Abstract
The basic helix-loop-helix (bHLH) proteins are a large family of transcription factors that control various developmental processes in eukaryotes, but the biological roles of most bHLH proteins are not very clear, especially in tomato. In this study, a PRE-like atypical bHLH gene was isolated and designated as SlPRE2 in tomato. SlPRE2 was highly expressed in immature-green fruits, moderately in young leaves, flowers, and mature-green fruits. To further research the function of SlPRE2, a 35 S:PRE2 binary vector was constructed and transformed into wild type tomato. The transgenic plants showed increased leaf angle and stem internode length, rolling leaves with decreased chlorophyll content. The water loss rate of detached leaves was increased in young transgenic lines but depressed in mature leaves. Besides, overexpression of SlPRE2 promoted morphogenesis in seedling development, producing light-green unripening fruits and yellowing ripen fruits with reduced chlorophyll and carotenoid accumulation in pericarps, respectively. Quantitative RT-PCR analysis showed that expression of the chlorophyll related genes, such as GOLDEN 2-LIKE and RbcS, were decreased in unripening 35 S:PRE2 fruit, and carotenoid biosynthesis-related genes PHYTOENE SYNTHASE1A and ζ-CAROTENE DESATURASE in ripening fruit were also down-regulated. These results suggest that SlPRE2 affects plant morphology and is a negative regulator of fruit pigment accumulation.
Introduction
The basic helix-loop-helix (bHLH) proteins are a large family of transcription factors that control metabolic, physiological and developmental processes in all eukaryotic organisms. They were considered to have different function based on the distinction of bHLH domain1, 2. The bHLH protein consists of about 60 amino acids organized in the basic region and HLH region3. The HLH region at the C-terminal is involved in the homo- or hetero-dimerization with other protein while the basic region in N-terminal for DNA-binding. Based on the DNA-binding ability, the bHLH proteins are divided into two groups, “DNA-binding bHLH” and “atypical bHLH” with no DNA-binding ability1, 4. Recent studies have shown that typical bHLH transcription factors participate in various plant growth and development processes, such as light signaling5, hormone signaling6, anthocyanin biosynthesis7, 8, fruit development9 and stress responses10. For example, overexpression of PIF5 induces leaf senescence and chlorophyll degradation in dark-grown Arabidopsis 11. The SlPIF1a is characterized to regulate carotenoid biosynthesis during fruit ripening by a light-dependent mechanism in tomato12. A bHLH35 gene from populous euphratica could regulate photosynthesis in Arabidopsis 13. In addition, the atypical bHLH genes have been shown to play a role in the regulation of hormone signaling14, 15, light signaling16, vascular development17 and grain size18, such as PRE1 and KIDARI/PRE6 19, 20.
PRE1 and KIDARI belong to the paclobutrazol-resistant (PRE) family with 6 members in Arabidopsis. PRE1, one of these atypical bHLHs, is initially identified to be an activator of gibberellin responses19. The further researches show that PRE1 mediates brassinosteroid, auxin, and light signaling21–23. The PRE3 is a dominant suppressor of BR mutant bri1-301 and is involved in regulation of light signal transduction in Arabidopsis 24, 25. The PRE4/BNQ3 also function in light signaling, and bnq3 mutant have pale-green flower and reduced chlorophyll level26. KIDARI/PRE6 is a repressor of light signaling and affects photomorphogenesis by negatively regulating HFR1 activity20, 27. Overexpression of ILI1 (INCREASED LAMINA INCLINATION1), a homology of PRE1 in rice, increased cell elongation through a mechanism involved in brassinosteroid signalling21. BU1 (BRASSINOSTEROID UPREGULATED1) is involved in brassinosteroid signaling and controls lamina joint bending in rice28. Overexpression of PGL1 (POSITIVE REGULATOR OF GRAIN LENGTH 1) and PGL2 in lemma/palea could increase grain length and weight in transgenic rice18, 29. To date, only Slstyle2.1, a PREs-like gene in tomato, was identified to control floral style length and contribute to the evolution of self-pollination in cultivated variety30. Through our genome-wide analysis of PRE family, there are five members in tomato including SlSTYLE2.1. However, their biological function in tomato growth is still unknown, which undoubtedly need further study.
Tomato is considered to be one of the best systems to study fleshy fruit ripening. Here, a homologous gene of Arabidopsis PREs, which was named SlPRE2 along the PRE-like gene style2.1 30, was cloned. In this study, overexpression of SlPRE2 was performed to investigate the role of SlPRE2 in tomato development. Transgenic tomatoes showed alteration of plant morphology by affecting light signaling and repression of fruit pigment accumulation. Our results indicate that SlPRE2 affects plant morphogenesis, fruit chlorophyll and carotenoid accumulation probably through influencing the activity of bHLH proteins involved in light signaling.
Results
SlPRE2 isolation and transcription pattern analysis
Based on the BLAST analysis in SGN (Sol Genomics Network, https://solgenomics.net/), 5 AtPREs like genes were isolated and named SlPRE1 to SlPRE5 in tomato (Fig. S1a,b). With the transcriptome analysis in SGN, expression profile of these SlPREs was performed in Fig. S1c. SlPRE1, which has been functionally identified as STYLE2.1 in tomato30, was specifically expressed in anthesis flower. SlPRE2 was highly expressed in 10 days post anthesis fruits(DPA). SlPRE3 had low expression abundance. Moreover, SlPRE4 was highly expressed in hypocotyl and vegetative meristem, while SlPRE5 was performed expressed in multiple tissue. The SlPRE2 was chosen for further investigation since its high expression in IMG fruit. Based on the sequence in SGN (sequence ID: Solyc02g067380.2.1), the SlPRE2 was isolated from tomato with specific primers SlPRE2-F and SlPRE2-R. Gene sequence analysis showed that SlPRE2 encodes a putative bHLH protein consisting of 94 amino acids (Fig. 1a). SlPRE1 has been functionally identified as STYLE2.1 in tomato30. SlPRE3 and SlPRE4 were reported as SlbHLH103 and SlbHLH131 by Sun, respectively31. As shown in Fig. 1a, amino acid sequence alignment of SlPRE2 and homologous proteins showed that SlPRE2 is highly homologous to AtPREs, OsBU1 and OsPGLs, which were identified as atypical bHLH18, 20, 23, 24, 28, 29 (Fig. 1a). These results indicated that SlPRE2 encodes an atypical bHLH transcription factor with no DNA-binding activity.
