Abstract
Human papillomavirus (HPV) infection is the most important risk factor for cervical cancer development. In HeLa cell line, the HPV viral genome is integrated at 8q24 in one allele of chromosome 8. It has been reported that the HPV fragment integrated in HeLa genome can cis‐activate the expression of proto‐oncogene MYC, which is located at 500 kb downstream of the integrated site. However, the underlying molecular mechanism of this regulation is unknown. A recent study reported that MYC was highly expressed exclusively from the HPV‐integrated haplotype, and a long‐range chromatin interaction between the integrated HPV fragment and MYC gene has been hypothesized. In this study, we provided the experimental evidences supporting this long‐range chromatin interaction in HeLa cells by using Chromosome Conformation Capture (3C) method. We found that the integrated HPV fragment, MYC and 8q24.22 was close to each other and might form a trimer in spatial location. When knocking out the integrated HPV fragment or 8q24.22 region from chromosome 8 by CRISPR/Cas9 system, the expression of MYC reduced dramatically in HeLa cells. Interestingly, decreased expression was only observed in three from eight cell clones, when only one 8q24.22 allele was knocked out. Functionally, HPV knockout caused senescence‐associated acidic β‐gal activity in HeLa cells. These data indicate a long‐distance interaction of the integrated HPV fragment with MYC gene and 8q24.22 region, providing an alternative mechanism relevant to the carcinogenicity of HPV integration.
Keywords: cervical cancer, HPV, MYC, long‐distance interaction, integration
Short abstract
What's new?
Infection with human papillomavirus (HPV) is a well‐established risk factor for cervical cancer. The mechanism by which HPV integration into the host genome influences cervical oncogenesis, however, is not fully understood. Using chromosome conformation capture, the authors of this study provide evidence for carcinogenic involvement of a long‐range chromatin interaction between integrated HPV fragments, the proto‐oncogene MYC, and the 8q24.22 chromosomal region in HeLa cells. Knock‐out of either the integrated HPV fragment or the 8q24.22 region in HeLa cells significantly reduced MYC expression. The findings offer an alternative explanation for the carcinogenic mechanism of HPV viral genome integration.
Human papilloma virus (HPV) infection is the major causative factor of cervical carcinoma, which ranks the fourth most common cancer among women globally.1 The known carcinogenic mechanisms of HPV infection in cervical carcinoma include: chronic viral infection, deregulation of cell cycle and chromosomal instability caused by viral oncoprotein E6/E7, and the integration of HPV DNA in human genome.2, 3, 4, 5 Indeed, our previous study demonstrated that alteration of human genome and functional aberration of integration‐targeted genes by HPV integration might play an important role in cervical oncogenesis, which provided additional evidences supporting that integrated viral DNA fragments may affect the expression and function of host genes near integration sites, and thereby resulting in malignant transformation of cervical cells.6
HeLa cell is one of the most widely used cell model in biomedical research of cervical cancer. In HeLa cells, multi‐copies of incomplete HPV viral genome integrated at chromosome 8q24, about 500 kb upstream of proto‐oncogene MYC.7, 8 There are lots of viewpoints considered that the integrated HPV fragment could activate the expression of MYC.9, 10 Considering the long distance between the two elements, previous studies mainly focused on the effect of HPV E6 and E7 proteins on the expression of MYC gene.11, 12 However, a recent study demonstrated that the highly expressed MYC was almost exclusively from the HPV‐integrated haplotype (mean ratio, 95:1) by RNA‐seq data, which implicated that the integration of viral genome, as a strong activator of MYC expression, acted in cis rather than trans manor.7 Therefore, they hypothesized that there is maybe a long‐range chromatin interaction between integrated HPV fragment and the HPV‐integrated haplotype MYC gene, but it still needed further evidence.
Chromosomes existing in nucleus present a highly complex three‐dimensional structure which may bring widely separated functional elements into close spatial proximity and create long‐range interactions in these elements.13 Chromosome conformation capture (3C) is a traditional and powerful technique that allows for the generation of three‐dimensional organization of genome. The 3C technique is based on sequential formaldehyde cross linking of chromatin, followed by fragmentation and re‐ligation of the cross‐linked chromatin. The re‐ligated fragments were detected by PCR with locus‐specific primers.14 In this study, by using 3C technique we explored the spatial proximity between the integrated HPV fragment and MYC locus in HeLa cells, and investigated the influence of the long‐range chromatin interaction on the expression of MYC by knocking out HPV or 8q24.22 region from human genome through CRISPR/Cas9 system. Our observation of multi‐locus involvement in the long‐range interaction of HPV on MYC expression further refined the carcinogenic mechanism of HPV integration.
