Abstract
The objective of this investigation was to identify the lineages of MRSA and MSSA with reduced susceptibility to chlorhexidine in Kuwaiti hospitals. 121 clinical MRSA and 56 MSSA isolates were included in this study. Antimicrobial susceptibility testing was performed for a selection of agents including chlorhexidine and resistance genes were amplified and sequenced. PFGE, spa typing, and MLST were completed for a selection of isolates. The results showed SCCmec II, III, IV, and V were present in 0.8, 21.5, 69.4, and 8.3% of the MRSA isolates. agr-1Sa was the most prevalent type in both MSSA (48%) and MRSA (54%). Forty-five percentage of MRSA contained pvl and 39% contained lukE-lukD, however, as many as 86% of MSSA contained pvl and 96.4% contained lukE-lukD. qac A-C genes were identified in 12.3% of MRSA, norA was present in 82.6% and blaZ in 94.2%. Among MSSA only 5.4% harbored qacA, 83% contained norA, and 91% blaZ. Multi-drug resistant ST239/t945 lineage containing a qac gene was the most identified S. aureus. However, other lineages, including ST772-MRSA-V/t4867/pvl(+)qacC/smr and non-qac harboring lineages of ST217-MRSAIV/t3244/pvl(–), ST34-MSSA/t161/pvl(+), ST5-MSSA/t688/pvl(+), ST5-MSSA/t4867/norA(+), and ST672-MSSA/t003/pvl(–), also showed reduced susceptibility to chlorhexidine. The observed reduced susceptibility of non-qac dependent MSSA isolates to chlorhexidine suggests the involvement of other elements in promoting higher MBC (≥30 mg/L). Our results confirm that monitoring MSSA is essential as they may have the potential to survive low level biocide exposure.
Keywords: MRSA, MSSA, PFGE, MLST, spa typing, chlorhexidine
Introduction
Staphylococcus aureus (S. aureus) is a collection of multi-lineage Gram-positive cocci that is commensal to humans and is commonly found in the upper respiratory tract of 20~30% of general population. Yet S. aureus is a leading cause of bacteraemia (Stefani et al., 2012) in hospitals, and is the most common cause of necrotizing pneumonia, skin, and soft-tissue infections in community (Klevens et al., 2007; DeLeo and Chambers, 2009).
S. aureus contains mobile genetic elements (MGE), comprising of bacteriophages, pathogenicity islands, and transposons that carry genes encoding for antimicrobial resistance and virulence factors. Horizontal gene transfer of these mobile elements may result in strains that are increasingly pathogenic, epidemic, and highly resistant to antibiotics (Tong et al., 2015). It has been suggested that methicillin resistant S. aureus (MRSA) has evolved from methicillin-susceptible S. aureus (MSSA) via acquisition of Staphylococcal cassette chromosome mec (SCCmec types I–XI) (Baranovich et al., 2010; Petersdorf et al., 2015). Usually lineages that are important in hospital-acquired infections harbor SCCmec III element containing a large number of resistance genes (Ito et al., 1999). In contrast, SCCmec IV and V are frequently detected in isolates causing community acquired infections, although recently they have also been associated with hospital-acquired infections (Ito et al., 2001; Pereira et al., 2014).
According to the National Committee for Clinical Laboratory Standards (CLSI), the criteria for identifying MRSA is defined as S. aureus harboring mecA or mecC genes or phenotypically showing minimum inhibitory concentration (MIC) of oxacillin or cefoxitin ≥4 mg/L. The presence of mecA gene confers resistance to β-lactam antibiotics but not to penicillins.
MRSA and MSSA may contain blaZ gene expressing a β-lactamase enzyme that confers resistance to penicillins only. It is thought that the advantage of the presence of the β-lactam resistance mechanism mediated by mecA gene would keep the bla system active (Milheiriço et al., 2011). Other resistance genes that may be present in S. aureus include the multidrug efflux transporter norA that confers resistance to a broad spectrum of compounds, comprising of fluoroquinolones, quaternary ammonium compounds, efflux inhibitors such as reserpine, verapamil, and some dyes [ethidium bromide (EtBr), rhodamine, and acridines] (Roy et al., 2013). Also, proteins of both MFS (major facilitator superfamily) and SMR (the small multidrug resistant) family encode efflux-mediated resistance to a range of structurally unrelated cationic and lipophilic substrates across the cell membrane. In general, MFS encodes genes such as qacA, conferring resistance to a range of chemicals including ethidium bromide and chlorhexidine, while qacB confers resistance primarily to monovalent organic cations and some divalent compounds. Finally, smr genes consisting of qacC and qacD (Mayer et al., 2001) confer resistance to quaternary ammonium compounds (QACs) (Shamsudin et al., 2012) but not to chlorhexidine.
It is essential to use biocides and administer antimicrobial agents to reduce the bacterial load and decrease the probability of infections in hospitals. Chlorhexidine has been one of the most frequently used biocides in hospitals, at concentrations ranging from 0.5 to 4%, due to its broad spectrum of activity, tolerability, and safety record. It is commonly used as a skin antiseptic prior to clinical procedures, in dressings, hand disinfections and when bathing patients. The mechanism of action of chlorhexidine is known to be by binding to the negatively charged bacterial cell wall affecting the osmotic equilibrium of the cell.
While MIC and minimum bactericidal concentration (MBC) are commonly used to detect reduced susceptibility to chlorhexidine, there is, nevertheless, neither a defined standardized method nor consensus on the meaning of resistance to this agent (Horner et al., 2012). Recently, Morrissey et al. (2014) attempted to define breakpoints for chlorhexidine on the basis of normal distribution of MICs for a given bacterial species, known as the epidemiological cut-off value (ECOFF). ECOFF is described as the upper limit of the normal MIC distribution for chlorhexidine for a specific species and not the likelihood of treatment failure and clinical breakpoints as it is applicable for antibiotics. In general, the advised dose of chlorhexidine usage is several times higher than the MBC, yet, if chlorhexidine concentration reaches sub-lethal levels over time (Bloomfield, 2002), those isolates with reduced susceptibility to chlorhexidine will remain viable, survive, and possibly persist.
This study identifies the characteristics of lineages of MRSA and MSSA with reduced susceptibility to chlorhexidine in Kuwaiti hospitals and supports the notion that MSSA should be considered as an important agent of infection among hospitalized patients.
Materials and methods
Bacterial isolates
One hundred and twenty one MRSA and 56 MSSA were randomly obtained from Kuwaiti S. aureus Reference Laboratory in 2013. Tables 1, 2 show the origin of the isolates. Based on the data provided by the MRSA Reference Laboratory all isolates were non-duplicates from single patients. It is noteworthy to mention that our collection contained colonizing as well as infecting isolates. The isolates were confirmed in our laboratory as S. aureus by Gram stain, morphology, catalase, and coagulase tests. PBP2 was detected with a slide latex agglutination test (Oxoid, Besingstoke, UK).