Figure 1.
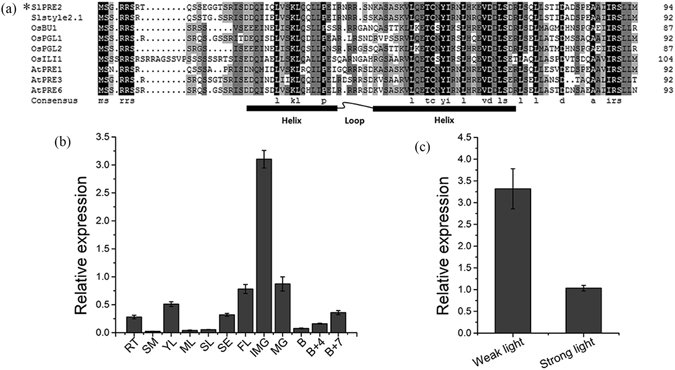
Multiple sequence alignment and expression profile of SlPRE2. (a) Multiple sequence alignment of SlPRE2 and other bHLH proteins. The SlPRE2 and Slstyle2.1 are proteins from tomato. OsBU1, OsPGL1, OsPGL2, OsILI1 are proteins from rice. AtPRE1–6 are proteins from Arabidopsis. Black and gray backgrounds indicate identical and similar amino acids. Convergence in structure is indicated by black box and curve. (b) The relative expression patterns of SlPRE2 in wild type tomato. RT, root; SM, stem; YL, young leaf; ML, mature leaf; SL, senescent leaf; SE, sepal; FL, flower; IMG, immature green fruit; MG, mature fruit; B, breaker fruit; B + 4, 4 days after breaker stage; B + 7, 7 days after breaker stage. (c) The relative expression levels in weak and strong light growth condition for 8 hours. Data are the mean ± SD of three biological replicates.
To extend our understanding of the role of SlPRE2 in tomato growth and development, quantitative PCR was performed to analyze the expression of SlPRE2 in various tissues, including roots, stems, leaves, flowers and fruits at different stages of development, from tomato cultivar Ailsa Craig (AC). The results showed that SlPRE2 was highly expressed in 15 DPA IMG fruits, moderately in young leaves, flowers, and mature-green fruits, and the lowest expression level was found in stems, mature and senescent leaves (Fig. 1b). Besides, a similar expression pattern of SlPRE2 in Solanum pimpinellifolium LA1589 was performed using Tomato Functional Genomics Database (http://ted.bti.cornell.edu/) (Table S1). Promoter analysis of SlPRE2 gene demonstrated that there are various light response elements, such as G-Box and ACE motif, located 800 bases upstream of the translation start site (ATG) (Table S2). To further explore its role in light signaling, tomato leaves exposed in weak (50 μmol·m−2·s−1) or strong light (800 μmol·m−2·s−1) for 8 hours were collected. Significantly suppressed expression of SlPRE2 was observed in strong light condition (Fig. 1c).
Overexpression of SlPRE2 alters leaf and stem morphology
To investigate the physiological function of SlPRE2 in tomato, transgenic lines overexpressing SlPRE2 were generated using constitutive CaMV 35 S promoter. Expression levels of SlPRE2 in independent 35 S:PRE2 lines were shown in Fig. S2. Compared with the wild type, the 35 S:PRE2 lines had various differences in plant morphology. First, 35 S:PRE2 lines had rolling mature leaves and the leaf rolling index was significantly increased (Fig. 2a,b, Fig. S3c), while the young leaves had no significant change (Fig. S3a,b). By scanning electron microscope analysis, the 35 S:PRE2 mature leaves had a significantly narrower stomatal aperture than that in wild type on the abaxial leaf surface, while there was no significant difference between the two groups in the stomatal pore length (Fig. 2c, Fig. S4). Furthermore, the transgenic plants had increased leaf angles and longer internodes (Fig. 2d,e). These results indicated that SlPRE2-overexpression affects the vegetative growth of tomato.
Figure 2.
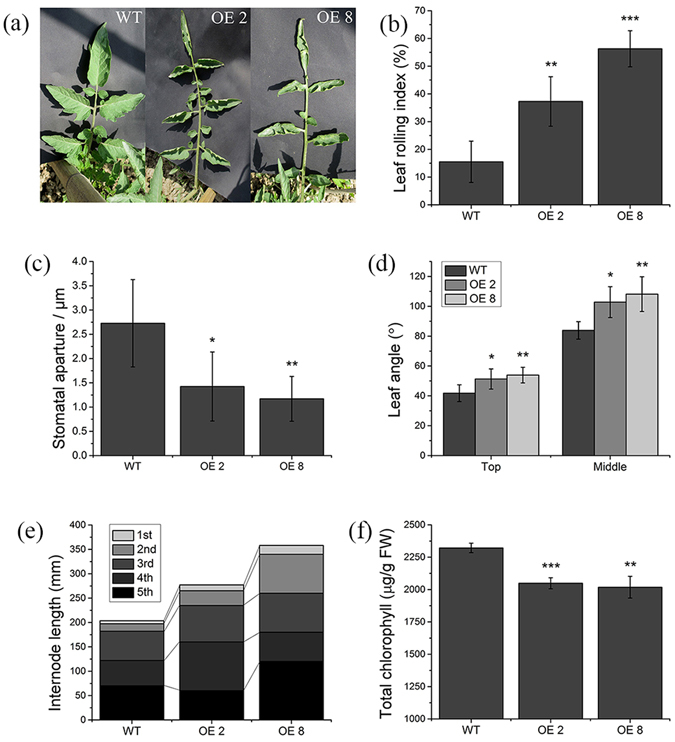
Phenotype of the SlPRE2 overexpression transgenic plants. (a) Leaf phenotype of wild type and transgenic lines. The 35 S:PRE2 transgenic lines showed rolling leaf trait. (b) Leaf rolling index (LRI) for the mature leaves. (c) Stomatal aperture of mature leaves on wild type and SlPRE2 overexpression lines. (d) Leaf tilt angle of the top young leaves and the middle mature leaves in wild type and transgenic lines. (e) The internode length for each of the top five internodes. (f) Chlorophyll content of leaves at the fifth from the top. Data are the mean ± SD of three biological replicates. *Significantly different from the wild type, P value t test < 0.05; ** for P < 0.01.
