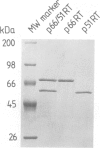
Abstract
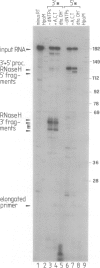
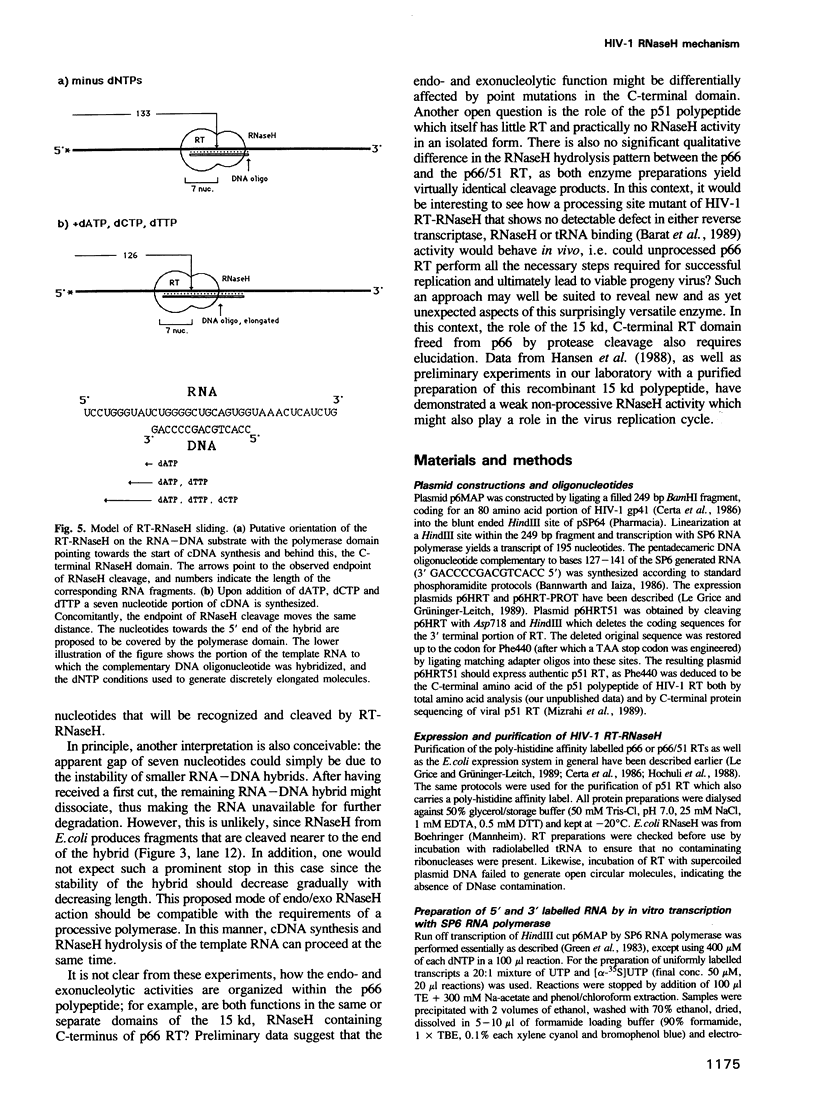
We have analysed the mechanism of ribonuclease H (RNaseH) induced cleavage of a defined RNA-DNA hybrid by human immuno-deficiency virus (HIV-1) reverse transcriptase (RT). An in vitro transcribed RNA labelled at the 3' end was hybridized to a pentadecameric DNA oligonucleotide complementary to an internal region of the RNA. Upon incubation of this RNA-DNA hybrid with recombinant p66 or p66/p51 HIV-1 reverse transcriptase, RT-RNaseH mediated cleavage is observed at most nucleotides within the short hybridized stretch, resulting in a spectrum of RNA fragments extending from the 3' label to this region and differing in length by one nucleotide. The same RNA, this time labelled at the 5' end, yields only one or two major cleavage products corresponding to RNA species extending from the 5' label to the middle of the hybridized region. Such a result can be explained by the action of both endonuclease and 3'----5' exonuclease activities inherent to the C-terminal domain of p66 RT. To investigate how RNaseH cleavage is coupled to reverse transcription, a combination of deoxynucleoside triphosphates was used which allowed controlled extension of the primer DNA. Concomitantly with the elongation of the oligonucleotide primer, RNaseH cleavage proceeds towards the 5' end of the RNA with identical increments, suggesting a simultaneous action of both activities.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bannwarth W., Iaiza P. A system for the simultaneous chemical synthesis of different DNA fragments on solid support. DNA. 1986 Oct;5(5):413–419. doi: 10.1089/dna.1986.5.413. [DOI] [PubMed] [Google Scholar]
- Barat C., Lullien V., Schatz O., Keith G., Nugeyre M. T., Grüninger-Leitch F., Barré-Sinoussi F., LeGrice S. F., Darlix J. L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989 Nov;8(11):3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certa U., Bannwarth W., Stüber D., Gentz R., Lanzer M., Le Grice S., Guillot F., Wendler I., Hunsmann G., Bujard H. Subregions of a conserved part of the HIV gp41 transmembrane protein are differentially recognized by antibodies of infected individuals. EMBO J. 1986 Nov;5(11):3051–3056. doi: 10.1002/j.1460-2075.1986.tb04605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., Gilboa E., Baltimore D. Mechanism of RNA primer removal by the RNase H activity of avian myeloblastosis virus reverse transcriptase. J Virol. 1984 Mar;49(3):686–691. doi: 10.1128/jvi.49.3.686-691.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Green M. R., Maniatis T., Melton D. A. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983 Mar;32(3):681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Hansen J., Schulze T., Mellert W., Moelling K. Identification and characterization of HIV-specific RNase H by monoclonal antibody. EMBO J. 1988 Jan;7(1):239–243. doi: 10.1002/j.1460-2075.1988.tb02805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizi A., McGill C., Hughes S. H. