Abstract
Background:
Chronic rhinosinusitis (CRS) is prevalent, morbid, and poorly understood. Extraction of electronic health record (EHR) data of patients with CRS may facilitate research on CRS. However, the accuracy of using structured billing codes for EHR-driven phenotyping of CRS is unknown. We sought to accurately identify CRS cases and controls using EHR data and to determine the accuracy of structured billing codes for identifying patients with CRS.
Methods:
We developed and validated distinct algorithms to identify patients with CRS and controls using International Classification of Diseases, Ninth Revision (ICD-9) and Current Procedural Terminology codes. We used blinded clinician chart review as the reference standard to evaluate algorithm and billing code accuracy.
Results:
Our initial control algorithm achieved a control positive predictive value (PPV) of 100% (i.e., negative predictive value of 100% for CRS). Our initial algorithm for CRS cases relied exclusively on billing codes and had a low case PPV (54%). Notably, ICD-9 code 471.x was associated with a case PPV of 85%, whereas the case PPV of ICD-9 code 473.x was only 34%. After multiple algorithm iterations, we increased the case PPV of our final algorithm to 91% by adding several requirements, e.g., that ICD-9 codes occur with 1 or more evaluations by a CRS specialist to enhance availability of objective clinical data for accurately phenotyping CRS.
Conclusion:
These algorithms are an important first step to identify patients with CRS, and may facilitate EHR-based research on CRS pathogenesis, morbidity, and management. Exclusive use of coded data for phenotyping CRS has limited accuracy, especially because CRS symptomatology overlaps with that of other illnesses. Incorporating natural language processing (e.g., to evaluate results of nasal endoscopy or sinus computed tomography) into future work may increase algorithm accuracy and identify patients whose disease status may not be ascertained by only using billing codes.
Keywords: Accuracy, billing code, chronic rhinosinusitis, Current Procedural Terminology, electronic health record, ICD-9, natural language processing, phenotyping, predictive value, sinusitis
Chronic rhinosinusitis (CRS) is characterized by persistent inflammation of the nasal and paranasal sinus mucosa, although its pathogenesis remains uncertain.1–4 Diagnosis of CRS requires indicative symptoms (e.g., nasal congestion or discharge) for ≥12 weeks' duration and objective confirmation by either nasal endoscopy or computed tomography (CT) of the sinuses, because CRS symptomatology overlaps with that of other upper airway disorders (including allergic and nonallergic rhinitis).1–3,5 CRS is divided into two types: CRS with nasal polyposis (CRSwNP) occurs in 20–30% of patients; however, most patients with CRS lack nasal polyps (CRSsNP).1,6 CRS is often associated with other airway diseases, including allergic rhinitis and/or asthma.1,2,6 Infrequently, CRS may occur as part of a multiorgan disease (e.g., cystic fibrosis, mucociliary dysfunction, or primary immunodeficiency1,2,6); in this setting, sinonasal inflammation is usually more severe than in single-organ disease.1,2,6
The prevalence of CRS is uncertain, but has been estimated as 13% of the U.S. population7—by this estimate, CRS was the second most common chronic condition in the United States.7,8 The burden of CRS is substantial: lower quality of life has been reported for CRS compared with heart disease or back pain,9 the annual costs of CRS exceed $8 billion in the United States (not including lost work or school days), and all major race/ethnic groups in the United States are affected.8,10,11 Few effective medical therapies for CRS are known; antibiotics are often prescribed but are of unproven benefit.1,2,12 Surgery is considered for recalcitrant cases, but postoperative recurrent disease is well described.1,8
Accurate identification of patients with CRS in the electronic health record (EHR) may accelerate understanding of the prevalence, pathophysiology, morbidity, and management of CRS, by combining EHR data with tools from epidemiology, bioinformatics, and health care quality research. To our knowledge, a validated method for identifying CRS in the EHR is not available. As part of a project to use EHR search algorithms to identify patients with CRS and controls in the NUgene Project13 (a biobank of DNA samples linked to EHR data at Northwestern University) and to investigate genetic determinants of CRS, the objective of this work was to develop and validate accurate algorithms to select patients with CRS and controls without CRS. We used structured billing codes to generate these EHR search algorithms, and we refined the algorithms using blinded chart review as the reference standard.
MATERIALS AND METHODS
This study was conducted using EHR data from the Northwestern Medicine Enterprise Data Warehouse (EDW).14 The EDW is an integrated repository of clinical and biomedical research data sources at Northwestern.14,15 Data for this study were accessed between March 1, 2012 and February 6, 2013, after obtaining approval from the Northwestern Feinberg School of Medicine Institutional Review Board with a waiver of informed consent.
