Abstract
The viruses circulating among Antarctic wildlife remain largely unknown. In an effort to identify viruses associated with Weddell seals (Leptonychotes weddellii) inhabiting the Ross Sea, vaginal and nasal swabs, and faecal samples were collected between November 2014 and February 2015. In addition, a Weddell seal kidney and South Polar skua (Stercorarius maccormicki) faeces were opportunistically sampled. Using high throughput sequencing, we identified and recovered 152 anellovirus genomes that share 63–70% genome-wide identities with other pinniped anelloviruses. Genome-wide pairwise comparisons coupled with phylogenetic analysis revealed two novel anellovirus species, tentatively named torque teno Leptonychotes weddellii virus (TTLwV) -1 and -2. TTLwV-1 (n = 133, genomes encompassing 40 genotypes) is highly recombinant, whereas TTLwV-2 (n = 19, genomes encompassing three genotypes) is relatively less recombinant. This study documents ubiquitous TTLwVs among Weddell seals in Antarctica with frequent co-infection by multiple genotypes, however, the role these anelloviruses play in seal health remains unknown.
Keywords: Anelloviridae, Weddell seal, South Polar skua, Ross Sea, Antarctica
1. Introduction
Infectious diseases are the leading causes of mass mortality in wildlife and the global uptick of emerging viral disease makes pathogen surveillance crucial for the protection of animal health (Dobson and Foufopoulos 2001; Morner et al. 2002; Blomström 2011). However, identifying viruses is difficult because traditional molecular techniques such as polymerase chain reaction (PCR) amplification and serology-based assays are only useful for the detection of known viruses and their close relatives (Wang et al. 2002; Symonds et al. 2009). Metagenomic approaches that enrich for viruses and use high throughput sequencing platforms are powerful approaches for revealing viral communities, including novel viruses, in animal populations (Edwards and Rohwer 2005; Delwart 2007; Rosario and Breitbart 2011).
Parts of Antarctica, a continent characterized by extreme climate and isolation, are inhabited by unique wildlife. The dense breeding grounds of some animals creates an increased risk of infectious diseases spreading amongst the population at an epidemic scale (Kerry et al. 1999). Similarly, increased human activity on the continent through tourism and research bases is thought to have exposed wildlife to diseases previously attributed to domestic animals. Such findings have led to concerns about pathogen introduction associated with anthropogenic activities (Austin and Webster 1993; Olsen et al. 1996; Gardner et al. 1997; Retamal et al. 2000; Torres 2000). Unfortunately, little is known about the pathogens associated with Antarctic animals.
Within the context of viral pathogens identified in Antarctic wildlife, the use of sequencing approaches has led to the identification of some viruses in Antarctic penguins, i.e. Adélie penguin (Pygoscelis adeliae), Chinstrap penguins (Pygoscelis antarctica) and Gentoo penguins (Pygoscelis papua), including an adenovirus, paramyxoviruses, orthomyxoviruses, a polyomavirus and a papillomavirus (Thomazelli et al. 2010; Hurt et al. 2014, 2016; Lee et al. 2014, 2016; Varsani et al. 2014, 2015). Similarly, a polyomavirus has been identified in sharp-spined notothen (Trematomus pennelii), an Antarctic fish (Buck et al. 2016), a parapoxvirus (Tryland et al. 2005) and a polyomavirus (Varsani et al. 2017) in Weddell seals (Leptonychotes weddellii), and an adenovirus has been identified in South Polar skua (Stercorarius maccormicki) (Park et al. 2012). In addition, serology-based assays have enabled the detection of a putative birnavirus and flavivirus (Morgan and Westbury 1981; Morgan et al. 1985; Gardner et al. 1997) in penguins (Adélie penguin, Blue penguin; Eudyptula minor and Emperor penguin; Aptenodytes forsteri), and a putative herpesvirus in Antarctic seals, namely Weddell seals and crabeater seals (Lobodon carcinophaga) (Harder et al. 1991; Stenvers et al. 1992).
A recent health assessment testing Weddell seals for antibodies to specific known bacterial and viral pathogens indicated that this population remains relatively naïve, leaving them potentially vulnerable to mass die-offs due to their close living proximity and lack of herd immunity (Yochem et al. 2009). Due to this vulnerability, it is important to identify viruses associated with these populations. Viral surveys will provide insight into the viral diversity that is currently associated with these seals, and will provide the genetic information necessary to develop new molecular assays to assess the prevalence of identified viruses and begin to elucidate their impact on animal health.
As part of an ongoing study on Weddell seals in the Ross Sea, we opportunistically sampled faeces, and took nasal and vaginal swabs to identify viruses associated with these animals. In these samples, as well as in a kidney sample from a deceased Weddell seal and a faecal sample from a South Polar skua (a bird that scavenges placenta and carcasses of seals), we identified a diversity of anelloviruses. Anelloviruses are non-enveloped, circular, negative sense, single-stranded DNA viruses that belong to the family Anelloviridae (Okamoto et al. 1998b; Biagini 2009). The first anellovirus, human torque teno virus (TTV), was discovered in a Japanese patient with posttransfusion hepatitis of unknown aetiology (Nishizawa et al. 1997). Since then, numerous anelloviruses have been characterized and grouped into 12 different genera, which have been found in a variety of hosts including pigs, wild boar, dogs, seals, sea lions, pine marteen, bats, horses, cats, sea turtles and a range of primates (Abe et al. 2000; Romeo et al. 2000; Okamoto et al. 2001; Martinez et al. 2006; Al-Moslih et al. 2007; Ng et al. 2009a,b, 2011; King et al. 2011; Nishiyama et al. 2014; Fahsbender et al. 2015). Despite their ubiquity and ability to cause persistent infections, the aetiology of anelloviruses remains unknown.
Most of what is known about anelloviruses is based on human TTV, which has a prevalence as high as 100% in some human populations (Ninomiya et al. 2008). Individuals frequently harbor multiple TTV genotypes (Niel et al. 2000; Nishiyama et al. 2014) and these have been identified throughout the body including in cervical secretions, nasal secretions, the umbilical cord, kidneys, blood, gastric tissue, and sweat (Spandole et al. 2015). There is no indication of tropism, but patterns of genotype compartmentalization, similar to human immunodeficiency virus, have been documented within the host (Maggi et al. 1999).
