Abstract
Objectives
Individualized treatment for bipolar disorder based on neuroimaging treatment targets remains elusive. To address this shortcoming, we developed a linguistic machine learning system based on a cascading Genetic Fuzzy Tree (GFT) design called the LITHium Intelligent Agent (LITHIA). Using multiple objectively defined fMRI and proton MRS (1H-MRS) inputs, we tested whether LITHIA could accurately predict lithium response in participants with first-episode bipolar mania.
Methods
We identified 20 subjects with first-episode bipolar mania who received an adequate trial of lithium over eight weeks and both fMRI and 1H-MRS scans at baseline pre-treatment. We trained LITHIA using 18 1H-MRS and 90 fMRI inputs over four training runs to classify treatment response and predict symptom reductions. Each training run contained a randomly selected 80% of the total sample and was followed by a 20% validation run. Over a different randomly selected distribution of the sample, we then compared LITHIA to eight common classification methods.
Results
LITHIA demonstrated nearly perfect classification accuracy and was able to predict post-treatment symptom reductions at 8 weeks with at least 88% accuracy in training and 80% accuracy in validation. Moreover, LITHIA exceeded the predictive capacity of the eight comparator methods and showed little tendency towards overfitting.
Conclusions
Results provide proof-of-concept that a novel GFT is capable of providing control to a multidimensional bioinformatics problem – namely prediction of lithium response – in a pilot data set. Future work on this, and similar machine learning systems, could help assign psychiatric treatments more efficiently, thereby optimizing outcomes and limiting unnecessary treatment.
Keywords: artificial intelligence, bipolar disorder, fMRI, fuzzy logic, genetic algorithm, lithium, machine learning, mania, region-of-interest, spectroscopy
Introduction
Bipolar disorder is a common, lifelong, and recurrent illness with lifetime prevalence rates up to 6% of the population when considering subthreshold mood symptoms.1,2 Although it is characterized by episodic mood dysregulation that fluctuates among manic, depressive, and euthymic (i.e., relatively symptom free) mood states, it is defined by the occurrence of mania. Patients experience an especially high degree of morbidity and mortality during manic episodes,3 highlighting the need to identify effective anti-manic treatments quickly.
Lithium is the oldest mood stabilizer and well established as a first-line anti-manic agent. Indeed, lithium is the only psychotropic agent specific to the treatment of bipolar disorder,3 making it uniquely suited for outcomes research targeting mania reduction. It is also particularly effective early in the course of illness.4 Nonetheless, many patients do not respond adequately to a trial of lithium monotherapy, and in the absence of reliable predictors of response, these patients are exposed to side effect risks without the benefit of eventual treatment response.
Despite the long history of lithium treatment for bipolar disorder, objective biological markers (i.e., blood or genetic markers) of potential treatment responsiveness to lithium and, indeed, all mood stabilizers have been lacking. Therefore, identifying an optimal treatment for an individual with bipolar disorder remains a time-consuming trial-and-error process.5 This approach is especially true after a first manic episode, which is necessary for the initial diagnosis of bipolar disorder, since there is little indication of which mood stabilizer class (e.g., lithium, second-generation antipsychotics or anticonvulsants) might be most beneficial in a given individual in the absence of a treatment response history.
With advances in neuroimaging, there has been considerable interest in brain markers of treatment effectiveness in serious mental illness. Yet, imaging research has failed to provide brain-based indicators of treatment response (or diagnosis) in bipolar disorder to date. One way forward has been to combine multiple clinical measures into a data-driven analytical approach to differentiate groups based on the pattern of relationships among measures.6 The data are then included as inputs in a bottom-up classification algorithm to find the best separation between groups. Traditionally, linear pattern classification has been conducted using inferential statistics, such as discriminant function analysis, or using support vector machines (SVMs), but these have yet to identify predictors for lithium response. Only more recently has machine learning applied non-linear solutions of large-scale bioinformatics problems, which is important for the study of bipolar disorder where relationships among neurocognitive measures and symptoms are often non-linear.7 Moreover, these newer algorithms are specifically tailored to situations in which the number of inputs vastly exceeds the sample size,8 as is often the case in neuroimaging research.
With the above considerations in mind, we examined whether a new machine learning system based on a cascading Genetic Fuzzy Tree (GFT) design could be used to predict lithium response in subjects with first-episode mania. Limitations inherent to standard fuzzy logic based machine learning systems stem, in part, from scalability issues. This problem derives from the need for a rule to be “learned” for every combination of values of membership functions for each input. The GFT method we employ for the present study mitigates these scalability concerns significantly by breaking the problem space into many sub-decisions. Thus, the GFT is vastly more scalable than traditional fuzzy logic based systems and maintains the base strengths of fuzzy logic; robustness to noise and subjectivity, adaptability of lessons learned from training, and high performance of final outputs. For this study, prediction is based on a set of objective pre-treatment neuroimaging parameters including relative functional magnetic resonance imaging (fMRI) activation and magnetic resonance spectroscopy (MRS) metabolite levels in various brain regions in a data-driven design. Enhanced classification accuracy using the GFT approach could help avoid unnecessary treatment, time, and expense in the subset of lithium non-responsive patients with bipolar disorder.
Methods
Participants
Twenty subjects who received open-label lithium to treat a first manic episode of bipolar I disorder were identified retrospectively from studies conducted within the University of Cincinnati Bipolar Imaging and Treatment Research Center (BITREC).9 Participants had been recruited during hospitalizations at the University of Cincinnati or Cincinnati Children’s Hospital Medical Center, provided informed consent (or assent with parental consent if < 18 years old), and completed both fMRI and MRS scans at a baseline scanning session. Fifteen subjects were less than 18 years old at the time of scan.
Subjects were included if they: (1) were 12 – 35 years old; (2) met DSM-IV criteria for bipolar I disorder, currently manic or mixed with a baseline Young Mania Rating Scale (YMRS)10 total score ≥ 20; (3) had no prior manic episodes and ≤ 2 prior depressive episodes; (4) had no previous psychiatric hospitalizations; (5) had < 3 months of lifetime psychotropic medication exposure, other than stimulants, including no active psychotropic medication in the two weeks prior to the index admission; and (6) no contraindication to taking lithium. Subjects were excluded by: (1) a history of substance dependence within three months prior to the index assessment; (2) any medical or neurological disorder that could affect fMRI assessments; (3) a history of significant developmental delays or estimated full-scale IQ score < 85; or (4) an MRI scan was contraindicated. We have recently reported imaging findings in a larger sample of both lithium and quetiapine treated patients, of which all patients in this present analysis were also included.9
Clinical Assessments
Diagnostic assessments were performed using the Structured Clinical Interview for DSM-IV, Patient version (SCID-P)11 or, for subjects under 18 years old, the Washington University Kiddie Schedule for Affective Disorders and Schizophrenia for School-Age Children - Present and Lifetime version (KSADS).12 Substance use assessments were augmented with the Addiction Severity Index (ASI).13 Manic symptoms were evaluated using the YMRS as noted. Changes in YMRS total scores between baseline and week 8 served as the treatment response measure of interest. Lithium response was defined as at least a 50% improvement between baseline pre-treatment and week 8 post-treatment YMRS scores.
Lithium Treatment Protocol
Subjects received open-label lithium for 8 weeks. Lithium was chosen because it is a first-line FDA-approved treatment for mania in children and adults with specific anti-manic effects. Open-label treatment was initiated during the index hospitalization and then continued following hospital discharge throughout the course of the study. Doses were adjusted by study clinicians based upon serum drug levels (target was 0.8 to 1.2 meq/L) and treatment response and tolerability. Adherence was verified by participant report, pill counts and serum levels when indicated.
