Abstract
The prognostic value of CpG island methylator phenotype (CIMP) in colorectal cancer remains unsettled. We aimed to assess the prognostic value of this phenotype analyzing a total of 1126 tumor samples obtained from two Norwegian consecutive colorectal cancer series. CIMP status was determined by analyzing the 5‐markers CAGNA1G, IGF2, NEUROG1, RUNX3 and SOCS1 by quantitative methylation specific PCR (qMSP). The effect of CIMP on time to recurrence (TTR) and overall survival (OS) were determined by uni‐ and multivariate analyses. Subgroup analyses were conducted according to MSI and BRAF mutation status, disease stage, and also age at time of diagnosis (<60, 60‐74, ≥75 years). Patients with CIMP positive tumors demonstrated significantly shorter TTR and worse OS compared to those with CIMP negative tumors (multivariate hazard ratio [95% CI] 1.86 [1.31‐2.63] and 1.89 [1.34‐2.65], respectively). In stratified analyses, CIMP tumors showed significantly worse outcome among patients with microsatellite stable (MSS, P < 0.001), and MSS BRAF mutated tumors (P < 0.001), a finding that persisted in patients with stage II, III or IV disease, and that remained significant in multivariate analysis (P < 0.01). Consistent results were found for all three age groups. To conclude, CIMP is significantly associated with inferior outcome for colorectal cancer patients, and can stratify the poor prognostic patients with MSS BRAF mutated tumors.
Keywords: Age of Onset, CIMP, Colon Cancer, DNA Methylation, Prognostic Factor
Short abstract
What's new?
As many as one‐fifth of colorectal cancers have a CpG island methylator phenotype (CIMP) involving widespread promoter DNA methylation. CIMP is associated with key factors related to disease outcome, including microsatellite instability and BRAF mutations. In this study, CIMP was found to be significantly associated with worse prognosis in colorectal cancer patients, particularly those with microsatellite stable (MSS) BRAF‐mutated tumors. In stratified analyses, trends toward worse survival were identified for CIMP‐positive stage III and stage IV patients in the MSS BRAF‐mutated group. The findings suggest that CIMP status should be included in prognostic analyses at time of diagnosis.
Abbreviations
- CIMP
CpG island methylator phenotype
- FFPE
formalin fixed paraffin embedded
- HR
hazard ratio
- MSI
microsatellite instability
- MSS
microsatellite stable
- mut
mutation
- OS
overall survival
- PMR
percent methylated reference
- TTR
time to recurrence
- qMSP
quantitative methylation specific polymerase chain reaction
- wt
wild type
Introduction
Colorectal cancer is one of the most common malignancies throughout the world, with almost 1.4 million new cases and close to 700 000 deaths each year.1 Clinicopathological staging according to the Tumor–Node–Metastases (TNM) system2 remains the most important prognostic factor. However, the TNM system fails to accurately predict the outcome of patients within stages,3 and robust markers to stratify these patients to improve prognostication and choice of therapy are clearly needed.
CpG islands are found in 50–70% of human gene promoters and are normally unmethylated. In cancer however, these islands frequently become hypermethylated, which is associated with aberrant silencing of several genes, including tumor suppressors.4 Approximately 15–20% of colorectal cancers are characterized by widespread promoter DNA methylation, referred to as the CpG island methylator phenotype (CIMP). CIMP positive tumors are associated with microsatellite instability (MSI), BRAF mutations, wild type TP53, poor differentiation, proximal location, and also female gender and older age.5, 6, 7
In addition to CIMP, there are two other well known molecular phenotypes of colorectal cancer, characterized by either MSI or chromosomal instability (CIN).8, 9, 10 Patients diagnosed with MSI generally have a good prognosis,11, 12, 13, 14 whereas patients with CIN have a much poorer prognosis,15 although with great stage dependent variation. In contrast, the prognostic effect of CIMP remains unsettled.16, 17, 18, 19, 20 Several studies fail to reach statistical significance due to insufficient sample size, and the effect of CIMP is often diminished after adjusting for other factors.21 We have analyzed the prognostic value of CIMP in two Norwegian population representative patient series (n > 1100), using a well‐established and validated CIMP panel,5, 22 adjusting for other molecular‐ and clinical variables, including stage, BRAF and MSI status.
Materials and Methods
Patient samples
Samples from a total of 1126 patients with stage I‐IV colorectal cancer were analyzed, obtained from two Norwegian series (Oslo 1,23, 24 and Oslo 225, 26). The Oslo 1 series comprised 762 formalin fixed paraffin embedded (FFPE) colorectal cancer tissue samples, collected from patients undergoing surgical resection at the Oslo University Hospital‐ Aker (OUH) in the period 1993–2003. The median age of the patients was 73 years (range 30–94 years). The Oslo 2 series included 364 fresh frozen samples from patients undergoing surgery at OUH in the period 2005–2011. The median age of the patients was 71 years (range 27–97 years).
The study was carried out according to the Helsinki declaration, and the research biobanks have been registered according to national legislation (numbers 2781 and 236–2005–16141). The study has been approved by the Regional Committee for Medical and Health Research Ethics (numbers 1.2005.1629 and S‐09282c 2009/4958), which requires that informed consent is obtained from patients enrolled in the study.