Table 1.
Clinical MRSA and MSSA isolates obtained from hospitals.
| Hospital | No. of MRSA | No. of MSSA |
|---|---|---|
| Adan | 10 | 0 |
| Amiri | 16 | 2 |
| Razi | 6 | 0 |
| Sabah | 30 | 11 |
| Armed Forces | 0 | 7 |
| Chest | 1 | 5 |
| Farwaniya | 24 | 3 |
| Ibn Sina | 0 | 1 |
| Jahra | 1 | 2 |
| Maternity | 13 | 4 |
| Mubaarak | 20 | 17 |
| Unknown | 0 | 4 |
| Total | 121 | 56 |
Table 2.
Sources of isolates in this study.
| Specimen | No. of MRSA | No. of MSSA |
|---|---|---|
| Abscess | 7 | 0 |
| Broncho Alveolar Lavage (BAL) | 0 | 1 |
| Blood | 7 | 8 |
| Ear | 5 | 3 |
| Eye | 4 | 2 |
| Fluid | 5 | 1 |
| Groin | 11 | 3 |
| High Vaginal Swab | 1 | 3 |
| Nasal | 26 | 8 |
| Catheter Tip | 2 | 1 |
| Pus | 14 | 2 |
| Rectal Swab | 1 | 0 |
| Skin | 12 | 4 |
| Sputum | 1 | 2 |
| Suction Tip | 0 | 2 |
| Swab | 6 | 1 |
| Thigh Abscess Pus | 0 | 1 |
| Tissue | 2 | 2 |
| Throat swab | 1 | 1 |
| Tracheal | 2 | 1 |
| Umbilical Swab | 1 | 1 |
| Urine | 2 | 0 |
| Wound | 10 | 5 |
| Unknown | 1 | 4 |
Susceptibility testing
Antibiotic susceptibility testing was performed by disc diffusion method, when applicable, following the Clinical Laboratory Standards Institute (2014) recommendations. The bacterial suspension (the final turbidity of a 0.5 McFarland Standard) was spread over the Mueller-Hinton agar uniformly and the antimicrobial discs were dispensed onto the agar plates using the disk dispenser and incubated overnight at 35°C. The antibiotic agents tested were penicillin (10 μg), cefoxitin (30 μg), ciprofloxacin (5 μg), clindamycin (2 μg) chloramphenicol (30 μg), erythromycin (15 μg), fusidic acid (10 μg), gentamicin (10 μg), kanamycin (30 μg), linezolid (10 μg), mupirocin (200 μg), rifampicin (5 μg), tetracycline (30 μg), teicoplanin (30 μg), tigecycline (15 μg), trimethoprim (5 μg). Inducible clindamycin resistance was detected with double-disk diffusion test (D-test). The diameter of zone of inhibition was measured (mm) and interpreted as recommended by EUCAST (2016) or CLSI guidelines (2014). MIC was determined by agar dilution method following CLSI recommendations for vancomycin (breakpoint >8 mg/L) and mupirocin (low-level resistance 8–64 mg/L and high-level resistance ≥512 mg/L). Bacterial growth above the breakpoint concentrations confirmed resistance to the relevant antibiotic. S. aureus ATCC 25923 was used as the control for disk diffusion and S. aureus ATCC 29213 for MIC assays. Isolates that showed resistance to at least three classes of antibiotic were considered as multi-drug resistant (MDR).
Minimum inhibitory concentrations (MIC) and minimum bactericidal concentration (MBC) of chlorhexidine
Reduced susceptibility to chlorhexidine by MIC and MBC was based on the method described by Morrissey et al. (2014) and Furi et al. (2013). In this study the MIC for chlorhexidine digluconate (CH-; 100 mg/mL in water; C9394; Sigma-Aldrich, St. Louis, MO, USA) was determined using broth microdilution method with doubling concentration of chlorhexidine and starting inocula of 1 × 105 CFU/mL. MBC was determined by subculturing 10 μL from each well without visible bacterial growth on Mueller-Hinton agar plates and incubating them at 37°C for 48 h. The first chlorhexidine dilution plate yielding three colonies or fewer was determined as MBC.
Interpretation of the results was based on the ECOFF, which is the upper limit of the normal MIC distribution for a given antimicrobial agent and a given species. For S. aureus and chlorhexidine MIC ≥ 4 mg/L and/or MBC ≥ 30 mg/L have been proposed.
DNA isolation
Total genomic DNA for PCR and sequencing was extracted using the DNeasy Blood & Tissue Kit (Qiagen Valencia, CA, USA) according to the manufacturer's instructions.
Detection of resistance and virulence genes
PCR was performed with HotStar Taq polymerase (Qiagen) according to the manufacturer's instructions and specific oligonucleotide primers for detection of the following genes: mecA, mecC, SCCmec, agr locus (agr-1Sa to agr-4Sa), lukE-lukD, lukS-PV, and lukF-PV, norA, qacA/B, qacC/qacD, qacG, qacH, blaZ, mupA, vanA, aac6′/aphD (Oligonucleotide primers are listed in Supplement 1). Amplified PCR products were purified with Qiagen purification kit (Qiagen Valencia, CA, USA) according to the manufacturer's instructions and both strands were sequenced by automated AB13100 DNA sequencer (Applied Biosystems, Carlsbad, CA, USA) system. The BLAST program of the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov) was used to search and compare databases for similar nucleic acid sequences.
spa typing
Based on the sequence analysis of polymorphic region X of protein A, a highly effective subtyping method for S. aureus is spa typing (Deurenburg et al., 2007). Amplification and sequencing of the variable region of the protein A gene, the sequence analysis and spa-type assignment were carried out as previously described (Harmsen et al., 2003). The X region of the spa gene was amplified by PCR with primers 1095F (5′-AGACGATCCTTCGGTGAGC-3′) and 1517R (5′-GCTTTTGCAATGTCATTTACTG-3′). DNA sequences were obtained with an ABI sequencer (Applied Biosystems). spa sequence types were determined with the database accessible via http://spa.ridom.de/ using BioNumerics version 7.1 (Applied Maths, Ghent, Belgium). The node distances between the spa types was calculated by minimum spanning tree method using Dice correlation (BioNumerics v.7.1.)
Multi-locus sequence typing (MLST)
Genomic DNA was extracted from overnight cultures and MLST was carried out according to the protocol for S. aureus on the MLST website (http://www.mlst.net). The fragments of seven housekeeping genes (Supplement 1) arcC, aroE, glpF, gmk, pta, tpi, and yqiL were amplified and sequenced. Nucleotide sequences were aligned and trimmed using Bionumerics 7.1 software (Applied Maths, Ghent, Belgium). Allele numbers and sequence types (STs) were assigned by submitting sequences to the MLST website. MLST was performed on the isolates that contained qacA/B/C and on those isolates that showed reduced susceptibility to chlorhexidine.
