Abstract
BACKGROUND
Inactivation processes can be classified into non‐thermal inactivation methods such as ethylene oxide and γ‐radiation, and thermal methods such as autoclaving. The ability of carbon dioxide enriched microbubbles to inactivate Pseudomonas putida suspended in physiological saline, as a non‐thermal sterilisation method, was investigated in this study with many operational advantages over both traditional thermal and non‐thermal sterilisation methods.
RESULTS
Introducing carbon dioxide enriched microbubbles can achieve ∼2‐Log reduction in the bacterial population after 90 min of treatment, addition of ethanol to the inactivation solution further enhanced the inactivation process to achieve 3, 2.5 and 3.5‐Log reduction for 2%, 5% and 10 %( v/v) ethanol, respectively. A range of morphological changes was observed on Pseudomonas cells after each treatment, and these changes extended from changing cell shape from rod shape to coccus shape to severe lesions and cell death. Pseudomonas putida KT 2440 was used as a model of gram‐negative bacteria.
CONCLUSION
Using CO2 enriched microbubbles technology has many advantages such as efficient energy consumption (no heat source), avoidance of toxic and corrosive reagents, and in situ treatment. In addition, many findings from this study could apply to other gram‐negative bacteria. © 2017 The Authors. Journal of Chemical Technology & Biotechnology published by John Wiley & Sons Ltd on behalf of Society of Chemical Industry.
Keywords: inactivation, cell lysis, CO2, microbubbles, Pseudomonas
INTRODUCTION
Traditionally, inactivation (sterilisation) methods can be classified into two main classes: thermal and non‐thermal sterilisation methods. Thermal methodologies, such as autoclaving and steam sterilisation, have been widely used to sterilise a variety of materials such as nutritional materials, medical devices and watery solutions. Other substances cannot be sterilised using these methodologies due to their heat‐sensitive compositions.1 Alternatively, non‐thermal methodologies have been applied on heat‐labile materials such as high protein foods as they can retain all their physical, mechanical and optical characteristics without denatured changes.1 Thermal methods such as steam sterilisation, however, have higher capital and operating costs than non‐thermal methods. On the other hand, thermal methods have slightly higher effectiveness than the non‐thermal methodologies.2 Therefore, there are increasing demands for non‐thermal sterilisation methods such as those using ethylene oxide, γ irradiation and supercritical CO2.2, 3, 4
However, these methods still have drawbacks, such as the toxicity and carcinogenicity of ethylene oxide,2 and changing the mechanical characteristics of biomaterials by γ‐irradiation.3 In contrast using carbon dioxide can avoid many of these drawbacks. Recently, CO2 has been used to preserve foodstuffs and to inactivate a wide range of microorganisms including yeasts and bacteria. Other potential applications for this methodology include in situ inactivation processes for both pretreatment slurry of lignocellulosic biomass after microbial pre‐treatment and contaminated algal culture. The primary action of using carbon dioxide as an effective inactivation technology is instigated by changing the balance of biological systems within the cells.4 The proposed targets for CO2 activity were reviewed and summarised previously.5
CO2 has a strong tendency to dissolve in water, which increases with hydrophobic solutions and at low temperatures. Interestingly, the CO2 solubility coefficient increases with decreasing temperature, and at 15 °C, the CO2 solubility coefficient is ∼1.6 The CO2 activity can be quantified using the survivor ratio, calculated using the following equation suggested previously:7
| (1) |
where N is the number of bacterial cells (colony forming units mL−1), N0 is the initial number of bacterial cells (colony forming units mL−1), k is the sterilisation rate constant min −1(D‐value) and t is the time (min).
The inactivation capability of CO2 depends on the dissolved CO2 concentration, while the pressure of pressurized CO2 might not have a direct effect on the microbial inactivation, it influences its dissolution of CO2.
Pseudomonas putida KT2440 is a gram‐negative, aerobic rod‐shaped bacterium, with a significant metabolic diversity. It is adapted to different environments such as soils, aquatic systems and rhizosphere.8 Its unique traits make it suitable as a bio‐control and growth‐promoting agent9 and therefore model organism for the current study.
The efficacy of this pre‐treatment process depends on the diffusion coefficient of carbon dioxide, which can be controlled by the contact time and interfacial area. The low rise velocity of microbubbles, coupled with their high surface area to volume ratio, provide superior mass transfer. To enhance mass transfer, CO2 needs to be introduced into the bulk liquid as microbubbles.10 In addition, microbubbles can generate free radicals during their collapse.11, 12 These radicals have a much higher standard redox potential (2.80 V) than oxidants such as ozone and hydrogen peroxide (2.07 and 1.77 V, respectively), and thus, they show instant and non‐selective reactivity with the majority of organic compounds present in solution.13, 14
pH influences not only the quantity of free radicals generated by the collapsed microbubbles but also the electrostatic profile of the microbubbles and the degree of ionization of organic compounds in aqueous solution.12 At pH above 4.5, the zeta (ζ) potential of microbubbles tends to have a negative sign, with the magnitude varying directly with increasing pH.15, 16 For values of pH below 4.5, the zeta potential has a positive value, and it increases with decreasing pH, resulting in a higher probability of negative charged compounds such as lipopolysaccharides (LPS) in the outer membrane of gram‐negative bacteria;17 which can approach the bubble interface and accelerate the reaction processes.12 For P. putida, the isoelectric point occurs at pH 3,18 and below this value, the net surface charge is positive, but negative otherwise.19 However, the pH dependent adhesion cannot be described as a linear function as shown previously.19 Based on P. putida average cell size: 0.5 to 0.6 µm in diameter and from 1.4 to 1.7 µm in length,20 it was calculated that one bacterium covers up to 1 µm2 of surface. However, the surface of each microbubble represents 7850 µm2, using 50 µm as the average microbubbles size. Thus, the adhesion intensity attained in the present experiments is sufficient to attach many bacterial cells, helping to direct diffusion of CO2 in aqueous phase via a bacterial membrane. CO2 can diffuse through the cell membrane of bacterial cells to accumulate in the phospholipid layer, leading to increased fluidity of plasma membrane and causing an anaesthesia effect for microbial cells.21
The aim of the present study is to explore the inactivation of P. putida using CO2 enriched microbubbles. Wild‐type cells were treated with CO2 microbubbles at low pressure and temperatures. Additives were used to enhance the activity of this process and to achieve a lower survivor ratio (greater log reduction [− Log]).
