Abstract
Calmodulin (CaM) functions depend on interactions with CaM‐binding proteins, regulated by . Induced structural changes influence the affinity, kinetics, and specificities of the interactions. The dynamics of CaM interactions with neurogranin (Ng) and the CaM‐binding region of /calmodulin‐dependent kinase II (CaMKII290−309) have been studied using biophysical methods. These proteins have opposite dependencies for CaM binding. Surface plasmon resonance biosensor analysis confirmed that and CaM interact very rapidly, and with moderate affinity ( ). Calmodulin‐CaMKII290−309 interactions were only detected in the presence of , exhibiting fast kinetics and nanomolar affinity ( ). The CaM–Ng interaction had higher affinity under ‐depleted ( and k −1 = 1.6 × 10−1s−1) than ‐saturated conditions ( ). The IQ motif of Ng (Ng27−50) had similar affinity for CaM as Ng under ‐saturated conditions ( ), but no interaction was seen under ‐depleted conditions. Microscale thermophoresis using fluorescently labeled CaM confirmed the surface plasmon resonance results qualitatively, but estimated lower affinities for the Ng ( ) and CaMKII290−309( ) interactions. Although CaMKII290−309 showed expected interaction characteristics, they may be different for full‐length CaMKII. The data for full‐length Ng, but not Ng27−50, agree with the current model on Ng regulation of /CaM signaling.
Keywords: calmodulin, calmodulin‐dependent kinase, surface plasmon resonance, microscale thermophoresis, neurogranin
1. INTRODUCTION
Biological processes are regulated via complex networks involving protein‐protein interactions complemented by protein interactions with nucleic acids, small organic ligands, and inorganic ions. The occurrence and localization of the proteins are regulated genetically while the concentration of small organic ligands and ions can be regulated by other mechanisms. The different regulatory mechanisms influence not only the localization of the effect but also how rapid the onset of the effect is as well as its duration and specificity. To better understand biological processes, we need new strategies that do not simply identify interacting proteins and their binding partners, their structures, and localization, but that also allow the interaction mechanism, affinities, and kinetics to be determined under various conditions reflecting the dynamics and specificities of their biological environment.
Calmodulin (CaM) is involved in many protein‐protein networks and thereby plays a central role in the regulation of physiological processes, including signaling and synaptic plasticity of the brain. It is regulated by variations in concentrations that influence its interactions with CaM‐binding proteins (CaMBP). They include enzymes, like /calmodulin‐dependent protein kinase II (CaMKII), and noncatalytic proteins, like neurogranin (Ng).1 Calmodulin interacts primarily in a ‐bound form, but the apo form of CaM can also interact with CaMBPs, giving CaM an active role over a broad range of concentration. The dynamics of the CaM interactome is not known as there is little information about the affinities of the interactions under different conditions and how it regulates the specificity of the interactions between CaM and CaMBPs. Here, we have devised biophysical methods that can provide both mechanistic and quantitative equilibrium‐based and kinetic data, i.e., affinity (K D) and association and dissociation rate constants (k 1 and k −1).
Calmodulin belongs to the helix‐loop‐helix motif (EF‐hand) family of ‐binding proteins and contains 2 pairs of canonical EF‐hands that are separated by an α‐helical linker.2 Binding of to apoCaM induces a conformational change from a compact to an open form. It exposes a hydrophobic pocket in both the N‐ and C‐terminal domains of CaM involved in interactions with a broad range of target proteins.2, 3, 4 Most proteins, like CaMKII, interact with /CaM (open form),5 while some proteins, like Ng and neuromodulin, have a preference toward apoCaM (compact form). 1, 6, 7, 8
Calmodulin‐dependent protein kinase II is one of the major signal transducers. The protein exists in various isoforms. The kinase subunit consists of 3 functional domains (catalytic, autoinhibitory, and self‐associated), assembled into dodecameric homooligomer and heterooligomer (reviewed in Lisman et al. and Hunter and Schulman9, 10). Excluding splicing variants, CaMKII occurs in 2 isoforms, namely, α and β. They are highly enriched in the brain, especially the hippocampus.11, 12 Both isoforms contain a CaM‐binding region, represented by the same polypeptide sequence (in human, aa290‐300 in CaMKIIα and aa291‐301 in CaMKIIβ). Calmodulin‐dependent protein kinase II is a key component for induction of long‐term potentiation in excitatory synapses. It acts by phosphorylation of Ser831 in GluA1‐containing α‐amino‐3‐hydroxy‐5‐methyl‐4‐isoxazole propionic acid receptors (AMPA receptors), thereby increasing single‐channel conductance.13 Furthermore, CaMKII leads to increased insertion of AMPA receptors into the postsynaptic membrane, which further increases synaptic strength.1 The interaction between /CaM and CaMKII leads to autophosphorylation and activation of the enzyme, which remains active even after it dissociates from /CaM.14
Neurogranin, together with neuromodulin and pep19, belongs to the calpacitin group of proteins that is primarily found in neurons. Expression of Ng has been shown to correlate with the onset of synaptogenesis in rats and mice in brain regions that display high levels of neuroplasticity, like the hippocampus.15 Neurogranin has a molecular weight of 7.8 kDa and has a sequence similar to neuromodulin in a region spanning 20 amino acids that contains an IQ motif (I/L/V)QXXXRXXXX(R/K)XX(F/I/L/V/W/Y).16 It corresponds to the protein kinase C phosphorylation and CaM‐binding site.6, 7 It has been shown that the interaction between Ng and CaM enhances synaptic strength.17 Recent data suggest that Ng slows down the diffusion of CaM and increases its concentration at dendritic spines, which, in turn, leads to enhanced sensitivitiy in the synapse.18
Proteins that are regulated by CaM display different levels of sensitivity. This is achieved by cooperativity between the CaM‐CaMBP complexes and the binding of . Positive cooperativity, characterized by an increase of affinity upon protein binding to calmodulin, has been described for the CaM‐troponin I and the CaM‐CaMKII complexes.19, 20, 21 An opposing effect on the affinity for to CaM has been observed for the CaM/Ng complex.8, 21 The modulation of CaM by different protein targets has recently been described according to the allosteric transition model of Monod, Wyman, and Changeux.22, 23
To better understand how CaM exerts its physiological effect, we were interested in developing informative assays that could provide information on the dynamics of the interactions between CaM and CaMBPs, and how the interactions are influenced by . We have previously used surface plasmon resonance (SPR) biosensor technology to distinguish the characteristics of the interactions of CaM and caldendrin with the scaffolding protein, AKAP79.24, 25 These studies revealed that although there were only minor differences in affinity, the interaction mechanisms translated into different kinetics and ‐dependencies, presumably leading to different functions in the synapse.
The interaction between CaM and Ng has been characterized in 2 independent studies by using isothermal titration calorimetry (ITC). They showed that Ng binds to CaM with nanomolar affinity in the absence and with low micromolar affinity in the presence6, 8 of . The present study aimed to shed additional light on this interaction by a time‐resolved approach using SPR biosensor analysis.
