Abstract
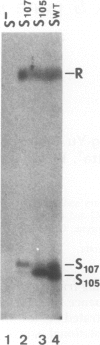
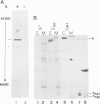
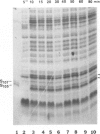
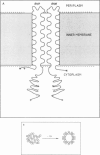
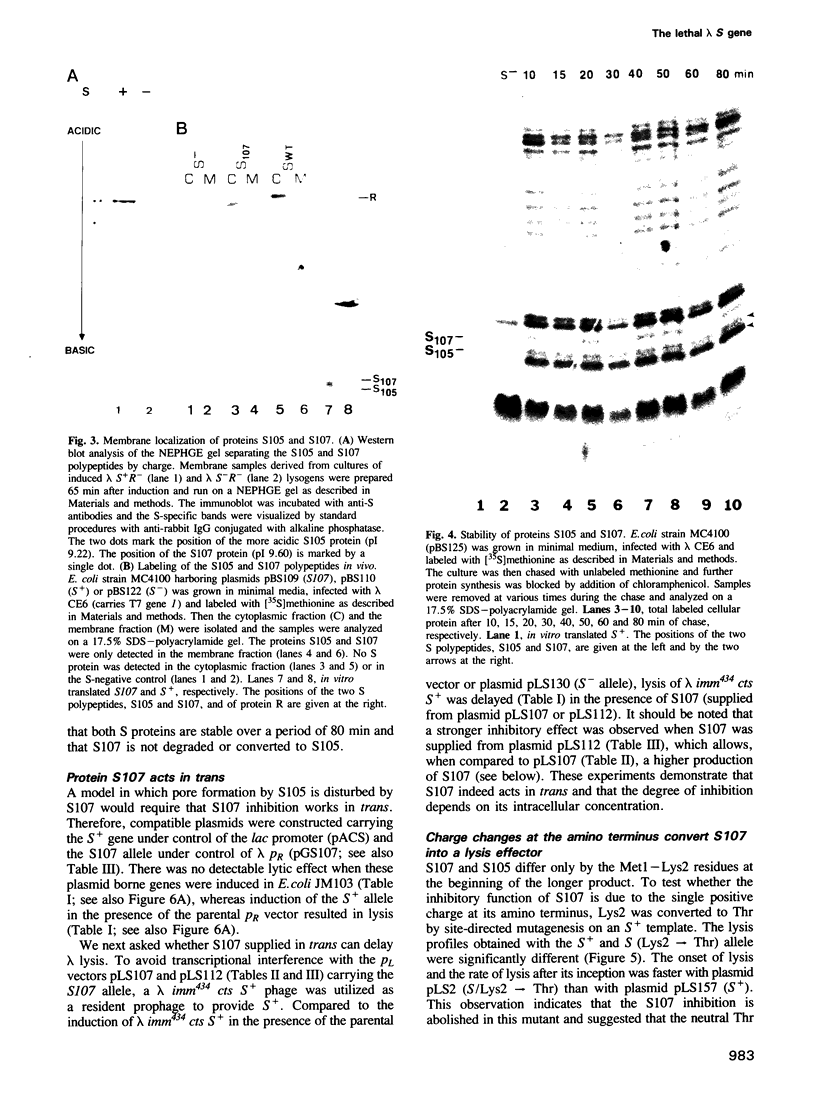
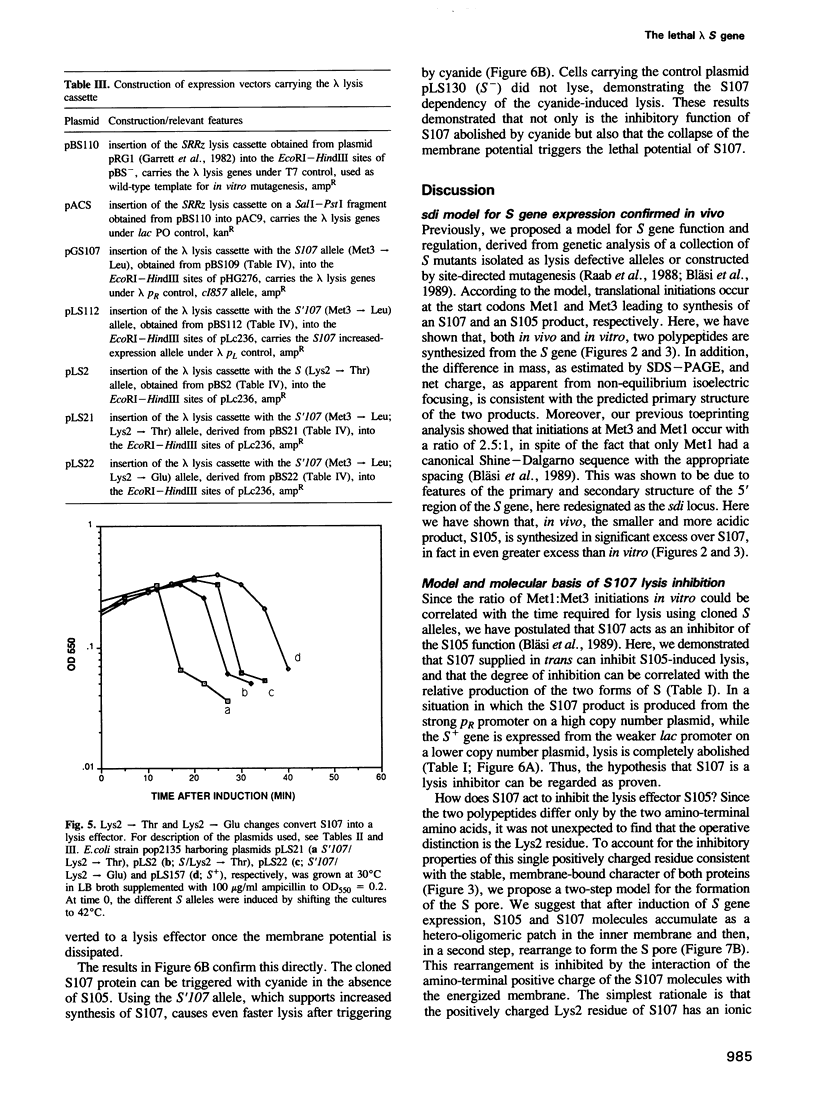
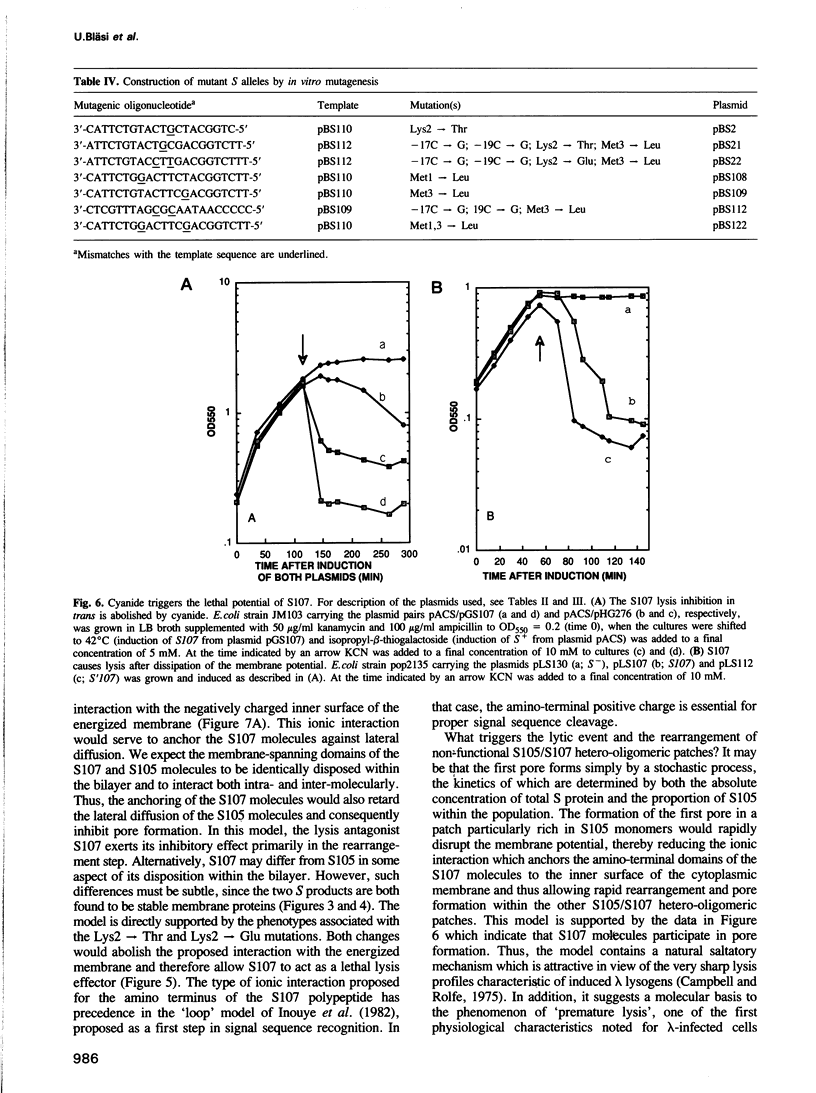
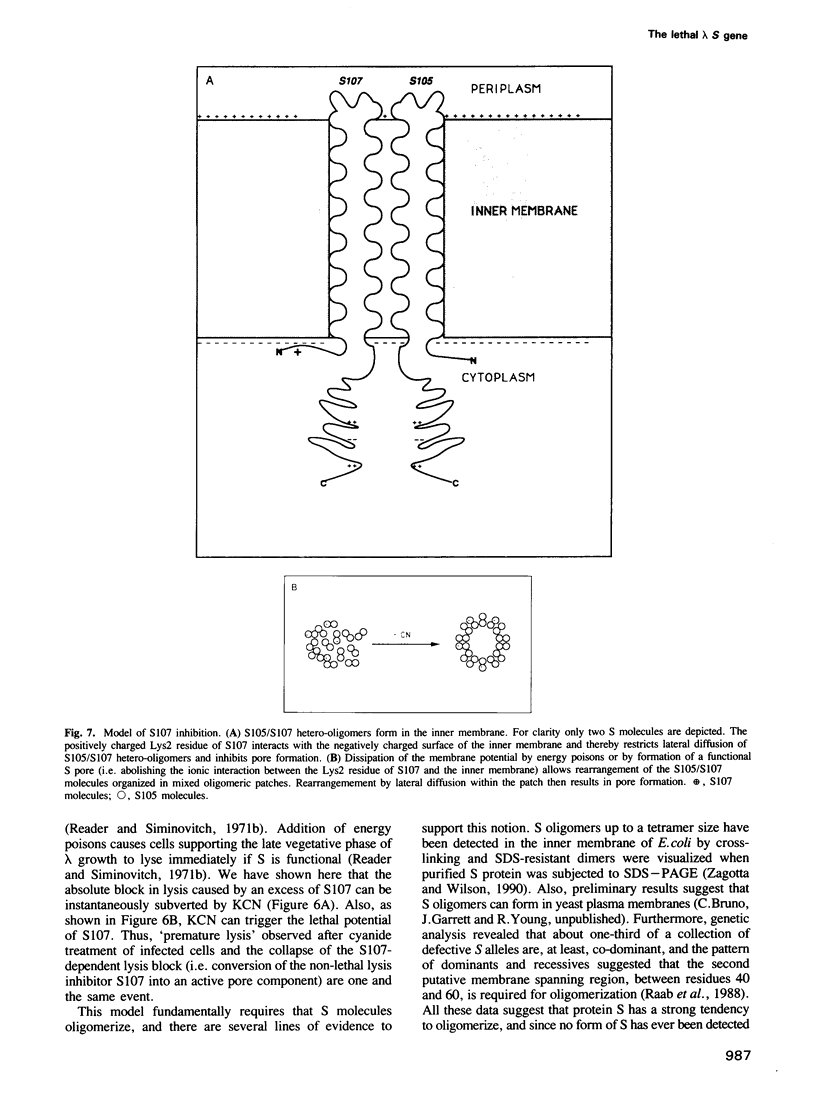
The 107 codon reading frame of the lambda lysis gene S begins with the codon sequence Met1-Lys2-Met3..., and it has been demonstrated in vitro that both Met codons are used for translational starts. Furthermore, the partition of initiation events at the two start codons strongly affects the scheduling of lysis. We have presented a model in which the longer product, S107, acts as an inhibitor of the shorter product, S105, the lethal lysis effector, despite the fact that the two molecules differ only in the Met-Lys residues at the amino terminus of S107. Using immunological and biochemical methods, we show in this report that the two predicted protein products, S105 and S107, are detectable in vivo as stable, membrane-bound molecules. We show that S107 acts as an inhibitor in trans, and that its inhibitory function is entirely defined by the positively charged Lys2 residue. Moreover, our data show that energy poisons abolish the inhibitory function of S107 and simultaneously convert S107 into a lysis effector. We propose a two step model for the