MATERIAL AND METHODS
Pseudomonas putida was cultivated at 30 °C for 24 h on the nutrient broth (Sigma–Aldrich, UK). Normal saline solution (0.85%) was prepared and refrigerated for 24 h at 6 °C. It is worth mentioning that normal saline is the generally favoured fluid as it is isotonic and less likely to interfere with the inactivation process. Diluted bacterial culture was prepared by mixing 900 mL of sterile cold saline with 100 mL of bacterial culture, which was grown for 18–24 h, and the final temperature set at 6 °C.
The reactor was connected to a CO2 (100%) gas cylinder (BOC, UK) through a diffuser (ceramic diffuser, point four systems Inc. UK), and the reactor has a final capacity 1.5 L (Fig. 1). These experiment sets were conducted for 90 min (time required to reach equilibrium state according to the preliminary studies), and samples were drawn every 15 min except the first sample, which was drawn after 7 mins. pH and temperature profiles were measured at the same time as drawing samples. Drawn samples were diluted with sterile normal saline and aseptically streaked on nutrient agar plates using an inoculation loop and incubated at 30 °C for 24 h, and thereafter; the grown colonies were counted using two software packages: ColonyCount, 2015 © Promega Corporation and HGColonyLT, 2014 © HyperGEAR Inc. Petri dishes with 30–300 colonies were counted, and colony‐forming unit per mL was calculated for each sample. In addition, a colony counter (Bio Spectrum 410 imaging system, UVP, UK) was also used to verify and correct the colony count. Survivor ratio was calculated and plotted using ln (N/N0) on Y‐axis and time on X‐axis, where N refers to the number of colony forming units for treated samples, N0 refers to the initial number of colony forming units before CO2 treatment.
Figure 1.
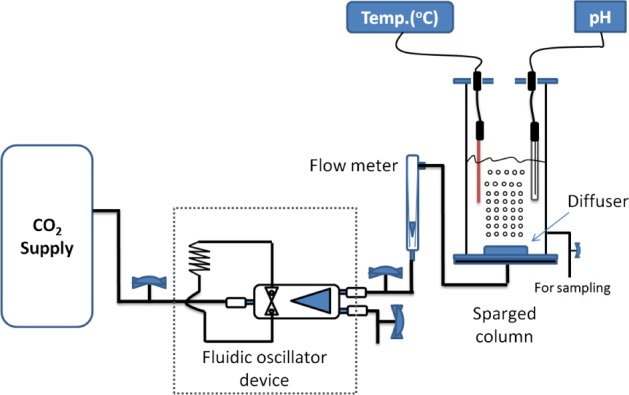
Schematic representation of the experimental set‐up. Pure CO2 gas (100%) is fed into the microbubbles diffuser.
In addition, the highest microbubble population fraction generated was of size less than 100 microns and the addition of ethanol to the treatment solution did not affect the bubble size significantly, as both chemicals were added at low concentrations and this has a very limited effect on the size of microbubbles.
Morphological examination of the bacterial cells
Combined microscopy
For each experiment, two slides for bacterial smear before and after the inactivation process were prepared and stained with gram stain according to the standard protocol described previously.22
Scanning electron microscopy
Microbial specimens were fixed chemically for examination with scanning electron microscopy (SEM). Initially, the specimens were kept in normal saline to avoid any changes as a result of osmolality as normal saline is isotonic solution. Thereafter, the specimens were mounted on 12.5 mm diameter stubs, attached with carbon‐sticky tabs, and then coated using an Edwards S150B sputter coater with approximately 25 nm of gold. The specimens were examined in a Philips XL‐20 scanning electron microscope at an accelerating voltage of 20 kV.
Determination of CO2 concentration
Kinetically, conversion of CO2 into carbonic acid is very slow, and ∼0.2% of CO2 can be converted to carbonic acid and its respective ions, while 99.8% of CO2 tends to remain as a dissolved gas, which can be shown in the dissociated equilibrium constraint:23
| (2) |
| (3) |
| (4) |
Carbonic acid is a diprotic acid, containing two hydrogen atoms ionizable in water and dissociates into bicarbonate and carbonate ions according to the following equations:
| (5) |
| (6) |
Obviously, it is possible to infer the concentration of dissolved CO2 from the pH assuming equilibrium and a well‐mixed system.
The equilibrium constraint for each dissociation reaction (5) and (6) are:
| (7) |
| (8) |
The system should satisfy the electroneutrality constraint, therefore
| (9) |
Dissociation of water to hydrogen and hydroxide ions with dissociation of CO2 can give five equations and the definitions of the pH relate with six unknowns, which are [H+], [OH−], [HCO3 −], [H2CO3], [CO3 2−] and [CO2 aq], which can be solved through a set of nonlinear algebraic equations (Table 1). Accordingly, CO2 concentration can be inferred from pH measurements as follows:
| (10) |
Table 1.
Chemical reactions with their corresponding reaction rate constants used to integrate the CO2 concentration equations in the current study
| Chemical reaction | Reaction rate constant | Unit | Reference | |
|---|---|---|---|---|
|
|
K f = 5.5 × 10− 6 | 1/s | 24, 25, 26 | |
|
|
K
r = 3 × 103
K eq = 1.8 × 10− 16 K w = 1 × 10− 14 |
1/mole/sec | ||
| CO 2+ | K f = 0.043 | 1/mole/sec | 27 | |
| CO 2 + H 2 O | K r = 14.98 | 1/s | ||
| K eq = 2.87 × 10− 3 | ||||
| HCO 3 + H | K f = 106.9 | |||
|
|
K r = 4.67 × 1010 | 1/mole/sec | ||
| K eq = 1.7 × 10− 4 | ||||
|
|
K eq = 5.62 × 10− 11 | 28 |
Therefore, the concentration of dissolved CO2 was measured using Equation (10) based on pH values measured by Mettler Toledo™ S220 (pH) meter. It is worth noting that addition of ethanol to the inactivation solution did not influence the pH of the solution and thus, the same Equation (10) was used to calculate the concentration of dissolved CO2 in the experimental sets with ethanol used as an additive.
RESULTS AND DISCUSSION
Inactivation of Pseudomonas putida using CO2 microbubbles with and without the ethanol addition
Pseudomonas putida cells were treated with CO2 microbubbles for 90 min at 100 mL min−1 flow rate and ∼1 bar. At equilibrium state, the inactivation efficiency was measured using two values, D‐value and L‐value. D‐value is defined as the time required for 1 log cycle reduction in microbial population, and calculated from the negative reciprocal of the slope of regression line from the straight part of the survivor curve 29 in Fig. 2(A). On the other hand, L value is the time during which the number of microbial cells remains constant before starting the inactivation of microorganisms.30 The D‐value for treatment with CO2 microbubbles was 64.8 min, while there was 2‐log reduction in P. putida population. In contrast, L value was 9.6 min. Two mechanisms are suggested to play a central role in this process: oxidative stress and the CO2 effects.
