Abstract
Background
Helicobacter pylori infection has been consistently associated with lack of access to clean water and proper sanitation, but no studies have demonstrated that the transmission of viable but nonculturable (VBNC) H. pylori can occur from drinking contaminated water. In this study, we used a laboratory mouse model to test whether waterborne VBNC H. pylori could cause gastric infection.
Materials and Methods
We performed five mouse experiments to assess the infectivity of VBNC H. pylori in various exposure scenarios. VBNC viability was examined using Live/Dead staining and Biolog phenotype metabolism arrays. High doses of VBNC H. pylori in water were chosen to test the “worst‐case” scenario for different periods of time. One experiment also investigated the infectious capabilities of VBNC SS1 using gavage. Further, immunocompromised mice were exposed to examine infectivity among potentially vulnerable groups. After exposure, mice were euthanized and their stomachs were examined for H. pylori infection using culture and PCR methodology.
Results
VBNC cells were membrane intact and retained metabolic activity. Mice exposed to VBNC H. pylori via drinking water and gavage were not infected, despite the various exposure scenarios (immunocompromised, high doses) that might have permitted infection with VBNC H. pylori. The positive controls exposed to viable, culturable H. pylori did become infected.
Conclusions
While other studies that have used viable, culturable SS1 via gavage or drinking water exposures to successfully infect mice, in our study, waterborne VBNC SS1 failed to colonize mice under all test conditions. Future studies could examine different H. pylori strains in similar exposure scenarios to compare the relative infectivity of the VBNC vs the viable, culturable state, which would help inform future risk assessments of H. pylori in water.
Keywords: Helicobacter pylori, infectivity, SS1, waterborne transmission
1. Introduction
Helicobacter pylori (H. pylori) is a gastrointestinal bacterium that causes gastritis, peptic ulcers and, over time, gastric adenocarcinoma.1, 2 Helicobacter pylori infection is hypothesized to be transmitted through multiple routes, including vertically from mother to child and through contaminated reservoirs like food and water.3, 4 A body of evidence suggests that contaminated water may be a source of H. pylori infection, with epidemiological studies consistently associating H. pylori infection with lack of access to potable drinking water and proper sanitation.3, 5, 6, 7, 8, 9 Furthermore, H. pylori has been detected in water using various molecular biology techniques, such as quantitative polymerase chain reaction (qPCR) and microscopy methods,5, 10, 11, 12, 13 and there are reports that it has been cultured from water.14, 15, 16, 17 Helicobacter pylori enters a viable but not culturable (VBNC) state within a few days after inoculation into water.18, 19, 20 This change is often accompanied by a morphological change from a spiral bacillus to a U‐shaped or coccoid form, and H. pylori has been found in the VBNC state in all these morphologies in the natural environment.18, 21 However, although H. pylori has been cultured from wastewater and drinking water, it is unclear whether this was due to the culturable form being present in the water or investigators being able to revert the VBNC form back to a culturable form using appropriate media.
The fact that H. pylori is present in both a culturable and VBNC state has not been accounted for when assessing risk associated with waterborne H. pylori. For example, a risk model of waterborne H. pylori infection using a quantitative microbial risk assessment methodology22 did not consider the VBNC form of H. pylori. Likewise, our recent study showing that constant exposure to the viable, cultural form of H. pylori in drinking water can infect mice did not account for exposure to the VBNC form.20 While previous studies found that VBNC H. pylori administered via gavage could cause infection in mice,19, 23 the gavage exposure method is not representative of exposure to drinking water. To fill this gap in the literature, we examined the infectivity of the VBNC form of H. pylori in water.