Material and Methods
Cell culture
HeLa and HEK293 cells were grown in Dulbecco's modified Eagle medium (DMEM), containing 10% fetal bovine serum (GIBCO), 100 IU/ml penicillin and 100 μg/ml streptomycin(GIBCO) at 37°C in 5% CO2 humidified incubator.
Chromosome conformation capture
Adherent cultured cells were made single‐cell suspension and fixed by adding formaldehyde to a final concentration of 2% followed by incubation for 10 min at room temperature. Cross‐linking was stopped by adding glycine to a final concentration of 1 M and incubating at room temperature for 5 min. The cells were then collected by centrifugation and washed twice with ice cold PBS. Cell lysis was performed by adding 1 ml of cell lysis buffer (10 mM Tris‐HCl, pH 7.5, 10 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 1 × protein inhibitor) and incubating for 30 min on ice. The cytoplasmic components were removed by centrifugation and the nuclear pellets were dissolved in 1× cutsmart buffer (NEB) containing 0.3% SDS. Then, 2% Triton X‐100 was added and incubated for an hour. Restriction enzyme TaqI (NEB) was used to digest large genome DNA to generate fragments by incubating at 65°C for 16 hrs, and then inactivated at 80°C for 30 min. The DNA fragments were end‐repaired by the use of T4 DNA ligase (NEB) and collected by phenol chloroform extraction. Finally, PCR amplification were performed by specific primers of HPV, MYC and 8q24.22. ACTB gene was used as a loading control for the 3C assay. Also, the ACTB‐MYC ligation was used as a specificity control for the 3C assay. The sequences of primers used in 3C assays are shown in Supporting Information Table S1.
The design and construction of CRISPR/Cas9 system
The CRISPR/Cas9 system was used to knock out HPV DNA and 8q24.22 from HeLa cells. To cleave HPV DNA from human genome, we designed two gRNAs which targeted at nucleotides of 5,790 and 2,057 nt of the HPV‐18 genome. Then the two gRNAs were cloned into pSpCas9(BB)‐2 A‐GFP (PX458), a plasmid which contains Cas9 from S. pyogenes with 2A‐EGFP and cloning backbone for sgRNA (Addgene, Cambridge, MA), respectively. Since the double strand breaks of HPV DNA cleaved by Cas9 nuclease would mainly repaired through the mutagenic non homologous end‐joining, co‐transfection of these two px458‐HPV‐gRNA plasmids into HeLa cells would result in the knock‐out of HPV fragment from human genome, approximately covering nucleotides from 5,790 to 2,057 nt of HPV‐18 genome. Similar strategy was used to design the CRISPR/Cas9 system for 8q24.22 knock‐out. Sequences of HPV and 8q24.22 gRNAs are listed in Supporting Information Table S1.
Establishment of stable HeLa cell lines with HPV or 8q24.22 knock out
The px458 vectors aimed at HPV or 8q24.22 region were transiently transfected into HeLa cells by using Lipofectamine® 2000 Transfection Reagent (Invitrogen). Forty‐eight hours post transfection, cells were trypsinized and seeded into 96‐well plates (1 cell/well). Three‐four weeks later, the visible cell colonies were detected by PCR to select the stable HeLa cell lines with HPV or 8q24.22 knock out. The primers used for PCR assay were listed in Supporting Information Table S1
RNA extraction and gene‐expression analysis
Total RNA of HeLa cells was extracted by using trizol reagent (Life Technologies) according to the manufacturer's instructions. Real time quantitative PCR was performed to measure the expression of MYC gene using Roche lightcycle 480 sequence detection system (Roche, Germany). The primers of MYC were listed in Supporting Information Table S1. The amplification protocol consisted of a pre‐denaturation step at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 20 sec, annealing at 60°C for 30 sec and extension at 72°C for 30 sec. The expression of MYC gene was determined using the comparative Ct method (2–ΔCt) after normalization to CTBP.