Figure 2.
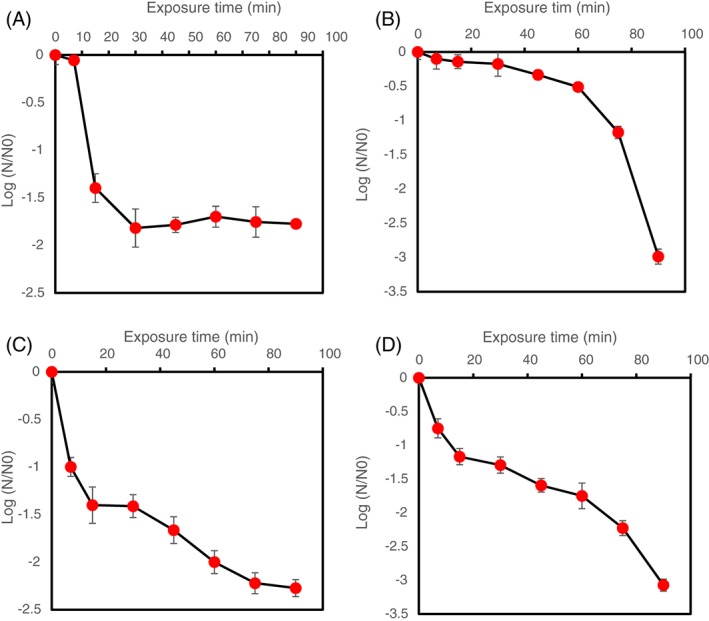
Survivor ratio of Pseudomonas putida after treatment with: (A) CO2 microbubbles; (B) CO2 microbubbles plus 2% (v/v) ethanol; (C) CO2 microbubbles plus 5% (v/v) ethanol; (D) CO2 microbubbles plus 10% (v/v) ethanol. Error bars depict standard deviation.
Regarding the oxidative stress, during the shrinkage and subsequent collapse of bubbles, some hydroxyl radicals are generated.12 In addition, the hydroxyl radicals generated are readily converted to the superoxide radicals and vice versa.31 Previously, P. putida was reported to go through an oxidative stress, when exposed to free radicals.32 Free radicals are reactive oxygen species (ROS), and are injurious species, reacting with different components of cellular systems such as lipids, proteins and DNA.33 Fundamentally, lipids seem to be the major targets for these species during an event of oxidative stress, and interestingly, the free radicals that are formed can directly react with the polyunsaturated fatty acids in the cell membrane, provoking lipid peroxidation. The latter reaction can change membrane properties and disrupt the membrane‐bound proteins.34 Amplification to this reaction occurs when more radicals are generated and more polyunsaturated fatty acids are broken down into other highly reactive products such as aldehydes. These highly reactive products can cause severe damage to vital compounds such as proteins.35
The mechanisms of the action of CO2 have been described in depth.5 CO2 is not a ‘natural product’ of the glucose metabolism pathway of P. putida, however, it can be produced during the catabolising of aromatic compounds by the β‐Ketoadipate pathway.36 All these factors work together to achieve the elevated log reduction observed in P. putida using CO2 microbubbles. In contrast, other microorganisms such as Zymomonas mobilis (another example of a gram‐negative bacteria) are expected to show a high degree of adaptation to CO2 as it is produced naturally during metabolism in both aerobic and anaerobic conditions.37, 38, 39 Figure 3 shows the temperature profile during the inactivation experiments.
Figure 3.
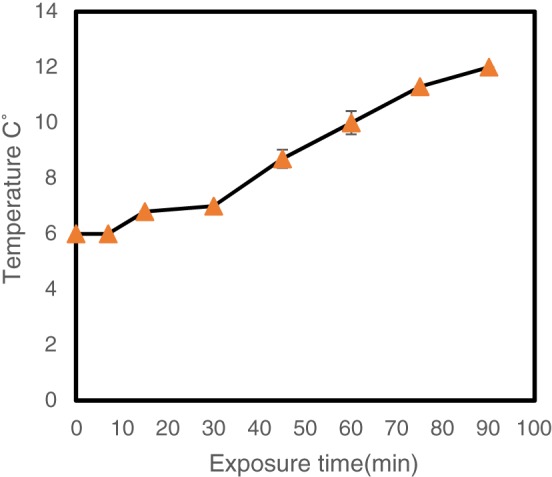
Temperature profile during CO2 enriched microbubbles inactivation process. Error bars depict standard deviation.
Low temperature was used to increase the CO2 solubility in order to enhance the inactivation activity. At 0 °C, the CO2 solubility is around 1.3 mol L−1, decreasing to ∼1 mol L−1 at 10 °C. Moreover, increasing the temperature to 20 °C will decrease the CO2 solubility to ∼0.7 mol L−1.40 Figure 3 shows there was a gradual increase in temperature of the solution from 6 °C to 12 °C.
To enhance the inactivation activity of CO2 microbubbles, organic solvents, are employed. These solvents are toxic to some microbial cells due to their tendency to partition preferentially in the cytoplasmic membranes, increasing the fluidity of the cell membrane, and ultimately causing an increase in the nonspecific permeabilization of the cytoplasmic membrane.41, 42 Moreover, the fatty acid composition in the cytoplasmic membrane of microbial cells can be changed significantly by these membrane‐active solvents.43, 44 Isomerisation is a mechanism employed by P. putida to adapt its cytoplasmic membrane to ethanol toxicity. In this mechanism, the Cis‐unsaturated fatty acids are isomerised to Trans‐unsaturated fatty acids, and the bacterial cells become much more robust to the ethanol stress.45 Figure 2(B) shows the survivor ratio of P. putida cells after treatment with CO2 microbubbles combined with 2 % (v/v) ethanol for 90 min at 100 mL min−1 flow rate and ∼1 bar.