Pulse-field gel electrophoresis (PFGE)
Clinical isolates were typed by Pulse-Field gel electrophoresis (PFGE) with the CHEF-DR II electrophoresis cell after digestion with SmaI restriction endonuclease enzyme (Bannerman et al., 1995). The running parameters were as follows: initial pulse 5 s, final pulse 40 s, at 6 V/cm for 20 h at 14°C. The gels were stained with ethidium bromide and scanned. Profiles were analyzed by the unweighted pair method with arithmetic average (UPGMA) using BioNumerics v.7.1. The development of the algorithms necessary for the comparison of fingerprinting profiles of isolates was based on the Dice correlation coefficient. The hierarchic Cluster analysis and phylogenetic trees were subsequently analyzed with an optimization of 1.5% and a tolerance of 1.5%. Isolates were considered to belong to the same PFGE clone if their Dice similarity index was ≥85%.
Qualitative real-time PCR amplification of qac genes
Real time qPCR was performed on qacA/B positive isolates. The total RNA was extracted using Qiagen RNeasy Kit (Qiagen Valencia, CA, USA) and cDNA was synthesized (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems) and used as a template for qPCR amplification using SYBR Green method (EvaGreen qPCR Mix Plus, Solis Biodyne) using the primers described previously (Furi et al., 2013). Two TaqMan probes with two different fluorophores at the 5 = end and a minor groove binder (MGB) at the 3 = end (Applied Biosystems, United Kingdom) were used in order to distinguish between qacA and qacB. Qualitative real-time PCRs were performed in a Light Cycler 480 system (Roche Diagnostics, Germany). Melt curve analysis was performed along with the amplification protocol to determine if non-specific products were amplified during the reaction. The Ct (cycle threshold) value and melting curve analysis was used to confirm the presence of one peak and one product. Further agarose gel analysis (3%) was performed to confirm the amplification of a single PCR product.
Statistical analysis
P-values were calculated using the t-test to determine whether differences between variants were significant using the unpaired two-tailed Mann-Whitney U (IBM SPSS version 23).
Results
Characterization of S. aureus isolates
All 121 MRSA isolates were mecA positive and consisted of 1 (0.8%) SCCmec II, 26 (21.5%) SCCmec III, 84 (69.4%) SCCmec IV, and 10 (8.3%) SCCmec V. mecC was not detected. Sixty-five (54%), 18 (15%), and 10 (8%) contained agr-1Sa, agr-2Sa, and agr-3Sa respectively, while 28 (23%) were agr negative. pvl was present in 45% (n = 55) while 39% (n = 47) contained lukE-lukD (Figure 1).
Figure 1.
The percentage of virulence genes detected in MSSA isolates is higher than MRSA.
Among MSSA, agr-1Sa, agr-2Sa, and agr-3Sa were present in 27 (48%), 14 (25%), and 15 (27%) of the isolates respectively. agr-4Sa was not detected. 86% (n = 48) contained pvl and 96.4% (n = 54) contained lukE-lukD (Figure 1).
Spa typing
Among the MRSA isolates, 36 known and two unknown spa types were identified: spa types t044 and t223 had the highest prevalence at 9.1% (n = 11) and 8.3% (n = 10) respectively. Among MSSA there were 37 known and 1 unknown spa types, t945 (9% n = 5 isolates) was the most prevalent. Altogether there was a two-fold variation in spa types among MSSA (38/56) vs. MRSA isolates (38/121) (68 vs. 31%) (p = 0.015). The common spa types shared among MRSA and MSSA were t002, t127, t223, t267, t304, t306, t688, t945, t3244, and t4867.
Figure 2 shows the node distances between spa types based on clustering by minimum spanning tree method using Dice correlation (BioNumerics v.7.1.). Node size is proportional to spa type frequency and line length is proportional to the number of mutational steps between the types.
Figure 2.
Relationship between spa types of MSSA and MRSA isolates in this study. Node size is proportional to spa type frequency and line length is proportional to the number of mutational steps between the types. The node distances between the spa types was calculated by minimum spanning tree method using Dice correlation (BioNumerics v.7.1.). Based on the default setting of 1%, distance intervals are created that are converted into distance units. For example two entries when having a similarity of 99.6% will have a distance of 0 (interval 100–99% = distance 0). Two entries that have a similarity of 98.7% will have a distance of 1 (interval 99–98% = distance 1).
MRSA- pvl (+) isolates belonging to spa types t044 (n = 7), t019 (n = 5), t852 (n = 5) were the most prevalent, followed by t105 (n = 3); t002, t005, t945, t2518, and t3107 (n = 2 each); t024, t032, t086, t127, t548, t657, t690, t902, t1347, t1560, t1752, t3244, t4867, and t7358 (n = 1 each).
pvl (+) MSSA isolates were t945 (n = 4), t267 (n = 3), and t4045 (n = 3) followed by t127, t304, t306, t359, t688, t3841, and t4867 (n = 2 each); and t002, t015, t021, t062, t084, t085, t166, t315, t346, t349, t376, t521, t701, t710, t1109, t1727, t2393, t2526, t2663, t3244, t3643, t7760, t9787 (n = 1 each).
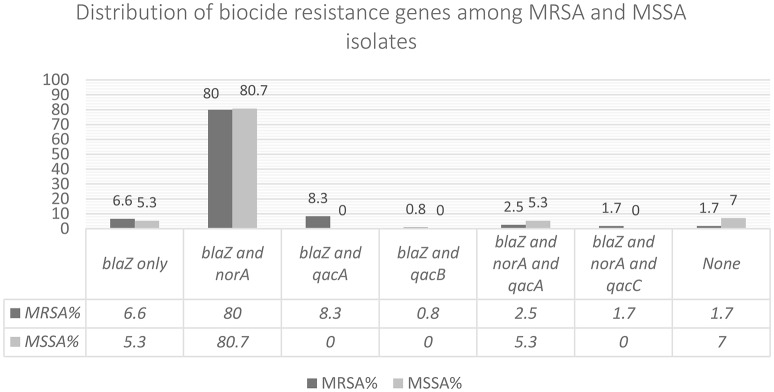
Detection of biocide resistance genes
Figure 3 shows the prevalence of combinations of biocide resistance gene observed in this study. qac genes were identified in 15 MRSA isolates (12.3%) of which 1 contained qacC (smr). Based on real time qPCR (Supplement 2), 13 isolates harbored qacA and 1 contained qacB. norA was present in 82.6% (n = 100), and blaZ in 94.2% (n = 114). Among MSSA, only 3 isolates (5.4%) harbored qacA gene. norA was present in 83% (n = 47), and blaZ in 91% (n = 51). We could not identify any statistically significant differences between the occurrences of either blaZ or norA in MRSA and MSSA isolates, however, MRSA was more likely to contain qacA gene than MSSA (p < 0.05).