DNA extraction, bisulfite treatment and determination of CIMP status
DNA from the fresh frozen colorectal tissue samples (Oslo 2) was extracted using either a standard phenol/chloroform protocol27 or magnetic beads (Maxwell 16, Promega). For the FFPE tissue samples (Oslo 1) the QIAmp DNA Mini kit (Qiagen, Hilden, Germany) was applied for DNA extraction as previously described.23 DNA from both series was bisulfite treated using the EpiTect Bisulfite kit (Qiagen) with 1.3 µg DNA as input, and purified using the QIAcube (Qiagen). For the FFPE tissue samples, an extra ethanol step was included during clean‐up. Quantitative methylation specific PCR (qMSP) was performed as previously described.28 In brief, the PCRs (95°C for 10 min, followed by 45 cycles of 95°C for 15 sec and 60°C for 60 sec) were carried out in triplicates in 384 well plates in the 7900HT Real‐Time PCR System (Life Technologies, Carlsbad, CA), and included 1xTaqMan Universal PCR Mastermix No AmpErase UNG (Life Technologies), 0.9 µM of each primer, 0.2 µM probe, and approximately 32.5 ng bisulfite treated template DNA. Water was included as template negative control, bisulfite treated DNA isolated from the whole blood of healthy individuals was included as methylation negative control, and bisulfite‐ converted in vitro methylated DNA (IVD, Chemicon; Millipore) was used as a methylation positive control. A standard curve was generated from IVD consisting of 1:5 serial dilutions (32.5 ng‐ 0.052 ng). To distribute template and master mix to the 384‐well plates, the EpMotion 5075 pipetting robot (Eppendorf, Hamburg, Germany) was used.
The threshold for censoring was cycle 35 and cycle 40 for the fresh frozen (Oslo 2) and FFPE (Oslo 1) samples, respectively. The median quantity value of the triplicates was used for data analysis. To normalize for DNA input the ALU‐C4 element was used as a control.29 The percent of methylated reference (PMR) values were calculated by dividing the median GENE:ALU ratio of the sample by the median GENE:ALU ratio of the positive control (IVD), and multiply by 100.
In accordance with the CIMP panel described by Weisenberger et al.5 including CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1, samples with a PMR value ≥10 for ≥3/5 markers were considered CIMP positive. PMR ≥ 10 was also used for scoring MLH1 methylation positive samples.30 CIMP and MLH1 status was successfully determined for 1122 samples, including all 364 fresh frozen samples and 758/762 = 99.5% of the FFPE samples.
Determination of MSI status
Of the 1122 samples successfully analyzed for CIMP, MSI status was available for 1113 samples, including 933 MSS and 180 MSI tumors.23, 24, 25 As previously described,23 MSI status was determined by fragment analyses of the five microsatellites BAT25, BAT26, D2S123, D5S346, and D17S250 (the Bethesda markers).
Mutation analyses of BRAF
BRAF exon 15 was amplified using the primers: sense 5’‐TCATAATGCTTGCTGTGATAGGA‐3'and antisense 5’‐GGCCAAAAATTTAATCAGTGGA‐3’. The PCR products were purified enzymatically by illustra ExoStar 1‐step (GE Healthcare Bio‐Sciences Corp., Piscataway, NJ, US) prior to sequencing using the BigDye Terminator v.1.1 Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA, US). The sequencing products were purified with Big Dye Xterminator (Applied Biosystems) or Sephadex (GE healthcare) and subjected to sequencing on an ABI 3730 DNA Sequencer (Applied Biosystems).
Statistical analysis
The statistical analyses were performed using IMB SPSS Statistics 21 and R version 3.2.2 together with the “survival”31 and “bootStepAIC”32 packages. χ2 or Fisher's exact tests (when expected frequencies were less than five) were applied to evaluate associations between CIMP and clinicopathological features (categorical variables). In survival analyses, the end points were five‐year time to recurrence (TTR) and five‐year overall survival (OS). TTR, defined according to Punt et al.33 was calculated from the date of surgery until the first of death from the same cancer, local recurrence or distal metastasis. Patients were censored at last follow‐up, death caused by other events than colorectal cancer and death due to postoperative complications (<3 months). OS was calculated from time of surgery until death of any cause, and cases were censored at last follow‐up. The effect of CIMP and BRAF were first independently evaluated in an univariate approach. Kaplan‐Meier method was used to estimate the survival curves and univariate Cox's proportional hazard models were fitted to the data. Hazard Ratios (HRs) and 95% confidence intervals (CIs) were derived from the model, and significance of the parameters was assessed using a Wald's test. The multivariate approaches were also based on Cox's proportional hazard model and preceded by a stepwise selection procedure by Akaike information criterion (AIC), in order to identify a subset of relevant predictor variables from the set of available clinicopathological data, both for the whole series and for the MSS tumors alone. To ensure robustness in the selection procedure, a Bootstrap approach with 1,000 iterations was implemented. A total of 1035 patients, including 872 with MSS tumors, had information on all predictor variables, and were included in the multivariate analyses. A χ2 test was first performed to evaluate whether the proportional hazards assumption for a Cox regression model fit was met. The significance of the variables included in the final model was then assessed by a Wald's test and p values <0.05 was considered statistically significant.
Finally, three alternative ways of stratifying patients according to age at diagnosis were tested, 1) <70 and ≥70, 2) <55, 55–74, ≥75, and 3) <60 years, 60–74 years, and ≥75 years. The third alternative achieved the lowest AIC value and was used for further analyses.
Results
CIMP and clinical‐ and molecular features
Among the 1122 samples successfully analyzed for CIMP status, 207 (18%) were CIMP positive, including sixty‐two of 933 (7%) MSS tumors and 143 of 180 (79%) MSI tumors. Associations between CIMP and clinical‐ and molecular features are shown separately for the MSS and MSI groups (Table 1), and across the whole series (Supporting Information Table 1). For all three groups, CIMP was significantly associated with BRAFV600E mutation, promoter methylation status of MLH1, right sided tumor localization, and female gender. In the MSS group, CIMP positive tumors were more frequently of advanced stages (III and IV) compared with CIMP negative tumors. An association between CIMP and age was only observed in the MSI group (Table 1). Associations between CIMP and clinical and molecular features stratified by age are summarized in Supporting Information Tables 2–4.
Table 1.