The study patient population included all patients with two or more office visits in the EDW between January 20, 1989 and February 6, 2013. Patients with only one visit in the EDW were excluded from this study because of concern that clinical information from the extant visit record might lack sufficient detail to determine phenotype (e.g., if a patient's only visit was for venipuncture or radiography).
Distinct EHR search algorithms were developed to identify patients with CRS (“cases”) or without CRS (“controls”) using International Classification of Diseases, Ninth Revision (ICD-9) diagnosis codes and Current Procedural Terminology (CPT) codes.16,17 The initial algorithm to identify CRS cases (Fig. 1) relied exclusively on CRS-related billing codes. Specifically, the initial CRS case algorithm selected patients with one or more clinical encounter (outpatient, inpatient, or emergency department) associated with any of the following: ICD-9 code for CRSwNP (471.x), CRS (473.x, which does not specify CRSwNP or CRSsNP), or CPT code for sinus surgery for CRS (Table 1).
Figure 1.
Initial Algorithm for the identification of subjects with CRS and controls without CRS. *Office visit includes outpatient scheduled office visits and urgent office visits. CRS, chronic rhinosinusitis; Dx, diagnosis; CT, computed tomography.
Table 1.
List of ICD-9 diagnosis and CPT codes used in the algorithm
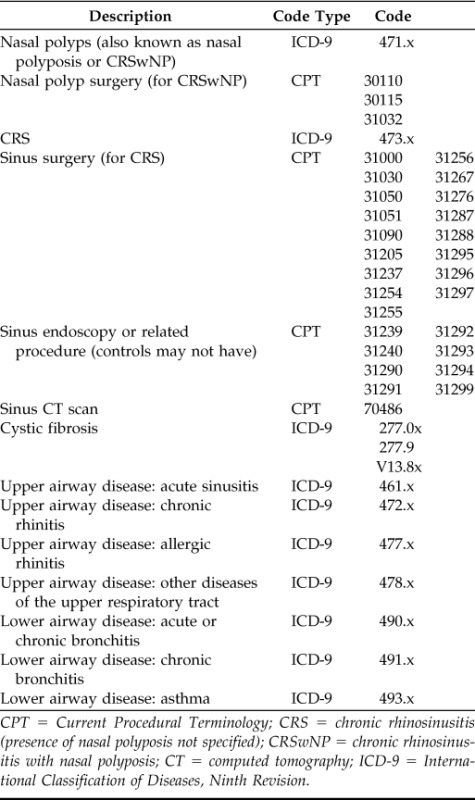
CPT = Current Procedural Terminology; CRS = chronic rhinosinusitis (presence of nasal polyposis not specified); CRSwNP = chronic rhinosinusitis with nasal polyposis; CT = computed tomography; ICD-9 = International Classification of Diseases, Ninth Revision.
The initial algorithm for controls excluded patients if their EHR contained any of the following: (1) any mention of CRS (including CRSwNP) by ICD-9 or CPT code, (2) order(s) for one or more sinus CT (regardless of whether CT was performed, because we assumed any order for sinus CT was prompted by active sinonasal symptoms that might signify CRS), or (3) ICD-9 codes associated with chronic upper or lower airway diseases (Fig. 1; Table 1). The latter two exclusion criteria were measures to decrease the likelihood of selecting patients with undiagnosed CRS. Chronic lower airway diseases were excluded in this control algorithm because lower airway dysfunction (e.g., asthma) often coincides with CRS.1,2,6
The reference standard to evaluate the algorithms was blinded clinician chart review. Two clinicians (J.H. and W.W.S.) reviewed charts of patients randomly selected by the algorithm. During chart review, patients were classified as CRS cases or not based on subjective and objective criteria, following guidelines for diagnosis of CRS.1,3 Patients were considered CRS cases if their charts contained evidence of CRS symptoms (e.g., nasal congestion or drainage) plus objective disease (e.g., pus or polyps during nasal endoscopy or sinus mucosal thickening, opacification, or air–fluid levels on CT). Operative notes for endoscopic sinus surgery (which uses nasal endoscopy, by definition) and office notes referencing prior sinus surgery were also used to adjudicate phenotype. If discrepancies arose between physicians' charted assessments and objective data (e.g., CRS appeared in a physician's clinical assessment but sinus CT results were normal), objective data prevailed.
Clinician-blinded review of randomly selected charts served as the reference standard to evaluate algorithm positive predictive value (PPV) and negative predictive value (NPV). Statistical analysis was performed using Stata/SE 10.0 (StataCorp. LP, College Station, TX).