2. Materials and Methods
2.1. Transmission and exposure groups
Our studies were carried out sequentially following our initial dosing experiments that examined the infectious dose of viable, culturable H. pylori in water.20 We performed four mouse experiments to assess the infectivity of VBNC H. pylori in various different exposure scenarios (Table 1). Concentrations of VBNC H. pylori were chosen based on previous studies19, 20, 23 and on the amounts of H. pylori found in sources of recreational and drinking water worldwide.24, 25 We first employed a classic single‐hit exposure model with waterborne VBNC H. pylori, examining whether a single day of water with a high dose of H. pylori could cause infection, choosing the high end of waterborne concentrations to test a “worst‐case” scenario and to try to ensure a higher chance of experimental infection; 4 weeks was chosen as the time to wait until euthanasia, given that She et al.19 had found slightly increased colonization rates at 4 weeks compared to 3 weeks. The sample size of 40 mice was chosen for consistency with our previous dosing experiments, in which each exposure group had 40 mice. When this failed to induce infection, we did two follow‐up experiments (Table 1, experiments 2 and 3). We increased the number of days of exposure (six instead of one), and also exposed severe combined immunodeficient mice to a single day of waterborne H. pylori, hypothesizing that more doses and immunocompromised hosts would be more likely to increase infection based on the results of our previous experiments.20 When these also failed to induce infection, we increased the exposure length again and increased the number of mice to 100 to increase the likelihood of seeing infection. In these experiments, we used a similar experimental design to our original dosing studies,20 exposing the mice to 56 days of contaminated water (experiment 4), and further decreasing the time until euthanasia. When this also failed to induce infection, we did a final follow‐up study in which we gavaged mice with four doses of ~2*108 cells of VBNC SS1 over 2 weeks. This, too, failed to induce infection.
Table 1.
Experimental overview of various drinking water exposure scenarios
| Experiment number | Exposure groups | Time to VBNC conversion | Exposure | Euthanized |
|---|---|---|---|---|
| 1 | 40 C57/BL6 mice (20 male, 20 female) | 2 d | Exposure to 1 d of 109 cells/L of VBNC Helicobacter pylori | 4 wk after final exposure |
| 2 | 40 C57/BL6 mice (20 male, 20 female) | 2 d | Exposure to 6 d of 109 cells/L of VBNC H. pylori a | 2 wk after final exposure |
| 3 | 10 C57/BL6 Severe Combined Immunodeficient mice (4 male, 6 female) | 2 d | Exposure to 1 d of 109 cells/L of VBNC H. pylori | 1 wk after final exposure |
| 4 | 100 C57/BL6 mice (50 male, 50 female) | 4 d | Consistent exposure to >109 cells/L of VBNC H. pylori over 56 d | 4 d after final exposure |
| Negative control | 10 C57/BL6 mice (4 male, 6 female) | N/A | Sterile, filtered tap water for 60 d | Day 60 |
| Positive control | 10 C57/BL6 mice (4 male, 6 female) | N/A | Consistent exposure to >109 cells/L of viable, culturable H. pylori over 56 d | 4 d after final exposure |
Mice were exposed to contaminated drinking water for 3 d, followed by 11 d of sterile water, and then another 3 d of contaminated water.
The mice were exposed to water contaminated with ~109 cells/L VBNC H. pylori (See Table 1). In experiments 1‐3, contaminated water was removed after 24 hours and replaced with either a bottle of freshly contaminated water or (when appropriate) sterilized, filtered tap water. Each exposure group had 20 cages, with two mice per cage per the Animal Care and Use Committee regulations. In experiment 4, water was changed twice per week, every 3‐4 days. As a negative control, 10 mice (five cages) were given sterile, filtered tap water for 9 weeks. As a positive control, 10 mice (five cages) were given sterile, filtered tap water inoculated with viable, culturable H. pylori for 9 weeks. All mice were housed at University Laboratory Animal Medicine facilities at the University of Michigan Medical School, and all experiments were approved by the Animal Care and Use Committee.
2.2. Bacterial strain
SS1 (Sydney Strain 1) was selected for this study for consistency with our previous studies,20 and because it colonizes mice more successfully than other H. pylori strains.26
2.3. H. pylori cultivation, counting, and inoculation
Helicobacter pylori cultivation was carried out as previously described.20 Briefly, SS1 was plated from stocks and grown at 37°C on 5% sheep blood tryptic soy agar II plates (BBL, Franklin Lakes, NJ, USA) in microaerobic conditions. After 3 days, colonies were collected and suspended in plates of Brucella broth (Remel, Columbus, OH, USA) supplemented with 10% heat‐inactivated fetal bovine serum (Fisher Scientific, Waltham, MA, USA). After shaking overnight in microaerobic conditions at 37°C, the broth was centrifuged at 1917 g and 4°C for 20 minutes. The supernatant was removed, and the pellet was suspended in 1× PBS. To confirm the concentration of H. pylori, the stock suspension was serially diluted onto 5% sheep blood tryptic soy agar II plates (BBL). Sterilized, filtered tap water was then inoculated with the stock suspension. After 4‐7 days of growth, the number of H. pylori colonies was counted and the stock solution concentration was back‐calculated.