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1218–1222. doi: 10.1073/pnas.85.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H. E., McCoy J. M., Seehra J. S., Richardson C. C. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J Biol Chem. 1989 Mar 15;264(8):4669–4678. [PubMed] [Google Scholar]
- Kane C. M. Renaturase and ribonuclease H: a novel mechanism that influences transcript displacement by RNA polymerase II in vitro. Biochemistry. 1988 May 3;27(9):3187–3196. doi: 10.1021/bi00409a010. [DOI] [PubMed] [Google Scholar]
- Keller W., Crouch R. Degradation of DNA RNA hybrids by ribonuclease H and DNA polymerases of cellular and viral origin. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3360–3364. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotewicz M. L., Sampson C. M., D'Alessio J. M., Gerard G. F. Isolation of cloned Moloney murine leukemia virus reverse transcriptase lacking ribonuclease H activity. Nucleic Acids Res. 1988 Jan 11;16(1):265–277. doi: 10.1093/nar/16.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug M. S., Berger S. L. Ribonuclease H activities associated with viral reverse transcriptases are endonucleases. Proc Natl Acad Sci U S A. 1989 May;86(10):3539–3543. doi: 10.1073/pnas.86.10.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. H., Verma I. M. Reverse transcriptase of RNA tumor viruses. V. In vitro proteolysis of reverse transcriptase from avian myeloblastosis virus and isolation of a polypeptide manifesting only RNase H activity. J Virol. 1978 Feb;25(2):652–663. doi: 10.1128/jvi.25.2.652-663.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder B., Purifoy D., Powell K., Darby G. AIDS virus reverse transcriptase defined by high level expression in Escherichia coli. EMBO J. 1987 Oct;6(10):3133–3137. doi: 10.1002/j.1460-2075.1987.tb02623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grice S. F., Ette R., Mills J., Mous J. Comparison of the human immunodeficiency virus type 1 and 2 proteases by hybrid gene construction and trans-complementation. J Biol Chem. 1989 Sep 5;264(25):14902–14908. [PubMed] [Google Scholar]
- Leis J. P., Berkower I., Hurwitz J. Mechanism of action of ribonuclease H isolated from avian myeloblastosis virus and Escherichia coli. Proc Natl Acad Sci U S A. 1973 Feb;70(2):466–470. doi: 10.1073/pnas.70.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoote M. M., Coligan J. E., Folks T. M., Fauci A. S., Martin M. A., Venkatesan S. Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus. J Virol. 1986 Nov;60(2):771–775. doi: 10.1128/jvi.60.2.771-775.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar C., Abbotts J., Broder S., Wilson S. H. Studies on the mechanism of human immunodeficiency virus reverse transcriptase. Steady-state kinetics, processivity, and polynucleotide inhibition. J Biol Chem. 1988 Oct 25;263(30):15657–15665. [PubMed] [Google Scholar]