Figure 3.
In this figure the combination of biocide resistance genes observed among the isolates has been shown.
Antimicrobial resistance patterns
Table 3 shows the percentage of resistant isolates to the tested antimicrobial agents. For MRSA all isolates were resistant to oxacillin and cefoxitin and susceptible to rifampicin, tigecycline, teicoplanin, and linezolid. There were two vancomycin resistant MRSA isolates with MIC = 16 mg/L, typed as: (R15): ST239-MRSA-III/t860/pvl(–) qacA (+) and (S52): ST772-MRSA-V/t4867/pvl(+)norA(+)qacC(+) (Table 4). PCR experiments using vanA primers did not yield any bands. All MSSA isolates were susceptible to rifampicin, mupirocin, linezolid, tigecycline, teicoplanin, oxacillin, and cefoxitin. All isolates that were resistant to both gentamicin and kanamycin contained aac6′/aphD gene encoding the aminoglycoside-modifying enzymes. 44.6% of MSSA and 62.8% of MRSA were MDR.
Table 3.
Percentage of resistance to each antimicrobial agent tested.
| Antimicrobial agent | No. resistant MRSA (%) | No. resistant MSSA (%) |
|---|---|---|
| Penicillin | 121 (100) | 47 (84) |
| Cefoxitin | 121 (100) | 0 |
| Ciprofloxacin | 40 (33) | 14 (25) |
| Chloramphenicol | 2 (1.7) | 0 |
| Clindamycin (C)a | 35 (29) | 16(29) |
| Clindamycin (I)b | 13 (11) | 0 |
| Erythromycin | 49 (40) | 16 (29) |
| Fusidic Acid | 24 (20) | 5 (9) |
| Gentamicin | 41 (34) | 38 (68) |
| Kanamycin | 52 (43) | 27 (48) |
| Mupirocin (HLR)c | 4 (3.3) | 0 |
| Mupirocin (LLR)d | 11 (9) | 0 |
| Tetracycline | 34 (28) | 16 (29) |
| Trimethoprim | 34 (28) | 7 (7) |
| Vancomycin | 2 (1.7) | 0 |
(C) Constitutive resistance to clindamycin;
(I) Inducible resistance to clindamycin;
(HLR) High level resistance to mupirocin;
(LLR) Low level resistance to mupirocin.
Table 4.
Typing characteristics and antimicrobial profile of qac(+) MRSA and MSSA and qac(–) isolates with reduced susceptibility to chlorhexidine.
| Key | ST | Spa | SCCmec | Specimen | Hospital | qac | norA | blaZ | pvl | LUKED | agr | CHX MBC(mg/L) | Antibiotic resistance |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R-3 | 22 | t4326 | IV | Groin | MUBARAK (ICU) | – | + | + | – | – | 1 | 30 | Pen, Gen, Kan, Ery, Cli I, Tri, Cip |
| R-4 | 22 | t852 | IV | Nasal | MUBARAK (ICU) | – | + | + | + | – | 1 | 30 | Pen, Gen, Kan, Ery, Cli C, Tri, Cip |
| R-12 | 22 | t852 | IV | Eye | MATERNITY | – | + | + | + | – | 1 | 30 | Pen, Gen, Kan, Tri |
| R-13 | 22 | t902 | IV | Nasal | SABAH (PICU) | – | – | + | + | – | 1 | 30 | Pen, Gen, Kan, Tri |
| R-15 | 239 | t860 | III | Groin | MUBARAK (Surgical) | A | – | + | – | + | 1 | 15 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip, Van |
| R-21 | 239 | t945 | III | Nasal | SABAH (ICU) | A | – | + | – | + | 1 | 30 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip,MupLLR |
| R-38 | 239 | t945 | III | Catheter Tip | SABAH (Internal) | A | – | + | – | – | 1 | 7.5 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip,MupLLR |
| R-45 | 239 | t860 | III | Groin | MUBARAK (Surgical) | A | – | + | – | + | 1 | 30 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip, Mup |
| R-53 | 239 | t860 | III | Blood | MATERNITY (NICU) | A | + | + | – | + | 1 | 30 | Pen, Gen, Kan, Ery, CliC, Tri, Fus, Cip,MupHLR |
| R-62 | 5 | t002 | II | Nasal | AMIRI (Internal) | – | + | + | – | + | 2 | 0.94 | Pen, Gen, Kan, Ery, CliC, Cip, MupHLR |
| R-67 | 80 | t044 | IV | Abscess | AMIRI | – | + | + | + | + | 1 | 1.875 | Pen, Kan, Tet, Fus, MupHLR |
| R-84 | 239 | t945 | III | Blood | SABAH (ICU) | A | + | + | + | + | 1 | 60 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip, MupLLR |
| R-87 | 80 | t044 | IV | Tissue | SABAH (PICU) | – | + | + | + | + | 1 | 1.875 | Pen, Gen, Kan, Ery, Cli I, Tri, Cip, MupLLR |
| R-101 | 239 | t945 | III | Skin | SABAH (Internal) | A | + | + | + | + | 1 | 30 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip |
| R-102 | 239 | t860 | III | Nasal | MUBARAK (Surgical) | A | – | + | – | + | 1 | 7.5 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip,MupLLR |
| R-103 | 239 | t860 | III | Pus | MUBARAK (Liver dialysis) | A | – | + | – | + | 1 | 0.94 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip,MupHLR |
| R-104 | 239 | t860 | III | Groin | MUBARAK (Surgical) | A | – | + | – | + | 1 | 0.94 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip,MupLLR, |
| R-105 | 239 | t945 | III | Groin | SABAH (SICU) | A | – | + | – | – | 1 | 0.94 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip,MupLLR |
| R-106 | 239 | t860 | III | Nasal | RAZI (Isolation Unit) | A | – | + | – | + | 1 | 0.94 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip,Mup LLR |
| R-107 | 239 | t945 | III | Tracheal | SABAH (Surgical) | B | – | + | – | + | 1 | 0.94 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip,MupLLR |