Continuous Performance Task with Emotional and Neutral Distractors (CPT-END)
During fMRI sessions, nonferromagnetic goggles were positioned on subjects to provide clear visualization of the CPT-END.9,14 This task is a visual oddball paradigm in which 70% of cues are colored squares, 10% are colored circles, 10% are emotionally neutral pictures, and 10% are emotionally unpleasant pictures. The neutral and emotional pictures were taken from the International Affective Picture System (IAPS; University of Florida) based upon the rating criteria developed by Yamasaki et al.15 Each visual cue required a response; the circles (targets) required a unique response (button 2), whereas the squares and pictures all required the same response (button 1). Imaging sessions consisted of two runs of 158 visual cues per run presented at three-second intervals for two seconds each. Emotional and neutral pictures were presented pseudorandomly. A fixation cross was presented for one second between cues.
Functional Magnetic Resonance Imaging (fMRI) Acquisition
Scanning was conducted at the University of Cincinnati Center for Imaging Research using a 4.0 Tesla Varian Unity INOVA Whole Body MRI/MRS system (Varian Inc., Palo Alto, CA, USA) following a protocol we have previously described.9,14,16 Briefly, to provide anatomic localization, a high-resolution, T1-weighted, 3-D brain scan was obtained followed by a multi-echo reference scan to correct for ghost and geometric distortions.17,18 For fMRI, whole-brain images (volumes) were acquired every three seconds using a T2*-weighted gradient-echo echoplanar imaging (EPI) pulse sequence [TR/TE = 3000/30 msec, FOV = 20.8 × 20.8 cm, matrix = 64 × 64 pixels, slice thickness = 5 mm, flip angle = 75°].
fMRI Image Processing
Functional MRI data were analyzed using AFNI (Analysis of Functional NeuroImages; http://afni.nimh.nih.gov/afni).19 Structural and functional images were co-registered based upon scanner coordinates. Functional images were corrected for motion using a six-parameter rigid body transformation.20 The maximum motion of any analyzed participant was < 5 mm. The average displacement between any successive TR pairs was < 0.1 mm. Additionally, each volume was inspected for signal artifacts using a semi-automated algorithm in AFNI and excluded from further analysis if uncorrectable head movement occurred. On average less than 16 volumes (10%) were removed from each run.
Anatomical and functional maps were transformed into stereotactic Talairach space using the ICBM452 template. Motion correction parameters were included as regressors of no interest and low frequency components of the signal were removed. Individual voxel-wise, event-related activation maps were created following standard AFNI procedures using an algorithm that compares the actual hemodynamic response to a canonical hemodynamic response function. Event-related response functions were calculated for the emotional pictures, neutral pictures and circles (targets). Squares provided the baseline against which hemodynamic responses were assessed.
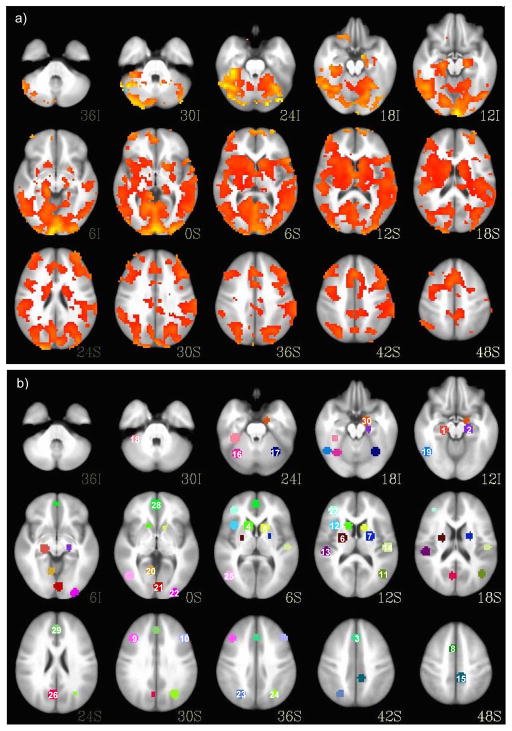
As shown in Figure 1, prior to treatment, fMRI regions-of-interest (ROIs) were established using voxel-wise contrasts between a larger sample of healthy (n = 41) and first-episode manic participants (n = 42) at baseline.9 Specifically, these analyses were completed in AFNI based upon Monte Carlo simulation using 10,000 iterations from which significant activation differences between groups were defined as p < .001 with a cluster of 30 voxels that resulted in a corrected threshold of p < .01.14 From this analysis, in order to define ROIs, spheres of 15 mm diameter were placed, centered on the voxel of maximum activation within identified clusters that differed between groups. A Gaussian blur with a full width at half maximum (FWHM) of 6 mm was applied. Percent signal change in activation served as the variable of interest. We chose this approach to reduce the number of machine learning inputs, reduce data dimensionality, and identify ROIs that may represent more likely targets of lithium treatment.
Figure 1.
Voxel-wise contrast showing regional activation differences between bipolar and healthy groups at baseline to define regions-of-interest (ROIs) for machine learning input (a). Fifteen mm spheres of different colors centered around local cluster maxima defining 30 functionally-derived ROIs including: 1 = L parahippocampal gyrus; 2 = R parahippocampal gyrus; 3 = B medial frontal cortex; 4 = L caudate; 5 = R caudate; 6 = L putamen; 7 = R putamen; 8 = B superior frontal cortex; 9 = L middle frontal cortex; 10 = R middle frontal cortex; 11 = R middle temporal gyrus; 12 = L anterior insula; 13 = L posterior insula; 14 = R insula; 15 = R precuneus; 16 = L cerebellar lobule VI/VIIa; 17 = R cerebellar lobule VI/VIIa; 18 = L cerebellar lobule VI; 19 = L fusiform gyrus; 20 = L lingual gyrus; 21 = R lingual gyrus; 22 = R inferior occipital gyrus; 23 = L parietal cortex; 24 = R parietal cortex; 25 = L inferior temporal gyrus; 26 = L dorsal posterior cingulate; 27 = L ventrolateral prefrontal cortex; 28 = B medial prefrontal cortex; 29 = B anterior cingulate cortex; 30 = R amygdala (b).
Proton Magnetic Resonance Spectroscopy (1H-MRS) Acquisition
A 1H-MRS scan was obtained immediately following the fMRI scan on the same system used for fMRI. A 1H TEM (Transverse ElectroMagnetic) head volume coil was used as a transmitter/receiver. A multi-slice scout image was initially acquired for positioning and 1H-MRS voxel placement. The scout image was followed by acquisition of a 3-D whole head MRI using a Modified Driven Equilibrium Fourier Transform (MDEFT) pulse sequence for tissue segmentation [TR = 13.1 ms, TE = 6 ms, TMD = 1.1 ms, data matrix = 256 × 192 × 192, FOV = 256 × 192 × 150 mm, slab thickness = 150 mm, axial orientation and 32 segments].17 For MRS data acquisition, the magnetic field homogeneity was optimized using a Fast Automatic Shimming Technique by Mapping Along Projections (FASTMAP). A typical line width at half maximum of the water signal was 10–12 Hz. The single-voxel 1H-MRS data were acquired in anterior cingulate cortex (ACC), and left and right ventrolateral prefrontal cortex (VLPFC) using a point resolved spectroscopy (PRESS) pulse sequence [20 × 20 × 20 (mm)3 voxel with TR = 2000 ms, TE = 23 ms, and 128 averages]. Water signal was suppressed by the VAPOR (variable pulse powers and optimizing relaxation delays) method.21 For computations of metabolite levels and eddy current correction, one reference spectrum without water suppression was collected at the same voxel positions using the same parameters except with four averages and reduced receiver gain.