Adding 2% ethanol enhanced the inactivation process, and caused ∼3‐log reduction in the microbial population after 90 min. Interestingly, the time during which the number of microbial cells remains constant‐‐L value‐‐ was∼30 min, ∼4 times longer than the previous experimental set while the D value was almost identical in both experiments. Enhancement of the inactivation activity can be attributed to mechanisms of action of both ethanol and CO2 on the cytoplasmic membrane of Pseudomonas cells and the combined action of both elements. Both CO2 and ethanol work mainly on the fatty acid composition of the membrane.
Changing the composition of the cytoplasmic membrane is not straightforward and a certain threshold needs to be reached before observing any changes in the membrane. The exact duration to reach this threshold has not been studied yet for both CO2 and ethanol. However, previous studies on the changes in the fatty acids profile are helpful to predict the duration. For example, Mejia et al.,46 reported changes in the fatty acids profile of Escherichia coli (gram‐negative bacterium) after exposure to heat shock stress for 30 min. Another example is the study by Boylan et al.,47 where Bacillus subtilis (gram‐positive bacterium) was exposed to environmental stresses such as salt stress for 15–30 min to accumulate β‐galactosidase.
During the first 30 min, the level of the Pseudomonas population was almost constant before the number of bacterial cells gradually decreased with time until the CO2 concentrations reached the equilibrium state.
In addition, it has been suggested that increasing the amount of ethanol used (the organic solvent) could intensify the inactivation process with CO2 microbubbles. Therefore, the concentration of ethanol was increased to 5% (v/v) to test this hypothesis. Figure 2(C) shows the survivor ratio of P. putida after treatment with CO2 enriched microbubbles plus 5% ethanol. Interestingly, there was almost a 2.5 log reduction of Pseudomonas population after treatment. Further, it can be noticed that the time during which the cell number remained constant was much less than in the previous experiments, as well as the D‐value, which reached around 46.8 min. This observation might not be expected but on the other hand, the L value was much lower than in the previous experiment. While decreasing the log reduction in this treatment might have resulted from the increase in bacterial tolerance to ethanol, the microbial exposure to a sub‐lethal level of ethanol might promote a cellular response to encounter this stress. For example, Vanbogelen et al.,48 and Waston49 reported induced expression of stress proteins as a result of exposure to sub‐lethal levels of ethanol and the same proteins were expressed as a result of heat shock. Therefore, there might be a cross‐protective response induced to face environmental stresses in the same manner. Indeed, this cross‐protective response was seen previously in Pseudomonas spp, on exposure to different aromatic compounds and heat. In all three cases, stress shock proteins were produced.50
The concentration of additive (ethanol) was increased to 10% (v/v) to verify the combined effects of ethanol and CO2 and to enhance the whole inactivation process. It can be observed from Fig. 2(D) that there was around 3.5 log reduction in the bacterial population after this treatment. In addition, the D‐value for this experimental set was 82.8 min, while the L‐value was around 4.8 min. Both of these values were higher than the previous experimental set, and consistent with the original hypothesis. Indeed, the magnitude of ethanol toxicity is associated with its concentration used.51 Increasing the ethanol concentration to 10% can cause chemical stress to the bacterial cells, and this stress might be analogous to other stresses.52 As mentioned above, P. putida can evolve adaptative mechanisms as a response to stresses.43, 48 Therefore, it was speculated that increasing the ethanol concentration above a certain threshold could exceed the ability of Pseudomonas cells to tolerate and respond to the elevated level of toxicity, causing serious injuries to the cytoplasmic membrane and eventually failure to keep the biological system balanced. Another important concept to be considered is chaotropicity. Ethanol is known as a chaotropic solute, resulting in water stresses in bacteria at concentrations similar to levels in the environment.53 Hallsworth et al.53 showed that ethanol did not affect cell turgor, but instead, perturbed macromolecule–water interactions and thereby destabilized cellular macromolecules, and inhibited growth. This bacterium responded to ethanol chaotropicity by specifically up‐regulating the synthesis of proteins involved in stabilizing protein structure, in lipid metabolism, and in membrane composition.53 However, destabilization of macromolecules in the biological system is an elastic process in comparison with the specific inactivation (such as inactivation with carbon dioxide). Macromolecules destabilization can be reversed up to a critical thermodynamic point by using kosmotropicity solutes that increase entropy and affect hydration of macromolecules, such as trehalose.53 These solutes tend to order water, and strengthen electrostatic interactions within organic macromolecules.54, 55, 56
CO2 concentrations during CO2‐enriched microbubbles sparging
The CO2 concentration was estimated using Equation (10). Figure 4(A), (B), (C) and (D) present CO2 concentrations at different pHs for all experimental sets. From Fig. 4, it can be noticed that the inactivation process was increased in conjunction with increase in CO2 concentration. This observation can be explained by the fact that the elevated CO2 concentration tends to accumulate in the phospholipid bilayers of cytoplasmic membrane, causing an increase in the penetrated CO2.21, 57 Also, increasing the dissolved concentration of CO2 was accompanied with decreasing survivor ratio of Pseudomonas population as the inactivation efficiency depends on CO2 concentration.58
Figure 4.
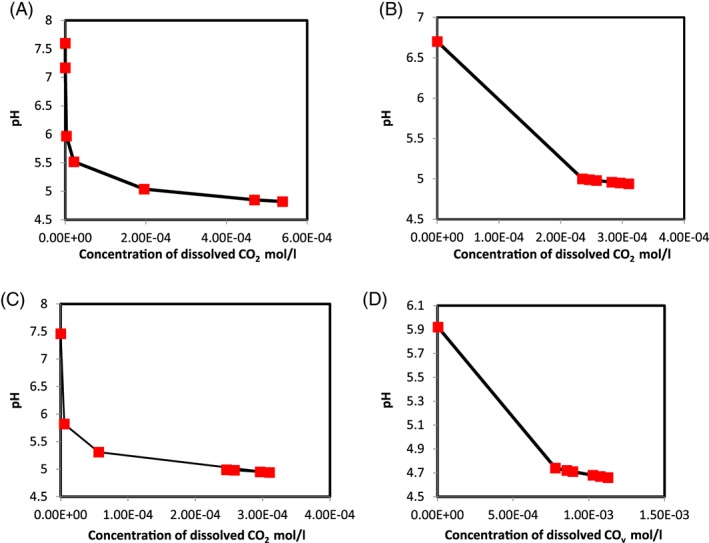
CO2 concentrations with different pHs observed during sparging. (A) CO2 microbubbles. (B) CO2 microbubbles plus 2% ethanol. (C) CO2 microbubbles plus 5% ethanol. (D) CO2 microbubbles plus 10% ethanol. Points are representative of triplicate results.