2.4. H. pylori viability in water
Previous methodologies for inducing the VBNC state have differed across studies. She et al.19 inoculated sterilized tap water with live H. pylori and stored it at 4°C for 40 days, defining cells as VBNC when they were in the coccoid state and did not grow when plated. Wang et al.23 incubated fresh H. pylori colonies in Ham's F12 medium with 10% calf serum for 3 days, then stored them at 4°C, defining cells as VBNC once they stopped growing. Cellini et al.27 inoculated a Brucella broth/2% fetal calf serum solution with fresh H. pylori and incubated it for 20 days until the cells no longer grew when plated. As we wanted to examine the infectivity of VBNC H. pylori in water, we chose to incubate H. pylori in water for the VBNC conversion.
Inoculated water was held for 2‐4 days at room temperature to ensure that the VBNC conversion had occurred before giving water to the mice. To check culturability, inoculated water was plated on 5% sheep blood tryptic soy agar II media and incubated for 7 days in microaerobic conditions at 37°C. Helicobacter pylori cells were checked for viability and morphology using microscopy at 60× magnification and Live/Dead BacLight Bacterial Viability Kit (Life Technologies, Eugene, OR, USA). To undertake viability and morphological analyses, 50 mL of water was centrifuged at 10 400 g for 3 minutes, the supernatant was removed, and cell pellets were suspended in BacLight Live/Dead dye. After incubating for a minimum of 15 minutes in the dark, the cell suspensions were examined in triplicate per the manufacturer's instructions.
Metabolic activity of VBNC H. pylori cells was examined using Biolog Phenotypic Microarray plates PM1, which contain 95 separate carbon sources which are commonly utilized by a variety of microbial species. All necessary reagents were purchased from Biolog (Hayward, CA, USA). Helicobacter pylori cells were grown on 5% Sheep blood Tryptic Soy Agar II media, then collected from the plates and suspended in sterile, autoclaved water. Cell suspensions were stored at room temperature for 0, 3, 4, 7, or 8 days. At each respective time point, cell suspensions were spun down at 1917 g for 20 minutes. The supernatant was discarded, and the resulting pellets were checked for metabolic activity using the PM1 plate. Briefly, pellets were re‐suspended in inoculating fluid IF‐0a GN/GP (1.2×), and then supplemented to a final concentration of 0.05% bovine serum albumin (BSA) and 1.25 mmol/L NaHCO3. (H. pylori has been shown to use more carbon sources and grow successfully in media containing BSA,28, 29 so it was included to ensure better visualization of metabolic activity in the VBNC state.) Dye mix D (Biolog) was then added to achieve a final concentration of 0.01%; 100 μL of this solution was pipetted into each well of the PM1 plate, which was then incubated in microaerobic conditions for 48 hours. Cells were considered to be metabolically active if they induced a color change in any of the wells containing nutrient sources, and the negative control had no color change. This was not measured in a quantitative way, but checked visually, as the purpose of this experiment was to examine H. pylori VBNC cell viability rather than examine the carbon sources used.
2.5. Dose estimation
To estimate the doses consumed by the mice, water bottles were weighed before being placed in cages and immediately after their removal. As water drips out of water bottles when they are placed in the cage and when the cages are moved, “dummy” bottles were filled with water and treated in the exact same way as experimental bottles. The amount of water lost from dummy bottles was averaged and that average was subtracted from the total water lost from each bottle. As mice were housed two per cage, the adjusted total per cage was then halved to provide the individual dose per mouse.
2.6. Mouse euthanasia, verification and quantification of infection
After exposure, the mice were euthanized and their stomachs were collected. The stomach was weighed, homogenized in 1× PBS, and serial dilutions of the homogenate were plated on H. pylori selective media (Columbia blood agar base with 10% horse blood, Dent Supplement, 300 mg/L urea, and 3500 U polymyxin B/L).30 Presumptive H. pylori isolates were counted and then checked for urease activity using urease indicator broth (0.33 mol/L urea, 0.2% Phenol Red, 0.02% NaN3, 0.01 mol/L pH 6.5 NaPO4 buffer). DNA was extracted from stomach homogenate using the QiaAMP DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) per the manufacturer's instructions. Extracted DNA was tested for the presence of the H. pylori VacA gene by PCR using the Takara PCR kit (Fisher, TAK RR001A) and primers VagA‐F (5‐CAATCTGTCCAATCAAGCGAG) and VagA‐R (5‐GCGTCAAAATAATTCCAAGG).31 PCR was run with an initial denaturation at 95°C for 3 minutes, followed by 35 cycles of 95°C for 1 minute, 52°C for 1 minute, 72°C for 1 minute, and a final extension at 72°C for 5 minutes. PCR products were visualized on a 1.5% agarose gel.