To supplement SPR experiments, we used microscale thermophoresis (MST) analysis as an orthogonal equilibrium‐based method to study CaM interactions. The technique is based on the motions of molecules and dispersed particles in a temperature gradient.26 Since thermodiffusion depends on the particle‐solvent interface, which changes upon a binding event, monitoring the dose‐response dependency of particles mobility allows the determination of equilibrium dissociation constant (K D).27 Another solution‐based label‐free method, UV fluorescence spectroscopy, was used as a third strategy to study the interactions between and CaM, validating the functionality of the designed assays. This method is commonly used for studies of CaM and monitors the changes in intrinsic tyrosine fluorescence in CaM upon ‐induced conformational changes.4, 28, 29
For the present study, we used a peptide of CaMKII including the CaM‐binding site (CaMKII290−309), and both full‐length Ng (Ngfl) and Ng‐derived peptide containing an IQ motif (Ng27−50, Figure 1).
Figure 1.

Primary structures of the studied polypeptides. The EF‐hand pairs of human calmodulin (CaM) (P62158) are highlighted (EF‐1 and EF‐2 in blue, EF‐3 and EF‐4 in green). The IQ motif of human neurogranin (Ng) (Q92686) is underlined
2. RESULTS
2.1. Biosensor surfaces for CaM interaction studies
Two types of sensor surfaces and experimental designs were used in the present study, one with CaM immobilized and the other with full‐lenth Ng (Ngfl) immobilized. The CaM surface was used for studies of CaM interactions with , CaMKII290−309, and Ng27−50. It was prepared by immobilizing CaM via amine coupling on SPR biosensor surfaces at surface densities of approximately 400 response units (RU). The Ngfl surface was used for interaction studies between CaM and Ngfl. The surface was prepared by capturing Ngfl containing a N‐terminal His‐tag to antipolyhistidine antibody‐coated sensor surfaces. An analysis cycle was then initiated by capturing Ngfl at levels of approximately 300 RU (Figure 2). After a stabilization period, CaM was injected to monitor the interaction. Before a new cycle was started, the antibody surface was completely regenerated by injecting 10 mM glycine, pH 1.5. The surface densities of captured Ngfl varied slightly between the different replicate experiments, but the relative variation in surface density within a replicate experiment was less than 1%, thereby providing comparable surface properties during the interaction analysis.
Figure 2.
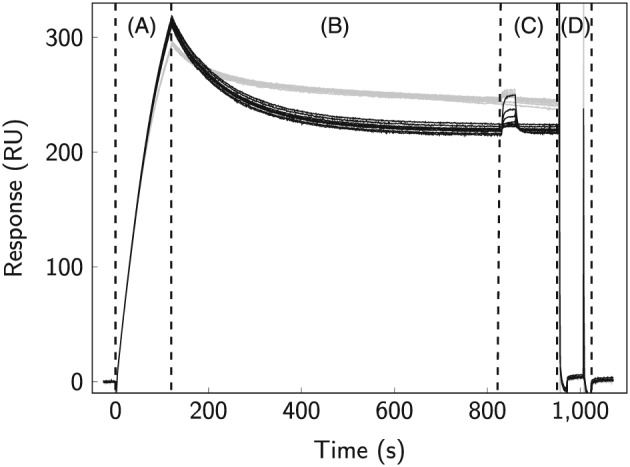
Assay setup for SPR‐based interaction studies between immobilized Ngfl and CaM. A, affinity capture of Ngfl to SPR biosensor surfaces by covalently immobilized polyhistidine antibody; B, stabilization period; C, injection of CaM under ‐saturated (gray) and ‐depleted conditions (black); D, surface regeneration
2.2. Validation of CaM interaction studies
Calmodulin interactions with and polypeptides were studied by several biophysical methods. The SPR, MST, and UV spectroscopic assays were all set up for analysis of the interaction between CaM and to characterize the basic system and validate the methods.
Surface plasmon resonance provided data for the interaction between and immobilized CaM, as shown in Figure 3A. The interaction was very rapid, so the association and dissociation rate constants could not be quantified. Steady‐state affinity analysis was performed, and the macroscopic K D was determined to K D = 2.8 μM at 25°C and K D = 3.2 μM at 22°C (Table 1).
Figure 3.
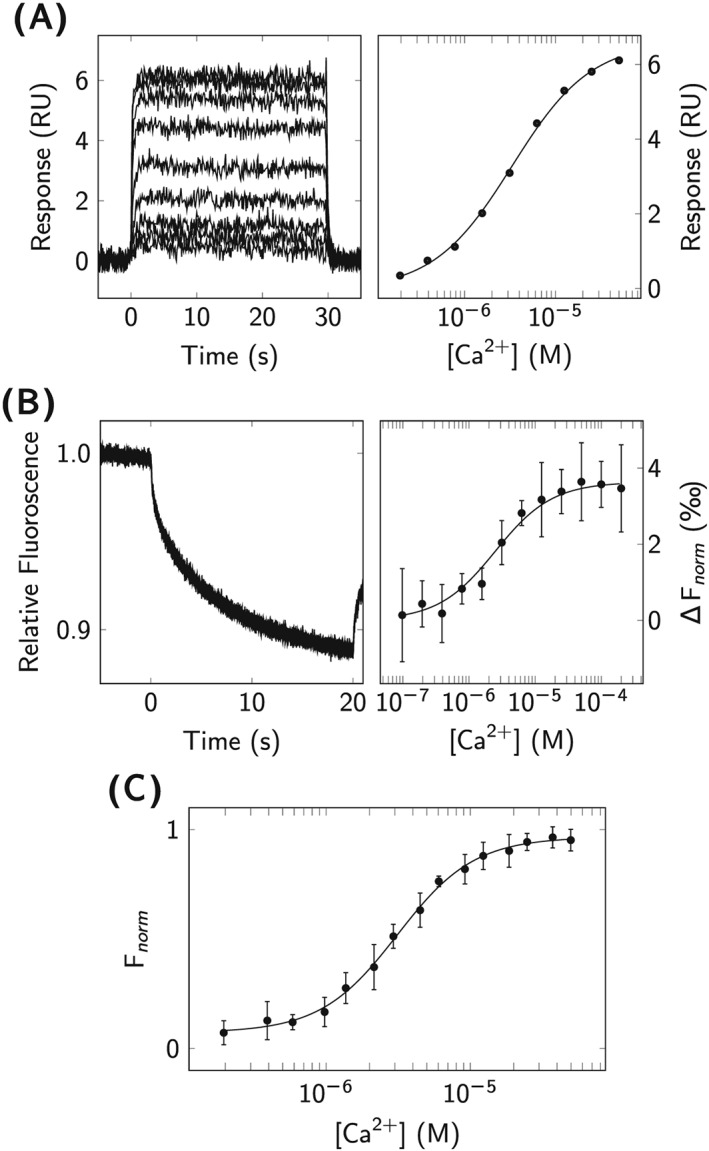
Biophysical analysis of ‐CaM interactions. A, SPR analysis of injected over immobilized CaM (left). The affinity was determined by fitting Equation 1 to the steady‐state signals extracted from the double‐referenced sensorgrams (right). B, MST analysis of fluorescently labeled CaM interaction with . Signals from the end of the MST traces (left) were plotted as a function of concentration (right) to determine the affinity using Equation 4. C, UV fluorescence spectroscopy of ‐CaM interaction: normalized intensities for the intrinsic tyrosine fluorescence of CaM (λ ex = 274 nm, λ em = 304 nm) were plotted as a function of concentration, and the E C 50 value was determined using Equation 3
Table 1.