Anelloviruses have been found to be highly diverse, even at the amino acid level of the coding open reading frames (ORFs), yet the genome organization remains relatively similar with at least two ORFs and a conserved untranslated region (UTR). ORF1 is the largest ORF and is predicted to encode the capsid protein, however, this has not been definitively confirmed (Kamahora et al. 2000; Okamoto et al. 2000). ORF2 encodes proteins thought to be involved in regulation of the innate and adaptive immune system, but the lack of an appropriate culture system has hindered the ability to determine the functionality of these proteins and TTV pathogenicity (Kakkola et al. 2007, 2009; Yu et al. 2007; Huang et al. 2012).
Here we analyze the genomes of the anelloviruses recovered from faeces, kidney, and vaginal and nasal swabs of Weddell seals and a faecal sample of a South Polar skua. Sequence analysis revealed two phylogenetically distinct anellovirus species that are prevalent in all sample types, indicating that anelloviruses are ubiquitous among Weddell seals.
2. Methods
2.1 Sample collection
As part of an ongoing diet study in the Ross Sea on Weddell seals, 42 Weddell seal faecal samples were opportunistically collected on the fast ice of McMurdo Sound (Antarctic) during the 2014/2015 field season. Even though there were tagged animals in the vicinity of the faeces, it was not possible to associate the sampled faeces to a particular tagged animal. In addition to these samples, we collected paired nasal and vaginal swabs from Nov 2014 to Feb 2015 25 adult female Weddell seals; for two additional animals, only nasal swabs were taken, and for three animals we also collected faecal samples (Supplementary Table 1). A subset of females sampled in Nov/Dec were resampled ∼60 days later in Jan/Feb, providing an additional eight nasal and nine vaginal samples. The nasal and vaginal swabs were refrigerated and stored in UTM™ Viral Transport Media (Copan). During the field season, a kidney was sampled from the carcass of a 14-year-old female (Flipper Tag# 8714A; specimen ID 17461) and frozen at −80 °C prior to analysis, see Varsani et al. (2017) for necropsy details. Finally, a South Polar skua faecal sample was collected off fresh snow at Cape Crozier, Ross Island.
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. Weddell seal samples were collected under National Marine Fisheries Service Marine Mammal permit #17411, Antarctic Conservation Act permit #2014-003, and University of Alaska Anchorage’s Institutional Animal Care and Use Committee approval #419971, with funding from the National Science Foundation grant ANT-1246463 to Jennifer M Burns. The skua faecal sample at Cape Crozier was collected under Animal Care and Use Permit #4130 through Oregon State University, Corvallis, OR, and Antarctic Conservation Act Permit #2006-010 from NSF through H.T. Harvey & Associates.
2.2 Sample processing
For each sample, ∼5 g of the faecal sample or tissue samples (in the case of the kidney) was resuspended in 20 ml of SM buffer (0.1 M NaCl, 50 mM Tris/HCl—pH 7.4, 10 mM MgSO4) and homogenized by vortexing for 30 s. The suspension was centrifuged at 10,000 × g for 10 min. Following this, the supernatant was sequentially filtered through 0.45 and 0.2 µm (pore size) syringe filters. Three grams of PEG 8000 (Sigma) was added to each of the filtrates and the solution was mixed gently to resuspend the PEG. The resulting suspension was incubated overnight at 4 °C to precipitate virions. The solution was centrifuged at 10,000×g for 20 min and the resulting pellet was resuspended in 2 ml of SM buffer.
Viral DNA was extracted using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics) from the resuspended virions (200 µl) from the faecal and kidney samples, and 200 µl of the UTM™ Viral Transport Media in which the swabs were stored. We used rolling-circle amplification (RCA) using the TempliPhi™ kit (GE Healthcare) to randomly amplify nucleic acids.
2.3 High throughput sequencing and sequence analysis
A 5-µl aliquot of the randomly amplified DNA from each of the Weddell seals faecal samples, nasal swabs and vaginal swabs was taken, pooled and labelled as WSP, WSN and WSV, respectively. The enriched DNA from the Weddell seal kidney sample was labelled as WSK and the faecal sample from the South Polar skua as SKP. The DNA samples WSP, WSN, WSV, WSK and SKP were then processed to generate ∼100-bp paired-end libraries for multiplex Illumina sequencing and sequenced on an Illumina 2500 (Illumina) platform at Macrogen Inc. (Korea). The resulting paired-end reads were de novo assembled using ABySS v1.9 (Simpson et al. 2009) with a k-mer of 64. Contigs of >750 nts were analyzed for viral-like sequences using BLASTx (Altschul et al. 1990) against a local viral sequence database.
In all of the WSP, WSN, WSV, WSK and SKP de novo assembled contigs, we identified sequences with similarities to anelloviruses. Based on these sequences we designed four pairs of abutting primers (Supplementary Table 2) to screen and recover the complete anellovirus genomes from each individual sample. The RCA product from each sample was used as a template for PCR amplification using Kapa HiFi Hotstart DNA polymerase with the following thermal cycling conditions: (95 °C for 3 min; 25 cycles of 98 °C for 20 s, 60 °C for 15 s, 72 °C for 2 min and a final extension of 72 °C for 3 min). The amplicons were resolved on a 0.7% agarose gel stained with SYBR Safe (ThermoFisher) and ∼2 kb size fragments were excised, gel purified and cloned into pJET1.2 plasmid vector (ThermoFisher). The resulting recombinant plasmids (five from each positive sample type) were Sanger sequenced by primer walking at Macrogen Inc. (South Korea).
To investigate the anellovirus diversity detected in seals, the pairwise identities of the anellovirus genomes and ORF1 sequences were determined using SDT v1.2 (Muhire et al. 2014). All anellovirus sequences with a detectable complete ORF1 were downloaded from GenBank (on the 18th of March 2017). 727 ORF1 sequences (including 152 from this study) were translated, aligned using MUSCLE (Edgar 2004) and then back translated. The resulting alignment was used to infer a Maximum likelihood phylogenetic tree using IQ-TREE (Nguyen et al. 2015) with GTR + I+G4 substitution model selected using ModelFinder (Kalyaanamoorthy et al. 2017). Branches with <60% bootstrap support (1,000 bootstrap iterations) were collapsed using TreeGraph2 (Stover and Muller 2010).
Evidence of recombination in the anelloviruses identified in this study was determined using RDP 4.58 (Martin et al. 2015) with default settings. Sequences were auto-masked for optimal recombination detection and only events detected with more than three different methods implemented in RDP 4.58 coupled with phylogenetic support for recombination and a P value of <0.05 were considered credible.