3. Results
3.1. Morphology, culturability, and metabolic activity of H. pylori in water
In all experiments, there was a complete loss of culturability of H. pylori 2‐3 days after initial inoculation into water. Despite being nonculturable, cells were still found to be membrane intact via Live/Dead staining 8 days after inoculation into water (Figure 1).
Figure 1.
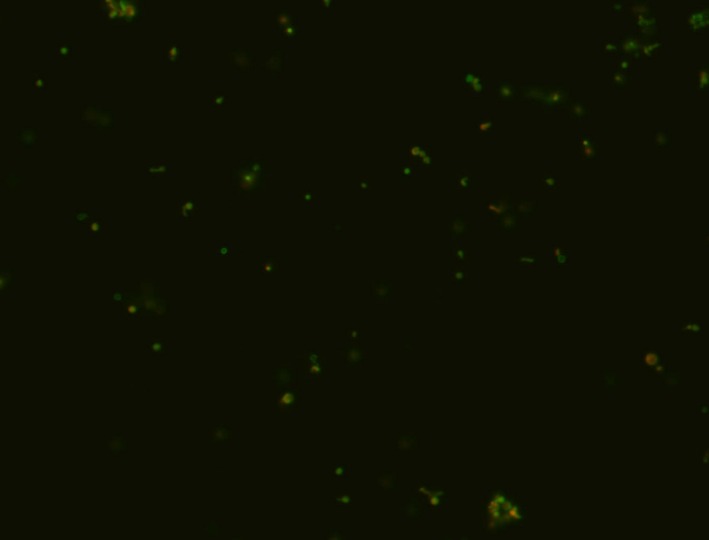
60× magnification of Helicobacter pylori suspension in water after 8 d of incubation at room temperature. Green cells are membrane intact, and red cells have membrane damage. The predominant form was coccoid
The VBNC H. pylori cells also induced color changes in the Biolog PM1 panels at each time point, respectively (Figure 2). This suggests that the cells were metabolically active, as they were metabolizing the carbon sources in each well. The cells in the viable, culturable state utilized many more carbon sources than any of the cells in the VBNC state. No differences in metabolic activity were seen between VBNC cells on days 3, 4, 7, and 8 (data not shown).
Figure 2.
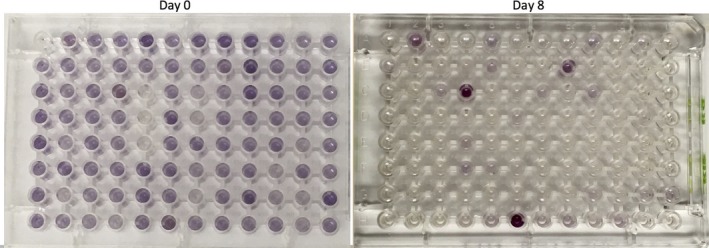
PM1 plates of Day 0 (viable and culturable) and Day 8 Helicobacter pylori cells (VBNC). Each well contains a different carbon source, and wells with a purple color change indicate that the carbon source was being used. Viable culturable H. pylori utilized a much wider variety of carbon sources than the VBNC H. pylori
3.2. Exposure to waterborne H. pylori
A cage was counted as infected if the following conditions were met: the quantitative culture plates had colonies with correct H. pylori morphology (small, round, and translucent), were positive for the rapid urease test, and were positive for PCR targeting the VacA gene. Cages were counted as positive if one or both mice in a cage were infected. If no mice were infected, then that cage was counted as negative. The results of the five exposure scenarios and the positive and negative controls are summarized in Table 2. Further, the mice dosed with SS1 via gavage were also not infected.
Table 2.