Associations between CIMP and clinical‐ and molecular features stratified by MSI status.
| MSS | MSI | |||||||
|---|---|---|---|---|---|---|---|---|
|
Total n |
CIMP− n (%) |
CIMP+ n (%) |
P value |
Total n |
CIMP− n (%) |
CIMP+ n (%) |
P value | |
| No. of patients | 933 | 871 (93) | 62 (7) | 180 | 37 (21) | 143 (79) | ||
| Gender | 0.004 | 0.024 | ||||||
| Male | 474 | 454 (96) | 20 (4) | 50 | 16 (32) | 34 (68) | ||
| Female | 459 | 417 (91) | 42 (9) | 130 | 21 (16) | 109 (84) | ||
| Age | 0.597 | <0.001 | ||||||
| <60 | 170 | 159 (94) | 11 (6) | 23 | 13 (57) | 10 (43) | ||
| 60–74 | 367 | 339 (92) | 28 (8) | 63 | 11 (17) | 52 (83) | ||
| ≥75 | 396 | 373 (94) | 23 (6) | 94 | 13 (14) | 81 (86) | ||
| Stage | <0.001 | 0.475 | ||||||
| I | 166 | 165 (99) | 1 (1) | 22 | 6 (27) | 16 (73) | ||
| II | 333 | 316 (95) | 17 (5) | 105 | 20 (19) | 85 (81) | ||
| III | 258 | 236 (91) | 22 (9) | 40 | 10 (25) | 30 (75) | ||
| IV | 173 | 151 (87) | 22 (13) | 13 | 1 (8) | 12 (92) | ||
| Localization | <0.001 | <0.001 | ||||||
| Right colon | 301 | 260 (86) | 41 (14) | 151 | 22 (15) | 129 (85) | ||
| Left colon | 329 | 314 (95) | 15 (5) | 18 | 7 (39) | 11 (61) | ||
| Rectum | 289 | 284 (98) | 5 (2) | 8 | 6 (75) | 2 (25) | ||
| BRAF | <0.001 | <0.001 | ||||||
| BRAF wt | 825 | 792 (96) | 33 (4) | 53 | 30 (57) | 23 (43) | ||
| BRAF mut | 55 | 28 (51) | 27 (49) | 111 | 3 (3) | 108 (97) | ||
| MLH1methylation | 0.024 | <0.001 | ||||||
| MLH1 unmeth | 929 | 869 (94) | 60 (6) | 35 | 25 (71) | 10 (29) | ||
| MLH1meth | 4 | 2 (50) | 2 (50) | 145 | 12 (8) | 133 (92) | ||
Meth, methylated; mut, mutation; No., number; unmeth, unmethylated; wt, wild type.
CIMP and survival analyses
Data from a total of 1118 patients (758 from Oslo 1 and 360 from Oslo 2) were available for survival analyses. Across all samples, CIMP alone was not significantly associated with OS or TTR (Table 2, and Supporting Information Table 5, respectively). In stage stratified analyses, there was a tendency towards worse survival for those with stage III CIMP positive tumors and significantly worse survival among patients with stage IV CIMP positive tumors, compared to those with CIMP negative tumors (Supporting Information Figure 1).
Table 2.
Univariate and multivariate Cox proportional hazard analyses with overall survival as endpoint.
| Patients, n | Events, n |
Univariate HR (95% CI) |
P value | Patients, n | Multivariate HR (95% CI) | P value | |
|---|---|---|---|---|---|---|---|
| Gender | |||||||
| Male | 525 | 227 | 1.00 (ref) | Not included | |||
| Female | 593 | 262 | 1.05 (0.88–1.26) | 0.575 | Not included | ||
| Age | |||||||
| <60 | 194 | 50 | 1.00 (ref) | 181 | 1.00 (ref) | ||
| 60–74 | 434 | 178 | 1.78 (1.30–2.44) | <0.001 | 403 | 1.97 (1.42–2.73) | <0.001 |
| ≥75 | 490 | 261 | 2.51 (1.85–3.40) | <.0001 | 451 | 3.51 (2.56–4.83) | <0.001 |
| Stage | |||||||
| I | 188 | 37 | 1.00 (ref) | 175 | 1.00 (ref) | ||
| II | 439 | 138 | 1.73 (1.20–2.49) | 0.003 | 408 | 1.43 (0.98–2.09) | 0.061 |
| III | 301 | 144 | 3.08 (2.14–4.42) | <0.001 | 275 | 2.75 (1.90–4.00) | <0.001 |
| IV | 188 | 169 | 11.96 (8.34–17.14) | <.0001 | 177 | 3.29 (1.90–5.68) | <0.001 |
| Localization | |||||||
| Right | 454 | 202 | 1.00 (ref) | Not included | |||
| Left | 350 | 164 | 1.06 (0.86–1.30) | 0.602 | Not included | ||
| Rectum | 298 | 116 | 0.81 (0.65–1.02) | 0.073 | Not included | ||
| CIMP | |||||||
| CIMP− | 912 | 392 | 1.00 (ref) | 846 | 1.00 (ref) | ||
| CIMP+ | 206 | 97 | 1.18 (0.94–1.47) | 0.144 | 189 | 1.89 (1.34–2.65) | <0.001 |
| MLH1methylation | |||||||
| MLH1 unmeth | 970 | 439 | 1.00 (ref) | 899 | 1.00 (ref) | ||
| MLH1 meth | 148 | 50 | 0.67 (0.50–0.90) | 0.008 | 136 | 0.35 (0.23–0.56) | <0.001 |
| MSI status | |||||||
| MSS | 930 | 423 | 1.00 (ref) | Not included | |||
| MSI | 179 | 61 | 0.69 (0.53–0.90) | 0.007 | Not included | ||
| BRAF | |||||||
| BRAF wt | 882 | 379 | 1.00 (ref) | 871 | 1.00 (ref) | ||
| BRAF mut | 167 | 78 | 1.17 (0.91–1.49) | 0.216 | 164 | 1.32 (0.92–1.9) | 0.128 |
| R status | |||||||
| R0 | 905 | 306 | 1.00 (ref) | 836 | 1.00 (ref) | ||
| R1 | 24 | 10 | 1.39 (0.74–2.61) | 0.304 | 24 | 1.28 (0.68–2.41) | 0.451 |
| R2 | 186 | 170 | 6.75 (5.55–8.21) | <0.001 | 175 | 4.30 (2.77–6.66) | <0.001 |
Meth, methylated; mut, mutation; unmeth, unmethylated; wt, wild type.
