Dear Editor
Salvia miltiorrhiza Bunge (Danshen) is a medicinal plant of the Lamiaceae family, and its dried roots have long been used in traditional Chinese medicine with hydrophilic phenolic acids and tanshinones as pharmaceutically active components (Xu et al., 2016; Zhang et al., 2014). The first step of tanshinone biosynthesis is the bicyclization of the general diterpene precursor (E, E, E)-geranylgeranyl diphosphate (GGPP) to copalyl diphosphate (CPP) by CPP synthases (CPSs), which is followed by cyclization or rearrangement reaction catalyzed by kaurene synthase-like enzymes (KSL). The resulting intermediate is usually an olefin, which requires the insertion of oxygen by cytochrome P450 mono-oxygenases (CYPs) for the final production of diterpenoids (Zi et al., 2014). While the CPS, KSL and several early acting CYPs (CYP76AH1, CYP76AH3 and CYP76AK1) for tanshinone biosynthesis have been identified in S. miltiorrhiza (Gao et al., 2009; Guo et al., in press; Guo et al., 2013; Zi and Peters, 2013), the majority of the overall biosynthetic pathway, as well as the relevant regulatory factors associated with tanshinone production, remains elusive (Figure 1B).
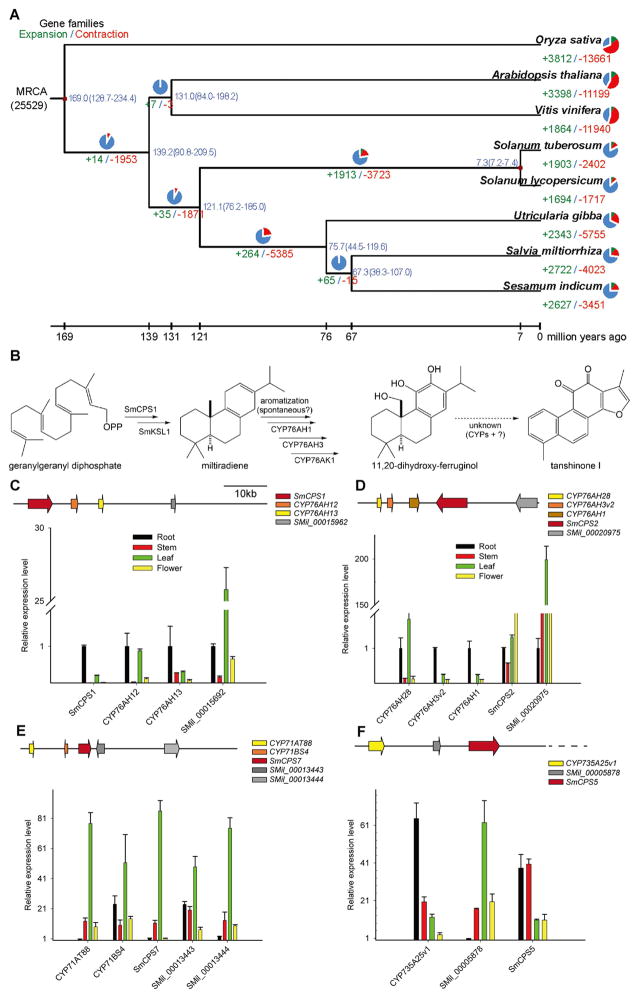
Figure 1. Evolutionary and functional analysis of S. miltiorrhiza genome.
(A) Phylogenetic analysis and divergence time estimation among eight plant species, along with gene-family dynamics for each branch. The tree was constructed based on 1,824 single-copy true orthologous genes. Divergence times between potato-tomato (7.2–7.4 MYA) and monocot-eudicot (128.7–234.4 MYA) were used as references for time calibration. Divergence times are indicated by the blue numbers beside the branching nodes. The number of gene-family contraction and expansion events is indicated by green and red numbers (respectively) below each species name.
(B) The predicted tanshinone biosynthetic pathway from the precursor GGPP to tanshinone I.
(C–F) Genomic configurations of four SmCPS/CYP gene clusters, and the expression profiles of these genes in different tissues. The scale bar was calculated from the relative expression levels of three qRT PCR repeats.