Affinities for interactions with CaM, studied by SPR, MST, and UV fluorescence spectroscopy. Data is presented as the average K D from the indicated number of replicate experiments (n) and the corresponding standard error (SE)
| Method | K D [M] (n) | T [°C] |
|---|---|---|
| SPR | (3.2±0.2)×10−6 (5) | 22 |
| SPR | (2.8±0.3)×10−6 (4) | 25 |
| MST | (3.0±0.5)×10−6 (4) | 25 |
| UV | (3.1±0.2)×10−6* (4) | 22 |
*E C 50 at total CaM concentration 1 μM, with the slope factor h=1.6
The interaction between and fluorescently labeled CaM was also analyzed at equilibrium by MST at 25°C (Figure 3B). The affinity was similar, with K D = 3.0 μM(Table 1).
In addition, the interaction between and CaM was measured at equilibrium by intrinsic tyrosine fluorescence, illustrated in Figure 3C. Experimental data, obtained at 22°C, were approximated using Equation 3, and the observable affinity was estimated to E C 50 = 3.1 μM at a CaM concentration of 1 μM, with a corresponding slope factor of h=1.6(Table 1). The determined slope factor of 1.6 indicates positive cooperativity of binding to the C‐lobe of CaM.
2.3. Analysis of CaM‐CaMKII290−309 interactions
Using SPR biosensor analysis, the interaction between CaMKII290−309 and immobilized CaM was observed at the presence of , while no interaction was detected under ‐depleted conditions. Figure 4A shows sensorgrams under ‐saturated (main graph) and depleted (insert) conditions.
Figure 4.
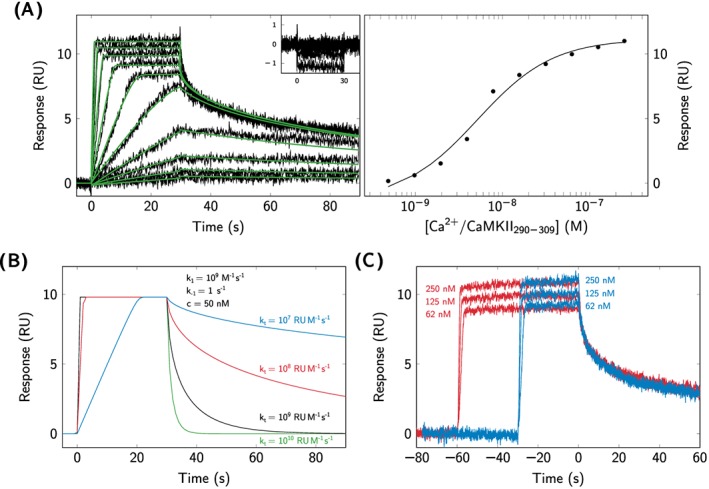
Kinetic analysis of CaM‐CaMKII290−309 interactions. A, CaMKII290−309 was injected over immobilized CaM under ‐saturated conditions (left) and ‐depleted conditions (shown in insert, left). A reversible 1‐step model (Scheme 1) was fitted to the double‐referenced sensorgrams (green solid lines). The affinity was determined by fitting Equation 1 to the steady‐signals extracted from the double‐referenced sensorgrams (right). B, Simulation of the effect of different values of the mass transport coefficient (k t) on the kinetics of the CaMKII290−309‐CaM interaction under ‐saturated conditions. C, Effect of varying injection times (blue: 30 s, red: 60 s) on interactions between CaMKII290−309 and CaM under ‐saturated conditions
The experimental data could be described by a reversible 1‐step model (Scheme 1), and kinetic parameters were very fast, initially approximated to 109 M−1 s−1 for the association rate constant and 1 s −1 for the dissociation rate constant. However, the instrument mass transport coefficient was 107 RU M−1 s−1, which, according to the empirical criterion by Karlsson (Equation 2),30 is indicative of mass transport–limited kinetic measurements. Simulations using the estimated rate constants and different mass transport coefficient (k t) showed that the sensorgrams were highly sensitive to variations in k t values (Figure 4B). Attempts to overcome the mass transport limitation by reducing the amount of immobilized CaM and increasing the flow rate to 90 μL min−1 were not successful. The kinetic parameters could therefore not be reliably estimated. For these reasons, the affinity for the CaM‐CaMKII290−309 interaction (K D = 7.1 nM, Table 2) was determined by steady‐state analysis, as shown in Figure 4A.
Table 2.
Affinities for interactions between CaM and CaMKII290−309 in the presence (2 mM CaCl2) of and in the absence of calcium. Data are presented as the average value from the indicated number of replicate experiments (n) and the corresponding standard error (SE)
| Interaction | K SPR D [M] (n) | K MST D [M] (n) |
|---|---|---|
| /CaM‐CaMKII290−309 | (7.1±2.5)×10−9 (5) | (1.9±0.7)×10−7 (3) |
| apoCaM‐CaMKII290−309 | nd (2) | nm |
nd ‐ no interaction was detected; nm ‐ not measured
Despite the difficulties in determining reliable kinetic rate constants, experiments with varying injection times were performed to obtain qualitative information about the interaction mechanism. Figure 4C shows that the sensorgrams from 3 different CaMKII290−309 concentrations injected for 30 s and 60 s, respectively, overlap very well. This indicates that CaMKII290−309 interacts with CaM according to a 1‐step model, as shown in Scheme 1.
The CaM‐CaMKII290−309 interaction under ‐saturated conditions was also evaluated by MST analysis, as shown in Figure 5A. The MST‐derived affinity was determined to K D = 190 nM, differing more than one order of magnitude from the SPR‐derived affinity (Table 2).
Figure 5.
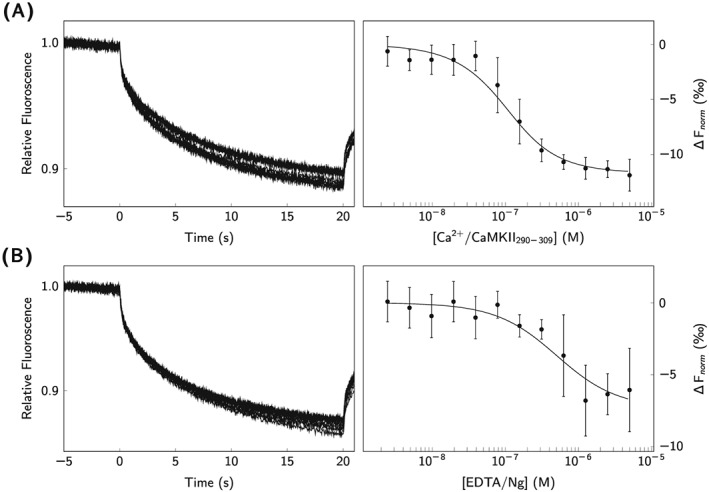
MST analysis of CaM‐CaMKII290−309 and CaM‐Ngfl interactions. Thermophoresis of fluorescently labeled CaM was measured over a range CaMKII290−309 and Ngfl concentrations. A, CaMKII290−309 in ‐saturating conditions; B, Ngfl under ‐depleted conditions. Extracted signals from the end of the Microscale thermophoresis traces were plotted as a function of concentration (black circles), and the affinities were determined by fitting Equation 4 to the data
2.4. Analysis of CaM‐neurogranin interactions
The interaction between CaM and Ng27−50, a peptide containing the CaM‐binding site of Ngfl and CaM, was initially studied in a similar manner as in the experiments with CaMKII290−309. Surprisingly, the interaction was only detected under ‐saturated conditions (Figure 6A). The relatively weak affinity was determined by steady‐state analysis to K D = 14 μM (Table 3). As a control experiment, Ng27−50 was immobilized by amine coupling, and CaM was injected as the analyte. Although Ng27−50 could be immobilized at high surface densities, theoretically sufficient for detection, an interaction with CaM was not detected (data not shown).