The multivariate models with OS and TTR as end points are shown in Table 2 and Supporting Information Table 5, respectively. Following the selection procedure and the coefficient testing CIMP was, in contrast to the univariate analysis, included and significant in both models after adjusting for stage, R status, age, MSI or MLH1 and BRAF mutation status.
Poor prognosis for patients with MSS CIMP positive tumors
Patients with CIMP positive MSS tumors displayed a significantly worse OS and shorter TTR compared to those with CIMP negative MSS tumors (Table 3 and Supporting Information Table 6, respectively. Visualized in Figure 1A). Further stratification by stage showed that CIMP was significantly associated with worse outcome among patients with MSS stage III and stage IV tumors (Figure 1C‐D).
Table 3.
Univariate and multivariate Cox proportional hazard analyses with OS survival as end point in patients with MSS tumors.
| Total, n | Events, n |
Univariate HR (95% CI) |
P value | Total, n |
Multivariate HR (95% CI) |
P value | |
|---|---|---|---|---|---|---|---|
| Gender | |||||||
| Male | 472 | 210 | 1.00 (ref) | Not included | |||
| Female | 458 | 213 | 1.10 (0.91–1.33) | 0.346 | Not included | ||
| Age | |||||||
| <60 | 170 | 45 | 1.00 (ref) | 163 | 1.00 (ref) | ||
| 60–74 | 365 | 161 | 1.92 (1.38–2.67) | <0.001 | 342 | 2.10 (1.49–2.95) | <0.001 |
| ≥75 | 395 | 217 | 2.57 (1.86–3.54) | <0.001 | 367 | 3.55 (2.54–4.96) | <0.001 |
| Stage | |||||||
| I | 165 | 34 | 1.00 (ref) | 156 | 1.00 (ref) | ||
| II | 333 | 111 | 1.78 (1.21–2.62) | 0.003 | 311 | 1.45 (0.98–2.16) | 0.064 |
| III | 257 | 123 | 2.94 (2.01–4.29) | <0.001 | 240 | 2.71 (1.83–4.00) | <0.001 |
| IV | 173 | 154 | 10.94 (7.51–15.92) | <0.001 | 165 | 2.72 (1.52–4.86) | <0.001 |
| Localization | |||||||
| Right | 301 | 152 | 1.00 (ref) | Not included | |||
| Left | 328 | 153 | 0.87 (0.69–1.09) | 0.217 | Not included | ||
| Rectum | 288 | 111 | 0.66 (0.52–0.85) | 0.001 | Not included | ||
| BRAF | |||||||
| BRAF wt | 822 | 359 | 1.00 (ref) | 818 | 1.00 (ref) | ||
| BRAF mut | 55 | 39 | 2.48 (1.78–3.45) | <0.001 | 54 | 1.57 (1.07–2.29) | 0.021 |
| CIMP | |||||||
| CIMP− | 868 | 379 | 1.00 (ref) | 813 | 1.00 (ref) | ||
| CIMP+ | 62 | 44 | 2.47 (1.80–3.37) | <0.001 | 59 | 1.84 (1.28–2.63) | <0.001 |
| MLH1methylation | |||||||
| MLH1 unmeth | 926 | 422 | 1.00 (ref) | Not included | |||
| MLH1 meth | 4 | 1 | 0.51 (0.07–6.60) | 0.497 | Not included | ||
| R status | |||||||
| R0 | 738 | 255 | 1.00 (ref) | 691 | 1.00 (ref) | ||
| R1 | 19 | 10 | 1.93 (1.02–3.62) | 0.042 | 19 | 1.55 (0.82–2.94) | 0.179 |
| R2 | 170 | 155 | 6.51 (5.28–8.01) | <0.001 | 162 | 4.92 (3.07–7.88) | <0.001 |
Meth, methylated; mut, mutation; unmeth, unmethylated; wt, wild type.
Figure 1.
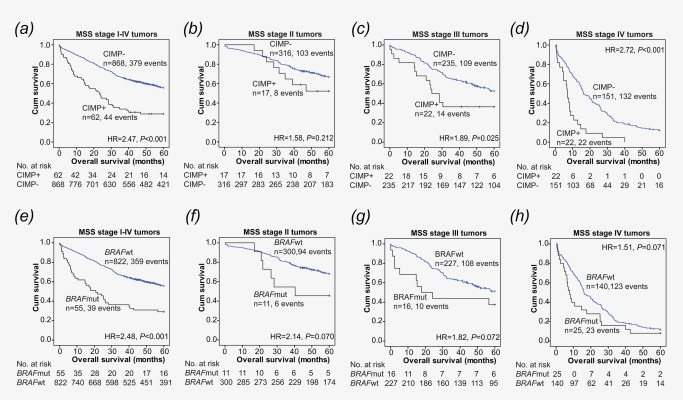
Kaplan Meier curves modeling the effect of CIMP (A‐D) and BRAF (E‐H) on OS among patients with MSS tumors.