Morphological changes on Pseudomonas putida cells using CO2 microbubbles with and without ethanol addition
The inactivated cells with CO2‐enriched microbubbles treatment were examined using microscopy to observe the morphological/numerical changes in the bacterial cells after treatment (Fig. 5). There was a clear reduction in the number in comparison with the untreated cells. Further observation (Fig. 6) revealed changes in the morphology such as shortening and shrinkage of the cells.
Figure 5.
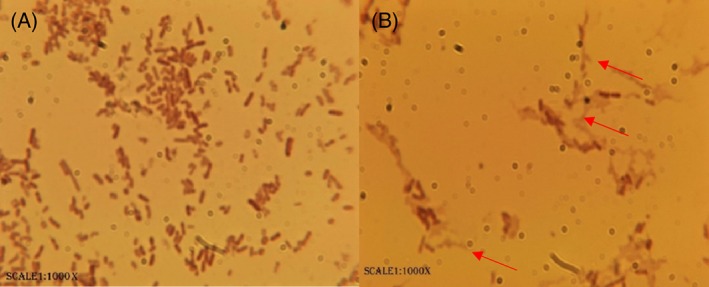
Numerical changes after treatment with CO2 microbubbles on 1000×. (A) Bacterial smears before sparging CO2. (B) Bacterial smear after treating with CO2‐enriched microbubbles for 90 min.
Figure 6.
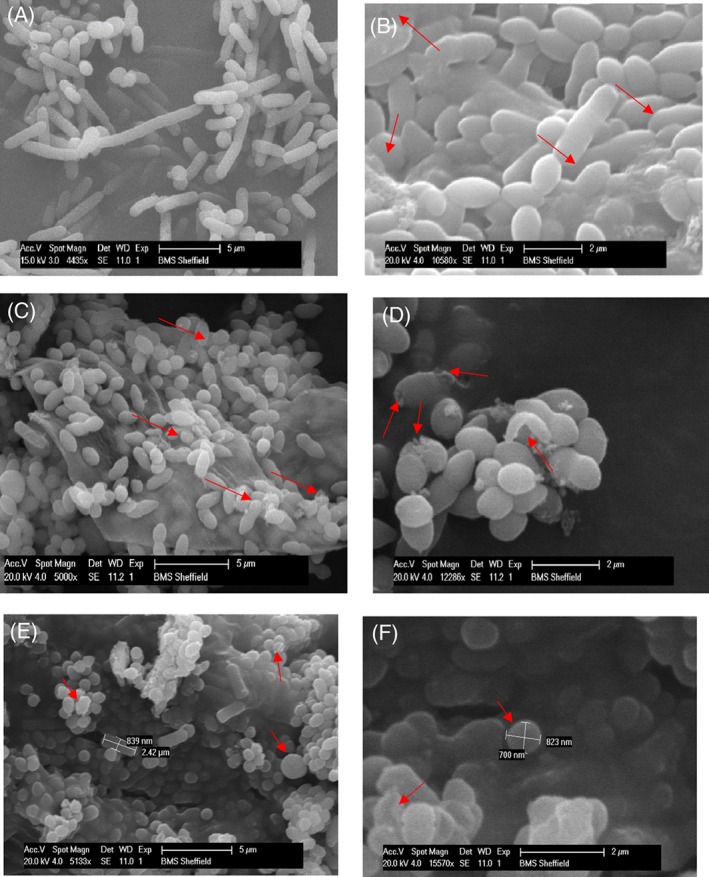
Morphological changes on (A) Pseudomonas cells before the inactivation process with CO2 microbubble plus 2% ethanol. (B) Pseudomonas cells after the inactivation process with CO2 microbubble plus 2% ethanol. (C) Pseudomonas cells before the inactivation process with CO2 microbubble plus 5% ethanol. (D) Pseudomonas cells after the inactivation process with CO2 microbubble plus 5% ethanol. (E) Pseudomonas cells before the inactivation process with CO2 microbubble plus 10% ethanol. (D) Pseudomonas cells after the inactivation process with CO2 microbubble plus 10% ethanol.
Much research has been done on using supercritical CO2 for the inactivation of microorganisms. For example, it was hypothesized that application of supercritical conditions can facilitate CO2 penetration into the cell membrane, consequently expanding the microbial cells and causing cell disruption.59 Indeed, the concept behind using supercritical CO2 was originally described by Fraser,60 when bacterial cells were burst after injecting CO2 under high pressure. Thereafter, this concept was used to recover some cellular constituents such as intracellular enzymes and proteins.61 While many of the previous studies on inactivation with CO2 were achieved under high pressure and elevated temperature (∼73 bars and 31.1 °C) to reach the supercritical state,62 the current study was achieved under relatively low‐pressure CO2 (∼1 bar) and low temperatures (6–12 °C). This suggests that high pressure and temperature are not the only factors affecting cell lysis by CO2 application.
Figure 6 shows morphological changes to the Pseudomonas cells after treatment with CO2 microbubbles plus ethanol as an additive. In Fig. 6(B), it is obvious that the Pseudomonas cells were aggregated after the treatment with CO2‐enriched microbubbles plus 2% ethanol. Many conditions can provoke microbial flocculation such as substrate acquisition, slow growth or starvation, physical and chemical stress and aggregation to protect against predation.63 Moreover, formation of microbial flocs can also help increase the metabolic activity of the stressed cells64 and enhance the microbial resistance to toxic compounds such as biocidal compounds.65, 66, 67 The changes in cell shapes can also be noticed in Fig. 6(B) in comparison with the normal bacterial cells in Fig 6(A) (the control), when Pseudomonas cells were observed to transition from rod cells to coccus cells. This transition might increase attachment of the bacterial cells, consequently leading to flocculation.68 Occasionally, the size reduction is accompanied by an increase in population under various environmental stresses.68, 69, 70, 71, 72, 73 Interestingly, the decrease in cell size was not accompanied with an increase in the bacterial population in the current study, as shown in Fig. 2(B). Changing the cells shape from rod to round shape increases the cell surface area to volume ratio; a useful metabolic response under starvation stress condition. Bacterial cells are known to adjust in order to effectively transport nutrients with minimum energy consumption.74 However, alternating the Pseudomonas cells shape from rod to round shape in the current study is likely a consequence of changes due to the environmental stresses mentioned above.