Figure 6.
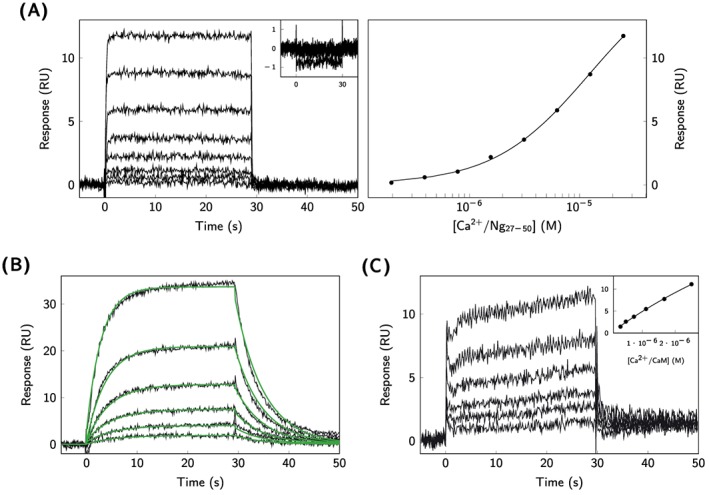
SPR analysis of CaM‐Ng interactions. A, Ng27−50 was injected over immobilized CaM under ‐saturated and ‐depleted (shown in insert) conditions (left). Equation 1 was fitted to the signals extracted from the end of the sensorgrams (right). B, CaM was injected under ‐depleted conditions over affinity captured Ngfl, and the kinetics rate constants were determined by fitting a reversible 1‐step model according to Scheme 1 (green solid lines). C, CaM was injected over affinity captured Ngfl under ‐saturated conditions, and the affinity was estimated using Equation 1
Table 3.
Kinetic rate constants and affinities for the interaction between CaM and Ng variants. Data is presented as as the average value from the indicated number of replicate experiments (n) and the corresponding standard error (SE)
| Interaction | k SPR 1 [M−1s−1] | k SPR −1 [s−1] | K SPR D [M] (n) | K MST D [M] (n) | |
|---|---|---|---|---|---|
|
|
nd | nd | (1.4±0.2)×10−5 (4) | nd (2) | |
| apoCaM‐Ng27−50 | nd | nd | nd (2) | nm | |
| apoCaM‐Ng | (3.4±0.2)×105 | (1.6±0.1)×10−1 | (4.8±0.4)×10−7(6) | (8.9±2.5)×10−7 (4) | |
| /CaM‐Ng | nd | nd | (1.9±0.2)×10−5(5) | nm |
nd ‐ parameter was not quantified or no interaction was detected; nm ‐ not measured
Since the results using Ng27−50 were not in accordance with the previous data,6, 8 experiments were performed also with Ngfl. For this, Ngfl was immobilized via antipolyhistidine antibody‐coated biosensor surfaces by affinity capture, as illustrated in Figure 2. Interactions of Ngfl with CaM were monitored both under ‐depleted (Figure 6B) and ‐saturated conditions (Figure 6C). In the absence of , the kinetics of the interaction could be described by a 1‐step interaction model (Scheme 1). The association rate constant was determined to k 1 = 3.4 × 105 M−1 s−1 and the dissociation rate constant to k −1 = 1.6 × 10−1 s−1, corresponding to an affinity of K D = 480 nM (Table 3). In the presence of , higher concentrations of CaM were required to obtain detectable signals, and the kinetic rate constants could not reliably be determined. By using steady‐state analysis, the affinity of Ngfl binding to Ca2+‐saturated CaM was estimated to K D = 19 μM(Table 3).
The interaction between Ngfl and CaM was further analyzed by MST, and Figure 5B illustrates the MST traces observed upon interaction. The affinity at equilibrium was determined to K D = 890 nM(Table 3). Because of the low affinity between Ng and CaM in the presence of , MST analysis would have required impractically high concentrations of Ng and was therefore not performed.
3. DISCUSSION
The CaM interactome, especially in synapses, is a complex and dynamic network of protein‐protein interactions. It is subjected to further modulation by variations of intracellular concentrations and by the spatial organization of the different binding partners. Because of its central role in mediating synaptic plasticity of the brain, it is of great interest to improve our understanding of the underlying interactions of CaM. In the present study, we pursued a purely in vitro based approach using complementary biophysical methodologies for kinetic and mechanistic interaction studies of two CaMBPs, Ng, and CaMKII. The 2 proteins differ not only in their nature, Ng being a noncatalytic protein of the calpacin family and CaMKII being an enzyme, but more importantly in the ‐dependency of their interactions with CaM. Developing information‐rich biophysical assays for studying such diverse proteins, with respect to structure, function, and ‐dependence is an important prerequisite to easily apply the methodology for studies of other CaMBPs.
Surface plasmon resonance biosensor technology is a powerful technique for kinetic studies of biomolecular interactions. The possibility to obtain kinetic information from the real‐time data allows one to gain more detailed insights into a given interaction than is provided by equilibrium‐based techniques. However, there are some issues that need to be considered in the design of an SPR assay. The preparation of a functional, sensitive, and stable sensor surface is a first critical step. The most commonly used technique to immobilize proteins onto biosensor surfaces is through amine coupling. This technique involves covalent attachment via primary amine groups of lysine residues in the target protein. This can potentially disturb interactions, especially if the lysine residues are located within the ligand‐binding site. Amine coupling was a suitable method for immobilizing CaM but did not result in a functional Ngfl surface. This was most likely due to the location of the majority of lysine residues within the IQ motif and therefore within the binding site for CaM, as shown in Figure 1. Affinity capture using an antibody targeted against the N‐terminal His residues of Ngfl was found to be a suitable alternative and allowed the extraction of reliable kinetic information (Figure 2). Analysis of the interactions between Ngfl and immobilized CaM, especially under ‐depleted conditions, provided similar results as with affinity‐captured Ngfl(data not shown). However, residual amounts of glycerol from the Ngfl stock gave rise to unwanted secondary effects that disturbed the analysis, especially during experiments under ‐saturated conditions that required higher concentration of Ngfl.
3.1. ‐CaM interactions studied by orthogonal biophysical assays
In the present study, different aspects of CaM interactions were analyzed. The primary aim was to characterize and compare the kinetics of Ng and CaMKII interactions with /CaM and apoCaM. As a first step, the ability to detect how CaM responded to concentrations was investigated by using SPR‐ and MST‐based analysis, complemented by fluorescence spectroscopy. For the SPR analysis, CaM was immobilized by amine coupling and the achieved surface density was sufficient to reliably monitor the interaction with adequate sensitivity. The macroscopic K D of 2.8 μM is in agreement with the half maximal concentration of the maximum response (K 1/2) that has been determined by the use of SPR biosensor analysis in a previous study. 31 From the biosensor analysis, it is not possible to distinguish whether binding is measured to an individual lobe of CaM or whether the macroscopic K D describes the overall affinity to the 4 EF‐hands of CaM.
interactions with CaM were additionally verified using MST analysis. The labeling chemistry involved in the MST experiments is essentially the same as the immobilization chemistry in the SPR‐based experiments, except of the probe nature — fluorescent labeling results in a covalent attachment of a bulky hydrophobic moiety rather than a flexible hydrophilic polymer in SPR experiments. However, MST represents a monophasic system, and the K D values acquired through the thermophoretic measurements are typically comparable with values obtained using other methods. It was found to be the case also for interactions with CaM.