1H-MRS Processing
Standard 1H-MRS spectra were obtained at each of three ROI locations and included NAA, mI, Glu, Glx (Glu + Gln), Cho, and Cr. To calculate each neurometabolite, localized spectra were curve fitted using LCModel (Linear Combination of Model spectra) using the reference of water signal in unsuppressed-water spectra.22 Data were corrected with T1 and T2 relaxation losses using previously published values.23 To clarify the influence of tissue heterogeneity, metabolite levels were corrected using tissue segmentation data. Metabolite levels were computed and presented as concentration (mM) with the water reference of the unsuppressed-water spectra. Differences in water concentrations, T1 and T2 relaxation times in gray matter, white matter and CSF were also taken into consideration for the computation. To determine the tissue contents within voxels, MDEFT images were processed using a contrast-driven algorithm executed with SPM2 (Statistical Parametrical Mapping; http://www.fil.ion.ucl.ac.uk/spm/).
Machine Learning Methodology
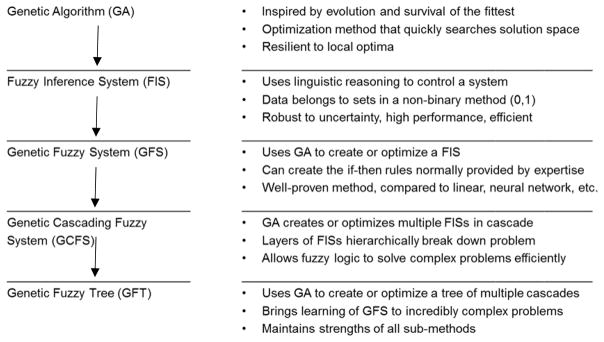
For this study, we developed a new linguistic machine learning system based on a cascading genetic fuzzy tree (GFT) design called the LITHium Intelligent Agent (LITHIA). Table 1 depicts the methodologies employed for LITHIA. As seen in Table 1, the methodology is an evolution of genetic algorithms24 and fuzzy systems.25 Our GFT methodology helps mitigate scalability concerns of standard genetic fuzzy systems by breaking the problem space into many sub-decisions. Variables that are directly coupled are placed in the same part of the controller while “slightly” coupled variables are, at least, placed in the same branch of the tree structure to offset any decrease in accuracy due to unaccounted for coupling between variables. Although any genetic algorithm could be utilized to train a system such as LITHIA in theory, for the reasons stated above, the EVE learning system was utilized presently [Psibernetix Inc., EVE System, http://www.psibernetix.com/services]. EVE is a patent-pending GFT with a fitness function for optimizing or training other similar GFT’s through recursive application.26–28
Table 1.
Methodologies underlying the final Genetic Fuzzy Tree (GFT) structure of LITHIA.
Initially, LITHIA was trained using the baseline YMRS total score, 18 MRS, 90 fMRI inputs and sex as inputs over four training runs. Each training run contained a randomly selected 80% of the total sample (n = 16) with the stipulation that it included at least four non-responders. The model was constrained to four lateral levels or “branches” including one each for MRS, fMRI circle (target), fMRI emotional and fMRI neutral data. It was also constrained to three inputs from a directly higher level, which resulted in up to 125 linguistic “if-then statements” controlling any given Fuzzy Inference System (FIS; this was done to limit the computational cost of any FIS in particular, not necessarily with respect to the cost of evaluating that FIS for a given set of inputs, but rather for the cost associated with training a FIS of such immense size). Each FIS classified states (i.e., activation and metabolite level) into a number of membership functions (i.e., very low, low, medium, high, very high). It then created if-then statements (e.g., if left amygdala response to emotional stimuli is very high, and right VLPFC response is low, then YMRS reduction is very high) and these logic-based rules were used to control the system. Over training, then, these rule bases were learned and the membership functions were tuned and optimized.
Neuroimaging values were provided to EVE, which was utilized to train LITHIA to identify each participant’s binary classification (yes or no), and continuous percent YMRS reduction accuracy (0–100%). For each of n patients the absolute value of % YMRS reduction was summed with either 1 or 0 if correctly classified as response given 50% reduction threshold. As a result, LITHIA was trained to minimize the following function:
| (1) |
Prediction accuracy, including sensitivity and specificity, was calculated.
Despite our data reduction approach for the fMRI ROIs, the number of input or predictor variables still greatly outnumbered the number of observations. While optimally a larger training set would be obtained, fuzzy logic has been demonstrated to be robust in training for complex problems with small sets of data or cases.29,30
Performance Evaluation
An 80/20 validation procedure was employed in each of five experimental runs. Each run included a different randomly selected distribution of 20 subjects with the stipulation that there be at least four non-responders in the training set (80%), and the remaining subjects in the validation set (20%). The first four experimental runs examined the consistency of results across different subject combinations in the training and validation sets. Performance was evaluated in training and validation sets using χ2 and dependent-sample t-tests. Good performance was defined by a greater likelihood of correct response classification relative to chance and similarity between estimated and actual symptom reduction.
The fifth and final randomly selected sample of the 20 subjects used the same constraints to compare LITHIA with eight common classification methods including: linear support vector regression, radial basis function support vector regression, stochastic gradient descent, least angle regression, lasso model, ridge regression, Bayesian ridge regression and decision tree. Performance was evaluated in the validation set using χ2 and dependent-sample t-tests. Good performance was defined by a greater likelihood of correct response classification for LITHIA relative to each of the other comparison methods and dissimilarity between symptom reduction for LITHIA and other methods, with LITHIA showing greater accuracy.
Results
Participant Characteristics and Clinical Response
We identified 20 subjects with bipolar disorder who had received 8 weeks of lithium treatment in addition to baseline fMRI and 1H-MRS scans. As seen in Table 2, within this group 15 (75%) subjects responded to lithium. There was no statistically significant difference between the responders and non-responders in age, sex, mood state, or presence of psychotic symptoms [ps > .05]. Both groups exhibited statistically similar YMRS total scores at baseline, but at week 8 responders had significantly lower scores than non-responders by definition [t(18) = 6.58, p < .001].
Table 2.
Demographic and clinical characteristic of bipolar I disorder subjects experiencing a first manic or mixed episode, divided between those who did and did not respond to lithium at 8 weeks of treatment.
| Characteristic | Bipolar I Disorder Subjects | |
|---|---|---|
|
| ||
| Responders (n=15) | Non-responders (n=5) | |
| Age, years | 17 (3) | 21 (6) |
| Sex, N, (%) male | 7 (47) | 3 (60) |
| Mood state, N (%) manic | 7 (50)a | 3 (60) |
| Psychosis present, N (%) | 3 (21)a | 3 (60) |
| Baseline YMRS, total score | 26 (5) | 22 (3) |
| 8-week YMRS, total scoreb | 6 (4) | 20 (6) |
Note.
One responder did not receive a DSM-IV diagnostic code, so beyond the diagnosis of first-episode bipolar I disorder, mood state and psychosis are not known in this case and n = 14.
Significant difference, responders v. non-responders (by definition), t(18) = 6.58, p < .001.
Values are mean (SD) unless otherwise stated. YMRS = Young Mania Rating Scale.
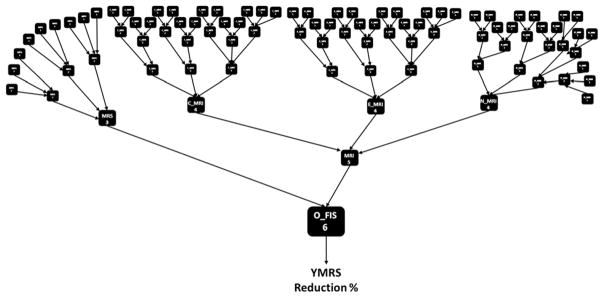
LITHIA Post-Training Model
Figure 2 depicts the post-training GFT of multiple cascades derived from the genetic algorithm described previously. As seen in Figure 2, there is a layered structure of FISs, each with different levels of connectivity and each utilizing many sub-decisions to reach the final control output (i.e., O_FIS). The model resulted in 93 individual FISs (961-digit string length), each exerting control of the system and leading to the final prediction of YMRS Reduction % with lithium treatment.