Overexpression of SlPRE2 inhibits chlorophyll accumulation and changes water losing rate in leaves
In addition to the changes in plant morphology, the leaves of 35 S:PRE2 lines also showed light green with decreased chlorophyll content (Fig. 2f). For understanding the molecular mechanism of chlorophyll breakdown, the photosynthesis and chloroplast development-related genes, GLK2, RbcS, Cab7, and DCL, were analyzed in wild type and 35 S:PRE2 mature leaves. Quantitative RT-PCR results displayed that the four genes were significantly down-regulated in 35 S:PRE2 lines (Fig. 3).
Figure 3.
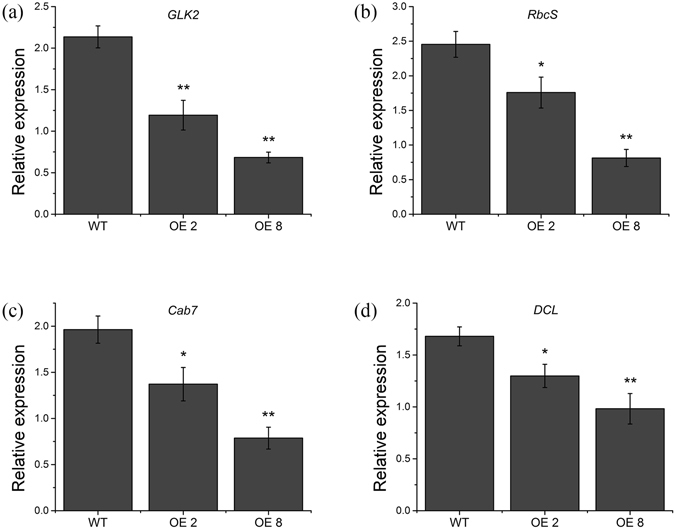
Expression levels of GLK2, RbcS, Cab7, and DCL in mature leaves of wild type and 35 S:PRE2 lines. Data are the mean ± SD of three biological replicates. *Significantly different from the wild type, P < 0.05; ** for P < 0.01.
The change of plant morphology could also affect the adaptation of plants to environment32. The rate of water loss of detached leaves was measured in young leaves and mature leaves from 35 S:PRE2 and wild type. As shown in Fig. 4, the young leaves of 35 S:PRE2 lines had a higher rate of water loss than wild type (Fig. 4a), however, the mature leaves of transgenic plants exhibited a lower rate of water loss (Fig. 4b).
Figure 4.
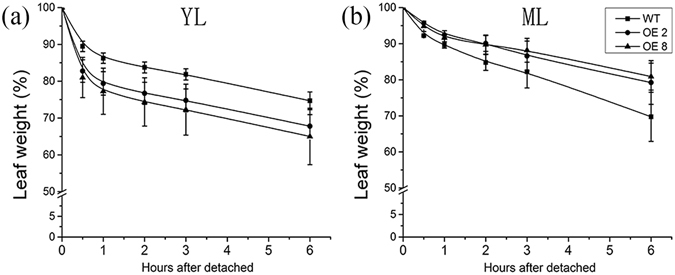
Rate of water loss from wild type and 35 S:PRE2 lines detached leaves at 25 °C room temperature. (a) and (b) represent rate of water loss in young leaves (YL) and mature leaves (ML), respectively. Data are the mean ± SD of three biological replicates.
Overexpression of SlPRE2 promotes hypocotyl elongation
To determine the roles of SlPRE2 in morphogenesis, the measurement of hypocotyl length was performed. Seeds of the wild type and SlPRE2 overexpression lines were germinated in dark or light for 8 days. Seedlings of transgenic lines grown in dark and light condition all showed an increase in hypocotyl growth compared with the wild type (Fig. 5a–d). To gain further information on the long hypocotyl phenotype of 35 S:PRE2 seedling, the transcription levels of HY5 and PIF4 were determined in 8-days-old dark-grown tomato seedlings. The HY5 has been reported to be a light signaling related gene in Arabidopsis and tomato33, 34. The PIF4 is a putative phytochrome interacting factor that may participate in light signaling in tomato. As shown in Fig. 5e and f, levels of HY5 and PIF4 mRNA were all decreased in 35 S:PRE2 seedlings.
Figure 5.
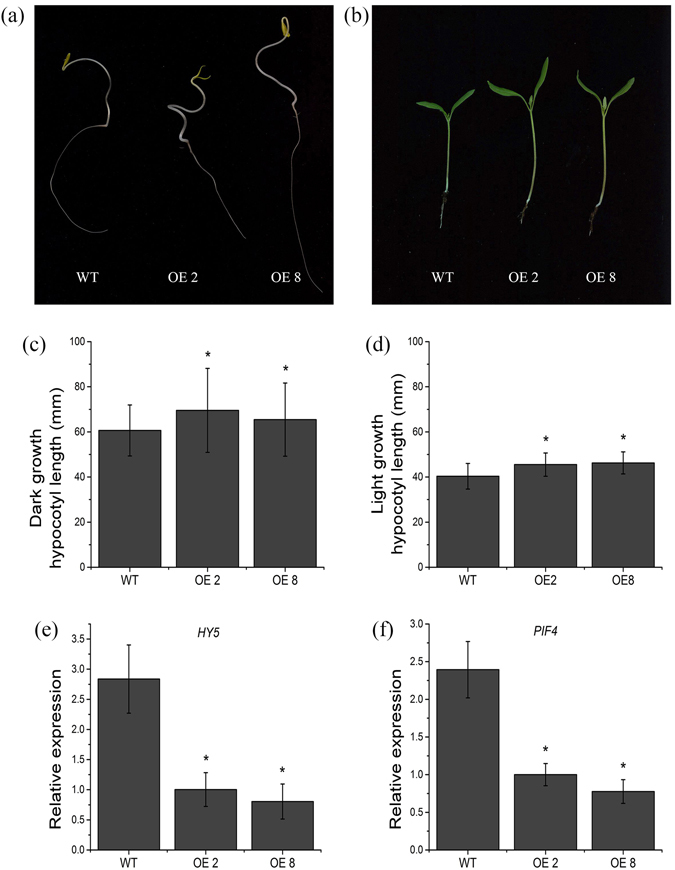
Phenotype of 35 S:PRE2 seedling and expression analysis of HY5 and PIF4. (a) and (b) photograph of seedling of two 35 S:PRE2 lines and wild type obtained at 8 days after sowing under dark and light condition, respectively. (c) and (d) hypocotyl length of two 35 S:PRE2 lines and wild type 8 days seedlings growing in dark and light condition, respectively. (e) and (f) respectively represent expression levels of HY5 and PIF4 in 8-days-old dark-grown seedlings. Data are the mean ± SD of three biological replicates. *Significantly different from the wild type, P < 0.05; ** for P < 0.01.