As a further complement to the biosensor and MST analyses, intrinsic tyrosine fluorescence spectroscopy was used. It is sensitive to binding to the C‐lobe of CaM. 32 The analysis indicated an E C 50 of 3.1 μM and positive cooperativity of binding within the C‐lobe of CaM. Thus, the labeling used in SPR and MST analyses did apparently not interfere with binding activity of CaM.
In summary, the results from SPR, MST, and UV fluorescence measurements show that neither the immobilization of CaM on biosensor surfaces nor the fluorescent labeling for MST analysis affected the apparent ‐binding properties, suggesting that these methods should provide reliable measurements of CaM interactions with various partners.
3.2. Qualitative kinetic analysis of CaM‐CaMKII290−309 interactions
Surface plasmon resonance and MST analysis both demonstrated that /CaM binds to CaMKII290−309 with nanomolar affinity (Figures 4A and 5A). The SPR‐based kinetic measurements showed that the interaction was fast and limited by diffusion of CaMKII290−309 to the CaM surface. The sensorgrams contained typical signs of limited mass‐transport, including (1) an initial linear association phase, (2) a sharp transition to steady state upon the delayed establishment of an equilibrium at the sensor surface, and (3) an apparently slow dissociation phase caused by rebinding of CaMKII290−309 before being washed off from the sensor surface.33, 34 Furthermore, evaluating the approximated kinetic rate constants for the /CaM‐CaMKII290−309 interaction showed that the association rate constant was lower than the mass‐transport coefficient, leading to a ratio based on the criterion introduced by Karlsson 30 that by far exceeded the empirically determined ratio of 5. When exceeding this ratio, mass transport effects have a strong effect on the determined parameters and lead to uncertainties in the determination of the kinetic parameters. For this reason, it needs to be emphasized that the apparently slow dissociation of CaMKII290−309 shown in Figure 4A does not correspond to the formation of a kinetically stable /CaM‐CaMKII complex. Simulations performed for the given interaction, using the approximated kinetic rate constants, showed high sensitivity toward variations of the mass transport coefficient, k t, resulting in rapid kinetics once the effect of k t on the association rate constant, k 1, became negligible (Figure 4B).
It is worth noting that the interaction could be completely broken up by a short (15 s) injection of HBS‐T‐EDTA (data not shown). The observed strength of the interaction can thus not be attributed to experimental artifacts due to unspecific binding, analyte precipitation, or other effects that do not correspond to a unspecific molecular recognition. Furthermore, practically identical dissociation curves for different injection times indicated that the interaction was not complex, and it was not meaningful to consider more complicated models than the simple 1‐step interaction model shown in 1.
Despite the inability to reliably quantify the kinetic rate constants for the /CaM‐CaMKII290−309 interaction, qualitatively, the analysis provided strong support for a very rapid binding event. This seems to be in agreement with results obtained for the interaction between dansylated CaM and both phosphorylated and unphosphorylated α‐CaMKII from the rat brain, where the association rate constants where determined to k 1 = 1.5 × 108 M−1 s−1 and k 1 = 0.5 × 108 M−1 s−1, respectively.35
Steady‐state analysis was not affected by the mass‐transport limitation, and the affinity was determined to K D = 7.1 nM. This is in agreement with results from fluorescence anisotropy measurements under ‐saturated conditions, where an affinity of less than 10 nM was reported for CaMKII290−309 5 and of K D = 45 nM for unphosphorylated α‐CaMKII.35 It is worth noticing that the affinity and kinetics of CaMKII290−309 to /CaM, although not fully resolved, seem to be in agreement with those obtained for unphosphorylated α‐CaMKII. Phosphorylated α‐CaMKII has a considerably slower dissociation rate constant (< 1 × 10−3 s−1) and therefore an affinity in the low picomolar range.35
The affinity for the /CaM‐CaMKII290−309 interaction determined by MST was more than 1 order of magnitude higher (Table 2) than determined with the SPR assay. Such a mismatch between MST‐ and SPR‐derived K D values for this interaction, under otherwise comparable conditions, might be caused by the introduction of a bulky hydrophobic group of the fluorescent label during the MST analysis, potentially leading to steric hindrance and a reduced affinity for this particular interaction.
In the absence of , the interaction of CaMKII290−309 with CaM was not detected within the measured concentration range. While the affinity under ‐depleted conditions has been reported to be in the mid micromolar range,5 determination of such a low affinity of CaMKII290−309 to apoCaM was not feasible in the given experimental setup. It would require high micromolar peptide concentrations, which would in turn likely lead to artifacts in the SPR biosensor assay. An affinity for the apoCaM‐CaMKII290−309 interaction is likely to be 4 to 5 orders of magnitude lower than for the /CaM‐CaMKII290−309. The relevance of such a weak interaction can be regarded as negligible, since CaMKII requires the initial activation by /CaM to mediate its physiological effects.
3.3. Biosensor analysis confirms Ng27−50 as a poor model system for full‐length neurogranin
When performing biochemical studies using recombinant proteins, a decision has to be made regarding the form of the investigated protein. Truncated proteins are commonly used to reduce the complexity and difficulties associated with studying full‐length proteins, like protein stability, solubility, and expression level. This was the case for CaMKII, which, as a wild‐type protein presents considerable challenges for in vitro biophysical interaction studies because of its size, domain organization, and supramolecular nature. The CaM‐binding domain of CaMKII has been mapped to the amino acids 290‐309 of the human full‐length protein. There is ample experimental data, both from direct interaction studies and competition experiments, showing that CaMKII290−309 binds to CaM with low nanomolar affinity and that the interaction practically exclusively occurs in the presence5, 36 of . This peptide is capable of substrate‐directed inhibition of full‐length CaMKII.36, 37 The ‐dependence of the substrate‐directed inhibition mimicked the behavior of the full‐length protein and the peptide, therefore, it is generally regarded as a valid model system to study the high‐affinity interaction between CaM and CaMKII. Furthermore, as discussed in Section 3.2, the binding data obtained for CaMKII290−309 seem to be in agreement with results for unphosphorylated CaMKII.
For Ng, the situation regarding the validity of using a truncated protein variant encompassing the IQ domain, which contains the CaM‐binding site of Ng, is not as clear as for CaMKII290−309. In 2 previous studies,6, 8 the role of the IQ domain of Ng (NgIQ) has been addressed experimentally, but the results were contradictory. In Kumar et al. 6, the authors conclude that Ng IQ‐motif containing peptide can be regarded as a valid model for the full‐length protein, while the results from Hoffman et al. 8 did not agree with this conclusion. The present results give support for concluding that the peptide is not a valid model system for full‐length Ng.