| R-110 | 239 | t945 | III | Sputum | RAZI (Surgery) | – | + | + | – | + | 1 | 0.94 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip, MupLLR |
| R-111 | 239 | t945 | III | Fluid | SABAH (Outpatient) | – | + | + | – | + | 1 | 0.94 | Pen, Gen, Kan, Ery, CliC, Tet, Fus, Cip, MupLLR |
| R-113 | 22 | t2518 | IV | Pus | FARWANIYA (Surgical) | A | – | + | + | – | 1 | 0.94 | Pen, Gen, Kan, Cip |
| S-7 | 239 | t945 | MSSA | Skin | SABAH (Internal) | A | + | + | + | + | 1 | 30 | Pen, Gen, Kan, Tob, Cip, Tri |
| S-19 | 1 | t127 | MSSA | High Vaginal Swab | MATERNITY | – | + | + | + | + | 3 | 30 | Pen, Kan |
| S-22 | 88 | t4045 | MSSA | Blood | MUBARAK | – | + | + | + | + | 3 | 60 | Pen, Kan, Tob |
| S-23 | 97 | t359 | MSSA | Blood | MUBARAK | – | + | + | + | + | 1 | 30 | Pen, Kan, Tob |
| S-24 | 5 | t4867 | MSSA | Ear | SABAH | – | + | + | + | + | 2 | 60 | Pen, Gen, Kan, Tob, Tri |
| S-25 | 1 | t127 | MSSA | Nasal | SABAH (Internal) | – | + | + | + | + | 3 | 60 | Pen, Gen, Kan, Tob |
| S-27 | 22 | t223 | MSSA | Eye | MUBARAK (Pediatric) | – | + | + | – | + | 1 | 30 | Pen, Tob, Kan |
| S-32 | 5 | t002 | MSSA | Skin | FARWANIYA (Pediatric) | – | + | + | + | + | 2 | 30 | Cip, Gen, Kan,Tob |
| S-35 | 15 | t084 | MSSA | High Vaginal Swab | MUBARAK | – | + | + | + | + | 2 | 30 | Pen, Gen, Kan, Tob |
| S-41 | 932 | t304 | MSSA | Sputum | SABAH (ICU) | – | + | + | + | + | 3 | 30 | Pen, Gen, Kan, Tob |
| S-42 | 672 | t003 | MSSA | Blood | FARWANIYA | – | – | + | – | + | 1 | >60 | Pen, Kan, Tob, Tri |
| S-43 | 965 | t062 | MSSA | Sputum | – | – | – | – | + | + | 2 | 30 | Pen, Gen, Kan, Tob |
| S-44 | 239 | t945 | MSSA | Groin | – | – | – | + | + | + | 1 | >60 | Pen, Gen, Kan, Tob, Ery, Cip |
| S-46 | 5 | t688 | MSSA | Nasal | ICU | – | – | – | + | + | 2 | 60 | Pen, Gen, Kan, Tob, |
| S-47 | 217 | t3244 | IV | Blood | AMIRI(ICU) | – | + | + | – | – | 1 | 60 | Pen, Gen, Kan, Tob, Ery, Cip, Tri |
| S-48 | 239 | t945 | MSSA | Groin | SABAH (Surgical) | – | – | + | + | + | 1 | 60 | Pen, Gen, Kan, Tob, Ery, Cip |
| S-49 | 97 | t359 | MSSA | High Vaginal Swab | MATERNITY | – | – | – | + | + | 1 | 30 | Kan, Tob, Tri |
| S-50 | 96 | t521 | MSSA | Suction tip | ARMED FORCES | – | – | + | + | + | 3 | 30 | Pen, Gen, Kan, Tob, Cip, Tri, |
| S-51 | 5 | t4867 | MSSA | Tracheal | SABAH (ICU) | – | + | + | + | + | 2 | 60 | Pen, Gen, Kan, Tob, Tri |
| S-52 | 772 | t4867 | V | Nasal Swab | SABAH (Surgical) | C | + | + | + | + | 2 | 30 | Pen, Gen, Kan, Tob, Cip, Tri, Van |
| S-53 | 34 | t166 | MSSA | Umbilical Swab | MUBARAK(NICU) | – | + | + | + | + | 3 | 30 | Pen, Kan, Tob, Tri |
| S-56 | 239 | t945 | MSSA | Groin | SABAH (Surgical) | A | + | + | + | + | 1 | 30 | Pen, Gen, Kan, Tob, Ery, Cip |
| S-57 | 239 | t945 | MSSA | Swab | SABAH (Surgical) | A | + | + | + | + | 1 | 7.5 | Pen, Gen, Kan, Tob, Cip |
Cip, Ciprofloxacin; Chl, Chloramphenicol; CHX MBC, Chlorhexidine Minimum Bactericidal Concentration; CliC, Constitutive resistance to clindamycin; CliI, Inducible resistance to clindamycin; Ery, Erythromycin; Fus, Fusidic Acid; Gen, Gentamicin; ICU, Intensive Care Unit; Kan, Kanamycin; MupHLR, High level resistance to mupirocin; MupLLR, Low level resistance to mupirocin; NICU, Neonatal Intensive Care Unit; Pen, Penicillin; PICU, Pediatric Intensive Care Unit; SICU, Surgical Intensive Care Unit; Tet, Tetracycline; Tri, Trimethoprim; Van, Vancomycin.
Mupirocin resistance in MRSA
We did not find any MSSA isolates resistant to mupirocin. However, 4 MRSA isolates showed high-level and 11 showed low-level resistance (Tables 3, 4). The presence of mupA gene was detected by PCR generating a 1.65 kb intragenic fragment for both resistant phenotypes. The four high-level mupirocin-resistant isolates were typed as (R-45, R-103): ST239-MRSAIII/t860/qacA(+), (R-53): ST239-MRSAIII/t860/qacA(+)/norA(+), and (R-62): ST5-MRSAII/t002/norA(+).
11 MRSA low-level mupirocin-resistant isolates were typed as (R-21, R-38, R-105): ST239-MRSAIII/t945/qacA(+), (R-84): ST239-MRSAIII/t945/norA(+)/qacA(+), (R-87): ST80-MRSAIV/t044/norA(+), (R-102, R-104, R-106): ST239-MRSAIII/t860/qacA(+), (R-107): ST239-MRSAIII/t945/qacB(+), (R-110, R-111) ST239-MRSAIII/t945/norA(+). These isolates were collected from a variety of specimens from different hospitals (Table 4).
qac genes, MLST, and chlorhexidine
For MRSA, MIC range was 0.5–2 mg/L with MIC50 = 1 mg/L and MIC90 = 2 mg/L, MBC range was 1–30 mg/L with MBC50 = 4 mg/L and MBC90 = 30 mg/L. For MSSA, MIC range was 0.5–60 mg/L with MIC50 = 1 mg/L and MIC90 = 2 mg/L. MBC range was from 4 to >60 mg/L with MBC50 = 30 mg/L and MBC90 = 60 mg/L. These results suggest that although more MRSA than MSSA isolates harbored qac genes (n = 15 vs. n = 3), MBC90 was higher for MSSA implying that the presence of qacA might not be significant in reducing susceptibility or prompting higher MBC to chlorhexidine.