CIMP status stratifies the poor prognosis group of patients with MSS and BRAF mutated tumors
BRAFV600 mutation was significantly associated with worse survival among patients with MSS tumors (univariate analysis; Table 3 and Supporting Information Table 6. Visualized in Figure 1E). The same trend was observed among patients with stage II, III or IV tumors (Figure 1F‐H).
To evaluate the effect of CIMP on BRAF, patients with MSS tumors were divided into four groups; 1: CIMP negative/BRAF wild type, 2: CIMP negative/BRAF mutated, 3: CIMP positive/BRAF wild type, and 4: CIMP positive/BRAF mutated. Results are presented in Figure 2 and Table 4, and show that patients with type 1, 2, or 3 tumors all had a significantly improved OS compared to patients with type 4 tumors (both CIMP positive and BRAF mutated). Although there were few patients within each stage, this finding was in general maintained in stage II‐III. For stage IV, no difference in survival was observed between patients with CIMP positive tumors with or without BRAF mutation (type 4 and type 3, respectively; Figure 2D).
Figure 2.
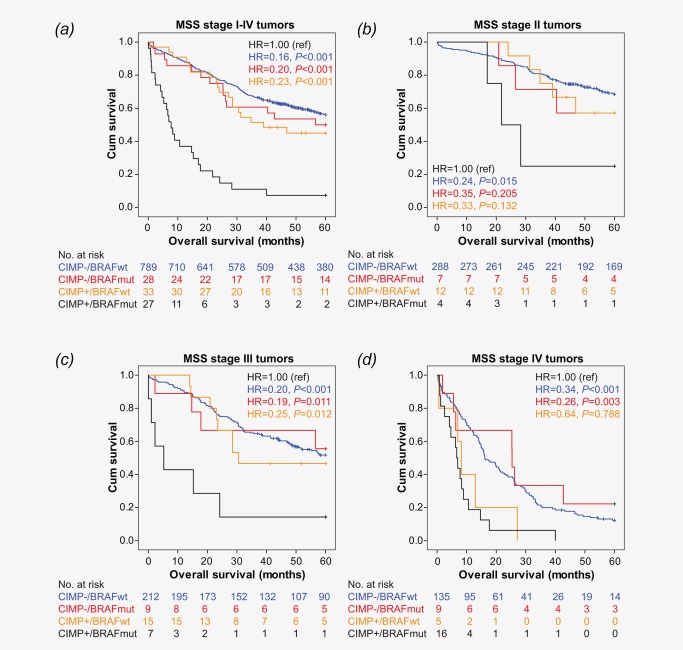
Effect of CIMP and BRAF on OS in patients with MSS tumors estimated by Kaplan Meier method.
Table 4.
Combined effect of CIMP and BRAF in MSS tumors.
| Overall survival | |||||||
|---|---|---|---|---|---|---|---|
| Total, n | Events, n | Univariate HR (95% CI) | P value | Total, n | Multivariate HR (95% CI) | P valuea | |
| CIMP‐BRAF | |||||||
| CIMP−/BRAFwt | 789 | 341 | 0.16 (0.11–0.24) | <0.001 | 785 | 0.28 (0.18–0.43) | <0.001 |
| CIMP−/BRAFmut | 28 | 14 | 0.20 (0.10–0.38) | <0.001 | 28 | 0.33 (0.17–0.64) | 0.001 |
| CIMP+/BRAFwt | 33 | 18 | 0.23 (0.12–0.42) | <0.001 | 33 | 0.40 (0.21–0.76) | 0.005 |
| CIMP+/BRAFmut | 27 | 25 | 1.00 (ref) | 26 | 1.00 (ref) | ||
Ajusted for age, stage, and R status. Mut, mutation; wt, wild type.
The worse survival for patients with CIMP positive BRAF mutated tumors remained significant after adjusting for stage, R status, and age (Table 4). Comparable findings were observed for TTR (not shown).
When looking specifically at the effect of CIMP in patients with MSS, BRAF mutated tumors, we observed that also within this poor prognostic group, CIMP was significantly associated with worse OS (HR = 4.39, 95% CI (2.21–8.70), P < 0.001; Figure 2) and shorter TTR (HR = 4.02, 95% CI (4.84–8.77), P < 0.001). The same trend was maintained among patients with stage II, III or IV tumors (Figure 2B‐D), showing that the worse survival observed was not only due to later stages.
From multivariate analyses, both CIMP and BRAF were included in the final model with OS as end point, suggesting that both variables have independent prognostic value. However, only CIMP was included in the model with TTR as endpoint (Supporting Information Table 6).
No effect of CIMP or BRAF among patients with MSI tumors
Among patients with MSI tumors, neither CIMP nor BRAF status was alone associated with survival (CIMP: OS, HR = 1.34, 95% CI (0.68–2.65); TTR, HR = 1.13, 95% CI (0.54–2.35); BRAF: OS, HR = 1.00, 95% CI (0.56–1.77); TTR, HR = 1.13, 95% CI (0.54–2.35)). Also when stratified by stage, no significant associations were seen, neither for CIMP nor for BRAF.
The effect of CIMP on survival is consistent across different age groups
The univariate effect of CIMP and BRAF on OS in the three analyzed age groups (<60, 60–74, ≥75) are summarized in Supporting Information Table 7 and Supporting Information Figure 2. The associations with survival were in general consistent across all three age groups.
Discussion
In this population representative series of colorectal cancer patients, we showed that patients with CIMP positive tumors had a significantly worse prognosis compared to patients with CIMP negative tumors after adjustment for age, stage, R status, and MSI or MLH1 methylation and BRAF mutation. The effect was strongest among patients with MSS tumors. Interestingly, CIMP status could further identify high risk patients among the poor prognosis group of patients with MSS and BRAF mutated tumors, a finding that remained significant after adjusting for stage, R status and age.