Figure 2.
The post-training Genetic Fuzzy Tree (GFT) of multiple cascades showing 93 individual Fuzzy Inference Systems (FISs) that help control the final output of Young Mania Rating Scale (YMRS) reduction % and lithium response classification.
LITHIA Training and Validation Results over Four Experimental Runs
Raw classification data for four separate 80% training and 20% validation runs appear in Appendix 1. In training, LITHIA correctly classified patients as responders or non-responders with 100% accuracy on each runs, which was significantly better than chance performance [χ2(1) = 10.67, p = .001]. Additionally, LITHIA estimated week-8 YMRS scores with between 87.9 – 91.2% mean accuracy, which was statistically similar to actual scores [ps > .05], despite having limited knowledge about the adequacy of inputs and limited training time. In validation, LITHIA also classified responders and non-responders 100% accurately [χ2(1) = 2.67, p = .10], with the exception of run four, which achieved 75% classification accuracy [p > .05]. There was only a slight decrease in the accuracy of week 8 symptom prediction in validation (range 80.4 – 90.4%) relative to training (range 87.9 – 91.2%).
LITHIA vs. Eight Deterministic Comparison Methods: Training Results
Over a different distribution of the 20 subjects, LITHIA was compared to eight common classification methods. In the interest of space, raw training data are not presented in tabular form here (data available from the corresponding author upon request), but instead overall classification and mean YMRS reduction accuracy is reported. In this training run, LITHIA again achieved 100% accuracy in classification and 89.8% mean accuracy predicting actual symptom score reductions. LITHIA compared favorably to the other methods, all of which also achieved perfect classification accuracy and > 80% accuracy predicting symptom reductions [linear support vector regression (classification = 100%, symptoms = 92.4%), radial basis function support vector regression (classification = 100%, symptoms = 92.3%), Stochastic gradient descent (classification = 100%, symptoms = 83.2%), least angle regression (classification = 100%, symptoms = 81.7%), lasso model (classification = 100%, symptoms = 81.0%), ridge regression (classification = 100%, symptoms = 97.0%), Bayesian ridge regression (classification = 100%, symptoms = 100%), and decision tree (classification = 100%, symptoms = 100%).
LITHIA vs. Eight Deterministic Comparison Methods: Validation Results
Raw validation data for LITHIA and the eight comparison methods appear in Appendix 2. In validation, LITHIA again achieved 100% classification accuracy and 91.8% mean accuracy predicting exact mania reduction, outperforming all other methods. Comparison methods validated over the same set, varied between 50% classification accuracy [χ2(1) = 2.67, p = .10] and 75% classification accuracy [p > .05], and between 61.0 and 83.9% accuracy predicting exact mania reduction [ps >.05]. None of the comparison methods classified the sole lithium non-responder accurately while LITHIA did. Note that the final three models (ridge regression, Bayesian ridge regression and decision tree, respectively) were likely substantially overfit, as the very high symptom prediction achieved in training (97.02%, 100% and 100%, respectively) was qualified by substantially lower (i.e., ≥ 20%) symptom prediction in validation (77.02%, 74.09% and 60.99%, respectively).
Discussion
This pilot study was conducted to obtain preliminary data and proof-of-concept that a novel machine learning system based on a genetic algorithm and fuzzy logic is capable of accurately classifying first episode manic subjects with bipolar disorder into groups that either responded or did not respond to lithium treatment. Preliminary results show strong predictive capability. Even with only 20 participants, LITHIA was able to apply control to the problem using linguistic rules.
The system demonstrated nearly perfect classification accuracy and was able to predict post-treatment symptom reductions at 8 weeks with at least 88% accuracy in training and 80% accuracy in validation. Moreover, LITHIA exceeded the predictive capacity of each of the eight comparison methods on average. Not only was it the only method to achieve 100% classification accuracy in the validation run, but it was also twice as accurate predicting average symptom reduction scores in validation, and showed very little tendency towards overfitting. Although LITHIA was not statistically superior to the comparator methods in validation, the small validation set (n = 4) employed for this pilot study may necessitate a more qualitative interpretation approach in light of low power. We are still working on visualization functionality, but unlike other classification methods, linguistic rules with weighted importance can be provided by LITHIA, along with the relative importance of inputs. LITHIA can directly explain why a prediction was given in English sentences, with no translation necessary, which will allow individuals who are untrained in intelligent systems to make use of the technology.
To our knowledge, these results are the first employing machine learning to predict treatment response in bipolar disorder. Indeed, there have been few machine learning studies to predict treatment response in the whole of psychiatry, probably due to the cost of incorporating brain indices in longitudinal treatment designs. In one prior study of patients with major depressive disorder, machine learning was employed to predict selective serotonin reuptake inhibitor (SSRI) treatment response using quantitative electroencephalogram (qEEG) to good effect (accuracy = 88%, sensitivity = 95%, specificity = 81%).31 However, in an early study using non-objective, observational data to predict fluoxetine treatment outcomes with an artificial neural network (ANN), the ANN was unable to predict clinical outcome.32 These findings support the notion that (1) newer machine learning applications may better predict outcome than their predecessors and (2) objective biomarkers are likely superior to subjective ones in bioinformatics applications.
In contrast to the dearth of treatment response studies, numerous diagnostic classification studies have employed Gaussian process classifier (GPC), support vector machine (SVM), and least absolute shrinkage and selection operator (LASSO) methods to predict diagnostic membership. However, research to classify different combinations of bipolar disorder, major depressive disorder, and schizophrenia patients based on objective neuroimaging parameters has met with mixed success. Obtained accuracy, sensitivity, and specificity indices have all ranged between 50–100%,33–45 suggesting that these methods are still limited. Future research building on newer methodologies incorporating genetic algorithms and GFTs, such as LITHIA, may help to overcome some of the shortcomings of prior generation classification methods.
As with all research, this study is limited. First, these data have not been validated using either a leave-one-out (LOO) cross-validation method or on an independent cohort of lithium-treated bipolar subjects, which is the gold-standard validation method. As a pilot study, the training cost for a LOO cross-validation is much higher than the base statistical methods used (seconds vs. hours) and the cost to identify a second, independent validation sample would have been prohibitive, but may be possible in the future. The likelihood of successful validation in a future sample would likely rely on its clinical composition being similar to the current sample, particularly with respect to young age. Heterogeneity in brain structure, function and chemistry across early adolescence and into adulthood may have a differential influence on LITHIA’s predictive ability relative to a neurologically mature validation cohort. Second, a larger sample size and additional training time would likely enhance statistical power and predictive ability. Although our sample was restricted to the retrospective data set we had on hand, we are currently conducting additional training in hopes of enhancing prediction. Third, it is possible that the training set was “overfit.” Although overfitting can be a problem in any machine learning application, the relatively good validation results suggest that overfitting, if present, was less of a problem for LITHIA than for the comparison models. Fourth, there is an apparent lack of clinical utility for the present data considering that fMRI and MRS measures, although objective in nature, are not routinely obtained in clinical settings. Moreover, lithium is a long-term prophylactic for which efficacy is determined over longer than eight weeks by means of both manic and depressive symptom reductions. Researchers should focus on incorporating objective measures derived from standard clinical imaging tests (e.g., structural MRI) and consider the long-term recurrence of depressive symptoms/episodes, in addition to mania, to enhance the clinical utility of prediction in future machine learning investigations. Fifth, without a control group, it is difficult to ascertain whether mania symptom reductions resulted from lithium treatment, natural episode resolution within the 8-week period, or other time-locked factors. Finally, although not specific to the present study, a lack of standardization of operating conditions (e.g., magnet strengths, number of head coil channels, software) can result in data that may not be comparable across sites.8
Despite these limitations, to our knowledge, this is the first study to apply linguistically based machine learning to the prediction of lithium response in bipolar disorder. As single neuroimaging parameters have largely failed the biomarker test in psychiatry, perhaps combining different measures in similar machine learning paradigms will yield better response prediction and diagnostic accuracy. Machine learning approaches are rapidly becoming capable of using high-dimensional data (e.g., thousands of voxels comprising brain scans) to identify combinations of inputs that optimally discriminate between groups. Current treatment and diagnostic prediction methods yield sensitivities and specificities ranging from 70–90%, which still leaves 3/10 patients with a mood disorder incorrectly diagnosed as healthy and ~1/10 healthy individuals incorrectly diagnosed as mood disordered.46 New and novel machine learning systems such as LITHIA offer promise for even better prediction in the near future.