lethal action of gene S: first, induction of the S gene results in the accumulation of S105 and S107 molecules in mixed oligomeric patches in the cytoplasmic membrane; second, S monomers rearrange by lateral diffusion within the patch to form an aqueous pore. The R gene product, a transglycosylase, is released through the pore to the periplasm, resulting in destruction of the peptidoglycan and bursting of the cell. According to this model, the lateral diffusion step is inhibited by the energized state of the membrane.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman E., Altman R. K., Garrett J. M., Grimaila R. J., Young R. S gene product: identification and membrane localization of a lysis control protein. J Bacteriol. 1983 Sep;155(3):1130–1137. doi: 10.1128/jb.155.3.1130-1137.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman E., Young K., Garrett J., Altman R., Young R. Subcellular localization of lethal lysis proteins of bacteriophages lambda and phiX174. J Virol. 1985 Mar;53(3):1008–1011. doi: 10.1128/jvi.53.3.1008-1011.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowska-Szewczyk K., Lipinska B., Taylor A. The R gene product of bacteriophage lambda is the murein transglycosylase. Mol Gen Genet. 1981;184(1):111–114. doi: 10.1007/BF00271205. [DOI] [PubMed] [Google Scholar]
- Bläsi U., Nam K., Hartz D., Gold L., Young R. Dual translational initiation sites control function of the lambda S gene. EMBO J. 1989 Nov;8(11):3501–3510. doi: 10.1002/j.1460-2075.1989.tb08515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. H., Rolfe B. G. Evidence for a dual control of the initiation of host-cell lysis caused by phage lambda. Mol Gen Genet. 1975 Aug 5;139(1):1–8. doi: 10.1007/BF00267990. [DOI] [PubMed] [Google Scholar]
- Fling S. P., Gregerson D. S. Peptide and protein molecular weight determination by electrophoresis using a high-molarity tris buffer system without urea. Anal Biochem. 1986 May 15;155(1):83–88. doi: 10.1016/0003-2697(86)90228-9. [DOI] [PubMed] [Google Scholar]
- Garrett J. M., Young R. Lethal action of bacteriophage lambda S gene. J Virol. 1982 Dec;44(3):886–892. doi: 10.1128/jvi.44.3.886-892.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett J., Fusselman R., Hise J., Chiou L., Smith-Grillo D., Schulz J., Young R. Cell lysis by induction of cloned lambda lysis genes. Mol Gen Genet. 1981;182(2):326–331. doi: 10.1007/BF00269678. [DOI] [PubMed] [Google Scholar]
- Inouye S., Soberon X., Franceschini T., Nakamura K., Itakura K., Inouye M. Role of positive charge on the amino-terminal region of the signal peptide in protein secretion across the membrane. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3438–3441. doi: 10.1073/pnas.79.11.