Already in 1993, we reported that patients with MSI showed an improved prognosis compared to those with MSS tumors.14 This finding was later confirmed in a meta‐analysis by Popat et al.12 Within the already good prognostic MSI group, CIMP does not seem to contribute with prognostic value,34, 35, 36 which is in agreement with our results. In the MSS group, however, several reports have demonstrated that patients with CIMP positive tumors have a significantly poorer survival compared to those with CIMP negative tumors.16, 17, 18 , 35, 36, 37, 38 With some exceptions, including the study by Barault et al 36 (n = 582 samples), the significant effect of CIMP is often lost in multivariate analyses. In this context a small sample size will be an obvious limitation, reducing the statistical power in subgroup analyses.21 In the present study, including >1000 patients, we confirmed that CIMP conferred a worse survival in the MSS subgroup. The inferior survival among patients with CIMP positive tumors remained significant in multivariate analyses, both across all samples and in the MSS subgroup.
Furthermore, patients with MSS BRAF mutated tumors have been shown to have a particularly poor prognosis.38, 39 We observed the same, and further demonstrated that CIMP was significantly associated with inferior survival among patients with MSS BRAF mutated tumors. CIMP thus defined a subgroup with worse prognosis among these patients. By analyzing 236 MSS tumors, Kim et al 17 also observed that patients with CIMP positive MSS BRAF mutated tumors had a particularly bad prognosis. CIMP was, however, not included in the final multivariate model, and they concluded that the inferior survival among patients with CIMP positive BRAF mutated tumors was attributed to the BRAF mutation.17 Samowitz et al 38 also looked at CIMP and BRAF among patients with MSS tumors. Although they found that patients with BRAF mutant CIMP positive tumors had a significantly worse survival compared to those with CIMP negative BRAF wild type tumors, they also concluded that the bad prognostic effect on survival was entirely due to the BRAF mutation. In their adjusted analysis of MSS tumors, CIMP was not significantly associated with OS or cancer specific survival.38 This is in contrast to our study, where CIMP is included in the final multivariate model and significant with both TTR and OS as end points. Furthermore, from our results it seems like it is the combination of CIMP and BRAF within MSS tumors that provides the worse survival. Patients with either a BRAF mutated or a CIMP positive tumor does not seem to have an inferior survival compared to patients with MSS tumors without any of these alterations. Interestingly, Phipps et al40 recently reported on the survival of 2050 colorectal cancer patients based on classification into five subgroups according to CIMP, MSI and BRAF and KRAS mutation status.41 Compared to patients with type four tumors (MSS, CIMP negative, BRAF/KRAS wild type), patients with type two tumors (MSS, CIMP positive, BRAF mutation) had the highest disease‐specific mortality.40 Although this stratification approach did not focus directly at the prognostic effect of CIMP within the MSS, BRAF mutated subgroup, it clearly underscores that patients with MSS, BRAF mutated CIMP positive tumors have a particularly poor outcome. We further show that this finding persisted in patients with stage II, III or IV disease.
We additionally observed that patients with stage IV CIMP positive tumors had a poorer prognosis, irrespective of BRAF and/or MSI status. This has also been observed by others,42, 43, 44 suggesting that CIMP may be used as a poor prognostic marker for advanced disease.
From a subset of the samples included in the Oslo 2 series, data indicated that the prognostic effect of CIMP might depend on the age of the patient. However, these findings were not replicated across a larger sample series (Oslo 1). Instead we observed that CIMP was associated with worse survival independent of age. Nevertheless, this is an interesting observation, and to our knowledge, few studies have looked at the prognostic effect of CIMP in younger (<60) versus old (>75) colorectal cancer patients. Approximately 40% of colorectal cancer patients are older than 75 years. Still, they are underrepresented in clinical trials of adjuvant therapy, mainly due to higher rates of co‐morbidities.45 This results in sparse data on how to best treat this group of patients, including whether they benefit from adjuvant therapy. Forty‐four percent of the patients included in the present study were 75 years or older. Worse survival of CIMP within the MSS and MSS BRAF mutated subgroup was also seen among these patients, indicating that CIMP status could be applicable to guide the choice of treatment in all colorectal cancer patients, including the elderly.
An apparent challenge within the CIMP literature is the high variation across studies when it comes to choice of gene panel, marker threshold and method to define CIMP, and also the lack of consistent stratification based on molecular and clinical features.46 These factors may influence the association between CIMP and survival, and also makes meta‐analyses challenging.47 To avoid potential biases introduced by qualitative methods such as methylation specific PCR (MSP),48 we have used quantitative MSP (qMSP) to analyze an accepted and validated CIMP panel.5, 22 In accordance with other publications,5, 16 , 49 we find that CIMP is commonly associated with the BRAFV600E mutation, MSI, MLH1 methylation, female gender, tumor localization, and older age, underscoring that the series used here is compatible to that of others. All patients included have control follow‐ups at the same hospital and the clinical data are continuously quality controlled and updated. The high number of samples included in this study combined with the high‐quality clinical data provides a unique opportunity for robust analyses, also within patient subgroups.
In the present study, we have implemented a selection procedure by AIC in order to determine the best‐fitting model from the initial set of clinicopathological data. This approach enables discarding of spurious variables, which may have added noise to the model and also helps preventing over‐fitting. We combined the selection procedure with a bootstrap re‐sampling method in order to guarantee the stability of the final model. By running the whole procedure independently for TTR and OS, stage, age, R status and CIMP were all systematically included in both models. In addition to stage, age and R status, which are variables known to be associated with survival, these results strongly suggest that CIMP is also a robust marker for survival among colorectal cancer patients, and should be considered in the prognosis of the disease.
In conclusion, we report that CIMP is associated with worse prognosis in colorectal cancer patients after adjusting for other factors, and specifically among patients with MSS and MSS BRAF mutated tumors.