Figure 6(D) shows the morphological changes that occurred in the Pseudomonas cells as a result of the treatment with CO2 enriched microbubbles plus 5% ethanol. It is apparent that the cells have lesions with loss of membrane integrity. Cell injury and death were previously observed, due to high pressure CO2 application.75 The current study, however, was conducted under low pressure (∼1 bar), and observing the same morphological changes on the bacterial cells is suggestive of an inactivation and subsequent cell lysis capability of CO2 according to the mechanisms reviewed previously.5 Using CO2 under high pressure can enhance the solubility of CO2 and consequently, accelerate cell injury and death.75 These changes are irreversible.4 Therefore, application of high pressure is a way to enhance the CO2 inactivation activity but by no means the only factor responsible for inactivation.
The morphological changes during treatment with the CO2‐enriched microbubbles plus 10% ethanol are presented in Fig. 6(E) and (F). Increasing the ethanol concentration can increase the membrane permeability of the bacterial cells, which is associated with chemical activities, inducing narcosis in biological systems.21 This effect is followed by leakage of protons and some vital ions such as potassium ions from the cytoplasmic membrane of the bacterial cells.76, 77 Interaction of these compounds with the phospholipid bilayers of the cytoplasmic membrane could cause substantial changes in the membrane structure. For example, lipophilic compounds tend to accumulate in the hydrophobic part of the cytoplasmic membrane, disturbing the interaction between acyl chains of the phospholipid bilayers as well as changing the fluidity of the membrane. Eventually swelling of the phospholipid bilayers of the bacterial cells can occur, resulting in a ball‐like shape.78
CONCLUSION
Pseudomonas Puitda KT2240 was used as a model for gram‐negative bacteria in the current study. Survivor ratio after each experiment was calculated after streaking the bacterial samples on nutrient agar plates. The initial log reduction with CO2 microbubbles alone was around 2 Log, while best Log reductions were achieved with 10%, 2% and 5% ethanol, respectively. Using microbubble technology for CO2 sparging caused both an oxidative stress and disturbance to the biological system of P. putida cells. Moreover, addition of ethanol amplified the activities of CO2 microbubbles and decreased the survivor ratio of P. putida. Several morphological changes were observed after each treatment, and these changes ranged from changing the cell shape from rod to round, lesions appearing on the bacterial cells, severe injurious signs and cell death.
ACKNOWLEDGEMENTS
ARM would like to thank the Iraqi Ministry of Higher Education and Scientific Research for the doctoral Scholarship. Many thanks also to Dr Mark S Thomas, Department of Infection and Immunity, the University of Sheffield, for donating Pseudomonas putida KT2440 strain. WZ would like to thank the EPSRC for support under grants EP/I019790/1 (microbubbles dynamics), EP/K001329/1(CO2 microbubbles), EP/N011511/1(in situ sterilisation using microbubbles during fermentation processes). JH would like to thank Plastok Ltd (UK) for microbubble aerator materials.
REFERENCES
- 1. Nair P, Currently practised sterilization methods‐some inadvertent consequences. J Biomater Appl 10:121–135 (1995). [DOI] [PubMed] [Google Scholar]
- 2. Dillow A, Dehghani F, Hrkach JS, Foster NR and Langer R, Bacterial inactivation by using near‐and supercritical carbon dioxide. Proc Nat Acad Sci 96:10344–10348 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Premnath V, Harris W, Jasty M and Merrill E, Gamma sterilization of UHMWPE articular implants: an analysis of the oxidation problem. Biomaterials 17:1741–1753 (1996). [DOI] [PubMed] [Google Scholar]
- 4. Hong S and Pyun Y, Inactivation kinetics of Lactobacillus plantarum by high pressure carbon dioxide. J Food Sci 64:728–733 (1999). [Google Scholar]
- 5. Garcia‐Gonzalez L, Geeraerd A, Spilimbergo S, Elst K, Van Ginneken L, Debevere J et al, High pressure carbon dioxide inactivation of microorganisms in foods: the past, the present and the future. Int J food Micro 117:1–28 (2007). [DOI] [PubMed] [Google Scholar]
- 6. Slaughter JC, The effects of carbon dioxide on yeasts, in Biotechnology Applications in Beverage Production , ed by Cantarelli C. and Lanzarini G. Elsevier Science Publishers, Netherland, 49–64 (1989). [Google Scholar]
- 7. Spilimbergo S and Bertucco A, Non‐thermal bacterial inactivation with dense CO2 . Biotechnol Bioeng 84:627–638 (2003). [DOI] [PubMed] [Google Scholar]
- 8. Timmis K, Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ Microbiol 4:779–781 (2002). [DOI] [PubMed] [Google Scholar]
- 9. Georgieva T, Nikolova D, Evstatieva Y, Licheva T and Savov V, Growth characteristics of Pseudomonas putida strains and effect of humic substances on cell density during batch cultivation. Bulgarian J Agricult Sci 20:82–86 (2014). [Google Scholar]
- 10. Zimmerman WB, Hewakandamby BN, Tesař V, Bandulasena HCH and Omotowa OA, On the design and simulation of an airlift loop bioreactor with microbubble generation by fluidic oscillation. Food Bioprod Process 87:215–227 (2009). [Google Scholar]
- 11. Li P and Tsuge H, Ozone transfer in a new gas‐induced contactor with microbubbles. J Chem Eng Japan 39:1213–1220 (2006). [Google Scholar]
- 12. Li P, Takahashi M and Chiba K, Enhanced free‐radical generation by shrinking microbubbles using a copper catalyst. Chemosphere 77:1157–1160 (2009). [DOI] [PubMed] [Google Scholar]
- 13. Hoigne J and Bader H, Rate constants of reactions of ozone with organic and inorganic compounds in water. Water Res 17:173–183 (1983). [Google Scholar]
- 14. Von Gunten U, Ozonation of drinking water: Part I . Oxidation kinetics and product formation. Water Res 37:1443–1467 (2003). [DOI] [PubMed] [Google Scholar]