Cell senescence analysis
Cell senescence was detected by using the Cell Senescence Assay (Sciencell) following the instructions based on the phenomenon with high β‐gal activity in senescent cells. Briefly, cells were seeded into six‐well plates and fixed by incubating with fixing solution for 15 min and then strained by staining solution in the dark for 24 hrs. After that, cells were rinsed with PBS and the blue stained cells were examined using a light microscope.
Southern blot
Restriction enzyme BamHI (NEB) was used to cut the whole genome DNA of wild HeLa cells and two HPV‐ko HeLa cell lines. Then the DNA was separated by gel electrophoresis and detected by the probe labeled with DIG which was a region from 727 to 1,376 nt in HPV genome.
Statistical analyses
The differences of expression levels between groups were analyzed by t test using GraphPad and a p values of <0.05 was considered significant.
Results
The integrated HPV fragment is close to MYC locus in spatial location
The 3C technique was used to explore the spatial relationship between integrated HPV fragment and MYC gene. The Schematic diagram of 3C procedure was shown in Figure 1 a. If the integrated HPV and MYC gene have close spatial proximity, the PCR assay of 3C by using specific primers of HPV and MYC could amplify four different products, when the crosslinked genome DNAs were used as the PCR template. As shown in Figure 1 b, we observed three PCR products which were amplified by primers HPV F+ MYC F(450 bp), HPV R + MYC F(500 bp) and HPV R + MYC R(260 bp) only in the crosslinked HeLa cells. Sequencing of the purified PCR products confirmed that all these three fragments contained both HPV and MYC genome sequences. To demonstrate the specificity of 3C assay, we added a housekeeping gene ACTB to the 3C experiments, which was not expected to interact with MYC. The PCR assays showed no amplification of ACTB‐MYC conjugated fragment by using primer pairs from ACTB and MYC. We also added a loading control to the 3C assays by using a primer pair amplifying a region within ACTB loci, which proved the reliability and the specificity of 3C assays. These results demonstrated the specificity of 3C assay and revealed a higher collision probability between HPV and MYC fragments digested by TaqI enzyme in reality than in random situation, suggesting that the integrated HPV fragment is close to MYC locus in physical location in HeLa cells.
Figure 1.
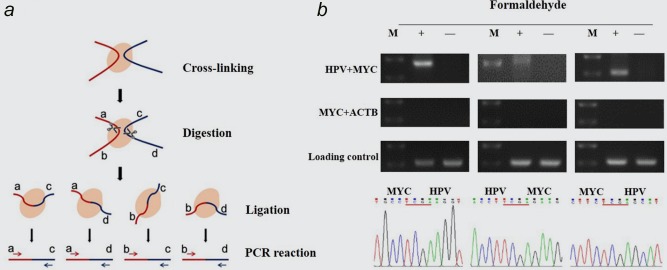
Physical proximity of HPV fragment and MYC gene detected by 3C assays. (a) Schematic diagram of 3C procedure. (b) HPV‐MYC ligation fragments detected by 3C assays. The PCR products were obtained from cross‐linked HeLa cells with three pairs of primers: HPV F + MYC F(410 bp), HPV F + MYC R(506 bp) and HPV R + MYC R(266 bp), respectively (upper). The sequencing of these fragments identified HPV‐MYC piecing sequences mediated by TaqI enzyme site (lower). ACTB‐MYC ligation(320 bp) and ACTB gene(277 bp) were used as specificity and loading control, respectively. Three independent experiments were performed for each 3C assay. M, DNA Marker. [Color figure can be viewed at wileyonlinelibrary.com]
The spatial location of integrated HPV fragment is close to 8q24.22 region
Next, we investigated whether there were other potential regions close to the integrated HPV fragment in three‐dimensional space except for MYC locus. Following the 3C protocol, we used primers of HPV genome together with Alu sequences, which were widely used to detect viral‐host junctions.15 We observed a unique PCR product in crosslinked HeLa cells by using HPV and Alu primers (Fig. 2 a, upper). The subsequent sequencing analysis demonstrated that this product was a fusion fragment consisted of HPV and partial human genome sequences mediated by TaqI enzyme digestion sites. When we searched the sequence of this fragment in the database of human genome (http://genome.ucsc.edu) for homologous sequence, we found that this fragment mapped its position to chromosome 8q24.22 region, a site about 3,300 kb down from the integrated HPV fragment. Next, to confirm the close location between the integrated HPV fragment and 8q24.22 region, we combined the specific primers of HPV with 8q24.22, and again found that the PCR products were consisted of HPV and 8q24.22 sequences (Fig. 2 a, lower). Therefore, these results indicated that the integrated HPV fragment located quite close to 8q24.22 region spatial distance by chromatin folding, although they were 3,300 kb away from each other in linear distance.