3. Results
3.1 Anellovirus identification and genome characterization
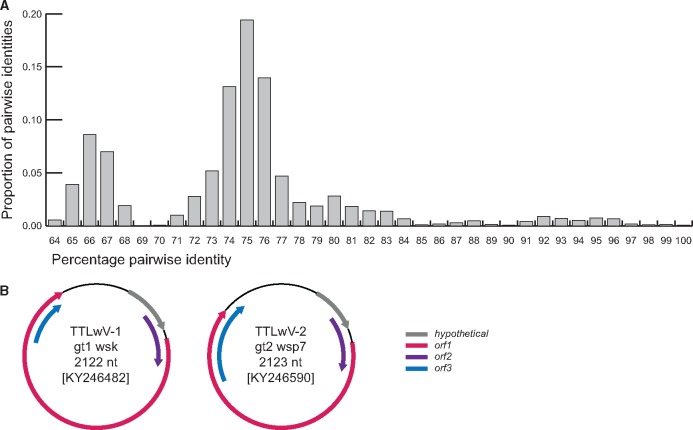
Analysis of the contigs from the de novo assembled reads from Illumina sequencing of WSP, WSN, WSV, WSK and SKP samples revealed high abundance of anellovirus-like sequences, polymavirus-like sequences from WSK which is reported in Varsani et al. (2017), circular replication-associated protein encoding viral-like sequences and various DNA bacteriophages. Given the high abundance of anellovirus-like sequences, we decided to focus on these for this report. Since anelloviruses have small circular genomes, PCRs using abutting primers were performed to recover 152 anellovirus genomes from the various samples, ranging in size from 2,105 to 2,212 nts. Analysis of the genome-wide pairwise identity of these revealed that the seal associated anellovirus genomes share >64% pairwise identity (Fig. 1). Of the 152 genomes identified, 74 genomes sharing >64% identity were recovered from seal faeces, 37 genomes sharing >70% identity from nasal swabs, 34 genomes sharing >72% identity from vaginal swabs, and four genomes sharing >73% identity from the kidney. In addition, three anellovirus genomes sharing >75% identity were recovered from South Polar skua faeces. These Antarctic anelloviruses share ∼63–70% genome-wide identities (Supplementary Data 1) with other pinniped anelloviruses from Pacific harbor seals (Phoca vitulina) sampled in USA (Pacific coast) and the Netherlands (HQ287751, KF373758, KF373760, KM262781, KM262785).
Figure 1.
(A) Distribution of genome-wide pairwise identities of TTLwVs from this study. (B) Schematic genome organizations of representatives from TTLwV-1 and TTLwV-2 which highlights three open reading frames and a hypothetical protein.
Based on species demarcation criteria of 35% divergence of the ORF1 amino acid sequences endorsed by the International Committee for the Taxonomy of Viruses (ICTV) (King et al. 2011), the anelloviruses identified in this study represent two species for which we propose the name torque teno Leptonychotes weddellii virus (TTLwV) -1 and -2. This species demarcation is also supported by the phylogenetic analysis of the ORF1 protein sequences of the TTLwVs (Fig. 2). The genome organization of representatives from the two phylogenetically distinct anellovirus species, TTLwV-1 and TTLwV-2, are illustrated in Fig. 1. Both genomes have the same organization, with three open reading frames and a hypothetical protein, and are approximately the same genome size, ∼2.1 kb. However, there are differences in ORF size and position, specifically for ORF3.
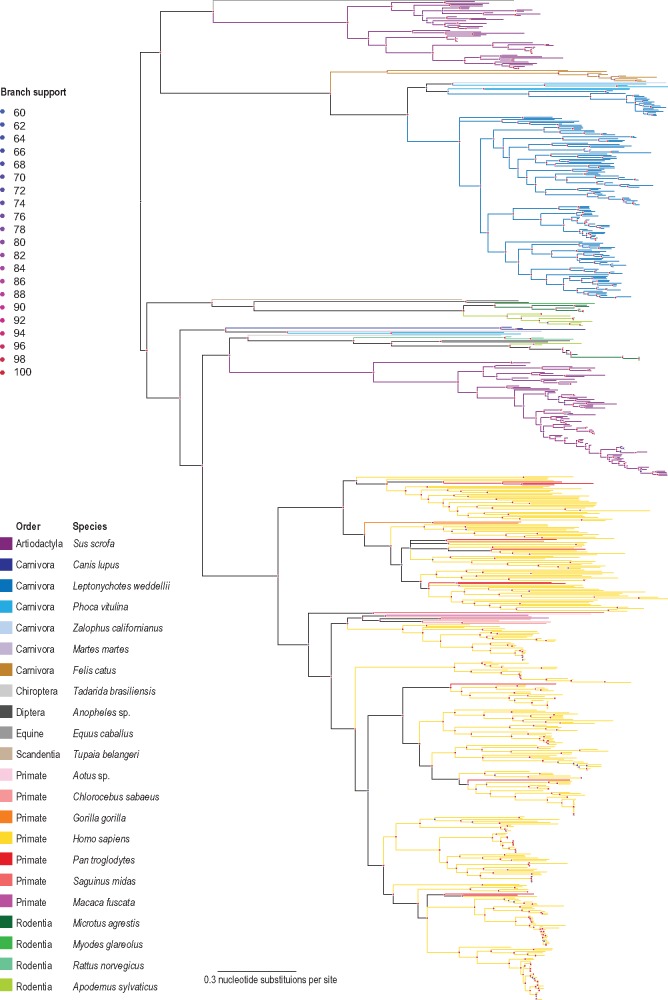
Figure 2.
Maximum-likelihood phylogenetic tree inferred from aligned ORF1 sequences of all publicly available anellovirus sequences together with those from this study. Branches with <60% bootstrap support have been collapsed.
A Maximum likelihood phylogenetic analysis of all available complete ORF1 nucleotide sequences from GenBank (n = 727) show that TTLwVs are related to other anelloviruses discovered in pinnipeds (Fig. 2), and most closely related to the Pacific harbor seal anelloviruses and torque teno Zalophus virus (Figs 2 and 3). In general, the ORF1 phylogenetic analysis (Supplementary Fig. S1) shows some level of host specificity and within lineages there appears to be a significant level of concordance between the phylogenies of the anelloviruses and their hosts (host phylogeny inferred with TimeTree; http://www.timetree.org/; Hedges et al. 2015; Kumar et al. 2017). Furthermore, it is clear that there are two lineages of porcine-associated, pinniped-associated and rodent-associated anelloviruses. Thus, it is highly likely that there were multiple diverse anelloviruses that were circulating amongst the most recent common ancestor (MRCA) of mammals. Within the hominoid-associated anelloviruses, those from chimpanzees (n = 10) and gorillas (n = 3) appear to be interspersed with those from humans (Fig. 2 and Supplementary Fig. S1). Within the primate lineage, given the depth of sampling of non-human primates, it is difficult to test for a coevolution hypothesis or infer any cross-species transmission events. The mosquito-associated anelloviruses are almost certainly derived from a vertebrate blood meal (see taxa marked with * in Supplementary Fig. S1).