Here we report the draft sequence and analysis of the S. miltiorrhiza genome by a hybrid assembly approach. Firstly, genomic DNA was extracted from S. miltiorrhiza line 99-3, a strain cultivated by IMPLAD, and 158.2 Gb Illumina data were generated on Hiseq 2000 platform (250-fold genome coverage, Supplementary Table 1) and assembled with Phusion2 (Mullikin and Ning, 2003), which resulted in a draft assembly of 558 Mb, with contig N50 of 2.47 Kb. Attempts with other assemblers, such as SOAPdenovo and Fermi, gave similar assembly metrics, suggesting intrinsic complexity of this plant genome. We then generated 8.19 Gb data with PacBio RS platform (3.74 Kb read length in average) and 8.65 Gb Roche/454 data (Supplementary Table 1). Celera Assembler (v7.0) was used for PacBio reads assembly after base-error correction with Roche/454 data, and the resultant contigs were combined with 454 reads for re-assembly. Finally, Illumina reads were mapped onto these contigs to correct single nucleotide polymorphisms (SNPs) and small insertions/deletions (indels) in homozygotes, which were presumably introduced by sequencing chemistry bias. This led to a final genome assembly of 538 Mb, with contig and scaffold N50 of 12.38 Kb and 51.02 Kb, respectively (Supplementary Table 2). Compared with the estimated genome size of 615 Mb by flow cytometry analysis (Supplementary Figure 1), the relatively small size of the assembled genome might result from the high repeat content of this species, as multiple copies of repetitive elements are presumably collapsed together.
By mapping the Illumina reads onto the draft assembly, 1,486,270 heterozygous SNPs (and 302,217 short indels) were identified, corresponding to 2.76 SNPs per Kb (Supplementary Table 3). This heterozygosity value was comparable to that of Populus (2.6 polymorphisms per kb) and grape (3.6 SNPs per kb).
Sequence annotation revealed that repetitive elements accounted for 54.44% of the genome (Supplementary Table 4), twice that of sesame, another species from the order Lamiales (Wang et al., 2014). Long terminal repeats were the most abundant, spanning 18.03% of the genome, while 55.58% of the repeats (30.26% of the genome) were unclassified, implying lineage-specific repeat expansion.
We predicted 30,478 protein-coding genes in the S. miltiorrhiza genome using ab initio and homology-based gene prediction methods (Supplementary Table 4), which were further validated by RNAseq data (Xu et al., 2015). Most of these genes (91.2%) had homologs in the non-redundant (nr) database at GenBank (E-value = 1e-5), and more than half (56.60%) could be assigned to KEGG pathways. Among them were 1,620 transcription factor (TF) genes, including 171 APETALA2, 139 bHLH, 291 MYB, and 78 WRKY family TFs (Supplementary Table 5). Several of these TFs have been previously associated with the biosynthesis of tanshinone and phenolic acid (Xu et al., 2016). In addition, 82 terpene synthase genes (TPS, Supplementary Table 6) involved in production of hemi-, mono-, sesqui- or di-terpenes, along with 437 CYPs (Supplementary Table 7) that catalyze various oxidation reactions, were identified.
Gene family evolution among eight plant species, including rice, Arabidopsis, grape, tomato, potato, bladderwort, sesame, and S. miltiorrhiza, was analyzed by CAFÉ (version 2.1). This suggests that gene family contraction outnumbered expansion along each lineage (Figure 1A). Intriguingly, families undergoing significant expansion in S. miltiorrhiza (P < 0.01) were primarily involved in stilbenoid, diarylheptanoid or gingerol biosynthesis (Ko00945), terpenoid biosynthesis (Ko00902), or steroid biosynthesis (Ko00100), which is consistent with the high production of tanshinones and phenolic acids by this medicinal plant. Phylogenomic analysis revealed that S. miltiorrhiza was most closely related to sesame, with an estimated divergence time of approximately 67 million years ago (Figure 1A).
Physical clustering of TPSs and CYPs is frequently associated with consecutive enzymatic actions in terpenoid biosynthesis (Boutanaev et al., 2015), and was investigated here. Four such TPS/CYP pairs were found in the draft S. miltiorrhiza genome (Figure 1, C–F). Three of these four CPSs have been previously characterized, with SmCPS1 and SmCPS2 involved in tanshinone biosynthesis in the roots and leaves, respectively, while SmCPS5 is required for gibberellin phytohormone metabolism (Cui et al., 2015). Interestingly, both SmCPS1 and SmCPS2 are flanked by genes from the CYP76AH sub-family. Notably, this includes the previously characterized CYP76AH1 (Guo et al., 2013). Even more strikingly, while this letter was in preparation it was reported that another of these CYP76AH sub-family members, CYP76AH3, was involved in tanshinone biosynthesis as well (Guo et al., in press), further validating the association of these biosynthetic gene clusters with tanshinone biosynthesis. Phylogenetic analysis suggests that the SmCPS1 and SmCPS2 clusters originated from duplication event of an ancestral CPS/CYP76AH pair (Supplementary Figure 2).