RESULTS
In this study, 996 charts were randomly selected and reviewed. One-quarter of these charts were randomly sampled to evaluate interobserver concordance, which was found to be 92%. Interobserver discordance was more likely to occur when objective evidence for CRS was embedded within physicians' written charts and not accessible from the “Imaging Results” section of the EHR (e.g., a CT scan from an outside facility may not have been found in the EHR section for imaging results, despite a written description of those results in a physician's note).
Our initial algorithm for controls was associated with a PPV of 100% for control subjects (i.e., NPV for CRS cases was 100%). In contrast, the CRS case PPV for our initial case algorithm was 54% ([136 true cases + 114 not cases]/250 predicted cases = 0.54; i.e., control NPV was 54%).
The largest source of error in our initial CRS case algorithm was the inability to classify patients with CRS because of absent objective data for distinguishing between CRS and other diseases. Of patients inaccurately selected by the initial CRS case algorithm, 88% had never been evaluated by a CRS specialist (i.e., otolaryngologist or allergist–immunologist at our center), but had received a presumptive diagnosis of CRS during generalist evaluation (and without CT or endoscopic examination). Patients who received CRS specialty care but were incorrectly identified as CRS cases often had EHRs that contained a CRS-associated ICD-9 code in one or more specialist visit(s) as a presumptive diagnosis, but these patients ultimately received alternate diagnoses after objective data were obtained (e.g., recurrent acute rhinosinusitis [three or more episodes of acute rhinosinusitis annually without intervening symptoms or objective abnormality], subacute rhinosinusitis [lasting >4 weeks but <12 weeks], nasal turbinate hypertrophy, deviated nasal septum, antrochoanal polyp, malignancy, glossopharyngeal neuralgia, oral antral fistula, or undifferentiated nasal lesion).
After multiple iterations of algorithm programming and chart review, we improved the accuracy of our CRS case algorithm (Table 2). For our final CRS case algorithm, PPV for CRS cases was 91% (i.e., NPV for controls was 91%). Our final CRS case algorithm contained several additional criteria (Fig. 2). We required specialty evaluation (i.e., otolaryngology or allergy–immunology) to increase the likelihood of available objective EHR data to confirm a diagnosis of CRS. Although unrestricted use of ICD-9 code 471.x identified CRS cases with fair accuracy (case PPV 85%), we often found ICD-9 code 471.1 (“polypoid sinus degeneration”) was associated with alternative diagnoses (e.g., antrochoanal polyp or undifferentiated nasal lesion), so 471.1 was excluded as a sufficient criterion to select for CRS. ICD-9 code 473.x was very inaccurate when used as the sole criterion to identify CRS (PPV 34%) because it often appeared as a working diagnosis; we discovered the accuracy of 473.x greatly improved when 473.x diagnoses were associated with two or more office visits in otolaryngology, because this pattern was most likely to indicate objective confirmation of CRS phenotype (i.e., by endoscopy and/or CT).
Table 2.
PPV of the final algorithm for CRS cases and controls
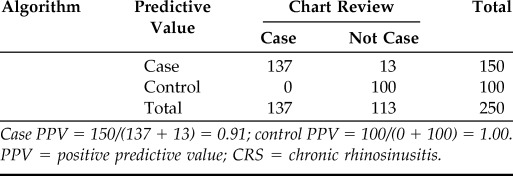
Case PPV = 150/(137 + 13) = 0.91; control PPV = 100/(0 + 100) = 1.00.
PPV = positive predictive value; CRS = chronic rhinosinusitis.
Figure 2.
Final algorithm for the identification of subjects with CRS and controls without CRS. *Office visit includes outpatient scheduled office visits and urgent office visits; **with otolaryngologist. 1,2,3All case diagnoses were entered by clinicians during encounters, including scheduled office visits, urgent office visits, emergency department visits, surgeries, and inpatient hospitalizations. The specific case diagnoses are as follows: 1Nasal polyp (i.e., CRSwNP) Dx: ICD-9 diagnosis codes 471.0, 471.8, or 471.9. 2Nasal polyp (i.e., CRSwNP) or CRS Dx: ICD-9 diagnosis codes 471.x or 473.x. 3CRS Dx: ICD-9 diagnosis code 473.x. See Table 1 for remaining ICD-9 diagnosis and CPT codes used in the remainder of the algorithm. Dx, diagnosis; CRS, chronic rhinosinusitis (presence of nasal polyposis not specified); CT, computed tomography; CRSwNP, CRS with nasal polyposis; ICD-9, International Classification of Diseases, Ninth Revision; CPT, Current Procedural Terminology.