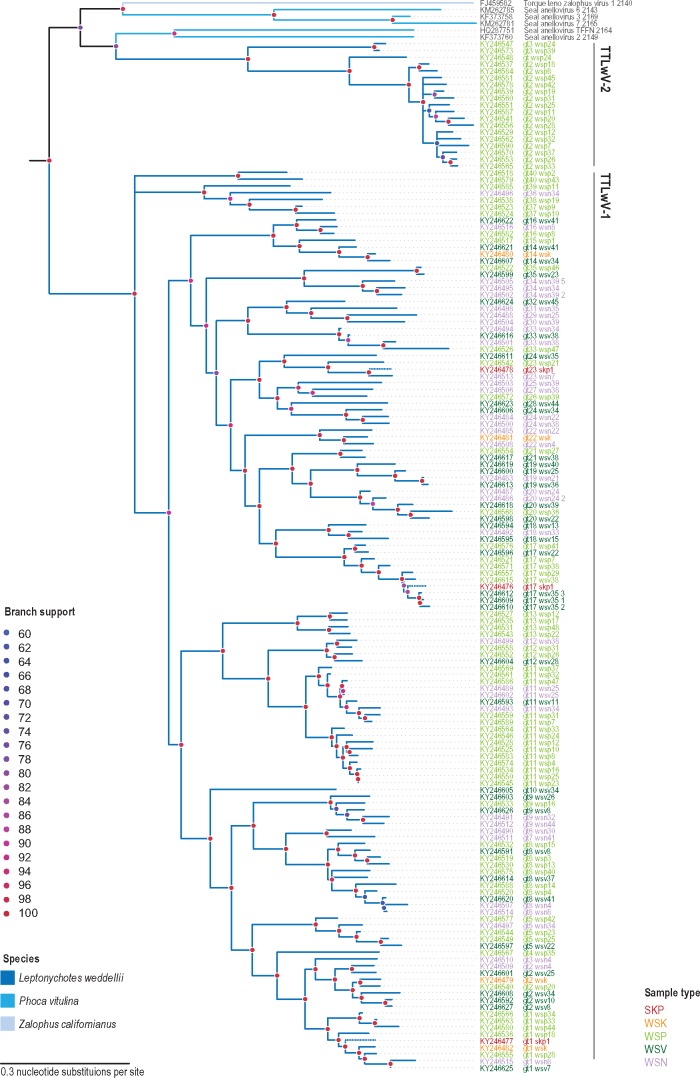
TTLwV-1 (n = 133) was identified in the South Polar skua faeces, as well as the kidney, nasal and vaginal swabs, and faeces from Weddell seals, while TTLwV-2 (n = 19) was exclusively found in seal faeces (Fig. 3). All of the genomes identified belong to the pinniped clade, indicating that the seal itself is the most probable host of all of TTLwVs.
Figure 3.
Maximum-likelihood phylogenetic tree inferred from aligned ORF1 sequences of TTLwVs and other closely related anelloviruses. Branches with <60% bootstrap support have been collapsed. TTLwV sequences from South Polar skua faeces (SKP) are shown with dotted branches.
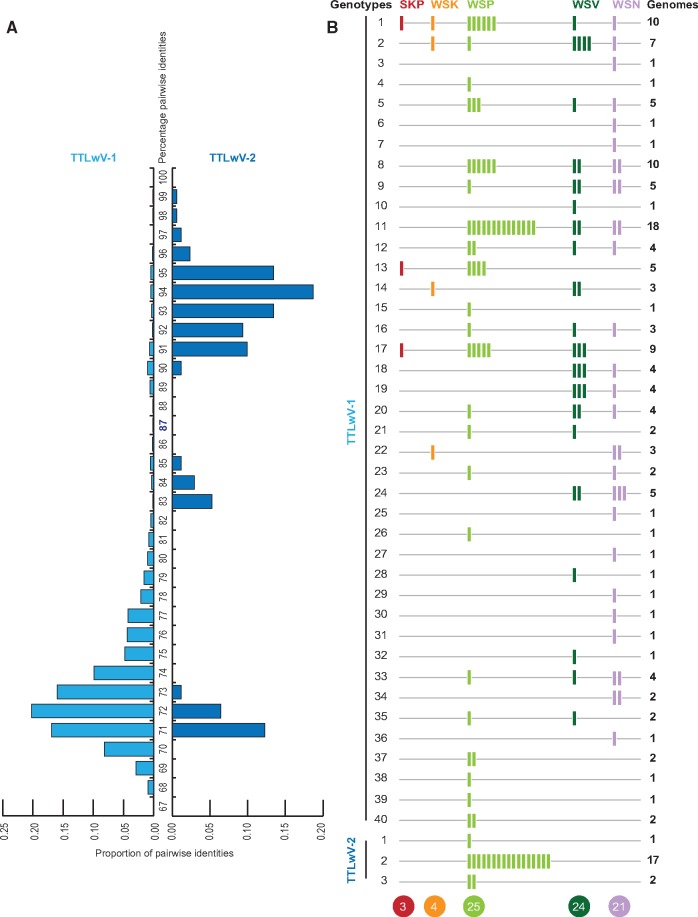
Based on the distribution of the pairwise identities of TTLwV-1 and TTLwV-2 ORF1 nucleotide sequences (Fig. 4), we established that genomes with ≥87% identity should be grouped into the same genotype. Accordingly, TTLwV-1 sequences were subdivided into a total of 40 genotypes, while TTLwV-2 was subdivided into three genotypes (Fig. 4).
Figure 4.
(A) Distribution of the genome-wide pairwise identities of TTLwV-1 and TTLwV-2 supporting 87% genotype demarcation for TTLwV-1 genome sequences. (B) Summary of genotypes found in different sample types. Each bar represents one genome. Total number of genotypes of TTLvW-1 and TTLvW-2 from each sample type are provided at the bottom.