Acknowledgments
This study was funded by an NIMH CIDAR award P50 MH077138 (Strakowski).
Appendix 1. Preliminary performance by LITHIA on four random training/validation splits (80/20%) with 20 total subjects
Run 1: Training
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 15.03 | 19.05% | 28.41% | 0 | 0 | Yes | 90.66% |
| 12 | 27 | 25 | 22.68 | 7.41% | 16.00% | 0 | 0 | Yes | 91.40% |
| 23 | 23 | 1 | 4.11 | 95.65% | 82.12% | 1 | 1 | Yes | 86.46% |
| 183 | 21 | 8 | 7.97 | 61.90% | 62.07% | 1 | 1 | Yes | 99.84% |
| 264 | 20 | 4 | 3.61 | 80.00% | 81.94% | 1 | 1 | Yes | 98.06% |
| 267 | 24 | 5 | 4.53 | 79.17% | 81.12% | 1 | 1 | Yes | 98.04% |
| 282 | 21 | 26 | 19.32 | −23.81% | 8.01% | 0 | 0 | Yes | 68.18% |
| 286 | 21 | 12 | 14.69 | 42.86% | 30.06% | 0 | 0 | Yes | 87.20% |
| 306 | 26 | 6 | 9.99 | 76.92% | 61.58% | 1 | 1 | Yes | 84.65% |
| 320 | 21 | 4 | 3.76 | 80.95% | 82.12% | 1 | 1 | Yes | 98.84% |
| 321 | 30 | 1 | 6.28 | 96.67% | 79.07% | 1 | 1 | Yes | 82.40% |
| 331 | 32 | 12 | 12.44 | 62.50% | 61.13% | 1 | 1 | Yes | 98.63% |
| 338 | 23 | 8 | 8.84 | 65.22% | 61.57% | 1 | 1 | Yes | 96.35% |
| 388 | 34 | 5 | 6.08 | 85.29% | 82.12% | 1 | 1 | Yes | 96.82% |
| 397 | 29 | 13 | 9.07 | 55.17% | 68.73% | 1 | 1 | Yes | 86.44% |
| 425 | 34 | 8 | 6.29 | 76.47% | 81.51% | 1 | 1 | Yes | 94.96% |
| Average: | 100% | 91.18% | |||||||
Run 1: Validation
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 189 | 20 | 4 | 6.28 | 80.00% | 68.61% | 1 | 1 | Yes | 88.61% |
| 197 | 20 | 21 | 15.80 | −5.00% | 21.00% | 0 | 0 | Yes | 74.00% |
| 248 | 21 | 5 | 5.27 | 76.19% | 74.89% | 1 | 1 | Yes | 98.70% |
| 398 | 31 | 9 | 11.91 | 70.97% | 61.57% | 1 | 1 | Yes | 90.60% |
| Average: | 100% | 87.98% | |||||||
Run 2: Training
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 17.22 | 19.05% | 18.01% | 0 | 0 | Yes | 98.96% |
| 12 | 27 | 25 | 22.14 | 7.41% | 18.01% | 0 | 0 | Yes | 89.40% |
| 23 | 23 | 1 | 7.19 | 95.65% | 68.73% | 1 | 1 | Yes | 73.08% |
| 189 | 20 | 4 | 6.27 | 80.00% | 68.65% | 1 | 1 | Yes | 88.65% |
| 197 | 20 | 21 | 16.40 | −5.00% | 18.01% | 0 | 0 | Yes | 76.99% |
| 264 | 20 | 4 | 3.77 | 80.00% | 81.13% | 1 | 1 | Yes | 98.87% |
| 282 | 21 | 26 | 17.22 | −23.81% | 18.01% | 0 | 0 | Yes | 58.18% |
| 286 | 21 | 12 | 13.75 | 42.86% | 34.52% | 0 | 0 | Yes | 91.67% |
| 306 | 26 | 6 | 8.18 | 76.92% | 68.53% | 1 | 1 | Yes | 91.61% |
| 320 | 21 | 4 | 5.32 | 80.95% | 74.68% | 1 | 1 | Yes | 93.73% |
| 321 | 30 | 1 | 7.66 | 96.67% | 74.48% | 1 | 1 | Yes | 77.81% |
| 331 | 32 | 12 | 8.03 | 62.50% | 74.90% | 1 | 1 | Yes | 87.60% |
| 338 | 23 | 8 | 7.19 | 65.22% | 68.73% | 1 | 1 | Yes | 96.49% |
| 388 | 34 | 5 | 6.23 | 85.29% | 81.67% | 1 | 1 | Yes | 96.37% |
| 398 | 31 | 9 | 10.95 | 70.97% | 64.68% | 1 | 1 | Yes | 93.71% |
| 425 | 34 | 8 | 8.65 | 76.47% | 74.56% | 1 | 1 | Yes | 98.09% |
| Average: | 100% | 88.20% | |||||||
Run 2: Validation
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 183 | 21 | 8 | 6.30 | 61.90% | 69.98% | 1 | 1 | Yes | 91.93% |
| 248 | 21 | 5 | 6.57 | 76.19% | 68.73% | 1 | 1 | Yes | 92.54% |
| 267 | 24 | 5 | 6.02 | 79.17% | 74.91% | 1 | 1 | Yes | 95.74% |
| 397 | 29 | 13 | 7.61 | 55.17% | 73.76% | 1 | 1 | Yes | 81.41% |
| Average: | 100% | 90.40% | |||||||
Run 3: Training
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 17.22 | 19.05% | 18.01% | 0 | 0 | Yes | 98.96% |
| 12 | 27 | 25 | 22.14 | 7.41% | 18.01% | 0 | 0 | Yes | 89.40% |
| 23 | 23 | 1 | 4.27 | 95.65% | 81.44% | 1 | 1 | Yes | 85.79% |
| 183 | 21 | 8 | 6.65 | 61.90% | 68.33% | 1 | 1 | Yes | 93.57% |
| 197 | 20 | 21 | 16.40 | −5.00% | 18.01% | 0 | 0 | Yes | 76.99% |
| 264 | 20 | 4 | 3.67 | 80.00% | 81.66% | 1 | 1 | Yes | 98.34% |
| 267 | 24 | 5 | 7.51 | 79.17% | 68.73% | 1 | 1 | Yes | 89.56% |
| 282 | 21 | 26 | 17.22 | −23.81% | 18.01% | 0 | 0 | Yes | 58.18% |
| 306 | 26 | 6 | 4.65 | 76.92% | 82.12% | 1 | 1 | Yes | 94.81% |
| 320 | 21 | 4 | 5.17 | 80.95% | 75.36% | 1 | 1 | Yes | 94.41% |
| 321 | 30 | 1 | 5.51 | 96.67% | 81.63% | 1 | 1 | Yes | 84.96% |
| 331 | 32 | 12 | 10.01 | 62.50% | 68.73% | 1 | 1 | Yes | 93.77% |
| 338 | 23 | 8 | 7.36 | 65.22% | 67.99% | 1 | 1 | Yes | 97.23% |
| 388 | 34 | 5 | 6.08 | 85.29% | 82.12% | 1 | 1 | Yes | 96.82% |
| 397 | 29 | 13 | 9.07 | 55.17% | 68.73% | 1 | 1 | Yes | 86.44% |
| 425 | 34 | 8 | 8.54 | 76.47% | 74.89% | 1 | 1 | Yes | 98.42% |
| Average: | 100% | 89.85% | |||||||
Run 3: Validation
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 189 | 20 | 4 | 3.67 | 80.00% | 81.64% | 1 | 1 | Yes | 98.36% |
| 248 | 21 | 5 | 6.64 | 76.19% | 68.39% | 1 | 1 | Yes | 92.20% |
| 286 | 21 | 12 | 16.36 | 42.86% | 22.11% | 0 | 0 | Yes | 79.25% |
| 398 | 31 | 9 | 5.54 | 70.97% | 82.12% | 1 | 1 | Yes | 88.85% |
| Average: | 100% | 89.67% | |||||||
Run 4: Training
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 12 | 27 | 25 | 22.14 | 7.41% | 18.01% | 0 | 0 | Yes | 89.40% |
| 23 | 23 | 1 | 5.91 | 95.65% | 74.31% | 1 | 1 | Yes | 78.65% |
| 183 | 21 | 8 | 5.20 | 61.90% | 75.25% | 1 | 1 | Yes | 86.66% |
| 189 | 20 | 4 | 5.03 | 80.00% | 74.83% | 1 | 1 | Yes | 94.83% |
| 197 | 20 | 21 | 16.40 | −5.00% | 18.01% | 0 | 0 | Yes | 76.99% |
| 248 | 21 | 5 | 5.38 | 76.19% | 74.39% | 1 | 1 | Yes | 98.20% |
| 264 | 20 | 4 | 3.73 | 80.00% | 81.33% | 1 | 1 | Yes | 98.67% |