While Ng27−50 interacted with /CaM with micromolar affinity (Table 3), the interaction with apoCaM was not detected. These findings were at first intriguing, especially considering results from ITC analysis reporting nanomolar affinity of Ng27−50 to apoCaM and micromolar affinity to /CaM.6 However, in a more recent study, where a similar peptide was used (Ng26−49 instead of Ng27−50), these results were not confirmed. Instead, fluorescence spectroscopy using Ng26−49 indicated positive heterotropic cooperativity for binding to CaM, while Ngfl indicated negative heterotropic cooperativity. 8 Furthermore, structural studies using nuclear magnetic resonance have shown that Ng26−49 forms contacts with residues of CaM only within the C‐lobe, while Ngfl forms contacts both with residues in the C‐ and N‐lobe.8 Such structural differences can be expected to result in considerable variations of the CaM‐binding properties of Ngfl and the derived IQ‐motif containing peptide.
In conclusion, the present data from SPR biosensor analysis confirms that Ng27−50 is not a suitable model system for Ngfl for studying CaM interactions.
3.4. Quantitative kinetic analysis of CaM interaction with Ng
Having confirmed that Ng27−50 is a poor model system for Ng, kinetic analysis was performed with the full‐length protein. As discussed above, for the SPR analysis, Ngfl was immobilized to SPR biosensor surfaces by affinity capture. Calmodulin was injected as analyte in solution both in its apo‐ and Ca2+‐saturated forms.
The influence of on the kinetics of the CaM‐Ngfl interaction was clearly seen (Figures 6B and 6C). For the apoCaM‐Ngfl interaction, both the association and dissociation rate constants could be resolved and corresponded to an affinity of K D = 480 nM(Table 3). There was a strong agreement between the affinity obtained by SPR and MST analysis, thereby supporting the validity of the determined K D values (Table 3). Under ‐saturated conditions, the kinetic rate constants were too rapid to be determined by SPR analysis, but it was shown that the affinity decreased to approximately K D = 19 μM.
Compared with previous analysis on the CaM‐Ngfl interaction, there is a good agreement of the present SPR‐ and MST‐based results with previous ITC‐based results 8. In the presence of 150 mM potassium chloride (KCl), the affinity of the apoCaM‐Ngfl interaction of K D = 480 nM, as determined by SPR and MST, is in good agreement with the affinity of K D = 800 nM determined by ITC. 8 The interaction of Ngfl with /CaM could previously not be resolved in the presence of 150 mM KCl 8; therefore, a valid comparison with the present data cannot be made. However, on a qualitative level, previous results obtained with varying KCl concentrations showed a consistently higher affinity of Ngfl to apoCaM than to /CaM,8 which was observed in the current study as well. Minor differences in measured affinities between the 2 different studies might be caused by differences in the primary structure of the protein constructs used. In Hoffman et al., 8 human Ngfl was used containing a D2A point mutation. We used wild‐type human Ngfl containing an N‐terminal His‐tag in the present study. It should therefore be noted that, while there are differences in the absolute affinities in studies on the Ng‐CaM interactions, there is strong agreement on the qualitative level that Ngfl interacts with CaM with reasonable affinity both under ‐saturated and ‐depleted conditions.
3.5. Role of bimodal neurogranin interaction with apo and /CaM
Based on the present and previous binding data, 8 it is clear that Ng binds with reasonable affinity to CaM both under ‐depleted and ‐saturated conditions. Other proteins, like CaMKII and AKAP79, whose interaction with CaM that we recently studied using similar methodology,25 have a clear preference for /CaM. This ability might reflect the physiological role of Ng in targeting and concentrating CaM at postsynaptic dendrites 18 that has been described as being a biochemical “capacitor” that releases /CaM either gradually or rapid in response to the strength and duration of pulses. 38 Under ‐depleted conditions, Ng is the predominant postsynaptic protein that binds apoCaM.18, 39 The relatively slow kinetics of the apoCaM‐Ng interaction and the accompanying middle nanomolar affinity are sufficient to target apoCaM and increase its availability for /CaM signaling. It has been observed that CaM has a high level of mobility within dendritic spines, 18 and if the CaM‐Ng complex would immediately dissociate upon an increase in concentration, as previously suggested,38 CaM could potentially diffuse from the postsynaptic dendrites. This would render the initial concentration of CaM within the dendrites unnecessary. Instead, by having a low affinity in the micromolor range, Ng is able to hold CaM in place until it is competed out by other CaMBPs, for instance CaMKII or AKAP79, that have faster association rate constants and affinities than Ng.25, 35
4. CONCLUSIONS
The current project has demonstrated how a biophysical strategy can be applied to elucidate the dynamic details of the calmodulin interactome and its regulation via . By studying the interactions of the isolated components directly and using a combination of real‐time and equilibrium‐based techniques in solution and on sensor surfaces, it was possible to extract details of the interactions. This has revealed distinctive features of the interactions between 2 proteins that interact with CaM in the apo form and the ‐bound form. Specifically, it adds a further dimension to the understanding of how Ng regulates CaM dynamics in synapses.
5. MATERIALS AND METHODS
5.1. Proteins and peptides
Calmodulin from the human brain was purchased from Genway Biotech Inc. (San Diego, California), and recombinant bovine CaM was purchased from Merck Millipore (Billerica, Massachusetts). The human and bovine orthologs are structurally identical. Full‐length recombinant human Ng (Ngfl, aa 1–78) with a His‐tag at the N‐terminus was purchased from Acris Antibodies (Herford, Germany). The IQ‐motif containing peptide of Ng (Ng27−50) and a peptide corresponding to the CaM‐binding region of CaMKIIα(CaMKII290−309) were purchased from Bachem (Bubendorf, Switzerland). An antipolyhistidine antibody was purchased from R&D Systems (Minneapolis, Minnesota). The amino acid sequences of the peptides and proteins used in the experiments are shown in Figure 1.
5.2. Surface plasmon resonance biosensor‐based interaction studies
Surface plasmon resonance (SPR) biosensor‐based interaction studies were performed on a Biacore S51 instrument using CM5 sensor chips (GE Healthcare, Uppsala, Sweden). For the interaction studies, sensor surfaces were prepared by immobilizing CaM or Ng.
Calmodulin was immobilized by amine coupling at a temperature of 25°C using a running buffer consisting of 10 mM HEPES, 150 mM NaCl, 0.05% Tween‐20, pH 7.4 (HBS‐T). The surface was activated by injecting 200 mM 1‐ethyl‐3‐(3‐dimethylaminopropyl)carbodiimide hydrochloride (EDC) and 50 mM N‐hydroxysuccinimide (NHS) (10 min, 20 μL min−1), followed by injecting 50 μg mL−1 CaM (diluted into 10 mM Na‐acetate, pH 4.0 (GE Healthcare)) for 20 min (2 μL min−1). The surface was deactivated by injecting 1 M ethanolamine, pH 8.5 (7 min, 25 μL min−1).
Full‐length neurogranin was immobilized by affinity capture via the N‐terminal His‐tag. The antipolyhistidine antibody was first immobilized by amine coupling at a concentration of 10 μg mL−1 (diluted into 10 mM Na‐acetate, pH 5.5, 10 min injection, 5 μL min−1). Subsequently, Ngfl, diluted to 25 μg mL−1 in either HBS‐T‐EDTA (HBS‐T supplemented with 1 mM EDTA) or HBS‐T‐CaCl2(HBS‐T supplemented with 2 mM CaCl2), was injected over the antibody surface (2 min, 10 μL min−1). During the experiments, the surface was regenerated after every injection of CaM by two 20 s pulses of 10 mM glycine, pH 1.5 (30 μL min−1).
interactions were measured at both 25°C and 22°C using HBS‐T as running and sample buffer. was injected in 2‐fold concentration series (200 nM − 50 μM) for 30 s at a flow rate of 90 μL min−1. Interaction studies of CaMKII290−309 and Ng27−50 with immobilized CaM were performed at 25°C both in the presence (HBS‐T‐CaCl2) and absence of (HBS‐T‐EDTA). CaMKII290−309(0.5 nM − 250 nM) and Ng27−50(200 nM − 25 μM) were injected in 2‐fold concentration series for 30 s at a flow rate of 90 μL.