- Mitra S. W., Chow M., Champoux J., Baltimore D. Synthesis of murine leukemia virus plus strong stop DNA initiates at a unique site. J Biol Chem. 1982 Jun 10;257(11):5983–5986. [PubMed] [Google Scholar]
- Mizrahi V., Lazarus G. M., Miles L. M., Meyers C. A., Debouck C. Recombinant HIV-1 reverse transcriptase: purification, primary structure, and polymerase/ribonuclease H activities. Arch Biochem Biophys. 1989 Sep;273(2):347–358. doi: 10.1016/0003-9861(89)90493-1. [DOI] [PubMed] [Google Scholar]
- Moelling K. Characterization of reverse transcriptase and RNase H from friend-murine leukemia virus. Virology. 1974 Nov;62(1):46–59. doi: 10.1016/0042-6822(74)90302-x. [DOI] [PubMed] [Google Scholar]
- Mous J., Heimer E. P., Le Grice S. F. Processing protease and reverse transcriptase from human immunodeficiency virus type I polyprotein in Escherichia coli. J Virol. 1988 Apr;62(4):1433–1436. doi: 10.1128/jvi.62.4.1433-1436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer C. A., Faras A. J. Mechanism of release of the avian rotavirus tRNATrp primer molecule from viral DNA by ribonuclease H during reverse transcription. Cell. 1982 Oct;30(3):797–805. doi: 10.1016/0092-8674(82)90284-7. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Fiore D. Ordered interstrand and intrastrand DNA transfer during reverse transcription. Science. 1988 Aug 26;241(4869):1064–1069. doi: 10.1126/science.2457948. [DOI] [PubMed] [Google Scholar]
- Repaske R., Hartley J. W., Kavlick M. F., O'Neill R. R., Austin J. B. Inhibition of RNase H activity and viral replication by single mutations in the 3' region of Moloney murine leukemia virus reverse transcriptase. J Virol. 1989 Mar;63(3):1460–1464. doi: 10.1128/jvi.63.3.1460-1464.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz O., Cromme F. V., Grüninger-Leitch F., Le Grice S. F. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989 Nov 6;257(2):311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- Starnes M. C., Cheng Y. C. Human immunodeficiency virus reverse transcriptase-associated RNase H activity. J Biol Chem. 1989 Apr 25;264(12):7073–7077. [PubMed] [Google Scholar]
- Tanese N., Goff S. P. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma I. M. Studies on reverse transcriptase of RNA tumor viruses III. Properties of purified Moloney murine leukemia virus DNA polymerase and associated RNase H. J Virol. 1975 Apr;15(4):843–854. doi: 10.1128/jvi.15.4.843-854.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Marzo Veronese F., Copeland T. D., DeVico A. L., Rahman R., Oroszlan S., Gallo R. C., Sarngadharan M. G. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science. 1986 Mar 14;231(4743):1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]