| 267 | 24 | 5 | 6.04 | 79.17% | 74.83% | 1 | 1 | Yes | 95.66% |
| 282 | 21 | 26 | 17.22 | −23.81% | 18.01% | 0 | 0 | Yes | 58.18% |
| 286 | 21 | 12 | 17.22 | 42.86% | 18.01% | 0 | 0 | Yes | 75.15% |
| 306 | 26 | 6 | 4.65 | 76.92% | 82.12% | 1 | 1 | Yes | 94.81% |
| 320 | 21 | 4 | 3.76 | 80.95% | 82.09% | 1 | 1 | Yes | 98.86% |
| 338 | 23 | 8 | 6.91 | 65.22% | 69.98% | 1 | 1 | Yes | 95.24% |
| 388 | 34 | 5 | 8.53 | 85.29% | 74.92% | 1 | 1 | Yes | 89.63% |
| 397 | 29 | 13 | 7.14 | 55.17% | 75.38% | 1 | 1 | Yes | 79.79% |
| 398 | 31 | 9 | 7.78 | 70.97% | 74.90% | 1 | 1 | Yes | 96.07% |
| Average: | 100% | 87.92% | |||||||
Run 4: Validation
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 17.22 | 19.05% | 18.01% | 0 | 0 | Yes | 98.96% |
| 321 | 30 | 1 | 20.23 | 96.67% | 32.57% | 1 | 0 | No | 35.90% |
| 331 | 32 | 12 | 8.13 | 62.50% | 74.60% | 1 | 1 | Yes | 87.90% |
| 425 | 34 | 8 | 8.37 | 76.47% | 75.39% | 1 | 1 | Yes | 98.92% |
| Average: | 75% | 80.42% | |||||||
Appendix 2. Validation performance of LITHIA relative to eight deterministic comparison methods
LITHIA
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 17.82 | 19.05% | 15.15% | 0 | 0 | Yes | 96.10% |
| 189 | 20 | 4 | 6.27 | 80.00% | 68.67% | 1 | 1 | Yes | 88.67% |
| 248 | 21 | 5 | 6.99 | 76.19% | 66.72% | 1 | 1 | Yes | 90.53% |
| 320 | 21 | 4 | 5.73 | 80.95% | 72.73% | 1 | 1 | Yes | 91.78% |
| Average: | 100% | 91.77% | |||||||
Linear Support Vector Regression
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 2.19 | 19.05% | 89.55% | 0 | 1 | No | 29.50% |
| 189 | 20 | 4 | 2.86 | 80.00% | 85.70% | 1 | 1 | Yes | 94.30% |
| 248 | 21 | 5 | 5.90 | 76.19% | 71.92% | 1 | 1 | Yes | 95.73% |
| 320 | 21 | 4 | 4.74 | 80.95% | 77.43% | 1 | 1 | Yes | 96.48% |
| Average: | 75% | 79.00% | |||||||
Radial Basis Function Support Vector Regression
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 5.33 | 19.05% | 74.64% | 0 | 1 | No | 44.41% |
| 189 | 20 | 4 | 3.53 | 80.00% | 82.35% | 1 | 1 | Yes | 97.65% |
| 248 | 21 | 5 | 5.34 | 76.19% | 74.59% | 1 | 1 | Yes | 98.40% |
| 320 | 21 | 4 | 5.06 | 80.95% | 75.89% | 1 | 1 | Yes | 94.94% |
| Average: | 75% | 83.85% | |||||||
Stochastic Gradient Descent
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 3.08 | 19.05% | 85.34% | 0 | 1 | No | 33.71% |
| 189 | 20 | 4 | 2.37 | 80.00% | 88.17% | 1 | 1 | Yes | 91.83% |
| 248 | 21 | 5 | 11.41 | 76.19% | 45.67% | 1 | 0 | No | 69.48% |
| 320 | 21 | 4 | 1.99 | 80.95% | 90.51% | 1 | 1 | Yes | 90.44% |
| Average: | 50% | 71.36% | |||||||
Least Angle Regression
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 5.31 | 19.05% | 74.73% | 0 | 1 | No | 44.32% |
| 189 | 20 | 4 | 4.55 | 80.00% | 77.27% | 1 | 1 | Yes | 97.27% |
| 248 | 21 | 5 | 6.14 | 76.19% | 70.78% | 1 | 1 | Yes | 94.59% |
| 320 | 21 | 4 | 5.64 | 80.95% | 73.13% | 1 | 1 | Yes | 92.18% |
| Average: | 75% | 82.09% | |||||||
Lasso Model
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 5.46 | 19.05% | 74.02% | 0 | 1 | No | 45.03% |
| 189 | 20 | 4 | 5.20 | 80.00% | 74.02% | 1 | 1 | Yes | 94.02% |
| 248 | 21 | 5 | 5.46 | 76.19% | 74.02% | 1 | 1 | Yes | 97.83% |
| 320 | 21 | 4 | 5.46 | 80.95% | 74.02% | 1 | 1 | Yes | 93.07% |
| Average: | 75% | 82.49% | |||||||
Ridge Regression
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 1.11 | 19.05% | 94.73% | 0 | 1 | No | 24.32% |
| 189 | 20 | 4 | 2.94 | 80.00% | 85.30% | 1 | 1 | Yes | 94.70% |
| 248 | 21 | 5 | 4.13 | 76.19% | 80.35% | 1 | 1 | Yes | 95.84% |
| 320 | 21 | 4 | 5.43 | 80.95% | 74.16% | 1 | 1 | Yes | 93.21% |
| Average: | 75% | 77.02% | |||||||
Bayesian Ridge Regression
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 0.68 | 19.05% | 96.78% | 0 | 1 | No | 22.27% |
| 189 | 20 | 4 | 2.99 | 80.00% | 85.07% | 1 | 1 | Yes | 94.93% |
| 248 | 21 | 5 | 2.79 | 76.19% | 86.71% | 1 | 1 | Yes | 89.48% |
| 320 | 21 | 4 | 6.17 | 80.95% | 70.63% | 1 | 1 | Yes | 89.68% |
| Average: | 75% | 74.09% | |||||||
Decision Tree
| Subject | Baseline YMRS |
Week 8 YMRS Actual |
Week 8 YMRS Estimated |
% Reduction Actual |
% Reduction Estimated |
Remission Actual |
Remission Estimated |
Correctly Classified? |
% Reduction Accuracy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 17 | 0.70 | 19.05% | 96.67% | 0 | 1 | No | 22.38% |
| 189 | 20 | 4 | 2.94 | 80.00% | 85.29% | 1 | 1 | Yes | 94.71% |
| 248 | 21 | 5 | 19.44 | 76.19% | 7.41% | 1 | 0 | No | 31.22% |
| 320 | 21 | 4 | 3.09 | 80.95% | 85.30% | 1 | 1 | Yes | 95.65% |
| Average: | 50% | 60.99% | |||||||
Footnotes
Conflicts of Interest Statement
Dr. Strakowski serves as DSMB chair for Sunovion and a consultant to Roche and Procter and Gamble. Dr. Adler has received research support through the University of Cincinnati from Johnson & Johnson, Merck, Forest, Otsuka, Purdue, Takeda, Pfizer, Shire, Sunovion and SyneuRx. He is a consultant to Sunovion. Dr. DelBello receives research support from Otsuka, Lundbeck, Shire, Sunovion and Pfizer. She is a consultant to Pfizer, Lundbeck, Sunovion, Otsuka, Supernus, Janssen and Neuronetics. Dr. Ernest is CEO of Psibernetix, Inc. We do not believe any of these relationships influence the reported results, but we report them for transparency. The remaining authors declare no conflicts.