Supporting information
Supporting Information Table 1. Associations between CIMP and clinical‐ and molecular features across all colorectal cancers analyzed
Supporting Information Table 2. Associations between CIMP and clinical‐ and molecular features stratified by age
Supporting Information Table 3. MSS tumors. Associations between CIMP and clinical‐ and molecular features stratified by age
Supporting Information Table 4. MSI tumors. Associations between CIMP and clinical‐ and molecular features stratified by age
Supporting Information Table 5. Univariate and multivariate Cox proportional hazard analyses with time to recurrence as endpoint.
Supporting Information Table 6. Univariate and multivariate Cox proportional hazard analyses among patients with MSS tumors with overall survival as end point.
Supporting Information Table 7. Univariate effect of CIMP and BRAF on overall survival stratified by age.
Supporting Information Figure 1. Effect of CIMP on OS modelled by Kaplan Meier plots in patients with (A) stage III, and (B) stage IV tumors.
Supporting Information Figure 2. Effect of CIMP and BRAF mutation on OS among patients with MSS tumors (A) <60 years, (B) 60‐74 years, and (C) ≥75 years.
Contributors Conception and design: RAL, GEL; Acquisition of data: HMV, SAD, MM, HH, GKP, MH, ME, OHS, AS, AN; Analyses and interpretation of data: HMV, MJ, SAD, MM, AN, RAL, GEL; Drafting of the manuscript: HMV, GEL; Study supervision: GEL; All authors were involved in revision of the manuscript and have approved the final version.
Funding This work was supported by grants from the South‐Eastern Norway Regional Health Authority (to G.E. Lind), Stiftelsen Kristian Gerhard Jebsen, the Norwegian Cancer Society (grant 72190 PR‐2006‐0442 to R.A. Lothe), the Research Council of Norway (project number 239961, to G.E. Lind; project number 218241 to R.A. Lothe through Norwegian Cancer Genomics Consortium, funding S.A. Danielsen as staff scientist), and the Centre of Excellence funding scheme (project number 179571).
Competing interests The authors declare no conflict of interests.
Reference List
- 1. Ferlay J, Soerjomataram I, Ervik M et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon,France: International Agency for Research on Cancer; 2013. Available from http://globocan.iarc.fr, accessed on 06/06/2016.
- 2. Edge SB, Byrd DR, Compton CC et al. AJCC cancer staging manual, 7th ed New York: Springer; 2010. [Google Scholar]
- 3. Bertagnolli MM, Redston M, Compton CC, et al. Microsatellite instability and loss of heterozygosity at chromosomal location 18q: prospective evaluation of biomarkers for stages II and III colon cancer–a study of CALGB 9581 and 89803. J Clin Oncol 2011; 29 3153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Issa JP, CpG island methylator phenotype in cancer. Nat Rev Cancer 2004; 4 988–93. [DOI] [PubMed] [Google Scholar]
- 5. Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006; 38 787–93. [DOI] [PubMed] [Google Scholar]
- 6. Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population‐based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 2005; 129 837–45. [DOI] [PubMed] [Google Scholar]
- 7. Issa JP, Shen L, Toyota M, CIMP, at last. Gastroenterology 2005; 129 1121–24. [DOI] [PubMed] [Google Scholar]
- 8. Kudryavtseva AV, Lipatova AV, Zaretsky AR, et al. Important molecular genetic markers of colorectal cancer. Oncotarget 2016; 7 53959–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kocarnik JM, Shiovitz S, Phipps AI, Molecular phenotypes of colorectal cancer and potential clinical applications. Gastroenterology Report 2015; 3 269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colussi D, Brandi G, Bazzoli F, et al. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. International Journal of Molecular Sciences 2013; 14 16365–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diep CB, Thorstensen L, Meling GI, et al. Genetic tumor markers with prognostic impact in Dukes' stages B and C colorectal cancer patients. J Clin Oncol 2003; 21 820–29. [DOI] [PubMed] [Google Scholar]
- 12. Popat S, Hubner R, Houlston RS, Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 2005; 23 609–18. [DOI] [PubMed] [Google Scholar]
- 13. Guastadisegni C, Colafranceschi M, Ottini L, et al. Microsatellite instability as a marker of prognosis and response to therapy: A meta‐analysis of colorectal cancer survival data. European Journal of Cancer 2010; 46 2788–98. [DOI] [PubMed] [Google Scholar]
- 14. Lothe RA, Peltomaki P, Meling GI, et al. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res 1993; 53 5849–52. [PubMed] [Google Scholar]
- 15. Walther A, Houlston R, Tomlinson I, Association between chromosomal instability and prognosis in colorectal cancer: a meta‐analysis. Gut 2008; 57 941–50. [DOI] [PubMed] [Google Scholar]
- 16. Hawkins N, Norrie M, Cheong K, et al. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology 2002; 122 1376–87. [DOI] [PubMed] [Google Scholar]
- 17. Kim JH, Shin SH, Kwon HJ, et al. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch 2009; 455 485–94. [DOI] [PubMed] [Google Scholar]
- 18. Lee S, Cho NY, Choi M, et al. Clinicopathological features of CpG island methylator phenotype‐positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathol Int 2008; 58 104–13. [DOI] [PubMed] [Google Scholar]
- 19. Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 2009; 58 90–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zanutto S, Pizzamiglio S, Lampis A, et al. Methylation status in patients with early stage colon cancer: a new prognostic marker?. Int J Cancer 2012; 130 488–89. [DOI] [PubMed] [Google Scholar]