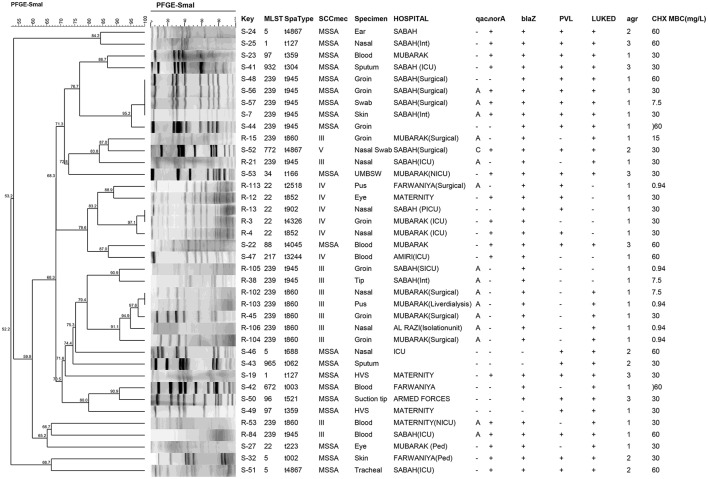
MLST and PFGE for qac(+) and those MRSA and MSSA isolates with MBC ≥ 30 mg/L are shown in Figure 4 and Table 4. Based on the suggested ECOFF, the reduced susceptibility to chlorhexidine did not exclusively depend on the presence of qac genes, however, MBC of chlorhexidine was higher for those qacA(+) isolates obtained from blood. For chlorhexidine, the median of MBC was statistically greater than the median of MIC (p-value < 0.0001). The median of MIC and MBC for qacA(+) isolates were higher than qacA(–) isolates (p-value 0.36 and 0.6, respectively). Results were analyzed using the unpaired two-tailed Mann-Whitney U (IBM SPSS version 23).
Figure 4.
PFGE profiles of qac(+) MRSA and MSSA and qac(–) isolates with reduced susceptibility to chlorhexidine. The hierarchic Cluster analysis and phylogenetic tree after digestion with SmaI is shown. PFGE banding patterns were analyzed using DICE/UPGMA with an optimization of 1.5% and a tolerance of 1.5% generated by BioNumerics software (v.7.1). The percentage of similarities between the clusters are shown.
Moreover, there were qacA(+) isolates that did not display high MBC (MBC ≤ 7.5 mg/L) (Table 4) as well as qacA(–) MSSA isolates with reduced susceptibility to chlorhexidine (including S-53: ST34-MSSA/t161 isolated from an infected umbilical cord of a neonate, S-48: ST239-MSSA/t945, S-46: ST5-MSSA/t688, S-51: ST5-MSSA/t4867, S-42:ST672-MSSA/t003, S-19:ST1-MSSA/t127, S-35:ST15-MSSA/t084, S-22:ST88/t4045, S-50: ST-96/t521, S-49: ST97/t359, S-41: ST932-MSSA/t304 and S-43: ST965-MSSA/t062). Likewise qacA(–) MRSA isolates including ST22-MRSA-IV (with variable spa types), ST239-MRSA-III and ST217-MRSA-IV showed reduced susceptibility to chlorhexidine (Table 4, Figure 4).
The only qacB(+) isolate (R-107) in our collection was susceptible (MBC ≤ 1 mg/L) whereas the only qacC(+) isolate (S-52) showed reduced susceptibility to chlorhexidine (MBC = 30 mg/L).
PFGE analysis
PFGE results for isolates with reduced susceptibility to chlorhexidine and those containing qac genes are shown in Figure 4. Cluster analysis was performed and phylogenic tree was prepared using dice similarities and unweighted matched-pair group which demonstrated diversity among the isolates. It also showed that none of the MSSA and MRSA isolates shared identical PF patterns.
Discussion
This is the first study to identify heterogeneous genotypes of qac and non-qac harboring MRSA and MSSA with reduced susceptibility to chlorhexidine in Kuwait. The MBC90 of chlorhexidine for MSSA isolates was higher than that of MRSA, even though the number of isolates containing qac genes was higher in the MRSA group (n = 15 vs. n = 3). These results suggest that elements other than qac genes may be important for reducing susceptibility to chlorhexidine. One factor to consider would be the expression of the agr quorum-sensing system which is already known to influence virulence factor production but which may also affect antimicrobial resistance and metabolism through its interaction with other staphylococcal gene regulators (Tseng et al., 2015). In our study virulence factors were generally more prevalent in MSSA. Forty-five percentage MRSA and 86% of MSSA contained pvl while 39% of MRSA and 96.4% of MSSA contained lukE-lukD genes. Similar to a previous report (Warren et al., 2016) we also observed qacA(+) MRSA isolates were more likely to exhibit mupirocin resistance.
In this study the predominant resistance gene was blaZ which was present in 94.2% of MRSA and 91% of MSSA usually in combination with other biocide genes. Thirteen MRSA and only 3 MSSA isolates (5.4%) harbored qacA and 16 out of 18 (89%) qac(+) isolates were typed as ST239. ST239-MRSA-SCCmecIII, also known as the hospital-acquired Brazilian/Hungarian clone, is one of the most successful lineages in the world owing its high rate of prevalence to an enhanced ability to form biofilm and a tendency to acquire genes that confer resistance to different classes of antimicrobials (Amaral et al., 2005). The most common reported spa type associated with ST239-MRSA is the ancestral spa type t037 (the plesiomorphic state) (Harris et al., 2010), but other spa types have also been linked to ST239. In this study spa types t945 and t860 were common between ST239-MRSA and ST239-MSSA however their PFGE banding patterns were dissimilar. In addition to ST239, other common lineages between MSSA and MRSA were ST5 and ST22. Similar observation but with different STs have been noted before (Crisóstomo et al., 2001). To determine whether these strains lost or gained Sccmec over time will require more genetic analysis.
Other MRSA lineages with reduced susceptibility to chlorhexidine were ST217, a single-locus variant of EMRSA-15, which is prevalent in Europe and India (Qi et al., 2005; Vignaroli et al., 2014; Bouchiat et al., 2015) and has been also detected from food-stuffs of animal origin (Lozano et al., 2009). Also the highly transmissible ST772-MRSA-V/t4867 (the Bengal clone) (Afroz et al., 2008) which has been detected in different countries (Fuentes et al., 2005; Nadig et al., 2012; Shore et al., 2014) including Kuwait (Boswihi et al., 2016). However, this is the first time qacC and reduced susceptibility to chlorhexidine has been identified in this infectious clone. qacC is a mobile gene that can be transferred between plasmids without the aid of insertion sequences or transposases also known as “DSO-gene-SSO” element (Wassenaar et al., 2016).
We also observed high MBC for chlorhexidine among MSSA lineages including ST34 and ST5; commonly linked to mother-to-infant transmission (Achermann et al., 2015; Benito et al., 2015), ST88-MSSA, a prevalent colonizer in Portugal and China (Qiao et al., 2014), ST97 (associated with food producing animals) (Lozano et al., 2011) and among other less reported lineages: ST96, ST932, and ST965.