References
- 1.Angst J, Gamma A, Benazzi F, Ajdacic V, Eich D, Rossler W. Toward a redefinition of subthreshold bipolarity: epidemiology and proposed criteria for bipolar-II, minor bipolar disorders and hypomania. J Affect Disord. 2003;73:133–146. doi: 10.1016/s0165-0327(02)00322-1. [DOI] [PubMed] [Google Scholar]
- 2.Judd LL, Akiskal HS. The prevalence and disability of bipolar spectrum disorders in the US population: re-analysis of the ECA database taking into account subthreshold cases. J Affect Disord. 2003;73:123–131. doi: 10.1016/s0165-0327(02)00332-4. [DOI] [PubMed] [Google Scholar]
- 3.Goodwin FK, Jamison KR. Manic-Depressive Illness: Bipolar Disorders and Recurrent Depression. 2. New York, NY: Oxford University Press; 2007. [Google Scholar]
- 4.Swann AC, Bowden CL, Calabrese JR, Dilsaver SC, Morris DD. Differential effect of number of previous episodes of affective disorder on response to lithium or divalproex in acute mania. Am J Psychiatry. 1999;156:1264–1266. doi: 10.1176/ajp.156.8.1264. [DOI] [PubMed] [Google Scholar]
- 5.Strakowski SM, Fleck DE, Maj M. Broadening the diagnosis of bipolar disorder: Benefits vs. risks. World Psychiatry. 2011;2011:181–186. doi: 10.1002/j.2051-5545.2011.tb00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linden DE. The challenges and promise of neuroimaging in psychiatry. Neuron. 2012;73:8–22. doi: 10.1016/j.neuron.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 7.Fleck DE, Shear PK, Strakowski SM. Manic distractibility and processing efficiency in bipolar disorder. In: Wood S, Allen N, Pantelis C, editors. Handbook of Neuropsychology and Mental Illness. Cambridge: Cambridge University Press; 2009. pp. 365–377. [Google Scholar]
- 8.Vidyasagar M. Identifying predictive features in drug response using machine learning: opportunities and challenges. Annu Rev Pharmacol Toxicol. 2015;55:15–34. doi: 10.1146/annurev-pharmtox-010814-124502. [DOI] [PubMed] [Google Scholar]
- 9.Strakowski SM, Fleck DE, Eliassen JC, Weber W, Durling M, Komoroski RA, Chu W-J, Norris M, Dudley JA, Welge JA, Blom T, Klein C, Strawn JR, DelBello MD, Lee J-H, Adler CM. fMRI brain activation changes following treatment of a first manic episode. Bipolar Disord. 2016 doi: 10.1111/bdi.12426. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young RC, Biggs JT, Ziegler VE, Meyer DA. A rating scale for mania: Reliability, validity and sensitivity. Br J Psychiatry. 1978;133:429–435. doi: 10.1192/bjp.133.5.429. [DOI] [PubMed] [Google Scholar]
- 11.First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV Axis I Disorders- Patient Edition (SCID-I/P) Biometrics Research Department. New York State Psychiatric institute; 722 West 168th Street, New York, NY 10032: 1995. [Google Scholar]
- 12.Kaufman J, Birmaher B, Brent D, Rao U, Flynn C, Morecit P, et al. Schedule for Affective Disorders and Schizophrenia for School-Age Children-Present and Lifetime Version (K-SADS-PL): initial reliability and validity data. J Am Acad Child Adolesc Psychiatry. 1997;36:980–988. doi: 10.1097/00004583-199707000-00021. [DOI] [PubMed] [Google Scholar]
- 13.McClellan AT, Kushner H, Metzger D, Peters R, Smith I, Grissom G, et al. The fifth edition of the Addiction Severity Index. J Sub Ab Treatment. 1992;9:199–213. doi: 10.1016/0740-5472(92)90062-s. [DOI] [PubMed] [Google Scholar]
- 14.Strakowski SM, Eliassen JC, Lamy M, Cerullo MA, Allendorfer JB, Madore M, et al. Functional magnetic resonance imaging brain activation in bipolar mania: evidence for disruption of the ventrolateral prefrontal-amygdala emotional pathway. Biol Psychiatry. 2011;69:381–388. doi: 10.1016/j.biopsych.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamasaki H, LaBar KS, McCarthy G. Dissociable prefrontal brain systems for attention and emotion. Proc Natl Acad Sci USA. 2002;99:11447–11451. doi: 10.1073/pnas.182176499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerullo MA, Fleck DE, Eliassen JC, Smith MS, DelBello MP, Adler CM, Strakowski SM. A longitudinal functional connectivity analysis of the amygdala in bipolar I disorder across mood states. Bipolar Disord. 2012;14:175–184. doi: 10.1111/j.1399-5618.2012.01002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Garwood M, Menon R, Adriany G, Andersen P, Truwit CL, Ugurbil K. High contrast and fast three-dimensional magnetic resonance imaging at high fields. Magn Reson Med. 1995;34:308–312. doi: 10.1002/mrm.1910340305. [DOI] [PubMed] [Google Scholar]
- 18.Schmithorst VJ, Dardzinski BJ, Holland SK. Simultaneous correction of ghost and geometric distortion artifacts in EPI using a multiecho reference scan. IEEE Trans Med Imaging. 2001;20:535–539. doi: 10.1109/42.929619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox RW. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 20.Cox RW, Jesmanowicz A. Real-time 3D image registration for functional MRI. Magn Reson Med. 1999;42:1014–1018. doi: 10.1002/(sici)1522-2594(199912)42:6<1014::aid-mrm4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 21.Tkac I, Staruck Z, Choi IY, et al. In vivo 1H NMR spectroscopy of rat brain at 1ms echo time. Magn Reson Med. 1999;41:649–656. doi: 10.1002/(sici)1522-2594(199904)41:4<649::aid-mrm2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 22.Provencher S. Estimation of metabolite concentration from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 23.Hetherington HP, Mason GF, Pan JW, et al. Evaluation of cerebral gray and white matter metabolite differences by spectroscopic imaging at 4.1T. Magn Reson Med. 1994;32:565–571. doi: 10.1002/mrm.1910320504. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg D. Genetic Algorithms in Search, Optimization, and Machine Learning. New York: Addison-Wesley; 1989. [Google Scholar]
- 25.Ross T. Fuzzy Logic with Engineering Applications. New York: Wiley & Sons; 1995. [Google Scholar]
- 26.Ernest N, Carroll D, Schumacher C, Clark M, Cohen K, Lee G. Genetic fuzzy based artificial intelligence for unmanned combat aerial vehicle control in simulated air combat missions. Journal of Defense Management. 2016 Epub ahead of print. [Google Scholar]