- 21. Jia M, Gao X, Zhang Y, et al. Different definitions of CpG island methylator phenotype and outcomes of colorectal cancer: a systematic review. Clin Epigenetics 2016; 8 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ogino S, Kawasaki T, Kirkner GJ, et al. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population‐based sample. J Mol Diagn 2007; 9 305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Merok MA, Ahlquist T, Royrvik EC, et al. Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: results from a large, consecutive Norwegian series. Ann Oncol 2013; 24 1274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bruun J, Kolberg M, Ahlquist TC, et al. Regulator of Chromosome Condensation 2 Identifies High‐Risk Patients within Both Major Phenotypes of Colorectal Cancer. Clin Cancer Res 2015; 21 3759–70. [DOI] [PubMed] [Google Scholar]
- 25. Berg M, Danielsen SA, Ahlquist T, et al. DNA sequence profiles of the colorectal cancer critical gene set KRAS‐BRAF‐PIK3CA‐PTEN‐TP53 related to age at disease onset. PLoS One 2010; 5 e13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sveen A, Agesen TH, Nesbakken A, et al. ColoGuidePro: a prognostic 7‐gene expression signature for stage III colorectal cancer patients. Clin Cancer Res 2012; 18 6001–10. [DOI] [PubMed] [Google Scholar]
- 27. Berg M, Agesen TH, Thiis‐Evensen E, et al. Distinct high resolution genome profiles of early onset and late onset colorectal cancer integrated with gene expression data identify candidate susceptibility loci. Mol Cancer 2010; 9 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andresen K, Boberg KM, Vedeld HM, et al. Novel target genes and a valid biomarker panel identified for cholangiocarcinoma. Epigenetics 2012; 7 1249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005; 33 6823–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Levine AJ, Phipps AI, Baron JA, et al. Clinicopathologic Risk Factor Distributions for MLH1 Promoter Region Methylation in CIMP‐Positive Tumors. Cancer Epidemiol Biomarkers Prev 2016; 25 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Springer, New York: 2000. [Google Scholar]
- 32. Rizopoulos D. bootStepAIC: Bootstrap stepAIC. In. http://CRAN.R-project.org/package=bootStepAIC: 2009.
- 33. Punt CJ, Buyse M, Kohne CH, et al. Endpoints in adjuvant treatment trials: a systematic review of the literature in colon cancer and proposed definitions for future trials. J Natl Cancer Inst 2007; 99 998–1003. [DOI] [PubMed] [Google Scholar]
- 34. Ward RL, Cheong K, Ku SL, et al. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol 2003; 21 3729–36. [DOI] [PubMed] [Google Scholar]
- 35. Dahlin AM, Palmqvist R, Henriksson ML, et al. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin Cancer Res 2010; 16 1845–55. [DOI] [PubMed] [Google Scholar]
- 36. Barault L, Charon‐Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population‐based series of 582 cases. Cancer. Res 2008; 68 8541–46. [DOI] [PubMed] [Google Scholar]
- 37. Shiovitz S, Bertagnolli MM, Renfro LA, et al. CpG Island Methylator Phenotype Is Associated With Response to Adjuvant Irinotecan‐Based Therapy for Stage III Colon Cancer. Gastroenterology 2014; 147 637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite‐stable colon cancers. Cancer Res 2005; 65 6063–69. [DOI] [PubMed] [Google Scholar]
- 39. Kakar S, Deng G, Sahai V, et al. Clinicopathologic characteristics, CpG island methylator phenotype, and BRAF mutations in microsatellite‐stable colorectal cancers without chromosomal instability. Arch Pathol Lab Med 2008; 132 958–64. [DOI] [PubMed] [Google Scholar]
- 40. Phipps AI, Limburg PJ, Baron JA, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 2015; 148 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jass JR, Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 2007; 50 113–30. [DOI] [PubMed] [Google Scholar]
- 42. Ogino S, Meyerhardt JA, Kawasaki T, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch 2007; 450 529–37. [DOI] [PubMed] [Google Scholar]
- 43. Lee MS, McGuffey EJ, Morris JS, et al. Association of CpG island methylator phenotype and EREG/AREG methylation and expression in colorectal cancer. Br J Cancer 2016; 114 1352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cha Y, Kim KJ, Han SW, et al. Adverse prognostic impact of the CpG island methylator phenotype in metastatic colorectal cancer. Br J Cancer 2016; 115 164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dienstmann R, Salazar R, Tabernero J, Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol 2015; 33 1787–96. [DOI] [PubMed] [Google Scholar]
- 46. Hughes LA, Khalid‐de Bakker CA, Smits KM, et al. The CpG island methylator phenotype in colorectal cancer: progress and problems. Biochim Biophys Acta 2012; 1825 77–85. [DOI] [PubMed] [Google Scholar]
- 47. Juo YY, Johnston FM, Zhang DY, et al. Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta‐analysis. Ann Oncol 2014; 25 2314–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut 2006; 55 1000–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hinoue T, Weisenberger DJ, Lange CP, et al. Genome‐scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 2012; 22 271–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Table 1. Associations between CIMP and clinical‐ and molecular features across all colorectal cancers analyzed
Supporting Information Table 2. Associations between CIMP and clinical‐ and molecular features stratified by age
Supporting Information Table 3. MSS tumors. Associations between CIMP and clinical‐ and molecular features stratified by age
Supporting Information Table 4. MSI tumors. Associations between CIMP and clinical‐ and molecular features stratified by age
Supporting Information Table 5. Univariate and multivariate Cox proportional hazard analyses with time to recurrence as endpoint.
Supporting Information Table 6. Univariate and multivariate Cox proportional hazard analyses among patients with MSS tumors with overall survival as end point.
Supporting Information Table 7. Univariate effect of CIMP and BRAF on overall survival stratified by age.
Supporting Information Figure 1. Effect of CIMP on OS modelled by Kaplan Meier plots in patients with (A) stage III, and (B) stage IV tumors.
Supporting Information Figure 2. Effect of CIMP and BRAF mutation on OS among patients with MSS tumors (A) <60 years, (B) 60‐74 years, and (C) ≥75 years.