Overexpression of SlPRE2 reduces pigment accumulation in fruit
For most fruits, the color will change during the ripening process. Tomato fruit ripening is characterized by a shift in color from green to red, which is induced by the breakdown of chlorophyll and the formation of carotenoids. In this study, the 35 S:PRE2 fruits showed light green pericarp at IMG, MG, and B stages, and yellowing pericarp at B + 4 and B + 7 stages compared to the wild type fruits (Fig. 6a). For chlorophyll content analysis, the tomato was cut into five sections along the vertical axis. The chlorophyll contents had a gradient descent from stem end to stylar end and the transgenic fruits have less chlorophyll than the WT at MG and B stages, especially in the stylar end (Fig. 6b,c). Transmission electron microscopy (TEM) revealed that the transgenic MG fruits display smaller chloroplasts and significantly fewer grana thylakoids as well as plastoglobule per chloroplast than the wild type (Fig. 6d). These results suggested that SlPRE2 is involved in the regulation of chloroplast development and chlorophyll accumulation in tomato. Besides, carotenoids that accumulated in ripening tomato fruit include lycopene, β-carotene, and lutein. Among them, lycopene is the main carotenoid and responsible for the red color of red tomatoes. Thus, the carotenoid contents in B + 4 and B + 7 fruits were determined by spectrophotometer. Total carotenoid contents were reduced by 10% to 25% in B + 4 and B + 7 fruits of transgenic lines (Fig. 6f). Meanwhile, the lycopene contents were decreased by 18% to 40% in transgenic B + 4 and B + 7 fruits (Fig. 6e).
Figure 6.
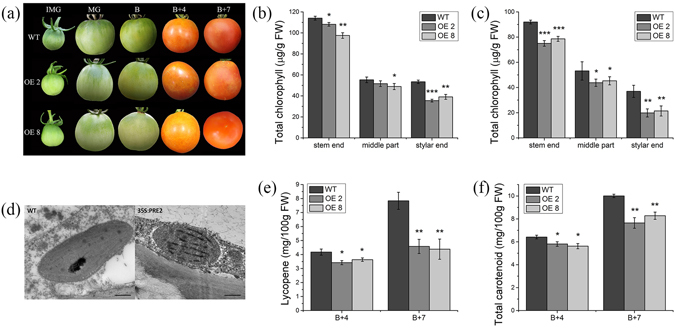
Phenotype and physiology in transgenic tomato fruits. (a) Fruits phenotype of wild type (top) and 35 S:PRE2 lines at IMG, MG, B, B + 4 and B + 7 stages (middle and bottom). (b) and (c) Chlorophyll contents of three latitudinal sections from wild type and 35 S:PRE2 fruits in MG and B stage. (d) Transmission electron microscopy images of MG fruit outer pericarp chloroplasts from wild type and 35 S:PRE2 lines. Bars = 0.5 μm. (e) and (f) Lycopene and total carotenoid content of fruit flesh in B + 4 and B + 7 stages. MG, mature fruit; B, breaker fruit; B + 4, 4 days after breaker stage; B + 7, 7 days after breaker stage. FW, fresh weight. Data are the mean ± SD of three biological replicates. *Significantly different from the wild type, P value t test < 0.05; ** for P < 0.01.
Transcriptional analysis of pigment-related genes in 35 S: SlPRE2 fruit
To further study the underlying causes of the decreased pigments in 35 S:PRE2 fruits, qRT-PCR was used to measure the transcript levels of genes involved in chlorophyll accumulation and carotenoid biosynthesis. The transcription levels of GLK2, RbcS, Cab7 and HY5 genes were notably down-regulated in 35 S:PRE2 fruits (Fig. 7). Moreover, GLK2 expression showed a descending trend from stem end to style end, which was consistent with the previous report described by Nguyen35, and its transcript accumulation in transgenic fruits was lower than the wild type in MG and B (Fig. S5). In addition, the carotenoid biosynthesis genes, PSY1 and ZDS, were markedly down-regulated at B + 4 and B + 7 stages, and the PDS was slightly increased in B + 4 fruit, but significantly decreased in B + 7 fruit (Fig. 8).
Figure 7.
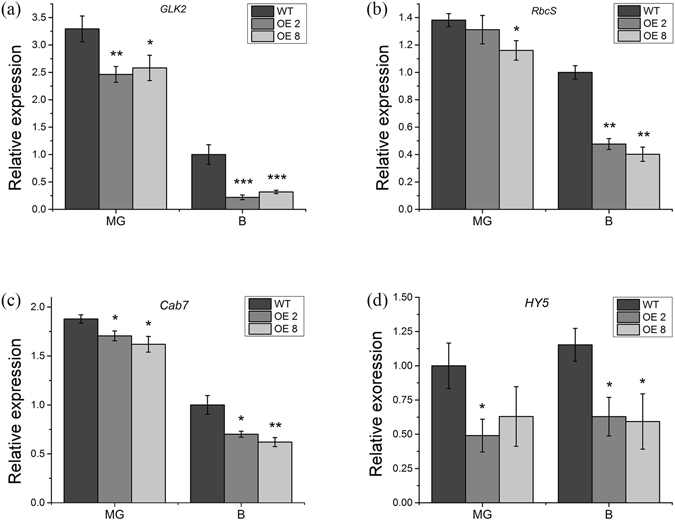
Chlorophyll metabolism related genes expression in wild type and 35 S:PRE2 lines. (a) to (d) respectively represent expression levels of GLK2, RbcS, Cab7, and HY5 in different stage fruits of wild type and 35 S:PRE2 lines. MG, mature fruit; B, breaker fruit. Data are the mean ± SD of three biological replicates. *Significantly different from the wild type, P < 0.05; ** for P < 0.01.