- 15. Yang C, Dabros T, Li D, Czarnecki J and Masliyah JH, Measurement of the Zeta potential of gas bubbles in aqueous solutions by microelectrophoresis method. J Colloid Interface Sci 243:128–135 (2001). [Google Scholar]
- 16. Takahashi M, ζ potential of microbubbles in aqueous solutions: electrical properties of the gas‐water interface. J Phys Chem 109:21858–21864 (2005). [DOI] [PubMed] [Google Scholar]
- 17. Bononi I, Balatti V, Gaeta S and Tognon M, Gram‐negative bacterial lipopolysaccharide retention by a positively charged new‐generation filter. Appl Environ Microbiol 74:6470–6472 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rochex A, Lecouturier D, Pezron I and Lebeault JM, Adhesion of a Pseudomonas putida strain isolated from a paper machine to cellulose fibres. Appl Microbiol Biotechnol 65:727–733 (2004). [DOI] [PubMed] [Google Scholar]
- 19. Nikolajeva V, Griba T and Petriņa Z, Factors influencing adhesion of Pseudomonas putida on porous clay ceramic granules. Environ Experiment Biol 10:77–80 (2012). [Google Scholar]
- 20. Hwang G, Park SR, Lee C‐H, Ahn I‐S, Yoon Y‐J and Mhin BJ, Influence of naphthalene biodegradation on the adhesion of Pseudomonas putida NCIB 9816–4 to a naphthalene‐contaminated soil. J Hazard Mater 172:491–493 (2009). [DOI] [PubMed] [Google Scholar]
- 21. Isenschmid A, Marison I and Stockar U Von, The influence of pressure and temperature of compressed CO2 on the survival of yeast cells. J Biotechnol 39:229–237 (1995). [DOI] [PubMed] [Google Scholar]
- 22. Harley JP, Laboratory Exercises in Microbiology, 5th edn McGraw‐Hill, Boston, MA, 1–12 (2002). [Google Scholar]
- 23. Al‐Mashhadani MKH, Bandulasena HCH and Zimmerman WB, CO2 mass transfer induced through an airlift loop by a microbubble cloud generated by fluidic oscillation. Ind Eng Chem Res 51:1864–1877 (2012). [Google Scholar]
- 24. Chatterjee A, Magee JL and Dey SK, The role of homogeneous reactions in the radionlisys of water. Radiat Res 96:1–19 (1983). [Google Scholar]
- 25. Zhang X and Houk KN, Acid/base catalysis by pure water: the aldol reaction. J Org Chem (70):9712–9716 (2005). [DOI] [PubMed] [Google Scholar]
- 26. Garrett BC, Dixon DA, Camaioni DM, Chipman DM, Johnson MA, Kimmel GA et al, Role of water in electron‐initiated processes and radical chemistry: issues and scientific advances. Chem Rev 105:355–389 (2005). [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y, Geochemical Kinetics. Princeton University Press, Princeton and Oxford: (2008). [Google Scholar]
- 28. Khalilitehrani M, An investigation on Modeling and Simulation of Chilled Ammonia Process using VOF Method. Master Thesis, Göteborg, Sweden: (2011). [Google Scholar]
- 29. Watanabe T, Furukawa S, Hirata J, Koyama T, Ogihara, H and Yamasaki M, Inactivation of Geobacillus stearothermophilus spores by high‐pressure carbon dioxide treatment. Appl Environ Microbiol 69:7124–7129 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oulé M, Tano K, Bernier A‐M and Arul J, Escherichia coli inactivation mechanism by pressurized CO2 . Can J Microbiol 52:1208–1217 (2006). [DOI] [PubMed] [Google Scholar]
- 31. Ragnar M, Eriksson T and Reitberger T, Radical formation in ozone reactions with lignin and carbohydrate model compounds. Holzforschung 53:292–298 (1999). [Google Scholar]
- 32. Kim J and Park W, Oxidative stress response in Pseudomonas putida . Appl Microbiol Biotechnol 98:6933–6946 (2014). [DOI] [PubMed] [Google Scholar]
- 33. Imlay JA, Pathways of oxidative damage. Annual Rev Microbiol 57:395–418 (2003). [DOI] [PubMed] [Google Scholar]
- 34. Cabiscol E, Tamarit J and Ros J, Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8 (2000). [PubMed] [Google Scholar]
- 35. Sheu KFR and Blass JP, The β‐ketoglutarate dehydrogenase complex. Annals New York Academy Sci 893:61–78 (1999). [DOI] [PubMed] [Google Scholar]
- 36. Ornston LN and Stanier RY, The conversion of catechol and protocatechuate to g‐ketoadipate by Pseudomonas putida . J Biol Chem 241:3776–3788 (1966). [PubMed] [Google Scholar]
- 37. Nipkow A, Sonnleitner B and Fiechter A, Effect of carbon dioxide on growth of Zymomonas mobilis in continuous culture. Microbiol Wiki 21:287–291 (1985). [Google Scholar]
- 38. Veeramallu UK and Agrawal P, The effect of CO 2 ventilation on kinetics and yields off cell‐mass and ethanol in the batch cultures of Zymomonas mobilis . Biotechnol Lett 8:811–816 (1986). [Google Scholar]
- 39. Conway T, The Entner‐Doudoroff pathway: history, physiology and molecular biology. FEMS Microbiol Rev 103:4465–4470 (1992). [DOI] [PubMed] [Google Scholar]
- 40. Carroll J, Slupsky J and Mather A, The solubility of carbon dioxide in water at low pressure. J Phys Chem Ref Data 20:1201–1209 (1991). [Google Scholar]
- 41. Heipieper HJ, Keweloh H and Rehm HJ, Influence of phenols on growth and membrane‐permeability of free and immobilized Escherichia coli . Appl Environ Microbiol 57):1213–1217 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heipieper HJ, Weber FJ, Sikkema J, Keweloh H and De Bont JAM, Mechanisms of resistance of whole cells to toxic organic solvents. Trends Biotechnol 12:409–415 (1994). [Google Scholar]
- 43. Ingram LO, Adaption of membrane lipids to alcohols. J Bacteriol 125:670–678 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ingram LO, Changes in lipid composition of Escherichia coli resulting from growth with organic solvents and with food changes in lipid composition of Escherichia coli resulting from growth with organic solvents and with food additives. Microbiology 33:1233–1236 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heipieper HJ and Meulenbeld G, Effect of environmental factors on the trans / cis ratio of unsaturated fatty acids in Pseudomonas putida S12. Appl Environ Microbiol 62:2773–2777 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mejia RI, Gomez‐Eichelmann MC and Fernandez MS, Fatty acid profile of Escherichia coli during the heat shock response. Biochem Molec Biol Int 47:835–844 (1999). [DOI] [PubMed] [Google Scholar]