Nevertheless in Staphylococci, qac genes may be associated with ecologically successful genetic lineages to retain a high level of endemicity and or epidemicity. In environments where biocide delivery is compromised, whether in the community or in hospitals, the possession of biocide resistance genes (qac) may contribute to clonal expansion (Cooper et al., 2012). Yet, high rate of the putative virulent pvl gene in MSSA clones with reduced susceptibility to chlorhexidine should be monitored with vigilance as they may lead to poor prognosis in patients.
Our overall conclusion from this study is that clinical MSSA isolates with reduced susceptibility to chlorhexidine are genetically diverse. Since MSSA infections are important globally and are not just a local problem, the typing information can be used to evaluate the significance of distribution of these lineages and their clonal relationships in other countries.
Author contributions
LV, AD, and EU conceived and designed the study. LV and FM acquired the data. LV, FM, and EU analyzed the data. LV, AD, FM, and EU drafted and critically evaluated the manuscript. All authors approved the final version of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Miss Qudsiya Electrowala and the staff of Kuwait MRSA reference laboratory for their assistance in providing the isolates and assistance in performing some of the preliminary identification experiments. The authors would also like to acknowledge the Research Unit for Genomics, Proteomics, and Cellomics Studies (OMICS) of the Health Sciences Centre, Kuwait University (Project No. SRUL02/13) for assisting in DNA sequencing.
Footnotes
Funding. This study was funded by Kuwait University Research Grant number NM02/12.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01359/full#supplementary-material
References
- Achermann Y., Seidl K., Kuster S. P., Leimer N., Durisch N., Ajdler-Schäffler E., et al. (2015). Epidemiology of methicillin-susceptible Staphylococcus aureus in a neonatology ward. Infect. Control Hosp. Epidemiol. 36, 1305–1312. 10.1017/ice.2015.184 [DOI] [PubMed] [Google Scholar]
- Afroz S., Kobayashi N., Nagashima S., Alam M. M., Hossain A. B., Rahman M. A., et al. (2008). Genetic characterization of Staphylococcus aureus isolates carrying Panton-Valentine leukocidin genes in Bangladesh. Jpn. J. Infect. Dis. 61, 393–396. [PubMed] [Google Scholar]
- Amaral M. M., Coelho L. R., Flores R. P., Souza R. R., Silva-Carvalho M. C., Teixeira L. A., et al. (2005). The predominant variant of the Brazilian epidemic clonal complex of methicillin-resistant Staphylococcus aureus has an enhanced ability to produce biofilm and to adhere to and invade airway epithelial cells. J. Infect. Dis. 192, 801–810. 10.1086/432515 [DOI] [PubMed] [Google Scholar]
- Bannerman T. L., Hancock G. A., Tenover F. C., Miller J. M. (1995). Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33, 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovich T., Zarake H., Shabana I. I., Nevzorova V., Turcutyuicov V., Suzuki H. (2010). Molecular characterization and susceptibility of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from hospitals and the community in Vladivostok, Russia. Clin. Microbiol. Infect. 16, 575–582. 10.1111/j.1469-0691.2009.02891.x [DOI] [PubMed] [Google Scholar]
- Benito D., Lozano C., Jiménez E., Albújar M., Gómez A., Rodríguez J. M., et al. (2015). Characterization of Staphylococcus aureus strains isolated from faeces of healthy neonates and potential mother-to-infant microbial transmission through breastfeeding. FEMS Microbiol. Ecol. 91:fiv007. 10.1093/femsec/fiv007 [DOI] [PubMed] [Google Scholar]
- Bloomfield S. F. (2002). Significance of biocide usage and antimicrobial resistance in domiciliary environments. J. Appl. Microbiol. 92, 144S–157S. 10.1046/j.1365-2672.92.5s1.15.x [DOI] [PubMed] [Google Scholar]
- Boswihi S. S., Udo E. E., Al-Sweih N. (2016). Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait hospitals: 1992-2010. PLoS ONE 11:e0162744. 10.1371/journal.pone.0162744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchiat C., El-Zeenni N., Chakrakodi B., Nagaraj S., Arakere G., Etienne J. (2015). Epidemiology of Staphylococcus aureus in Bangalore, India: emergence of the ST217 clone and high rate of resistance to erythromycin and ciprofloxacin in the community. New Microbes New Infect. 14, 15–20. 10.1016/j.nmni.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2014). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement. Document M100-S21. Wayne, PA: CLSI. [Google Scholar]
- Cooper B. S., Kypraios T., Batra R., Wyncoll D., Tosas O., Edgeworth J. D. (2012). Quantifying type-specific reproduction numbers for nosocomial pathogens: evidence for heightened transmission of an Asian sequence type 239 MRSA clone. PLoS Comput. Biol. 8:e1002454. 10.1371/journal.pcbi.1002454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisóstomo M. I., Westh H., Tomasz A., Chung M., Oliveira D. C., de Lencastre H. (2001). The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. U.S.A. 98, 9865–9870. 10.1073/pnas.161272898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo F. R., Chambers H. F. (2009). Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J. Clin. Invest. 119, 2464–2474. 10.1172/JCI38226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenburg R. H., Vink C., Kalenic S., Friedrich A. W., Bruggeman C. A., Stobberingh E. E. (2007). The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect Dis. 13, 222–235. 10.1111/j.1469-0691.2006.01573.x [DOI] [PubMed] [Google Scholar]
- EUCAST (2016). European Committee on Antimicrobial Susceptibility Testing. European society of clinical microbiology and infectious diseases. Available online at: http://www.eucast.org
- Fuentes D. E., Navarro C. A., Tantaleán J. C., Araya M. A., Saavedra C. P., Pérez J. M., et al. (2005). The product of the qacC gene of Staphylococcus epidermidis CH mediates resistance to beta-lactam antibiotics in gram-positive and gram-negative bacteria. Res. Microbiol. 156, 472–477. 10.1016/j.resmic.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Furi L., Ciusa M. L., Knight D., Di Lorenzo V., Tocci N., Cirasola D., et al. (2013). Evaluation of reduced susceptibility to quaternary ammonium compounds and bisbiguanides in clinical isolates and laboratory-generated mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 3488–3497. 10.1128/AAC.00498-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen D., Claus H., Witte W., Rothgänger J., Claus H., Turnwald D., et al. (2003). Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41, 5442–5448. 10.1128/JCM.41.12.5442-5448.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. R., Feil E. J., Holden M. T., Quail M. A., Nickerson E. K., Chantratita N., et al. (2010). Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474. 10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner C., Mawer D., Wilcox M. (2012). Reduced susceptibility to chlorhexidine in staphylococci: is it increasing and does it matter? J. Antimicrob. Chemother. 67, 2547–2559. 10.1093/jac/dks284 [DOI] [PubMed] [Google Scholar]