We also revised the CPT codes used in our final CRS case algorithm. After surveying our center's otolaryngologists about their coding practices during patient care, we removed CPT codes nonspecific for CRS (31239, 31240, 31290, 31291, 31292, 31293, 31294, and 31299; Table 1) and added others used by these physicians specifically for patients with CRS (31295, 31296, 31297, 31000, 31030, 31050, 31051, 31205, and 31090). Finally, our final algorithm for CRS cases and controls excluded patients with any ICD-9 code associated with cystic fibrosis, because we intended this algorithm for the study of CRS independent of known monogenic diseases (e.g., cystic fibrosis).
Finally, we applied our final algorithms to the EDW to estimate the number of patients with and without CRS at our center. Out of 799,682 patients with two or more office visits in the EDW, our final algorithms identified 5983 CRS cases and 593,602 controls. These algorithms were unable to determine disease status (CRS case versus control) for 25% of this EDW population (n = 200,097).
DISCUSSION
In this study, we developed and tested the accuracy of EHR search algorithms to identify patients with or without CRS at our center, using blinded clinician chart review as the reference standard. Our initial algorithm for CRS cases had poor accuracy for identifying CRS, but after multiple algorithm iterations, our final CRS case algorithm PPV was >90%. Our algorithm to select patients without CRS was highly accurate (control PPV = 100%).
Although claims-derived, electronic diagnosis of acute upper respiratory illness has been associated with >95% sensitivity and specificity,18 much less is known regarding chronic upper airway disease, despite its high prevalence and antibiotic use.1,2 Prior work by Bhattarchayya et al.10 used ICD-9 and CPT codes to identify patients with CRS in a national insurance claims database. Our results agree with their finding that unfettered use of coding data was nonspecific for identifying CRS and thus overestimated CRS prevalence.10 These investigators found CRS prevalence decreased from 20 to 2.3% when their algorithms required orders for objective testing: they stipulated patients could only be classified with CRS if their EHR included CPT coding affiliated with sinus CT or endoscopy.10 Requiring CPT codes associated with objective testing was a strength of this study. However, these algorithms did not distinguish between results that were normal and results consistent with CRS; furthermore, a reference standard to evaluate algorithm accuracy was not performed.10 For example, if a patient met ICD-9 criteria for CRS, presence of CPT coding for sinus CT would have classified that patient with CRS even if CT results were negative for disease.
Our study is not the first to highlight the limitations of problem list terminologies,19 but we believe this work makes several contributions. To our knowledge, our study was the first to evaluate the accuracy of ICD-9 and CPT codes for CRS using a reference standard. We refined our EHR search algorithms through modification and iteration and, consequently, improved algorithm accuracy for identifying patients with CRS. We evaluated the accuracy of CRS-associated ICD-9 codes (i.e., 471.x and 473.x) and discovered the specificity of algorithm criteria varied by ICD-9 code, with 473.x requiring the most restrictive specifications to achieve reasonable accuracy. Our algorithm to identify patients without CRS was very accurate, which may be valuable for secondary uses of EHR data (e.g., epidemiology and bioinformatics) that require well-phenotyped control subjects.
Our study was not without limitations. Currently, our algorithms are center-specific, and our final CRS algorithm requirement for specialist evaluation may limit generalizability. Variation in clinical use of ICD-9 and CPT codes by center, specialty, and individual may further decrease algorithm generalizability. Our algorithms must be updated for use with ICD-10 codes. At this time, our CRS algorithm does not contain temporal criteria for the diagnosis of CRS (i.e., ≥12 weeks of symptoms)1–3; this could be incorporated into future work. We were unable to evaluate algorithm sensitivity and specificity because resource constraints precluded us from randomly selecting a new set of charts from the EHR to reapply our algorithms. Last but not least, these algorithms would benefit from addition of natural language processing (NLP) to evaluate results of objective tests for CRS (e.g., sinus CT findings, either in an EHR's Results section or in the written physician's chart) and clinical assessments by CRS specialists (e.g., in encounter note assessment sections). NLP may help determine presence of CRS in patients whose disease status can not be ascertained solely via billing codes. Finally, NLP could be very useful when the number of potential cases is too large for manual chart review to be practical. For our genetics study, because the number of genotyped patients identified with our final CRS algorithm was ∼200, we manually reviewed encounter notes and sinus CT results for CRS cases that were identified using ICD-9 code 473.x. This took two reviewers a total of 40 hours for roughly 200 subjects, so manual chart review would not scale for a larger study.
CONCLUSION
In conclusion, we developed and validated accurate algorithms to identify patients with CRS and controls from our EHR, using chart review as a reference standard. This work is an important first step to accurately identify patients with this morbid, costly, and poorly understood disease and may help facilitate research regarding the prevalence, pathogenesis, morbidity, and care of CRS.