- 47. Boylan SA, Redfield AR, Brody MS and Price CW, Stress‐induced activation of the sigma B transcription factor of Bacillus subtilis . J Bacteriol 175:7931–7937 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vanbogelen RA, Kelley PM and Neidhardt FC, Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli . J Bacteriol 169:26–32 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Watson K, Microbial stress proteins. Adv Microbiol Physiol 31:184–223 (1990). [DOI] [PubMed] [Google Scholar]
- 50. Park SH, Oh KH and Kim CK, Adaptive and cross‐protective responses of Pseudomonas sp. DJ‐12 to several aromatics and other stress shocks. Current Microbiol 43:176–181 (2001). [DOI] [PubMed] [Google Scholar]
- 51. Mrozik A, Piotrowska‐Seget Z and Łabuzek S, Cytoplasmatic bacterial membrane responses to environmental perturbations. Polish J Environ Studies 13:487–494(2004). [Google Scholar]
- 52. Ingram LO, Microbial tolerance to alcohols: role of the cell membrane. Trends Biotechnol 4:40–44 (1986). [Google Scholar]
- 53. Hallsworth JE, Heim S and Timmis KN, Chaotropic solutes cause water stress in Pseudomonas putida . Environ Microbiol 5:1270–1280 (2003). [DOI] [PubMed] [Google Scholar]
- 54. Mansure JJC, Panek AD, Crowe LM and Crowe JH, Trehalose inhibits ethanol effects on intact yeast cells and liposomes. Biochimica Biophysica Acta ‐ Biomembranes 1191:309–316 (1994). [DOI] [PubMed] [Google Scholar]
- 55. Shah D, Johnston TP and Mitra AK, Thermodynamic parameters associated with guanidine HCl‐ and temperature‐induced unfolding of bFGF . Int J Pharmaceut 169:1–14 (1998). [Google Scholar]
- 56. Cray JA, Stevenson A, Ball P, Bankar SB, Eleutherio ECA, Ezej TC et al, Chaotropicity: a key factor in product tolerance of biofuel‐producing microorganisms. Current Opin Biotechnol 33:228–259 (2015). [DOI] [PubMed] [Google Scholar]
- 57. Spilimbergo S, Elvassore N and Bertucco A, Microbial inactivation by high‐pressure. J Supercrit Fluids 22:55–63 (2002). [Google Scholar]
- 58. Kobayashi F, Ikeura H, Odake S and Hayata Y, Inactivation of Saccharomyces cerevisiae by equipment pressurizing at ambient temperature after generating CO2 microbubbles at lower temperature and pressure. LWT ‐ Food Sci Technol 56:543–547 (2014). [Google Scholar]
- 59. Darani K and Mozafari MR, Supercritical fluids technology in bioprocess industries: a review. J Biochem Technol 2:144–152 (2010). [Google Scholar]
- 60. Fraser D, Bursting bacteria by release of gas pressure. Nature 167:33–34 (1951). [DOI] [PubMed] [Google Scholar]
- 61. Lin H and Chen L, Method for recovery of intracellular material by disruption of microbial cells with carbon dioxide under pressure. US patent 5306637, West Lafayette, Ind., Indiana (1994).
- 62. Díaaz‐Reinoso B, Moure A, Domiänguez H and Parajoä JC, Supercritical CO2 extraction and purification of compounds with antioxidant activity. J Agric Food Chem 54:2441–2469 (2006). [DOI] [PubMed] [Google Scholar]
- 63. Bossier PA and Verstraete W, Triggers for microbial aggregation in activated sludge? Appl Microbiol Biotechnol 45:1–6 (1996). [Google Scholar]
- 64. McFeters GA, Egli T, Wilberg E, Schneider R, Suozz M and Giger W, Activity and adaptation of nitrilotriacetate (NTA)‐degrading bacteria: field and laboratory studies. Water Res 24:875–881 (1990). [DOI] [PubMed] [Google Scholar]
- 65. Costerton JW, Cheng KJ, Greesy GG, Ladd TI, Nickel JC, Dasgupta M et al, Bacterial biofilms in nature and disease. Annu Rev Microbiol 41:435–464 (1987). [DOI] [PubMed] [Google Scholar]
- 66. Giwercman B, Jensen ET, Hoiby N, Kharazmi A and Costerton W, Induction of beta‐lactamase production in Pseudomonas aeruginosa biofilm. Antimicrob Agents Chemother 35:1008–1010 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Anwar H, Strap JL and Costerton JW, Establishment of aging biofilms: possible mechanism of bacterial resistance to antimicrobial therapy. Antimicrob Agents Chemother 36:1347–1351 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fakhruddin ANM and Quilty B, The response of Pseudomonas putida CP1 cells to nutritional, chemical and environmental stresses. World J Microbiol Biotechnol 22:507–514 (2006). [Google Scholar]
- 69. Novitsky JA and Morita RY, Morphological characterization of small cells resulting from nutrient starvation in a psychrophilic marine Vibrio . Appl Environ Microbiol 32:619–622 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Amy PS and Morita RY, Starvation‐survival patterns of sixteen freshly isolated open‐ocean bacteria. Appl Environ Microbiol 45:1109–1115 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Givskov M, Eberl L, Moller S, Poulsen LK and Molin S, Responses to nutrient starvation in Pseudomonas‐Putida Kt2442 – analysis of general cross‐protection, cell‐shape and macromolecular content. J Bacteriol 176:7–14 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mueller RF, Bacterial transport and colonization in low nutrient environments. Water Res 30:2681–2690 (1996). [Google Scholar]
- 73. Makarov AA, Dorofeev AG and Panikov NS, Cell shape and size of starving microorganisms as determined by computer image analysis. Microbiology 67:264–273 (1998). [Google Scholar]
- 74. Sanin SL, Sanin FD and Bryers JD, Effect of starvation on the adhesive properties of xenobiotic‐degrading bacteria. Process Biochem 38:909–914 (2003). [Google Scholar]
- 75. Hong SI, Park WS and Pyun YR, Inactivation of Lactobacillus sp. from Kimchi by high‐pressure carbon dioxide. Food Sci Technol 30:681–685 (1997). [Google Scholar]
- 76. Leão C and Van Uden N, Effects of ethanol and other alkanols on passive proton influx in the yeast Saccharomyces cerevisiae . Biochimica Biophysica Acta 774:43–48 (1984). [DOI] [PubMed] [Google Scholar]
- 77. Cartwright CP, Jurosze J‐R, Beavan MJ, Ruby FMS and De Moraisi SMF, Ethanol dissipates the proton‐motive force across the plasma membrane of Saccharomyces cerevisiae . Microbiology 132:369–377 (1986). [Google Scholar]
- 78. Sikkema J, de Bont JA and Poolman B, Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev 59:201–222 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]