To further investigate the role of these clusters in tanshinone biosynthesis, the tissue specific expression of the genes was analyzed using RNA-Seq data. Much as previously reported (Cui et al., 2015), SmCPS1 and SmCPS2 are most highly expressed in the roots and leaves/flowers, respectively. However, the expression patterns of the CYP76AH sub-family members do not simply follow that of the co-clustered CPSs. Instead, despite being clustered with the root specific SmCPS1, CYP76AH12 is equally expressed in both the roots and leaves, although the linked CYP76AH13 is more specifically expressed in roots. In addition, despite being clustered with the more aerial tissue specific SmCPS2, CYP76AH1 and CYP76AH3 are quite specifically expressed in roots, although the linked CYP76AH28P is more highly expressed in the leaves. All of these expression patterns were validated by qRT-PCR (Figure 1, C–F). Taken together, it seemed to imply that the decoupling of expression between CPSs and their flanking CYPs had occurred after gene cluster duplication event.
The SmCPS7/CYP cluster contains two members of CYP71 family (Fig. 1E), CYP71AT88 and CYP71BS4. Given that a number of CYPs from the CYP71 family are involved in (di)terpenoid biosynthesis (Zi et al., 2014), this raises the possibility that this cluster might participate in a common diterpenoid biosynthetic pathway.
For the SmCPS5/CYP cluster (Fig. 1F), previous work had suggested that SmCPS5 is involved in gibberellin metabolism (Cui et al., 2015), while CYP735A25v1 has no known role in such phytohormone metabolism. Thus, this particular pair of enzymes seems unlikely to operate together in a common pathway.
We then compared the tissue-specific expression patterns of all 437 annotated CYP genes with that of SmCPS1. Thirty-two CYPs exhibited similar expression patterns to SmCPS1 across different organs examined (R2 > 0.85) (Supplementary Table 8). As expected, this includes CYP76AH1, whose role in tanshinone biosynthesis was first suggested on the basis of similar co-expression analysis (Guo et al., 2013), as well as CYP76AH3 and CYP76AK1, whose recently reported roles in tanshinone biosynthesis were discovered by the same approach (Guo et al., in press). Hence, the remaining co-regulated CYPs provide additional candidates for investigation of the tanshinone biosynthetic pathway.
The traditional use of Danshen involves decoction with water, indicating an important role for the hydrophilic phenolic acids. These include rosmarinic acid (RA), salvianolic acid and lithospermic acid B, whose biosynthesis involves both general phenylpropanoid metabolism and the more specific tyrosine-derived pathway. As previously reported, the genome contains 29 genes from 9 families potentially involved in S. miltiorrhiza phenolic acid biosynthesis. Notably, most families had multiple genes with distinct expression patterns, implying diversified roles for these natural products. In addition, from the 80 laccases genes, 5 were identified as potentially involved in the conversion of RA to salvianolic acid, based on their specific expression in the root phloem and xylem tissues. Thus, the genome sequence reported here is providing important insights into the biosynthesis of these water-soluble natural products as well.
While the S. miltiorrhiza plant that was used for genome sequencing has purple flowers, the white-flowered landrace S. miltiorrhiza is known for better medical quality. To evaluate the genetic differences between these varieties, a white-flowered plant was selected for sequencing and comparative analysis. The number of homozygous SNPs (1,719,024) was roughly twice that of heterozygotes, corresponding to the fixed polymorphism level of 3.87 SNPs per Kb. Overall, 49,521 non-synonymous SNPs were identified, among which 580 protein-coding genes were affected through the formation of premature stop codons. Nine KEGG pathways were significantly enriched with non-synonymous amino acid changes, including pathways for diterpenoid, flavonoid and phenylpropanoid biosynthesis, as well as those for Toll-like receptor signaling and plant-pathogen interactions (Supplementary Figure 3).
While the average sequencing depth for the white-flower plant was 42X, more than 10% of the genome had no coverage at all. For further investigation, 28.6 Mb genomic regions longer than 1 Kb with no mapping coverage were analyzed. Interestingly, only 12.68% of these regions were comprised of repetitive sequences, a much lower proportion than the genome average (54.44%). In total, these regions contained 107 genes, which appear to have been lost in the white-flower landrace, including 11 disease-resistance genes, 4 CYPs, and 13 transcription factors. At least some of this intergenomic diversity is hypothesized to contribute to the phenotypic differences between these two varieties, such as flower coloration, and tanshinone content, which will be the subject of future investigations.