Overview of experimental results
| Experiment number | Average number of VBNC Helicobacter pylori cells per liter drinking water (range) | Average cumulative ingested dose per mouse (range) | Number of infected cages n/N (%) | Total number of infected mice n/N (%) | VacA PCR‐positive results n/N (%) |
|---|---|---|---|---|---|
| Experiment 1 | 109 | 106 | 0/20 (0%) | 0/40 (0%) | 0/40 |
| Experiment 2 | 2.14E9 (1.15E9‐3.42E9) | 5.33E7 (4.09E7‐6.91E7) | 0/20 (0%) | 0/40 (0%) | 0/40 |
| Experiment 3 | 2.22E9 | 5.44E6 (4.20E6‐6.19E6) | 0/5 (0%) | 0/10 (0%) | 0/10 |
| Experiment 4 | 7.49E9 (9.30E8‐2.04E10) | 2.30E9 (1.75E9‐3.83E9) | 0/50 (0%) | 0/100 (0%) | 0/100 |
| Negative control | 0 | 0 | 0/5 (0%) | 0/10 (0%) | 0/10 |
| Positive control | 4.80E9 (2.42E8‐2.04E10) | 1.07E9 (8.45E8‐1.64E9) | 5/5 (100%) | 8/10 (80%) | 8/10 (80%) |
Results from viable but nonculturable H. pylori dosing experiments. While the positive control showed consistent levels of infection with previous studies, mice exposed to VBNC H. pylori showed no signs of infection.
The negative controls showed no signs of infection, and confirmed H. pylori cultures were recovered from 8 of 10 positive controls. None of the mice exposed to VBNC H. pylori showed any sign of infection, either via culture or PCR.
4. Discussion
We were unable to cause infection in mice with the VBNC form of SS1, either in drinking water or via gavage. Our inability to cause infection was surprising, given the known capacity of this strain to successfully infect mice,26, 32 our wide range of exposure scenarios, and our previously published study that showed that SS1 in water could infect mice in a dose‐dependent manner.20 In our previous study, 4 weeks of exposure to water spiked with 109 CFU/L, 108 CFU/L 107 CFU/L, and 106 CFU/L of H. pylori caused infection in 39 of 40, 33 of 40, four of 38, and one of 40 mice, respectively. The ingested cumulative doses are two‐ to 2000‐fold lower than those used in this current experiment, showing that SS1 is less infectious (or completely noninfectious) in the VBNC state than when viable and culturable. This suggests that H. pylori strains may be less infectious than when viable and culturable.
However, there are few dosing experiments in the literature that examine this phenomenon. She et al.19 found that 11 of 16 mice gavaged with VBNC H. pylori were infected compared to 14 of 16 gavaged with the same dose of viable, culturable H. pylori. Also using gavage to administer doses, Cellini et al.27 showed that eight of 20 mice were infected from VBNC H. pylori compared to nine of 20 with viable, culturable H. pylori. Both studies used strains that were recently isolated from clinical biopsies of patients with ulcers. Combined with our results from drinking water and gavage exposure to SS1, this suggests that different strains may differ in their ability to infect mice when in the VBNC state.
Our inability to cause infection could be due in part to the drinking water exposure route, which may affect the dose that reaches the stomach compared to gavage methods. Gavage directly inoculates the stomach with a large bolus of bacteria, while drinking water contains comparatively lower doses and must go through the mouth and esophagus before reaching the stomach, which may result in bacterial losses along the way. While this may affect our results, our total cumulative doses—especially in experiment 5—were comparable to (or higher than) the doses reported in previous studies (108‐4*108 CFU/dose). Further, our gavage experiments showed no signs of infection either. Finally, our previous study in which we administered viable, culturable H. pylori to mice in drinking water found relatively similar dose/response rates as other studies that were carried out with gavage.20
4.1. Limitations and public health implications
As with any animal study, we cannot be certain that our results accurately reflect what would occur with human exposure. As H. pylori is a human pathogen, it is possible that the VBNC form is more infectious in humans than in mice. Further, we only exposed mice to one strain of H. pylori, and it is possible that other strains would be more infectious in the VBNC state than SS1, as has been seen in other published papers in the literature.19, 27 Despite our large sample sizes and high doses, our inability to infect mice with VBNC H. pylori via drinking water suggests that VBNC SS1 in water is not infectious in mice. This may reflect the strain that we used, the route of exposure, or may simply mean that we did not account for some crucial piece of the puzzle that is yet unknown about the transmission of H. pylori via water. The genetic variability of H. pylori strains is vast,33 so it may be possible that some strains lack the capability to persist in water, but instead are transmitted only via other exposures, such as person‐to‐person or fecal‐oral routes.4
4.2. Future directions
Examining different strains of VBNC H. pylori in these exposure scenarios would give insight into the trade‐offs of survival and infectivity associated with the VBNC state. Further, investigating the distributions of VBNC vs viable, culturable H. pylori populations in the natural environment would provide a better understanding of the infectivity of the various forms of H. pylori. Such experiments would allow for more accurate risk assessments of H. pylori in water, as it is very likely that multiple strains and forms of H. pylori are present in contaminated drinking or surface water sources.