Calmodulin interactions with immobilized Ngfl were studied at 25°C at a flow rate of 30 μL min−1. Calmodulin was injected for 30 s in 2‐fold dilution series (250 nM − 8 nM) in HBS‐T‐EDTA and in 1.5‐fold dilution series (2.5 μM − 300 nM) in HBS‐T‐CaCl2.
Experimental data was double‐referenced by subtracting the response from an untreated reference surface and the average response from 2 blank samples. Biacore T200 Evaluation v1.0 Software (GE Healthcare) was used for steady‐state and kinetic analysis. For the steady‐state analysis, average signals from a 5 s interval were extracted from the end of the association phase of the sensorgrams and analyzed as a function of ligand concentration using Equation 1. The parameters for the responses when saturation is reached, R max, and for the offset, m, were fitted locally, while the K D was fitted as a global parameter, [L] corresponds to the ligand concentration.
| (1) |
For kinetic analysis, a 1:1 interaction model (Scheme 1) was globally fitted to the double‐referenced data to determine the association rate constant, k 1, the dissociation rate constant, k −1, and the corresponding K D value.
The effect of limited mass‐transport on kinetic measurements was evaluated using Equation 2 introduced in Karlsson.30 Simulation of the influence of different values of mass‐transport coefficient, k t, on the kinetics of CaMKII290−309 was performed using BIAevaluation v3.0.2 (GE Healthcare).
| (2) |
5.3. Ultraviolet fluorescence spectroscopy
Fluorescence measurements were performed on a Fluoromax‐4 Spectrofluorometer (Horiba, Kyoto, Japan) at 22°C using a 0.5 mL 4‐side polished quartz cuvette (Hellma Analytics, Müllheim, Germany) with a 10 mm light path. Intrinsic tyrosine fluorescence of CaM was measured at an excitation wavelength of λ ex = 274 nm and an emission wavelength of λ em = 304 nm using slit widths of 1 nm and 10 nm, respectively.
Two‐fold serial dilutions of starting from 40 mM CaCl2 in CaM stock solution (10 mM HEPES, 150 mM NaCl, 10 μM (micromoles) apoCaM, pH 7.4) were prepared. For determination of the affinity, 5 μL aliquots from each dilution point were titrated into CaM stock solution.
Fluorescence intensities within every titration series were normalized to span a range from 0 to 1. Equation 3 was fitted to the data using nonlinear regression in the BIAevaluation software 3.0.2. F min and F max correspond to the normalized fluoresence intensity of ‐free and ‐saturated CaM, respectively, E C 50 to the observable K D at the given protein concentration, [ ], to the total concentration, and h to the slope factor that indicates degree of the binding cooperativity.
| (3) |
5.4. Affinity analysis using microscale thermophoresis
Microscale thermophoresis–based interaction analysis was performed on a Monolith NT. Automated MST instrument (NanoTemper Technologies GmbH, Munich, Germany) using standard treated AK002 capillaries at a temperature of 25°C. Excitation and MST power were set to 20%. The initial state was recorded for 5 s, thermophoresis for 20 s and the back diffusion for 1 s.
For MST experiments, CaM was labeled with Cyanine5 NHS‐activated ester (Lumiprobe GmbH, Hannover, Germany). The dye was dissolved in 100% dimethyl sulfoxide to a final concentration of 540 μM, and the labeling reaction was performed by incubating 2.5 μM of CaM and 5 μM of the fluorophore in HBS‐T supplemented with 1% dimethyl sulfoxide for 30 min at room temperature in the dark. The reaction was stopped by buffer exchange to HBS‐T using spin desalting columns with a molecular weight cutoff of 7 kDa (Thermo Scientific, Rockford, USA) according to the instructions of the manufacturer.
Thermophoretic measurements were performed at a constant CaM concentration of 75 nM. Ligands were prepared in 2‐fold dilution series (concentrations specified under Results). Interaction of with CaM was analyzed in HBS‐T, CaMKII290−309 with CaM in HBS‐T‐CaCl2. Analysis of the interaction between apoCaM and Ngfl was performed in HBS‐T‐EDTA. Data from at least 3 independent replicate experiments were analyzed using the NTAnalysis software (NanoTemper GmbH). The macroscopic dissociation constant (K D) was determined using Equation 4:
| (4) |
where [B 0] corresponds to the total concentration of target binding sites, [L 0] to the concentration of titrated ligand, and [B L] to the concentration of formed complex between ligand and target binding sites. 27
ACKNOWLEDGMENTS
This project was supported by the SynSys Project (FP7 Health project 242167) and the Swedish Research Council (VR, no. D0571301) (HD). The authors thank Mari Kullman Magnusson (Beactica AB, Uppsala, Sweden) for assistance during the development of the SPR assay and Dr Nicolas Le Novere and Dr Massimo Lai (The Babraham Institute, UK) for discussion. The authors further thank the SciLifeLab Drug Discovery and Development platform at Uppsala University for providing access to the MST instrument.