In summary, we present a draft assembly of the S. miltiorrhiza genome using long reads from the PacBio RS platform to supplement short Illumina reads, which resulted in significant improvement of the assembly quality. This hybrid approach proved effective for the highly repetitive and complex genome of S. miltiorrhiza, enabling assembly of sufficiently large enough scaffolds for the identification of potential biosynthetic gene clusters. The four CPS/CYP gene clusters revealed here, along with other genes potentially encoding biosynthetic enzymes (e.g., in tashinone biosynthesis - Supplementary Table 9), provide a strong foundation for understanding the biochemical diversity and pharmaceutical qualities of S. miltiorrhiza. Moreover, access to the genome sequence is further expected to enable molecular breeding with this important traditional medicinal herb.
Acknowledgments
FUNDING
This work was supported by the National Natural Science Foundation of China (81130069, 81573398, 31400278), the National Key Technology R&D Program (2012BAI29B01), the Key Project of Chinese National Programs for Fundamental Research and Development (2013CB127000) and the US National Institutes of Health (GM109773).
Footnotes
ACCESSION NUMBERS
Raw Illumina Hiseq 2000, the Roche/454 and PacBio sequencing reads of S. miltiorrhiza line 99-3 and Raw Illumina Hiseq 2000 sequencing reads of The white-flowered landrace S. miltiorrhiza have been submitted to the NCBI Sequence Read Archive database (SRP051524, SRP051564, SRP028388). All of the data generated in this project, including those related to genome assembly, gene prediction, gene functional annotations, and transcriptomic data, may also be downloaded from our web portal at http://www.ndctcm.org/shujukujieshao/2015-04-23/27.html.
SUPPLEMENTARY INFORMATION
Supplementary Information is available at Molecular Plant Online.
AUTHOR CONTRIBUTIONS
SC, XQ, and JS conceived the study. QL, YL, JX, JQ and YuZ sequenced the genome. HX and ZN assembled the genome. Annotation and evolutionary analysis of the genome were performed by CSo and YuZ. HL, YiZ, WS and RP analyzed the putative gene clusters. HL, BW, XZ, AJ, ZX, XWL, XEL, LH, DN, HY and GC performed the experiments. SC, XQ, JS, CL, JL, XL, JW, CSu and YW coordinated the project. HX, JS, HL, YuZ, JX, CSo, GS, AH, ZN, RJP, XQ and SC wrote the paper. All of the authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boutanaev AM, Moses T, Zi J, Nelson DR, Mugford ST, Peters RJ, Osbourn A. Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci USA. 2015;112:E81–88. doi: 10.1073/pnas.1419547112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Duan L, Jin B, Qian J, Xue Z, Shen G, Snyder JH, Song J, Chen S, Huang L, et al. Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza Bunge. Plant Physiol. 2015;169:1607–1618. doi: 10.1104/pp.15.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett. 2009;11:5170–5173. doi: 10.1021/ol902051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Ma X, Cai Y, Ma Y, Zhan Z, Zhou YJ, Liu W, Guan M, Yang J, Cui G, et al. Cytochrome P450 promiscuity leads to a bifurcating biosynthetic pathway for tanshinones. New Phytol. doi: 10.1111/nph.13790. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Zhou YJ, Hillwig ML, Shen Y, Yang L, Wang Y, Zhang X, Liu W, Peters RJ, Chen X, et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci USA. 2013;110:12108–12113. doi: 10.1073/pnas.1218061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullikin JC, Ning Z. The phusion assembler. Genome Res. 2003;13:81–90. doi: 10.1101/gr.731003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yu S, Tong C, Zhao Y, Liu Y, Song C, Zhang Y, Zhang X, Wang Y, Hua W, et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014;15:R39. doi: 10.1186/gb-2014-15-2-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Ji A, Zhang X, Song J, Chen S. Biosynthesis and regulation of a ctive constituents in medicinal model plant Salvia miltiorrhiza. Chin Herbal Med. 2016;8:3–11. [Google Scholar]
- Xu Z, Peters RJ, Weirather J, Luo H, Liao B, Zhang X, Zhu Y, Ji A, Zhang B, Hu S, et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 2015;82:951–961. doi: 10.1111/tpj.12865. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yan YP, Wu YC, Hua WP, Chen C, Ge Q, Wang ZZ. Pathway engineering for phenolic acid accumulations in Salvia miltiorrhiza by combinational genetic manipulation. Metab Eng. 2014;21:71–80. doi: 10.1016/j.ymben.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Zi J, Mafu S, Peters RJ. To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu Rev Plant Biol. 2014;65:259–286. doi: 10.1146/annurev-arplant-050213-035705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi J, Peters RJ. Characterization of CYP76AH4 clarifies phenolic diterpenoid biosynthesis in the Lamiaceae. Org Biomol Chem. 2013;11:7650–7652. doi: 10.1039/c3ob41885e. [DOI] [PMC free article] [PubMed] [Google Scholar]