ACKNOWLEDGMENTS
The authors acknowledge Dr. Bruce K. Tan, Dr. Rakesh K. Chandra, Dr. David B. Conley, and Dr. Robert C. Kern (all from the Department of Otolaryngology, Northwestern University Feinberg School of Medicine) for their helpful discussions regarding coding practices.
Footnotes
Funded by the American Academy of Allergy, Asthma, and Immunology ARTrust Mini Grant, Ernest Bazley Grant, U01 HG006388, UL1RR025741, R01 AI082984, P01 AI106683, and T32 AI083216
The authors have no conflicts of interest to declare pertaining to this article
REFERENCES
- 1. Fokkens WJ, Lund VJ, Mullol J, et al. European position paper on rhinosinusitis and nasal polyps 2012. Rhinol Suppl 1–298, 2012. [PubMed] [Google Scholar]
- 2. Slavin RG, Spector SL, Bernstein IL, et al. The diagnosis and management of sinusitis: A practice parameter update. J Allergy Clin Immunol 116:S13–S47, 2005. [DOI] [PubMed] [Google Scholar]
- 3. Meltzer EO, Hamilos DL, Hadley JA, et al. Rhinosinusitis: Developing guidance for clinical trials. J Allergy Clin Immunol 118:S17–S61, 2006. [DOI] [PubMed] [Google Scholar]
- 4. Georgy MS, Peters AT. Chapter 8: Rhinosinusitis. Allergy Asthma Proc 33(suppl 1):S24–S27, 2012. [DOI] [PubMed] [Google Scholar]
- 5. Leung R, Chaung K, Kelly JL, Chandra RK. Advancements in computed tomography management of chronic rhinosinusitis. Am J Rhinol Allergy 25:299–302, 2011. [DOI] [PubMed] [Google Scholar]
- 6. Hsu J, Peters AT. Pathophysiology of chronic rhinosinusitis with nasal polyp. Am J Rhinol 25:285–290, 2011. [DOI] [PubMed] [Google Scholar]
- 7. Pleis JR, Ward BW, Lucas JW. Summary health statistics for U.S. adults: National Health Interview Survey, 2009. Vital Health Stat 10:1–207, 2010. [PubMed] [Google Scholar]
- 8. Hamilos DL. Chronic rhinosinusitis: Epidemiology and medical management. J Allergy Clin Immunol 128:693–707, 2011. [DOI] [PubMed] [Google Scholar]
- 9. Gliklich RE, Metson R. The health impact of chronic sinusitis in patients seeking otolaryngologic care. Otolaryngol Head Neck Surg 113:104–109, 1995. [DOI] [PubMed] [Google Scholar]
- 10. Bhattacharyya N. Incremental health care utilization and expenditures for chronic rhinosinusitis in the United States. Ann Otol Rhinol Laryngol 120:423–427, 2011. [DOI] [PubMed] [Google Scholar]
- 11. Soler ZM, Mace JC, Litvack JR, Smith TL. Chronic rhinosinusitis, race, and ethnicity. Am J Rhinol Allergy 26:110–116, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meltzer EO, Hamilos DL. Rhinosinusitis diagnosis and management for the clinician: A synopsis of recent consensus guidelines. Mayo Clin Proc 86:427–443, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc 19:212–218, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Northwestern Medical Enterprise Data Warehouse. http://informatics.northwestern.edu/blog/edw/
- 15. Pacheco JA, Avila PC, Thompson JA, et al. A highly specific algorithm for identifying asthma cases and controls for genome-wide association studies. American Medical Informatics Association (AMIA) annual symposium proceedings/AMIA symposium, November 14–18, 2009, San Francisco, CA 497–501, 2009. [PMC free article] [PubMed] [Google Scholar]
- 16. Centers for Medicare & Medicaid Services ICD-9 Code Lookup. http://www.cms.gov/medicare-coverage-database/staticpages/icd-9-code-lookup.aspx [PubMed]
- 17. American Medical Association CPT Code/Value Search. https://ocm.ama-assn.org/OCM/CPTRelativeValueSearch.do?submitbutton=accept
- 18. Linder JA, Bates DW, Williams DH, et al. Acute infections in primary care: Accuracy of electronic diagnoses and electronic antibiotic prescribing. J Am Med Inform Assoc 13:61–66, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fung KW, McDonald C, Srinivasan S. The UMLS-CORE project: A study of the problem list terminologies used in large healthcare institutions. J Am Med Inform Assoc 17:675–680, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]