5. Conclusions
We found that mice exposed to VBNC SS1 H. pylori via drinking water were not infected, despite the various exposure scenarios (immunocompromised, high doses) that might have promoted infection with VBNC bacteria. While other studies that have used viable, culturable SS1 to successfully infect mice via gavage and drinking water, our results suggest that VBNC SS1 is either not infectious (or potentially has greatly reduced infectivity). Future studies could examine different H. pylori strains in similar exposure scenarios to compare the relative infectivity of the VBNC vs the viable, culturable state, which would help inform future risk assessments of H. pylori in water.
Disclosures of Interests
The authors declare no conflict of interests.
Acknowledgements
This study was supported partially by the Dow Sustainability Fellowship to K.F.B., the Rackham Research Fellowship to K.F.B, the Graham Institute of Sustainability Integrated Assessment Grant to M.V. and C.X., and also by a generous donation from Dr. Laurence Baker, D.O. (Collegiate Professor of Cancer Developmental Therapeutics; Professor, Departments of Internal Medicine and Pharmacology; University of Michigan School of Medicine). Many thanks to Lisa Marchlewicz, Kari Neier, and Joseph Kochmanski for their assistance.
Boehnke KF, Eaton KA, Fontaine C, et al. Reduced infectivity of waterborne viable but nonculturable Helicobacter pylori strain SS1 in mice. Helicobacter. 2017;22:e12391 https://doi.org/10.1111/hel.12391
References
- 1. Parsonnet J, Friedman GD, Vandersteen DP, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;324:232‐235. [DOI] [PubMed] [Google Scholar]
- 2. Parsonnet J, Hansen S, Rodriguez L, et al. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267‐1271. [DOI] [PubMed] [Google Scholar]
- 3. Goh K‐L, Chan W‐K. Epidemiology of Helicobacter Pylori and public health implications. Helicobacter. 2011;16:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kusters JG, van Vliet AHM, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449‐490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hulten K, Han S, Enroth H, et al. Helicobacter pylori in the drinking water in Peru. Gastroenterology. 1996;110:1031‐1035. [DOI] [PubMed] [Google Scholar]
- 6. Soto G, Bautista CT, Roth DE, et al. Helicobacter pylori reinfection is common in Peruvian adults after antibiotic eradication therapy. J Infect Dis. 2003;188:1263‐1275. [DOI] [PubMed] [Google Scholar]
- 7. Ramirez‐Ramos A, Gilman RH, Leon‐Barua R, et al. Rapid recurrence of Helicobacter pylori infection in Peruvian patients after successful eradication. Clin Infect Dis. 1997;25:1027‐1031. [DOI] [PubMed] [Google Scholar]
- 8. Klein PD, Opekun AR, Smith EO, et al. Water source as risk factor for Helicobacter pylori infection in Peruvian children. Lancet. 1991;337:1503‐1506. [DOI] [PubMed] [Google Scholar]
- 9. Aziz RK, Khalifa MM, Sharaf RR. Contaminated water as a source of Helicobacter pylori infection. J Adv Res. 2015;6(4):539‐547. https://doi.org/10.1016/j.jare.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moreno Y, Piqueres P, Alonso JL, Jiménez A, González A, Ferrús MA. Survival and viability of Helicobacter pylori after inoculation into chlorinated drinking water. Water Res. 2007;41:3490‐3496. [DOI] [PubMed] [Google Scholar]
- 11. Moreno‐Mesonero L, Moreno Y, Alonso JL, Ferrús MA. DVC‐FISH and PMA‐qPCR techniques to assess the survival of Helicobacter pylori inside Acanthamoeba castellanii . Res Microbiol. 2016;167(1):29‐34. https://doi.org/10.1016/j.resmic.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 12. Nayak AK, Rose JB. Detection of Helicobacter pylori in sewage and water using a new quantitative PCR method with SYBR green. J Appl Microbiol. 2007;103:1931‐1941. [DOI] [PubMed] [Google Scholar]
- 13. Santiago P, Moreno Y, Ferrús MA. Identification of viable Helicobacter pylori in drinking water supplies by cultural and molecular techniques. Helicobacter. 2015;20(4):252‐259. https://doi.org/10.1111/hel.12205. [DOI] [PubMed] [Google Scholar]