- 27.Ernest N, Cohen K, Garcia E, Schumacher C, Casbeer D. Multi-agent cooperative decision making using genetic cascading fuzzy systems. AIAA SciTech Conference; Kissimmee, FL. 2015. [Google Scholar]
- 28.Ernest N, Cohen K, Schumacher C, Casbeer D. Learning of intelligent controllers for autonomous unmanned combat aerial vehicles by genetic cascading fuzzy methods. SAE Aerospace Systems and Technology Conference; Cincinnati, OH. 2014. [Google Scholar]
- 29.Cordon O, Herrera F, et al. Genetic fuzzy systems evolutionary tuning and learning of fuzzy knowledge bases. World Scientific; 2001. [Google Scholar]
- 30.Ernest N. Electronic Dissertation. University of Cincinnati; 2015. Genetic fuzzy trees for intelligent control of unmanned combat aerial vehicles. [Google Scholar]
- 31.Khodayari-Rostamabad A, Reilly JP, Hasey GM, de Bruin H, Maccrimmon DJ. A machine learning approach using EEG data to predict response to SSRI treatment for major depressive disorder. Clin Neurophysiol. 2013;124:1975–1985. doi: 10.1016/j.clinph.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 32.Winterer G, Ziller M, Linden M. Classification of observational data with artificial neural networks versus discriminant analysis in pharmacoepidemiological studies: Can outcome of fluoxetine treatment be predicted? Pharmacopsychiatry. 1998;31:225–231. doi: 10.1055/s-2007-979333. [DOI] [PubMed] [Google Scholar]
- 33.Costafreda SG, Fu CH, Picchioni M, Toulopoulou T, McDonald C, Kravariti E, Walshe M, Prata D, Murray RM, McGuire PK. Pattern of neural responses to verbal fluency shows diagnostic specificity for schizophrenia and bipolar disorder. BMC Psychiatry. 2011;11:18. doi: 10.1186/1471-244X-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grotegerd D, Suslow T, Bauer J, Ohrmann P, Arolt V, Stuhrmann A, Heindel W, Kugel H, Dannlowski U. Discriminating unipolar and bipolar depression by means of fMRI and pattern classification: a pilot study. Eur Arch Psychiatry Clin Neurosci. 2013;263:119–131. doi: 10.1007/s00406-012-0329-4. [DOI] [PubMed] [Google Scholar]
- 35.Koutsouleris N, Meisenzahl EM, Borgwardt S, Riecher-Rössler A, Frodl T, Kambeitz J, Köhler Y, Falkai P, Möller HJ, Reiser M, Davatzikos C. Individualized differential diagnosis of schizophrenia and mood disorders using neuroanatomical biomarkers. Brain. 2015;138:2059–2073. doi: 10.1093/brain/awv111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mourão-Miranda J, Almeida JR, Hassel S, de Oliveira L, Versace A, Marquand AF, Sato JR, Brammer M, Phillips ML. Pattern recognition analyses of brain activation elicited by happy and neutral faces in unipolar and bipolar depression. Bipolar Disord. 2012;14:451–460. doi: 10.1111/j.1399-5618.2012.01019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mourão-Miranda J, Oliveira L, Ladouceur CD, Marquand A, Brammer M, Birmaher B, Axelson D, Phillips ML. Pattern recognition and functional neuroimaging help to discriminate healthy adolescents at risk for mood disorders from low risk adolescents. PLoS One. 2012;7:1–9. doi: 10.1371/journal.pone.0029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mwangi B, Wu MJ, Bauer IE, Modi H, Zeni CP, Zunta-Soares GB, Hasan KM, Soares JC. Predictive classification of pediatric bipolar disorder using atlas-based diffusion weighted imaging and support vector machines. Psychiatry Res. 2015;234:265–271. doi: 10.1016/j.pscychresns.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mwangi B, Wu M-J, Cao B, Passos IC, Lavagnini L, Keser Z, Zunta-Soares GB, Hasan KM, Kapczinski F, Soares JC. Individualized prediction and clinical staging of bipolar disorder using neuroanatomical biomarkers. Bio Psych CNNI. 2016;1:186–194. doi: 10.1016/j.bpsc.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redlich R, Almeida JJ, Grotegerd D, Opel N, Kugel H, Heindel W, Arolt V, Phillips ML, Dannlowski U. Brain morphometric biomarkers distinguishing unipolar and bipolar depression. A voxel-based morphometry-pattern classification approach. JAMA Psychiatry. 2014;71:1222–1230. doi: 10.1001/jamapsychiatry.2014.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocha-Rego V, Jogia J, Marquand AF, Mourao-Miranda J, Simmons A, Frangou S. Examination of the predictive value of structural magnetic resonance scans in bipolar disorder: A pattern classification approach. Psychol Med. 2014;44:519–532. doi: 10.1017/S0033291713001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnack HG, Nieuwenhuis M, van Haren NE, Abramovic L, Scheewe TW, Brouwer RM, Hulshoff Pol HE, Kahn RS. Can structural MRI aid in clinical classification? A machine learning study in two independent samples of patients with schizophrenia, bipolar disorder and healthy subjects. Neuroimage. 2014;84:299–306. doi: 10.1016/j.neuroimage.2013.08.053. [DOI] [PubMed] [Google Scholar]
- 43.Serpa MH, Ou Y, Schaufelberger MS, Doshi J, Ferreira LK, Machado-Vieira R, Menezes PR, Scazufca M, Davatzikos C, Busatto GF, Zanetti MV. Neuroanatomical classification in a population-based sample of psychotic major depression and bipolar I disorder with 1 year of diagnostic stability. Biomed Res Int. 2014;2014:1–9. doi: 10.1155/2014/706157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu MJ, Mwangi B, Bauer IE, Passos IC, Sanches M, Zunta-Soares GB, Meyer TD, Hasan KM, Soares JC. Identification and individualized prediction of clinical phenotypes in bipolar disorders using neurocognitive data, neuroimaging scans and machine learning. Neuroimage. 2016 doi: 10.1016/j.neuroimage.2016.02.016. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu MJ, Wu HE, Mwangi B, Sanches M, Selvaraj S, Zunta-Soares GB, Soares JC. Prediction of pediatric unipolar depression using multiple neuromorphometric measurements: A pattern classification approach. J Psychiatr Res. 2015;62:84–91. doi: 10.1016/j.jpsychires.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Savitz JB, Rauch SL, Drevets WC. Clinical application of brain imaging for the diagnosis of mood disorders: the current state of play. Mol Psychiatry. 2013;18:528–539. doi: 10.1038/mp.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]