3.2 Evidence of recombination
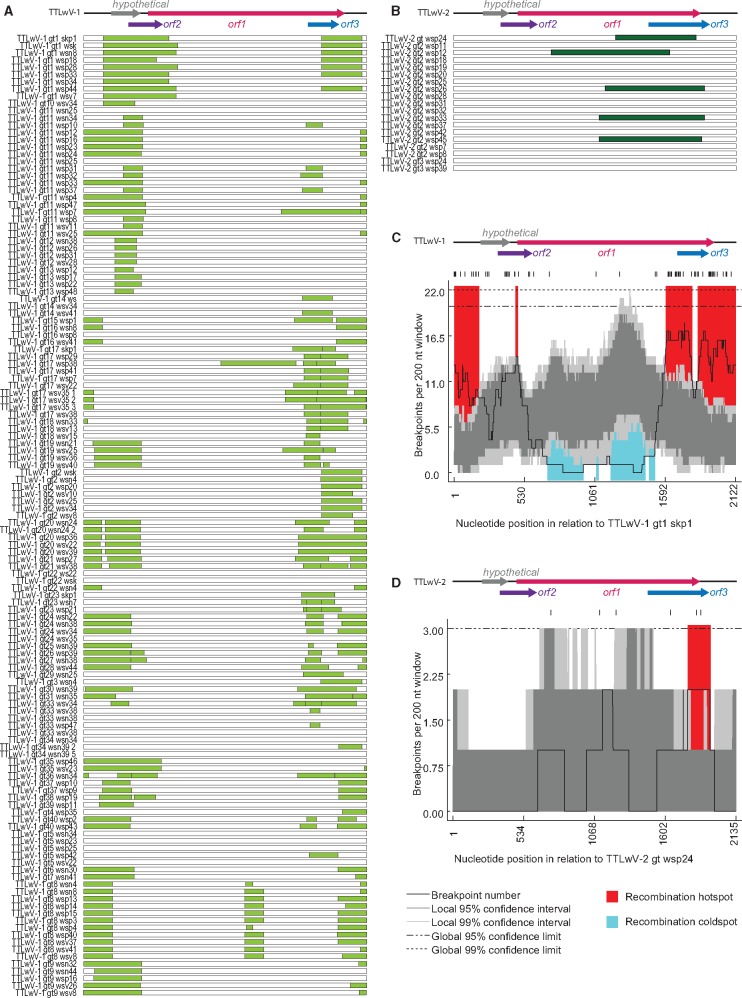
Evidence of recombination within the genomes was detected by analyzing each species individually with RDP 4.58 (Martin et al. 2015). TTLwV-1 and TTLwV-2 genomes have differing recombination patterns. About 89% of TTLwV-1 sequences had at least one recombination event, while only 26% of TTLwV-2 sequences had a recombination event (Fig. 5). The majority of the recombination events in TTLwV-1 were located in the highly conserved translated region (UTR), which is similar to the recombination hotspots found in human TTV and in a global anellovirus analysis (Worobey 2000; Lefeuvre et al. 2009). In contrast with the cold spots in ORF1 of the TTLwV-1 genome, TTLwV-2 has recombination hotspots in the coding regions, ORF1 and ORF3. Recombination may be a driving force of anellovirus diversity, especially in TTLwV-1 (Fig. 5).
Figure 5.
Summary of recombinant regions identified in (A) TTLwV-1 (light green bars) and (B) TTLwV-2 (dark green bars). Recombination breakpoint distribution plots for (C) TTLwV-1 and (D) TTLwV-2, with breakpoint hot-spots indicated in red and cold-spots in blue. The dark and light grey areas of the plots indicate 95% and 99% confidence intervals, respectively. Detectable breakpoint positions are indicated by vertical lines at the top of the graphs. The thick black line represents the plot of the number of breakpoints detected within the 200-nt window region (window was moved along each of the represented alignments 1 nt at a time).
3.3 Prevalence of TTLwV in Weddell seals
Paired nasal and vaginal swabs were collected from 25 seals and 76% (19/25) of these tested positive for TTLwV-1 in at least one of the paired samples. When parsed individually, 64% of the nasal swabs and 72% of the vaginal swabs were TTLwV-1 positive. None of these samples tested positive for TTLwV-2. Of the 45 faecal samples collected, all were TTLwV positive, with 98% positive for TTLwV-1 and 40% for TTLwV-2.
The number of genotypes per type of sample, and the prevalence of each genotype, illustrated in Fig. 4 and Supplementary Fig. S2, show the most common genotypes of TTLwV-1 are 1, 8, and 11, while genotype 2 completely dominates TTLwV-2. Notably TTLwV-1 genotype 1 was also recovered from the South Polar skua faeces. The seal faeces, vaginal, and nasal swabs had comparable diversity with 25, 24, and 21 characterized genotypes, respectively. Although a single kidney sample was analyzed, four genotypes were identified in this organ. Additionally, while most seals harbored one or two TTLwV-1 genotypes, a few seals harbored many more (maximum n = 9 genotypes seal ID 16603; Supplementary Fig. S2).
There is no clear distribution pattern of the 40 genotypes of TTLwV-1, yet TTLwV-2 was only found in the seal faeces (40% of the faecal samples). Since TTLwV-2 is related to other anelloviruses isolated from pinnipeds, it is likely to also be infecting seals, but may have a different and more specific tropism than TTLwV-1, which seems to have pan-tropism. The overlapping genotypes of TTLwV-1 found in the faeces, nasal, and vaginal swabs suggest possible faecal-oral transmission of this virus, and indicates that future studies may be able to test the faeces alone to capture anellovirus diversity in seals.
3.4 TTLwV-1 identification in South Polar skua faeces
TTLwV-1 was recovered from South Polar skua faeces that was opportunistically sampled. South Polar Skua faeces contained three TTLwV-1 genotypes (1, 13, 17; Figs 3 and 4), which were also identified in the seal kidney, faeces, nasal, and vaginal swabs. The presence of various TTLwV-1 genotypes in South Polar skua faeces may reflect viruses that are dietary in origin since these predatory birds feed on Weddell seal placenta and scavenge seal carcasses, which may contain TTLwV, hence the likely explanation for presence of this in its faeces.
4. Conclusion
The advent of NGS technology has proven to be a powerful tool for virus discovery and has changed the field of virology. Here we took advantage of this technology to investigate viral presence and diversity in Antarctic wildlife, which remains largely unknown. Previous studies of Weddell seals have focused on wildlife exposure to known viruses, limiting our understanding of the myriad of viruses present in this unique environment.