Figure 8.
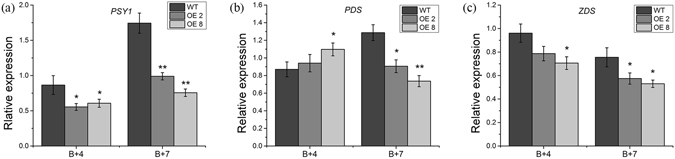
Carotenoid biosynthesis genes expression in fruits from 35 S:PRE2 lines and wild type. (a) to (c) respectively represent expression levels of PSY1, PDS, and ZDS in different stage fruits of wild type and 35 S:PRE2 lines. B + 4, 4 days after breaker stage; B + 7, 7 days after breaker stage. Data are the mean ± SD of three biological replicates. *Significantly different from the wild type, P < 0.05; ** for P < 0.01.
Discussion
Light is an important factor for plant growth, which affects various development processes, such as de-etiolation, cell elongation and shade avoidance through the regulation of many genes. Previous studies have demonstrated that PREs involve in plant growth regulation and light signal transduction14, 20, 25. In Arabidopsis, ATBS1/PRE3 suppresses the dwarf phenotype of BR mutants by an inhibition of negative BR signaling components, and it is involved in light signaling pathway by affecting the expression of genes related to light signaling, including PIF3 and HY5 24, 25. With activation tagging technology, the PRE1 activation-tagged mutant showed long hypocotyl and slightly narrow pale green leaves, and the hypocotyls length were all increased in transgenic Arabidopsis with overexpression of PRE2-PRE5 genes19. Mara et al. showed that mutant in PRE4/BNQ3 induced a pale-green flower and declined chlorophyll content in Arabidopsis 26. KIDARI was identified as a repressor of light signaling and photomorphogenesis in Arabidopsis, and co-expression of KIDARI suppressed HFR1-ox phenotypes and showed relatively long hypocotyls20, 27. Along with PRE1, these atypical bHLH transcription factors interact with HFR1 and PARs and rescue PIFs activity in light signaling23. Rice plants with overexpression of BU1 and ILI1 all showed increased bending of lamina joint respectively, while RNAi plants had erect leaves21, 28.
In our study, overexpression of SlPRE2 resulted in various plant morphological variation. Firstly, although the shape of young leaves had no difference between the wild type and transgenic lines, the young leaves in transgenic lines had higher water loss rate than the wild type, and the mature leaves of the 35 S:PRE2 plant were curled upwardly with reduced chlorophyll content and water loss rate (Figs 2f and 4). The phenotype of pale green leaves in 35 S:PRE2 lines are consistent with the character performed in PRE1 activation-tagged mutant leaves and PRE4/BNQ3 mutant sepals and carpels19, 26. It is well known that the degree of leaf rolling are linearly related to leaf water potential and plants with curly leaves also had an increased resistance to water stress36. Leaf rolling reduced effective leaf area and transpiration, thus leading to the closure of stomata, which increases drought avoidance37. Our results revealed that overexpression of SlPRE2 elevates the rate of water loss of young leaves. Thus, we can speculate that mature leaves rolling may be one way to hold water in leaves.
Secondly, the leaf angle and stem internode length were increased in 35 S:PRE2 lines (Fig. 2d,e), which is in line with phenotypes displayed in ILI1- and BU1-overexpression rice plants21, 28. Plants under shade avoidance also showed increased stem internode and leaf angle to get more light for photosynthesis38, 39. Given that the effective area of curly mature leaves of 35 S:PRE2 plant was decreased, this morphological variation may be a strategy for light competition by increasing effective area of leaf for receiving light and improving photosynthetic efficiency. Besides, promoter analysis of the SlPRE2 gene showed that there are several light responsiveness cis-acting regulatory elements in the 800 bp upstream of the start codon (Table S2). Meanwhile, SlPRE2 was significantly inhibited in strong light (Fig. 1c), and 35 S:PRE2 seedlings showed long hypocotyls along with the remarkably repressed expressions of light signaling related genes PIF4 and HY5 (Fig. 5e,f). These results indicated that SlPRE2 affects plant morphology probably through influencing light signaling transduction.
Fruit ripening is a complex process, including loss of green color and accumulation of carotenoid, soften pericarp with cell wall degradation and metabolism change with flavor and nutrient accumulation. Tomato fruit ripening is influenced by multiple factors, such as development signal, phytohormones, nutrient status, temperature, and light. Among them, ethylene is a major trigger for fruit ripening in tomato. Besides, many genes affect fruit ripening through controlling chlorophyll metabolism, photosynthesis, and light signaling. For example, the UNIFORM RIPENING (U) encodes an MYB transcription factor Golden 2-like (SlGLK2) which controls chlorophyll accumulation and distribution in developing tomato fruit, and the uniform ripening (u) mutant has a light green fruit phenotype, while overexpression of SlGLK2 results in dark-green fruits40. The SlHY5 is not only an important light signaling related gene but also a positive regulator of fruit pigmentation. Down-regulation of SlHY5 by RNA interference shows light green immature fruits with a reduced chlorophyll accumulation34. In addition, chlorophyll a/b-binding protein (CAB) and ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (RbcS) are reported to be essential to photosynthesis rate in photosynthetic tissues41, 42. In this study, overexpression of SlPRE2 resulted in reduced chlorophyll content in unripe fruit (Fig. 6), which phenotype is consistent with that in 35 S:PRE2 leaves we identified (Fig. 2f). Moreover, the transcript levels of GLK2, HY5, RbcS, and Cab7 in 35 S:PRE2 leaves and fruits were significantly decreased (Figs 3, 7). Transmission electron microscopy (TEM) showed that the transgenic MG fruits displayed defective chloroplast (Fig. 6d). Together with the expression pattern of SlPRE2 in fruits of different period, it is possible that SlPRE2 might regulate the balance between frequent fruit cell expansion and chlorophyll accumulation processes in immature fruit. However, these results indicated that SlPRE2 is a repressor of chlorophyll accumulation and chloroplast development.