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. Z., Goodman H. M., O'Farrell P. H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977 Dec;12(4):1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- Raab R., Neal G., Garrett J., Grimaila R., Fusselman R., Young R. Mutational analysis of bacteriophage lambda lysis gene S. J Bacteriol. 1986 Sep;167(3):1035–1042. doi: 10.1128/jb.167.3.1035-1042.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab R., Neal G., Sohaskey C., Smith J., Young R. Dominance in lambda S mutations and evidence for translational control. J Mol Biol. 1988 Jan 5;199(1):95–105. doi: 10.1016/0022-2836(88)90381-6. [DOI] [PubMed] [Google Scholar]
- Reader R. W., Siminovitch L. Lysis defective mutants of bacteriophage lambda: genetics and physiology of S cistron mutants. Virology. 1971 Mar;43(3):607–622. doi: 10.1016/0042-6822(71)90286-8. [DOI] [PubMed] [Google Scholar]
- Reader R. W., Siminovitch L. Lysis defective mutants of bacteriophage lambda: on the role of the S function in lysis. Virology. 1971 Mar;43(3):623–637. doi: 10.1016/0042-6822(71)90287-x. [DOI] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Stewart G. S., Lubinsky-Mink S., Kuhn J. pHG276: a multiple cloning site pBR322 copy number vector expressing a functional lac alpha peptide from the bacteriophage lambda PR promoter. Plasmid. 1986 May;15(3):182–190. doi: 10.1016/0147-619x(86)90036-3. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]