Samples tested from Weddell seals from the Ross Sea led to the discovery of 152 anellovirus genomes. Sequencing complete genomes enabled the recognition of two new species, TTLwV-1 and TTLwV-2 and the role recombination plays in driving TTLwV diversity. TTLwV is phylogenetically related to the other pinniped-associated anelloviruses that were recovered from the brain and lungs of the Pacific harbor seal, the lungs, liver, lymph nodes and tonsils of a California sea lion (Zalophus californianus), and the faeces from subantarctic fur seals (Arctocephalus tropicalis) and South American fur seals (Arctocephalus australis) (Ng et al. 2009b, 2011; Bodewes et al. 2013; Kluge et al. 2016). However, this is the first time anelloviruses have been characterized in Weddell seals and the first time they have been described in Antarctic vertebrates.
Although anelloviruses cause persistent infections and are ubiquitous among humans and various animal species, their aetiology remains a mystery (Spandole et al. 2015). This was the first time a prevalence study showed anelloviruses to be ubiquitous within a pinniped population, with TTLwVs present in 100% of the seal faecal samples. TTLwV-1 genomes from the seal faeces were related to those recovered from the vaginal and nasal swabs and kidney, indicating that these were shedding from the seal itself and not infecting seal food sources. The fact that the same TTLwV-1 genotypes were found within the seal tissues and faeces may also suggest that this anellovirus species is transmitted through the faecal-oral route, which is a hypothesis proposed for the transmission of TTV in humans (Okamoto et al. 1998a; Ukita et al. 1999). Although TTLwV-2 was only detected in faecal samples, phylogenetic analysis indicates that this species may also infect seals. Failure to detect TTLwV-2 in the seal tissues tested here suggests that this species has more specific tropism than TTLwV-1.
Nevertheless, sampling of Weddell seal faeces alone could give a broad perspective of anellovirus diversity circulating within this Antarctic pinniped population. Sampling faeces may therefore be a valuable, non-invasive sampling tool for capturing the diversity and prevalence of pinniped anelloviruses. Previous prevalence studies of pinniped-associated anelloviruses screened the serum and lungs, which may have greatly underestimated the prevalence of anelloviruses in pinniped populations (Ng et al. 2011; Fahsbender et al. 2015).
Future studies focusing on the anelloviruses in Antarctica will provide insight into their presence in other species and how they are transmitted through the food web. The South Polar skua faeces derived TTLwVs are most likely acquired from scavenging Weddell seal placenta or carcasses. Therefore, detection of TTLwV in South Polar skua faeces and Weddell seal samples may provide an example of a situation in which viruses could be used as proxies for trophic interactions (Dayaram et al. 2016; Godinho et al. 2017). Additionally, it remains to be determined whether TTLwV is present in Antarctic surface waters, as has been shown for some anelloviruses in Italy and Japan (Haramoto et al. 2005; Verani et al. 2006).
More work needs to be done to determine the role of TTLwVs in Weddell seal health. TTLwVs are diverse and pervasive in this population, with individuals infected with multiple genotypes. The health effects of infection by specific genotypes or co-infection with different genotypes remain unknown. With the exception of the dead Weddell seal from which the kidney sample infected with a polyomavirus (Varsani et al. 2017) was obtained, all other animals were in apparent good health, with most sighted months and years following handling. Determining the viral load of TTLwV may prove to be important for providing clues as to the strength of seal immune systems since anellovirus loads are thought to be good indicators of immunosuppression (Hofer 2014).
Supplementary Material
Acknowledgements
D.G.A. and G.B. are supported by the US National Science Foundation (NSF; ANT-0944411). All the samples were collected with logistics supplied from the US Antarctic Program. S.K. was supported by NSF ANT-0944747. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. E.F. and M.B. were supported by the US National Science Foundation’s Assembling the Tree of Life Program Grant DEB-1239976. A.K. and R.B. were supported by Institutional Development Awards (IDeA) Networks of Biomedical Research Excellence Assistantships (grant number P20GM103395) from the National Institute of General Medical Sciences of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIH. All the wet bench molecular work and sequencing was supported by personal funds of A.V.
Data availability
All sequence data reported in this study has been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers KY246476–KY246627.
Supplementary data
Supplementary data are available at Virus Evolution online.
Conflict of interest: None declared.
References
- Abe K. et al. (2000) ‘TT Virus Infection in Nonhuman Primates and Characterization of the Viral Genome: Identification of Simian TT Virus Isolates’, Journal of Virology, 74: 1549–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Moslih M. I., Perkins H., Hu Y. W. (2007) ‘Genetic Relationship of Torque Teno Virus (TTV) between Humans and Camels in United Arab Emirates (UAE)’, Journal of Medical Virology, 79: 188–91. [DOI] [PubMed] [Google Scholar]
- Altschul S. F. et al. (1990) ‘Basic Local Alignment Search Tool’, Journal of Molecular Biology, 215: 403–10. [DOI] [PubMed] [Google Scholar]
- Austin F. J., Webster R. G. (1993) ‘Evidence of Ortho- and Paramoxyoviruses in Fauana from Antarctica’, Journal of Wildlife Diseases, 29: 568–71. [DOI] [PubMed] [Google Scholar]
- Biagini P. (2009) ‘Classification of TTV and Related Viruses (Anelloviruses)’, in Villiers E.-M., Hausen H. (eds.) TT Viruses, pp. 21–33. Springer Berlin Heidelberg. [DOI] [PubMed] [Google Scholar]
- Blomström A.-L. (2011) ‘Viral Metagenomics as an Emerging and Powerful Tool in Veterinary Medicine’, Veterinary Quarterly, 31: 107–14. [DOI] [PubMed] [Google Scholar]
- Bodewes R. et al. (2013) ‘Novel B19-Like Parvovirus in the Brain of a Harbor Seal’, PloS One, 8: e79259.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck C. B. et al. (2016) ‘The Ancient Evolutionary History of Polyomaviruses’, PLoS Pathogens, 12: e1005574.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayaram A. et al. (2016) ‘Diverse Circular Replication-Associated Protein Encoding Viruses Circulating in Invertebrates Within a Lake Ecosystem’, Infection, Genetics and Evolution, 39: 304–16. [DOI] [PubMed] [Google Scholar]