Carotenoids are one of the most diverse classes of natural compound. It was also accumulated in leaves, flowers, fruit of plants. They are essential for plant photosynthesis and protection cell from photo-oxidation. Moreover, they furnish flowers and fruits with distinct colors that are designed to attract animals to disperse seeds. So far, the carotenoid biosynthesis pathway is very clear in higher plant. Generally, the first dedicated compound in the carotenoid pathway, 15-cis-phytoene, is formed through the catalyzes of phytoene synthase1 (PSY1), a key enzyme for carotenoid biosynthetic pathway43. After that, phytoene desaturase (PDS) and ζ-carotene desaturase (ZDS) catalyze the synthesis of 9,9′-di-cis-ζ-carotene and all-trans-lycopene44, 45. Absent function of PSY1 in tomato fruit results in yellow flesh phenotype43. In this study, we analyzed the expression of these tomato genes, PSY1, PDS, and ZDS. qPCR revealed that the transcript levels of these genes were significantly reduced in ripe fruits, especially in B + 7 transgenic fruits (Fig. 8), which were confirmed by the reduced lycopene and total carotenoid contents in 35 S:PRE2 fruits (Fig. 6e,f). These results suggested that SlPRE2 negatively regulates carotenoid accumulation during fruit ripening by repressing the expression of these carotenoid biosynthetic genes.
Materials and Methods
Plant materials and growth conditions
In this study, the tomato (Solanum lycopersicum Mill. cv. Ailsa Craig) was used as the wild type. Plants were grown in standard greenhouses with 16 h light (27 °C) and 8 h dark (19 °C) cycle. In wild type, the developmental stages of tomato fruits were divided into IMG (immature green at 15 DPA), MG (mature green at 32DPA), B (breaker with the color change from green to yellow), B + 4 (4 days after breaker) and B + 7 (7 days after breaker) stages according to the days post anthesis (DPA) and fruit color. For a test of gene expression under different light condition, plants were exposed in weak (50 μmol·m−2·s−1) or strong (800 μmol·m−2·s−1) light condition for 8 hours, respectively. All plant samples were immediately frozen in liquid nitrogen and stored at −80 °C.
Multiple sequence alignment analysis
Multiple sequence alignment of SlPRE2 with other PRE-like protein was conducted using the ClustalX 1.83 and DNAMAN version 7.0 programs. The peptide sequences were selected from Arabidopsis thaliana, Solanum lycopersicum and Oryza sativa according to their sequence similarity and functions reported. For phylogenetic analysis, peptide sequences of PREs proteins were selected, and the phylogenetic tree was constructed using MEGA 5 software.
Total RNA isolation and quantitative real-time PCR analysis
Total RNA was extracted from various tissues using RNAiso Plus (Takara). 2 μg RNA was treated with DNase I (Promega). The first-strand cDNA synthesis was performed with the M-MLV reverse transcriptase (Promega) using oligo(dT)20 primer. The synthesized cDNA was diluted three times with nuclease-free water for qRT-PCR analysis. Quantitative real-time PCR was performed using the GoTaq qPCR Master Mix (Promega). The condition for qPCR amplification was as follow: 95 °C for 3 min; 40 cycles of 95 °C for 30 s, 60 °C for 45 s and 72 °C for 30 s. Amplification was followed by a melting curve analysis with continual fluorescence data acquisition during the 60–95 °C melt. SlCAC and SlEF1α were used as internal reference genes for tomato development and abiotic stress studies, respectively46, 47. In addition, gene-specific primers for qPCR analysis are listed in Table S3.
Vector construction and plant transformation
To generate 35 S:PRE2 lines, the full-length coding sequence of the SlPRE2 gene was amplified by PCR from tomato (AC) cDNA using primers SlPRE2-F (5′ CGGGATCCTCAAAAGAATCATCTCAAAATA 3′) and SlPRE2-R (5′ CGAGCTCGTAAACATCAATACAAGCACAC 3′) which have been tailed with BamH I and Sac I restriction site at the 5′ end respectively. The amplified products were digested and linked into the plant binary vector pBI121 to produce the SlPRE2-overexpression vector pBI121-SlPRE2 driven by the CaMV 35 S promoter. After that, the pBI121-SlPRE2 vector was transferred into tomato cotyledon explants through Agrobacterium-mediated transformation method as described by Chen et al.48. The transgenic plants were selected on kanamycin medium and detected by PCR with primers NPT II-F (5′ GACAATCGGCTGCTCTGA 3′) and NPT II-R (5′ AACTCCAGCATGAGATCC 3′).
Leaf rolling index and leaf angle measurements
The measurements of leaf rolling index and leaf angle were taken during the 9:00 to 11:00 AM using ruler and protractor respectively. The natural width (Ln) and the greatest width (Lg) of the leaf blade were determined. Leaf rolling index was calculated according to the following equation49: Leaf rolling index (%) = (Lg − Ln)/Lg × 100. Leaf angle was determined as the adaxial angle between a stem and a leaf 50.