- 14. Moreno Y, Ferrús MA. Specific detection of cultivable Helicobacter pylori cells from wastewater treatment plants. Helicobacter. 2012;17:327‐332. [DOI] [PubMed] [Google Scholar]
- 15. Lu Y, Redlinger TE, Avitia R, Galindo A, Goodman K. Isolation and genotyping of Helicobacter pylori from untreated wastewater. Appl Environ Microbiol. 2002;68:1436‐1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bahrami AR, Rahimi E, Ghasemian Safaei H. Detection of Helicobacter pylori in city water, dental units' water, and bottled mineral water in Isfahan, Iran. Sci World J. 2013. https://doi.org/10.1155/2013/280510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Al‐Sulami A, Al‐Edani T, Al‐Abdula A. Culture method and PCR for the detection of Helicobacter pylori in drinking water in Basrah Governorate Iraq. Gastroenterol Res Pract. 2012. https://doi.org/10.1155/2012/245167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adams BL, Bates TC, Oliver JD. Survival of Helicobacter pylori in a natural freshwater environment. Appl Environ Microbiol. 2003;69:7462‐7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. She FF, Lin JY, Liu JY, Huang C, Su DH. Virulence of water‐induced coccoid Helicobacter pylori and its experimental infection in mice. World J Gastroenterol. 2003;9:516‐520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boehnke KF, Eaton KA, Valdivieso M, Baker LH, Xi C. Animal model reveals potential waterborne transmission of Helicobacter pylori infection. Helicobacter. 2015;20:326‐333. [DOI] [PubMed] [Google Scholar]
- 21. Oliver JD. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev. 2010;34:415‐425. [DOI] [PubMed] [Google Scholar]
- 22. Ryan M, Hamilton K, Hamilton M, Haas CN. Evaluating the potential for a Helicobacter pylori drinking water guideline. Risk Anal. 2014;34:1651‐1662. [DOI] [PubMed] [Google Scholar]
- 23. Wang X, Sturegård E, Rupar R, et al. Infection of BALB/c A mice by spiral and coccoid forms of Helicobacter pylori . J Med Microbiol. 1997;46:657‐663. [DOI] [PubMed] [Google Scholar]
- 24. Voytek MA, Ashen JB, Fogarty LR, Kirshtein JD, Landa ER. Detection of Helicobacter pylori and fecal indicator bacteria in five North American rivers. J Water Health. 2005;3:405‐422. [DOI] [PubMed] [Google Scholar]
- 25. Holman CB, Bachoon DS, Otero E, Ramsubhag A. Detection of Helicobacter pylori in the coastal waters of Georgia, Puerto Rico and Trinidad. Mar Pollut Bull. 2014;79:354‐358. [DOI] [PubMed] [Google Scholar]
- 26. Lee A, O'Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386‐1397. [DOI] [PubMed] [Google Scholar]
- 27. Cellini L, Allocati N, Angelucci D, et al. Coccoid Helicobacter pylori not culturable in vitro reverts in mice. Microbiol Immunol. 1994;38:843‐850. [DOI] [PubMed] [Google Scholar]
- 28. Testerman TL, Gee DJMC, Mobley HLT. Helicobacter pylori growth and urease detection in the chemically defined medium Ham ‘s F‐12 nutrient mixture. J Clin Microbiol. 2001;39:3842‐3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chouankam A, Lei X‐H, Bochner B. Phenotype MicroArray Determination of Metabolic Pathway Activities in Bacteria: Helicobacter pylori as an example; 2012.
- 30. Degnan AJ, Sonzogni WC, Standridge JH. Development of a plating medium for selection of Helicobacter pylori from water samples. Appl Environ Microbiol. 2003;69:2914‐2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Atherton JC, Cover TL, Twells RJ, Morales MR, Hawkey CJ, Blaser MJ. Simple and accurate PCR‐based system for typing vacuolating cytotoxin alleles of Helicobacter pylori . J Clin Microbiol. 1999;37:2979‐2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wanken AE, Conway T, Eaton KA. The Entner‐Doudoroff pathway has little effect on Helicobacter pylori colonization of mice. Infect Immun. 2003;71:2920‐2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. J Clin Invest. 2004;113:321‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]