- Delwart E. L. (2007) ‘Viral Metagenomics’, Reviews in Medical Virology, 17: 115–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A., Foufopoulos J. (2001) ‘Emerging Infectious Pathogens of Wildlife’, Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 356: 1001–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity’, BMC Bioinformatics, 5: 113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R. A., Rohwer F. (2005) ‘Viral Metagenomics’, Nature Reviews. Microbiology, 3: 504–10. [DOI] [PubMed] [Google Scholar]
- Fahsbender E. et al. (2015) ‘Development of a Serological Assay for the Sea Lion (Zalophus californianus) Anellovirus, ZcAV’, Scientific Reports, 5: 9637.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner H. et al. (1997) ‘Poultry Virus Infection in Antarctic Penguins’, Nature, 387: 245.. [DOI] [PubMed] [Google Scholar]
- Godinho M. T. et al. (2017) ‘Genome Sequence of Cauliflower Mosaic Virus Identified in Earwigs (Doru luteipes) through a Metagenomic Approach’, Genome Announcements, 5: e00043–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E. et al. (2005) ‘One-Year Monthly Monitoring of Torque Teno Virus (TTV) in Wastewater Treatment Plants in Japan’, Water Research, 39: 2008–13. [DOI] [PubMed] [Google Scholar]
- Harder T. C., Plötz J., Liess B. (1991) ‘Antibodies against European Phocine Herpesvirus Isolates Detected in Sera of Antarctic Seals’, Polar Biology, 11: 509–12. [Google Scholar]
- Hedges S. B. et al. (2015) ‘Tree of Life Reveals Clock-Like Speciation and Diversification’, Molecular Biology and Evolution, 32: 835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer U. (2014) ‘Microbiome: Anelloviridae Go Viral’, Nature Reviews. Microbiology, 12: 4–5. [DOI] [PubMed] [Google Scholar]
- Huang Y. W. et al. (2012) ‘Rescue of a Porcine Anellovirus (Torque Teno Sus Virus 2) from Cloned Genomic DNA in Pigs’, Journal of Virology, 86: 6042–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt A. C. et al. (2016) ‘Evidence for the Introduction, Reassortment, and Persistence of Diverse Influenza A Viruses in Antarctica’, Journal of Virology, 90: 9674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt A. C. et al. (2014) ‘Detection of Evolutionarily Distinct Avian Influenza a Viruses in Antarctica’, mBio, 5: e01098–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkola L. et al. (2009) ‘Replication of and Protein Synthesis by TT Viruses’, in de Villiers E.-M., Hausen H.Z. (eds.) TT Viruses: The Still Elusive Human Pathogens, pp. 53–64. Berlin, Heidelberg: Springer Berlin Heidelberg. [Google Scholar]
- Kakkola L. et al. (2007) ‘Construction and Biological Activity of a Full-Length Molecular Clone of Human Torque Teno Virus (TTV) Genotype 6’, The FEBS Journal, 274: 4719–30. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S. et al. (2017) ‘ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates’, Nature Methods, 14: 587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamahora T., Hino S., Miyata H. (2000) ‘Three Spliced mRNAs of TT Virus Transcribed from A Plasmid Containing the Entire Genome in COS1 Cells’, Journal of Virology, 74: 9980–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerry K., Riddle M., Clarke J. (1999) Diseases of Antarctic Wildlife. A report for SCAR and COMNAP. SCAR.
- King A. M. Q. et al. (2011) Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier. [Google Scholar]
- Kluge M. et al. (2016) ‘Metagenomic Survey of Viral Diversity Obtained from Feces of Subantarctic and South American Fur Seals’, PloS One, 11: e0151921.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. et al. (2017) ‘TimeTree: A Resource for Timelines, Timetrees, and Divergence Times’, Molecular Biology and Evolution, 34: 1812–9. [DOI] [PubMed] [Google Scholar]
- Lee S. Y. et al. (2014) ‘A Novel Adenovirus in Chinstrap penguins (Pygoscelis antarctica) in Antarctica’, Viruses, 6: 2052–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y. et al. (2016) ‘Genetic and Molecular Epidemiological Characterization of a Novel Adenovirus in Antarctic Penguins Collected between 2008 and 2013’, PloS One, 11: e0157032.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeuvre P. et al. (2009) ‘Widely Conserved Recombination Patterns among Single-Stranded DNA Viruses’, Journal of Virology, 83: 2697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi F. et al. (1999) ‘High Prevalence of TT Virus Viremia in Italian Patients, Regardless of Age, Clinical Diagnosis, and Previous Interferon Treatment’, The Journal of Infectious Diseases, 180: 838–42. [DOI] [PubMed] [Google Scholar]
- Martin D. P. et al. (2015) ‘RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes’, Virus Evolution, 1: vev003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez L. et al. (2006) ‘Torque Teno Virus (TTV) is Highly Prevalent in the European Wild Boar (Sus scrofa)’, Veterinary Microbiology, 118: 223–9. [DOI] [PubMed] [Google Scholar]
- Morgan I. R., Westbury H. A. (1981) ‘Virological Studies of Adelie Penguins (Pygoscelis adeliae) in Antarctica’, Avian Diseases, 25: 1019–26. [PubMed] [Google Scholar]
- Morgan I. R., Westbury H. A., Campbell J. (1985) ‘Viral Infections of Little Blue Penguins (Eudyptula minor) along the Southern Coast of Australia’, Journal of Wildlife Diseases, 21: 193–8. [DOI] [PubMed] [Google Scholar]
- Morner T. et al. (2002) ‘Surveillance and Monitoring of Wildlife Diseases’, Revue Scientifique Et Technique-Office International Des Epizooties, 21: 67–76. [DOI] [PubMed] [Google Scholar]
- Muhire B. M., Varsani A., Martin D. P. (2014) ‘SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation’, PloS One, 9: e108277.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. F. et al. (2009a) ‘Discovery of a Novel Single-Stranded DNA Virus from a Sea Turtle Fibropapilloma by Using Viral Metagenomics’, Journal of Virology, 83: 2500–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. F. et al. (2009b) ‘Novel Anellovirus Discovered from a Mortality Event of Captive California Sea Lions’, The Journal of General Virology, 90: 1256–61. [DOI] [PubMed] [Google Scholar]
- Ng T. F. et al. (2011) ‘Metagenomic Identification of a Novel Anellovirus in Pacific Harbor Seal (Phoca vitulina richardsii) Lung Samples and Its Detection in Samples from Multiple Years’, The Journal of General Virology, 92: 1318–23. [DOI] [PubMed] [Google Scholar]