Chlorophyll and carotenoid analysis
Chlorophyll contents were measured in the expanded leaves (mature leaves), IMG (25DPA), MG and B fruits. Tissues from the IMG, MG and B fruits were divided into five latitudinal sections along the vertical axis, and the stem end, middle part and stylar end were selected to determine the chlorophyll contents respectively35. The selected tissues (~0.5 g) were ground and dissolved in 10 mL of 80% acetone. The extract was centrifuged at 1500 g for 10 min, then the supernatant was transferred to a new lightproof tube, and its absorbance was measured at 645 nm and 663 nm with a spectrophotometer (lambda 900 UV/VIS/NIR, Perkin Elmer). Total chlorophyll contents were calculated with the following equation: total chlorophyll (mg/l FW) = 8.02A663 + 20.2A64551, 52. Carotenoids were measured in the B + 4 and B + 7 fruit. The selected tissues (~1.0 g) were ground and dissolved in 10 mL of 60:40 (v/v) hexane: acetone. The extract was centrifuged at 4000 g for 5 min and the absorbance of the supernatant was measured at 450 nm with a spectrophotometer. Total carotenoid contents were calculated with the following equation: total carotenoids (mg/100 g FW) = 4 × A450 × 1043, 53. For lycopene quantitation, 0.25 g pericarps from the B + 4 and B + 7 fruits were ground and dissolved in 8 mL of 2:1:1 (v/v/v) hexane: ethanol: acetone. The extract was shaken for 30 min then 1 mL distilled water was added and shaken for 5 minutes. The extracted solution was left for 15 min to separate into two phases (polar upper layer and nonpolar layer). The absorbance of the supernatant was measured at 503 nm by spectrophotometer. Lycopene contents were quantified using a molar extinction coefficient of 1.585 × 105 M−1 cm−1 54.
Transmission electron microscopy and scanning electron microscopy
Outer pericarp tissues from MG fruits were selected and fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer for 4 h and washed three times in the 0.1 M phosphate buffer for 15 min. Then tissues were postfixed in 1% OsO4 at 4 °C for 1 hour, and dehydrated in an ethanol series and infiltrated in spur resin. Ultrathin sections were viewed with Hitachi H-7500 transmission electron microscope. For analysis of stomatal characteristics, mature leaves of wild type and transgenic plants were fixed in 2.5% glutaraldehyde. After being fixed for 3 hours, leaf tissues were dehydrated through an ethanol series. Then the samples were dried and coated with gold for observation using Hitachi scanning electron microscope S-3000N. The stomatal size was measured using Image J software.
Statistical analysis
Data were analyzed by one-way analysis of variance (ANOVA) and the t-test. Differences were considered to be significant at a level of p < 0.05. The measurement values were displayed as means with standard deviation (SD) of three biological replicates.
Electronic supplementary material
Acknowledgements
This work was supported by National Natural Science Foundation of China (no. 30600044, 31572129), and the Natural Science Foundation of Chongqing of China (cstc2015jcyjA80026), and Chongqing University Postgraduates’ Innovation Project (CYB15027).
Author Contributions
Z.Z., Z.H., and G.C. designed the experiment. Z.Z. and W.Y. performed the research. Z.Z. and X.Y. carried out the gene clone and plasmid construction. Z.Z., X.G., and J.H. analyzed data. Z.Z., Z.H., and G.C. wrote the article. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04092-y
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Littlewood TD, Evan GI. Transcription factors 2: helix-loop-helix. Protein profile. 1995;2:621. [PubMed] [Google Scholar]
- 2.Seo PJ, Hong SY, Kim SG, Park CM. Competitive inhibition of transcription factors by small interfering peptides. Trends Plant Sci. 2011;16:541–549. doi: 10.1016/j.tplants.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Jones S. An overview of the basic helix-loop-helix proteins. Genome Biology. 2004;5:226–226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. plant physiology. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Li X, Li K, Liu H, Lin C. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 2013;9:e1003861. doi: 10.1371/journal.pgen.1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi Y, et al. bHLH transcription factors that facilitate K+ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci. Signal. 2013;6:ra48–ra48. doi: 10.1126/scisignal.2003760. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L-L, Shi M-Z, Xie D-Y. Regulation of anthocyanin biosynthesis by nitrogen in TTG1–GL3/TT8–PAP1-programmed red cells of Arabidopsis thaliana. Planta. 2012;236:825–837. doi: 10.1007/s00425-012-1674-2. [DOI] [PubMed] [Google Scholar]
- 8.Tominaga-Wada R, Nukumizu Y, Wada T. Tomato (Solanum lycopersicum) homologs of TRIPTYCHON (SlTRY) and GLABRA3 (SlGL3) are involved in anthocyanin accumulation. Plant signaling & behavior. 2013;8:e24575. doi: 10.4161/psb.24575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groszmann M, Paicu T, Smyth DR. Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis. The Plant Journal. 2008;55:40–52. doi: 10.1111/j.1365-313X.2008.03469.x. [DOI] [PubMed] [Google Scholar]
- 10.Feng H-L, et al. A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant physiology and biochemistry. 2013;73:309–320. doi: 10.1016/j.plaphy.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Zhang, Y., Liu, Z., Chen, Y., He, J. & Bi, Y. PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Science (2015). [DOI] [PubMed]
- 12.Llorente, B. et al. Tomato fruit carotenoid biosynthesis is adjusted to actual ripening progression by a light-dependent mechanism. The Plant Journal, n/a-n/a, doi:10.1111/tpj.13094 (2015). [DOI] [PubMed]
- 13.Dong Y, et al. A novel bHLH transcription factor PebHLH35 from Populus euphratica confers drought tolerance through regulating stomatal development, photosynthesis and growth in Arabidopsis. Biochem Biophys Res Commun. 2014;450:453–458. doi: 10.1016/j.bbrc.2014.05.139. [DOI] [PubMed] [Google Scholar]
- 14.Bai MY, Fan M, Oh E, Wang ZY. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell. 2012;24:4917–4929. doi: 10.1105/tpc.112.105163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malinovsky FG, et al. Antagonistic regulation of growth and immunity by the Arabidopsis basic helix-loop-helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bHLH1. Plant Physiol. 2014;164:1443–1455. doi: 10.1104/pp.113.234625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. Embo Journal. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohashi-Ito, K., Matsukawa, M. & Fukuda, H. An atypical bHLH transcription factor regulates early xylem development downstream of auxin. Plant and Cell Physiology, pct013 (2013). [DOI] [PubMed]
