y, et al. Epigenetic inactivation of the miR-34a in hematological malignancies. Carcinogenesis. 2010;31:745–750. doi: 10.1093/carcin/bgq033. [DOI] [PubMed] [Google Scholar]
- 11.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 13.McGinley L, McMahon J, Strappe P, et al. Lentiviral vector mediated modification of mesenchymal stem cells & enhanced survival in an in vitro model of ischaemia. Stem Cell Res Ther. 2011;2:12. doi: 10.1186/scrt53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cosme J, Liu PP, Gramolini AO. The cardiovascular exosome: Current perspectives and potential. Proteomics. 2013 doi: 10.1002/pmic.201200441. [DOI] [PubMed] [Google Scholar]
- 15.Gross JC, Chaudhary V, Bartscherer K, et al. Active Wnt proteins are secreted on exosomes. Nat Cell Biol. 2012;14:1036–1045. doi: 10.1038/ncb2574. [DOI] [PubMed] [Google Scholar]
- 16.Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hergenreider E, Heydt S, Treguer K, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 18.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajstura J, Rota M, Whang B, et al. Bone marrow cells differentiate in cardiac cell lineages after infarction independently of cell fusion. Circ Res. 2005;96:127–137. doi: 10.1161/01.RES.0000151843.79801.60. [DOI] [PubMed] [Google Scholar]
- 20.Boon RA, Iekushi K, Lechner S, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 21.Kooijmans SA, Vader P, van Dommelen SM, et al. Exosome mimetics: a novel class of drug delivery systems. Int J Nanomedicine. 2012;7:1525–1541. doi: 10.2147/IJN.S29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi YJ, Lin CP, Ho JJ, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13:1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai C, Whitesell L, Rogers AB, et al. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendillo ML, Santagata S, Koeva M, et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bu P, Chen KY, Chen JH, et al. A microRNA miR-34a-Regulated Bimodal Switch Targets Notch in Colon Cancer Stem Cells. Cell Stem Cell. 2013;12:602–615. doi: 10.1016/j.stem.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.