- Nguyen L. T. et al. (2015) ‘IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies’, Molecular Biology and Evolution, 32: 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niel C., Saback F. L., Lampe E. (2000) ‘Coinfection with Multiple TT Virus Strains Belonging to Different Genotypes is a Common Event in Healthy Brazilian Adults’, Journal of Clinical Microbiology, 38: 1926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya M. et al. (2008) ‘Development of PCR Assays with Nested Primers Specific for Differential Detection of Three Human Anelloviruses and Early Acquisition of Dual or Triple Infection During Infancy’, Journal of Clinical Microbiology, 46: 507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama S. et al. (2014) ‘Identification of Novel Anelloviruses with Broad Diversity in UK Rodents’, The Journal of General Virology, 95: 1544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa T. et al. (1997) ‘A Novel DNA Virus (TTV) Associated with Elevated Transaminase Levels in Posttransfusion Hepatitis of Unknown Etiology’, Biochemical and Biophysical Research Communications, 241: 92–7. [DOI] [PubMed] [Google Scholar]
- Okamoto H. et al. (1998a) ‘Fecal Excretion of a Nonenveloped DNA Virus (TTV) Associated with Posttransfusion Non-A-G Hepatitis’, Journal of Medical Virology, 56: 128–32. [PubMed] [Google Scholar]
- Okamoto H. et al. (1998b) ‘Molecular Cloning and Characterization of a Novel DNA Virus (TTV) Associated with Posttransfusion Hepatitis of Unknown Etiology’, Hepatology Research, 10: 1–16. [Google Scholar]
- Okamoto H. et al. (2001) ‘Genomic and Evolutionary Characterization of TT Virus (TTV) in Tupaias and Comparison with Species-Specific TTVs in Humans and Non-Human Primates’, The Journal of General Virology, 82: 2041–50. [DOI] [PubMed] [Google Scholar]
- Okamoto H. et al. (2000) ‘TT Virus mRNAs Detected in the Bone Marrow Cells from an Infected Individual’, Biochemical and Biophysical Research Communications, 279: 700–7. [DOI] [PubMed] [Google Scholar]
- Olsen B. et al. (1996) ‘Salmonella enteritidis in Antarctica: Zoonosis in Man or Humanosis in Penguins?’, Lancet (London, England), 348: 1319–20. [DOI] [PubMed] [Google Scholar]
- Park Y. M. et al. (2012) ‘Full Genome Analysis of a Novel Adenovirus from the South Polar Skua (Catharacta maccormicki) in Antarctica’, Virology, 422: 144–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retamal P. et al. (2000) ‘Detection of anti-Brucella Antibodies in Pinnipeds from the Antarctic Territory’, Veterinary Record, 146: 166–7. [DOI] [PubMed] [Google Scholar]
- Romeo R. et al. (2000) ‘High Prevalence of TT Virus (TTV) in Naive Chimpanzees and in Hepatitis C Virus-Infected Humans: Frequent Mixed Infections and Identification of New TTV Genotypes in Chimpanzees’, The Journal of General Virology, 81: 1001–7. [DOI] [PubMed] [Google Scholar]
- Rosario K., Breitbart M. (2011) ‘Exploring the Viral World Through Metagenomics’, Current Opinion in Virology, 1: 289–97. [DOI] [PubMed] [Google Scholar]
- Simpson J. T. et al. (2009) ‘ABySS: A Parallel Assembler for Short Read Sequence Data’, Genome Research, 19: 1117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandole S. et al. (2015) ‘Human Anelloviruses: An Update of Molecular, Epidemiological and Clinical Aspects’, Archives of Virology, 160: 893–908. [DOI] [PubMed] [Google Scholar]
- Stenvers O., Plotz J., Ludwig H. (1992) ‘Antarctic Seals Carry Antibodies Against Seal Herpesvirus’, Archives of Virology, 123: 421–4. [DOI] [PubMed] [Google Scholar]
- Stover B. C., Muller K. F. (2010) ‘TreeGraph 2: Combining and Visualizing Evidence from Different Phylogenetic Analyses’, BMC Bioinformatics, 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E. M., Griffin D. W., Breitbart M. (2009) ‘Eukaryotic Viruses in Wastewater Samples from the United States’, Applied and Environmental Microbiology, 75: 1402–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomazelli L. M. et al. (2010) ‘Newcastle Disease Virus in Penguins from King George Island on the Antarctic region’, Veterinary Microbiology, 146: 155–60. [DOI] [PubMed] [Google Scholar]
- Torres D. (2000) ‘Antarctic Territory’, The Veterinary Record, 146: 166–7. [DOI] [PubMed] [Google Scholar]
- Tryland M. et al. (2005) ‘Isolation and Partial Characterization of a Parapoxvirus Isolated from a Skin Lesion of a Weddell Seal’, Virus Research, 108: 83–7. [DOI] [PubMed] [Google Scholar]
- Ukita M. et al. (1999) ‘Excretion into Bile of a Novel Unenveloped DNA Virus (TT Virus) Associated with Acute and Chronic Non-A-G Hepatitis’, The Journal of Infectious Diseases, 179: 1245–8. [DOI] [PubMed] [Google Scholar]
- Varsani A. et al. (2017) ‘Identification of a Polyomavirus in Weddell Seal (Leptonychotes weddellii) from the Ross Sea (Antarctica)’, Archives of Virology, 162: 1403–7. [DOI] [PubMed] [Google Scholar]
- Varsani A. et al. (2014) ‘A Novel Papillomavirus in Adelie Penguin (Pygoscelis adeliae) Faeces Sampled at the Cape Crozier Colony, Antarctica’, The Journal of General Virology, 95: 1352–65. [DOI] [PubMed] [Google Scholar]
- Varsani A. et al. (2015) ‘Identification of an Avian Polyomavirus Associated with Adelie Penguins (Pygoscelis adeliae)’, The Journal of General Virology, 96: 851–7. [DOI] [PubMed] [Google Scholar]
- Verani M. et al. (2006) ‘One-Year Monthly Monitoring of Torque Teno Virus (TTV) in River Water in Italy’, Water Science and Technology, 54: 191.. [DOI] [PubMed] [Google Scholar]
- Wang D. et al. (2002) ‘Microarray-Based Detection and Genotyping of Viral Pathogens’, Proceedings of the National Academy of Sciences, 99: 15687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M. (2000) ‘Extensive Homologous Recombination among Widely Divergent TT Viruses’, Journal of Virology, 74: 7666–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem P. K. et al. (2009) ‘Health Assessment of Weddell Seals, Leptonychotesweddellii, in McMurdo Sound, Antarctica’, in Kerry K.R., Riddle M. (eds.) Health of Antarctic Wildlife: A Challenge for Science and Policy, pp. 123–138. Berlin, Heidelberg: Springer Berlin Heidelberg. [Google Scholar]
- Yu X. et al. (2007) ‘Genome Analysis and Epidemiological Investigation of Goose Circovirus Detected in Eastern China’, Virus Genes, 35: 605–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data reported in this study has been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers KY246476–KY246627.