Seeger C, Talibov VO, Danielson UH. Biophysical analysis of the dynamics of calmodulin interactions with neurogranin and Ca2+/calmodulin‐dependent kinase II. J Mol Recognit. 2017;30;e2621 https://doi.org/10.1002/jmr.2621
Abbreviations: CaM, calmodulin; MST, microscale thermophoresis; SPR, surface plasmon resonance; UV, ultraviolet spectrofluorimetry
REFERENCES
- 1. Xia Z, Storm DR. The role of calmodulin as a signal integrator for synaptic plasticity. Nat Rev Neurosci. 2005;6(4):267–276. [DOI] [PubMed] [Google Scholar]
- 2. Babu YS, Bugg CE, Cook WJ. Structure of calmodulin refined at 2.2 – resolution. J Mol Biol. 1988;204(1):191–204. [DOI] [PubMed] [Google Scholar]
- 3. Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax Ad. Solution structure of calcium‐free calmodulin. Nat Struct Biol. 1995;2(9):768–776. [DOI] [PubMed] [Google Scholar]
- 4. LaPorte DC, Wierman BM, Storm DR. Calcium‐induced exposure of a hydrophobic surface on calmodulin Biochemistry. 1980;19(16):3814–3819. [DOI] [PubMed] [Google Scholar]
- 5. Evans T, Shea MA. Energetics of calmodulin domain interactions with the calmodulin binding domain of CaMKII. Proteins: Structure, Function, and Bioinformatics. 2009;76(1):47–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kumar V, Chichili VPR, Zhong L, et al. Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin. Sci Rep. 2013;3:1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baudier J, Deloulme JCh, Van Dorsselaer A, Black D, Matthes HW. Purification and characterization of a brain‐specific protein kinase C substrate, neurogranin (p17). Identification of a consensus amino acid sequence between neurogranin and neuromodulin (gap43) that corresponds to the protein kinase C phosphorylation site and the calmodulin‐binding domain. J Biol Chem. 1991;266(1):229–237. [PubMed] [Google Scholar]
- 8. Hoffman L, Chandrasekar A, Wang X, Putkey JA, Waxham MN. Neurogranin alters the structure and calcium binding properties of calmodulin. J Biol Chem. 2014;289(21):14644–14655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci. 2002;3(3):175–190. [DOI] [PubMed] [Google Scholar]
- 10. Hunter T, Schulman H. CaMKII structure ‐ an elegant design. Cell. 2005;123(5):765–767. [DOI] [PubMed] [Google Scholar]
- 11. Brocke L, Chiang LW, Wagner PD, Schulman H. Functional implications of the subunit composition of neuronal CaM kinase II. J Biol Chem. 1999;274(32):22713–22722. [DOI] [PubMed] [Google Scholar]
- 12. Erondu NE, Kennedy MB. Regional distribution of type II /calmodulin‐dependent protein kinase in rat brain. The J Neurosci. 1985;5(12):3270–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Derkach V, Barria A, Soderling TR. /calmodulin‐kinase II enhances channel conductance of α‐amino‐3‐hydroxy‐5‐methyl‐4‐isoxazolepropionate type glutamate receptors. Proceedings of the National Academy of Sciences. 1999;96(6):3269–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lai Y, Nairn AC, Greengard P. Autophosphorylation reversibly regulates the /calmodulin‐dependence of /calmodulin‐dependent protein kinase II. Proceedings of the National Academy of Sciences. 1986;83(12):4253–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gerendasy DD, Sutcliffe JG. Rc3/neurogranin, a postsynaptic calpacitin for setting the response threshold to calcium influxes. Mol Neurobiol. 1997;15(2):131–163. [DOI] [PubMed] [Google Scholar]
- 16. Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. The FASEB Journal. 1997;11(5):331–340. [DOI] [PubMed] [Google Scholar]
- 17. Zhong L, Cherry T, Bies CE, Florence MA, Gerges NZ. Neurogranin enhances synaptic strength through its interaction with calmodulin. The EMBO Journal. 2009;28(19):3027–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petersen A, Gerges NZ. Neurogranin regulates cam dynamics at dendritic spines. Sci Rep. 2015;5:11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keller CH, Olwin BB, LaPorte DC, Storm DR. Determination of the free‐energy coupling for binding of calcium ion and troponin I to calmodulin. Biochemistry. 1982;21(1):156–162. [DOI] [PubMed] [Google Scholar]
- 20. Olwin BB, Storm DR. Calcium binding to complexes of calmodulin and calmodulin binding proteins. Biochemistry. 1985;24(27):8081–8086. [DOI] [PubMed] [Google Scholar]
- 21. Gaertner TR, Putkey JA, Waxham MN. Rc3/neurogranin and /calmodulin‐dependent protein kinase II produce opposing effects on the affinity of calmodulin for calcium. J Biol Chem. 2004;279(38):39374–39382. [DOI] [PubMed] [Google Scholar]
- 22. Lai M, Brun D, Edelstein SJ, Le Novère N. Modulation of calmodulin lobes by different targets: an allosteric model with hemiconcerted conformational transitions. PLoS Computational Biology. 2015;11(1):e1004063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Monod J, Wyman J, Changeux J‐P. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12(1):88–118. [DOI] [PubMed] [Google Scholar]
- 24. Gorny X, Mikhaylova M, Seeger C, Reddy PP, Reissner C, Schott BH, Danielson UH, Kreutz MR, Seidenbecher Constanze. AKAP79/150 interacts with the neuronal calcium‐binding protein caldendrin. J Neurochem. 2012;122(4):714–726. [DOI] [PubMed] [Google Scholar]
- 25. Seeger C, Gorny X, Reddy PP, Seidenbecher C, Danielson UH. Kinetic and mechanistic differences in the interactions between caldendrin and calmodulin with AKAP79 suggest different roles in synaptic function. J Mol Recognit. 2012;25(10):495–503. [DOI] [PubMed] [Google Scholar]
- 26. Duhr S, Braun D. Why molecules move along a temperature gradient. Proceedings of the National Academy of Sciences. 2006;103(52):19678–19682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein‐binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. [DOI] [PubMed] [Google Scholar]
- 28. Dedman JR, Potter JD, Jackson RL, Johnson JD, Means AR. Physicochemical properties of rat testis ‐dependent regulator protein of cyclic nucleotide phosphodiesterase. J Biol Chem. 1977;252:8415–8422. [PubMed] [Google Scholar]
- 29. Pundak S, Roche RS. Tyrosine and tyrosinate fluorescence of bovine testes calmodulin: calcium and pH dependence. Biochemistry. 1984;23(7):1549–1555. [DOI] [PubMed] [Google Scholar]
- 30. Karlsson R. Affinity analysis of non‐steady‐state data obtained under mass transport limited conditions using BIAcore technology. J Mol Recognit. 1999;12(5):285–292. [DOI] [PubMed] [Google Scholar]
- 31. Dell'Orco D, Sulmann S, Linse S, Koch K‐W. Dynamics of conformational ‐switches in signaling networks detected by a planar plasmonic device. Anal Chem. 2012;84(6):2982–2989. [DOI] [PubMed] [Google Scholar]
- 32. Richman PG, Klee CB. Specific perturbation by of tyrosyl residue 138 of calmodulin. J Biol Chem. 1979;254(12):5372–5376. [PubMed] [Google Scholar]
- 33. Schuck P. Kinetics of ligand binding to receptor immobilized in a polymer matrix, as detected with an evanescent wave biosensor. I. A computer simulation of the influence of mass transport. Biophys J. 1996;70(3):1230–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nico J, Plomp E, Fischer MJE, Ruijtenbeek R. Kinetic analysis of the mass transport limited interaction between the tyrosine kinase lck SH2 domain and a phosphorylated peptide studied by a new cuvette‐based surface plasmon resonance instrument. Anal Biochem. 2000;279(1):61–70. [DOI] [PubMed] [Google Scholar]
- 35. Meyer T, Hanson PI, Stryer L, Schulman H. Calmodulin trapping by calcium‐calmodulin‐dependent protein kinase. Science. 1992;256(5060):1199–1202. [DOI] [PubMed] [Google Scholar]
- 36. Payne ME, Fong YL, Ono T, Colbran RJ, Kemp BE, Soderling TR, Means AR. Calcium/calmodulin‐dependent protein kinase II. characterization of distinct calmodulin binding and inhibitory domains. J Biol Chem. 1988;263(15):7190–7195. [PubMed] [Google Scholar]
- 37. Colbran RJ, Schworer CM, Hashimoto Y, Fong YL, Rich DP, Smith MK, Soderling TR. Calcium/calmodulin‐dependent protein kinase II. Biochem J. 1989;258(2):313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gerendasy DD, Herron SR, Watson JB, Sutcliffe JG. Mutational and biophysical studies suggest RC3/neurogranin regulates calmodulin availability. J Biol Chem. 1994;269(35):22420–22426. [PubMed] [Google Scholar]
- 39. Zhabotinsky AM, Camp RN, Epstein IR, Lisman JE. Role of the neurogranin concentrated in spines in the induction of long‐term potentiation. The J Neurosci. 2006;26(28):7337–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]