- Ito T., Katayama Y., Hiramatsu K. (1999). Cloning and nucleotide sequence determination of the entire mec DNA of pre methicillin-resistant Stapylococcus aureus N315. Antimicrob. Agents Chemother. 43, 1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Katayama Y., Asada K., Mori N. (2001). Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45, 1323–1336. 10.1128/AAC.45.5.1323-1336.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., et al. (2007). Active Bacterial Core surveillance (ABCs) MRSA Investigators. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298, 1763–1771. 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- Lozano C., Gómez-Sanz E., Benito D., Aspiroz C., Zarazaga M., Torres C. (2011). Staphylococcus aureus nasal carriage, virulence traits, antibiotic resistance mechanisms, and genetic lineages in healthy humans in Spain, with detection of CC398 and CC97 strains. Int. J. Med. Microbiol. 301, 500–505. 10.1016/j.ijmm.2011.02.004 [DOI] [PubMed] [Google Scholar]
- Lozano C., López M., Gómez-Sanz E., Ruiz-Larrea F., Torres C., Zarazaga M.. (2009). Detection of methicillin-resistant Staphylococcus aureus ST398 in food samples of animal origin in Spain. J. Antimicrob. Chemother. 64, 1325–1326. 10.1093/jac/dkp378 [DOI] [PubMed] [Google Scholar]
- Mayer S., Boos M., Beyer A., Fluit A. C., Schmitz F. J. (2001). Distribution of the antiseptic resistance genes qacA, qacB, and qacC in 497 methicillin-resistant and –susceptible European isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 47, 896–897. 10.1093/jac/47.6.896 [DOI] [PubMed] [Google Scholar]
- Milheiriço C., Portelinha A., Krippahl L., de Lencastre H., Oliveira D. C. (2011). Evidence for a purifying selection acting on the β-lactamase locus in epidemic clones of methicillin-resistant Staphylococcus aureus. BMC Microbiol. 11:76. 10.1186/1471-2180-11-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey I., Oggioni M. R., Knight D., Curiao T., Coque T., Kalkanci A., et al. (2014). Evaluation of epidemiological cut-off values indicates that biocide resistant subpopulations are uncommon in natural isolates of clinically-relevant microorganisms. PLoS ONE 9:e86669. 10.1371/journal.pone.0086669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadig S., Velusamy N., Lalitha P., Kar S., Sharma S., Arakere G.. (2012). Staphylococcus aureus eye infections in two Indian hospitals: emergence of ST772 as a major clone. Clin. Ophthalmol. 6, 165–173. 10.2147/OPTH.S23878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira V. C., Riboli D. F., da Cunha Mde L. (2014). Characterization of the clonal profile of MRSA isolated in neonatal and pediatric intensive care units of a University Hospital. Ann. Clin. Microbiol. Antimicrob. 7:50 10.1186/s12941-014-0050-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersdorf S., Herma M., Rosenblatt M., Layer F., Henrich B. (2015). A novel staphylococcal cassette chromosome mec Type XI Primer for detection of mecC-harboring methicillin-resistant Staphylococcus aureus directly from screening specimens. J. Clin. Microbiol. 53, 3938–3941. 10.1128/JCM.02328-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W., Ender M., O'Brien F., Imhof A., Ruef C., McCallum N., et al. (2005). Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Zurich, Switzerland (2003): prevalence of type IV SCCmec and a new SCCmec element associated with isolates from intravenous drug users. J. Clin. Microbiol. 43, 5164–5170. 10.1128/JCM.43.10.5164-5170.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Ning X., Chen Q., Zhao R., Song W., Zheng Y., et al. (2014). Clinical and molecular characteristics of invasive community-acquired Staphylococcus aureus infections in Chinese children. BMC Infect. Dis. 7:582 10.1186/s12879-014-0582-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. K., Kumari N., Pahwa S., Agrahari U. C., Bhutani K. K., Jachak S. M., et al. (2013). NorA efflux pump inhibitory activity of coumarins from Mesua ferrea. Fitoterapia 90, 140–150. 10.1016/j.fitote.2013.07.015 [DOI] [PubMed] [Google Scholar]
- Shamsudin M. N., Alreshidi M. A., Hamat R. A., Alshrari A. S., Atshan S. S., Neela V. (2012). High prevalence of qacA/B carriage among clinical isolates of meticillin-resistant Staphylococcus aureus in Malaysia. J. Hosp. Infect. 81, 206–208. 10.1016/j.jhin.2012.04.015 [DOI] [PubMed] [Google Scholar]
- Shore A. C., Tecklenborg S. C., Brennan G. I., Ehricht R., Monecke S., Coleman D. C. (2014). Panton-Valentine leukocidin-positive Staphylococcus aureus in Ireland from 2002 to 2011: 21 clones, frequent importation of clones, temporal shifts of predominant methicillin-resistant S. aureus clones, and increasing multi resistance. J. Clin. Microbiol. 52, 859–870. 10.1128/jcm.02799-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani S., Chung D. R., Lindsay J. A., Friedrich A. W., Kearns A. M., Westh H., et al. (2012). Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int. J. Antimicrob. Agents 39, 273–282. 10.1016/j.ijantimicag.2011.09.030 [DOI] [PubMed] [Google Scholar]
- Tong S. Y., Holden M. T., Nickerson E. K., Cooper B. S., Köser C. U., Cori A., et al. (2015). Genome sequencing defines phylogeny and spread of methicillin-resistant Staphylococcus aureus in a high transmission setting. Genome Res. 25, 111–118. 10.1101/gr.174730.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng S. P., Lin Y. T., Tsai J. C., Hung W. C., Chen H. J., Chen P. F., et al. (2015). Genotypes and phenotypes of Staphylococcus lugdunensis isolates recovered from bacteremia. J. Microbiol. Immunol. Infect. 48, 397–405. 10.1016/j.jmii.2013.11.006 [DOI] [PubMed] [Google Scholar]
- Vignaroli C., Mancini A., Varaldo P. E. (2014). Composite SCCmec element in single-locus variant (ST217) of epidemic MRSA-15 clone. Emerging Infect. Dis. 20, 905–907. 10.3201/eid2005.130934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren D. K., Prager M., Munigala S., Wallace M. A., Kennedy C. R., Bommarito K. M., et al. (2016). Prevalence of qacA/B genes and mupirocin resistance among methicillin- resistant Staphylococcus aureus (MRSA) isolates in the setting of chlorhexidine bathing without mupirocin. Infect. Control Hosp. Epidemiol. 37, 1–7. 10.1017/ice.2016.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar T. M., Ussery D. W., Ingmer H. (2016). The qacC Gene has recently spread between rolling circle plasmids of Staphylococcus, indicative of a novel gene transfer mechanism. Front. Microbiol. 27:1528 10.3389/fmicb.2016.01528 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.