- 18.Heang D, Sassa H. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS One. 2012;7:e31325. doi: 10.1371/journal.pone.0031325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, et al. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:591–600. doi: 10.1093/pcp/pcj026. [DOI] [PubMed] [Google Scholar]
- 20.Hyun Y, Lee I. KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol. 2006;61:283–296. doi: 10.1007/s11103-006-0010-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhang LY, et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao Y, Oh E, Choi G, Liang Z, Wang ZY. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol Plant. 2012;5:688–697. doi: 10.1093/mp/sss011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castelain M, Le Hir R, Bellini C. The non-DNA-binding bHLH transcription factor PRE3/bHLH135/ATBS1/TMO7 is involved in the regulation of light signaling pathway in Arabidopsis. Physiol Plant. 2012;145:450–460. doi: 10.1111/j.1399-3054.2012.01600.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Zhu Y, Fujioka S, Asami T, Li J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell. 2009;21:3781–3791. doi: 10.1105/tpc.109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mara CD, Huang T, Irish VF. The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell. 2010;22:690–702. doi: 10.1105/tpc.109.065946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong SY, et al. A competitive peptide inhibitor KIDARI negatively regulates HFR1 by forming nonfunctional heterodimers in Arabidopsis photomorphogenesis. Mol Cells. 2013;35:25–31. doi: 10.1007/s10059-013-2159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka A, et al. BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 2009;151:669–680. doi: 10.1104/pp.109.140806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heang D, Sassa H. An atypical bHLH protein encoded by POSITIVE REGULATOR OF GRAIN LENGTH 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breeding science. 2012;62:133. doi: 10.1270/jsbbs.62.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen KY, Cong B, Wing R, Vrebalov J, Tanksley SD. Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science. 2007;318:643–645. doi: 10.1126/science.1148428. [DOI] [PubMed] [Google Scholar]
- 31.Sun H, Fan HJ, Ling HQ. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics. 2015;16:9. doi: 10.1186/s12864-014-1209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bohnert HJ, Nelson DE, Jensen RG. Adaptations to environmental stresses. The plant cell. 1995;7:1099. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 34.Liu YS, et al. Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9897–9902. doi: 10.1073/pnas.0400935101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen CV, et al. Tomato GOLDEN2-LIKE Transcription Factors Reveal Molecular Gradients That Function during Fruit Development and Ripening. Plant Cell. 2014;26:585–601. doi: 10.1105/tpc.113.118794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadioglu A, Terzi R. A dehydration avoidance mechanism: leaf rolling. The Botanical Review. 2007;73:290–302. doi: 10.1663/0006-8101(2007)73[290:ADAMLR]2.0.CO;2. [DOI] [Google Scholar]
- 37.Abd Allah AA. Genetic studies on leaf rolling and some root traits under drought conditions in rice (Oryza sativa L.) African Journal of Biotechnology. 2009;8:6241–6248. [Google Scholar]
- 38.Bongers FJ, Evers JB, Anten NP, Pierik R. From shade avoidance responses to plant performance at vegetation level: using virtual plant modelling as a tool. New Phytologist. 2014;204:268–272. doi: 10.1111/nph.13041. [DOI] [PubMed] [Google Scholar]
- 39.Keuskamp DH, Sasidharan R, Pierik R. Physiological regulation and functional significance of shade avoidance responses to neighbors. Plant signaling & behavior. 2010;5:655–662. doi: 10.4161/psb.5.6.11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powell ALT, et al. Uniform ripening Encodes a Golden 2-like Transcription Factor Regulating Tomato Fruit Chloroplast Development. Science. 2012;336:1711–1715. doi: 10.1126/science.1222218. [DOI] [PubMed] [Google Scholar]
- 41.Wanner LA, Gruissem W. Expression dynamics of the tomato rbcS gene family during development. The Plant Cell. 1991;3:1289–1303. doi: 10.1105/tpc.3.12.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffman NE, et al. A cDNA clone encoding a photosystem I protein with homology to photosystem II chlorophyll a/b-binding polypeptides. Proceedings of the National Academy of Sciences. 1987;84:8844–8848. doi: 10.1073/pnas.84.24.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fray RG, Grierson D. Identification and Genetic-Analysis of Normal and Mutant Phytoene Synthase Genes of Tomato by Sequencing, Complementation and Co-Suppression. Plant Molecular Biology. 1993;22:589–602. doi: 10.1007/BF00047400. [DOI] [PubMed] [Google Scholar]
- 44.Mann V, Pecker I, Hirschberg J. Cloning and characterization of the gene for phytoene desaturase (Pds) from tomato (Lycopersicon esculentum) Plant molecular biology. 1994;24:429–434. doi: 10.1007/BF00024111. [DOI] [PubMed] [Google Scholar]
- 45.Pecker I, Chamovitz D, Linden H, Sandmann G, Hirschberg J. A single polypeptide catalyzing the conversion of phytoene to zeta-carotene is transcriptionally regulated during tomato fruit ripening. Proceedings of the National Academy of Sciences. 1992;89:4962–4966. doi: 10.1073/pnas.89.11.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Exposito-Rodriguez, M., Borges, A. A., Borges-Perez, A. & Perez, J. A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. Bmc Plant Biology8 (2008). [DOI] [PMC free article] [PubMed]
- 47.Nicot N, Hausman JF, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany. 2005;56:2907–2914. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- 48.Chen GP, et al. Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. plant physiology. 2004;136:2641–2651. doi: 10.1104/pp.104.041608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi Z, et al. Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta. 2007;226:99–108. doi: 10.1007/s00425-006-0472-0. [DOI] [PubMed] [Google Scholar]
- 50.Li P-F, et al. Dryland Wheat Domestication Changed the Development of Aboveground Architecture for a Well-Structured Canopy. PloS one. 2014;9:e95825. doi: 10.1371/journal.pone.0095825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. plant physiology. 1949;24:1. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porra R, Thompson W, Kriedemann P. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 1989;975:384–394. doi: 10.1016/S0005-2728(89)80347-0. [DOI] [Google Scholar]
- 53.Forth D, Pyke KA. The suffulta mutation in tomato reveals a novel method of plastid replication during fruit ripening. J Exp Bot. 2006;57:1971–1979. doi: 10.1093/jxb/erj144. [DOI] [PubMed] [Google Scholar]
- 54.Lavecchia R, Zuorro A. Improved lycopene extraction from tomato peels using cell-wall degrading enzymes. European Food Research and Technology. 2008;228:153–158. doi: 10.1007/s00217-008-0897-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