Figure 2.
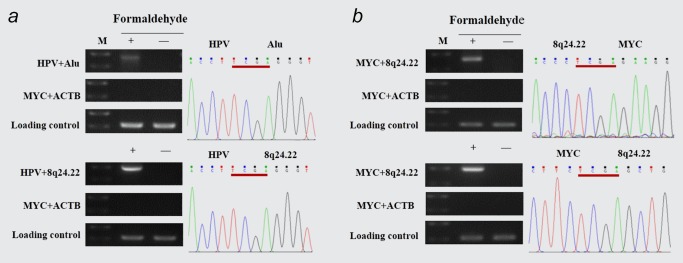
Physical proximity of HPV‐8q24.22 and MYC‐8q24.22 detected by 3C assays. (a) HPV‐8q24.22 ligation fragments were amplified and sequenced in cross‐linked HeLa cells using primer pairs of HPV F + Alu(430 bp) (upper) and HPV F + 8q24.22 F (450 bp) (lower). (b) MYC‐8q24.22 ligation fragments were amplified and sequenced as (A) by using primers of MYC R + 8q24.22 F (376 bp), MYC R + 8q24.22 R (476 bp). ACTB‐MYC ligation and ACTB gene were used as specificity and loading control, respectively. Three independent experiments were performed for each 3C assay. [Color figure can be viewed at wileyonlinelibrary.com]
The spatial locus of MYC gene is close to 8q24.22 region
Since the integrated HPV fragment is close to not only MYC locus but also 8q24.22 region in physical space, we wondered whether MYC locus is close to 8q24.22 region too. Specific primers of MYC and 8q24.22 were used in the 3C assay. As expected, we detected the presence of MYC and 8q24.22 genome fusion sequences linked by TaqI enzyme digestion sites in cross‐linked HeLa cells (Fig. 2 b), suggesting that MYC locus is also close enough to 8q24.22 region in spatial relationship, even though MYC locus is approximately 2,800 kb upstream from 8q24.22 region in linear condition.
The integrated HPV fragment, MYC locus and 8q24.22 region make up a trimer in spatial location
On the basis of the above results, it is of interest to explore whether the three elements, namely the integrated HPV fragment, MYC locus and 8q24.22 region, are spatially and temporally close to each other in HeLa cells. To detect this, we constructed HPV and 8q24.22 knock‐out HeLa cell strains by using CRISPR/Cas9 technology.16 It has been reported that the integrated HPV DNA in HeLa cells are partial copy of the HPV‐18 genome which interspersed with region of human chromosome 8q24.21. The integrated HPV contains only two‐thirds of the complete viral genome, including E6 and E7 viral genes which amplified to a copy number of approximately 12, but lacking a functional copy of E2, an inhibitor of E6 and E7 (Fig. 3 a). In addition, the integrated structure also contains a non‐coding regulatory region of HPV genome.7 Considering the complexity of the HPV integration, we deigned dual gRNAs which targeted HPV18 genome at nucleotide 5,790 and 2,057, respectively (NCBI Reference Sequence: NC_001357.1; Fig. 3 a). Using single clone isolation, we generated stable HPV knock‐out HeLa cell strains. The PCR and sequencing assays identified a 4,120 bp viral DNA fragment removed from HPV‐ko HeLa cells (Fig. 3 b). In consistent with this result, 3C method revealed that in contrast to the spatial co‐location of MYC locus and 8q24.22 region in the wild type HeLa cells, the physical proximity of MYC locus and 8q24.22 region disappeared in HPV‐ko HeLa cells. HEK293 cell line lacking HPV integration also showed no physical proximity of MYC locus and 8q24.22 region, which was used as a negative control of 3C assay (Fig. 3 c). This result indicated that the integrated HPV fragment is necessary for the formation of the spatial proximity between MYC locus and 8q24.22 region.
Figure 3.
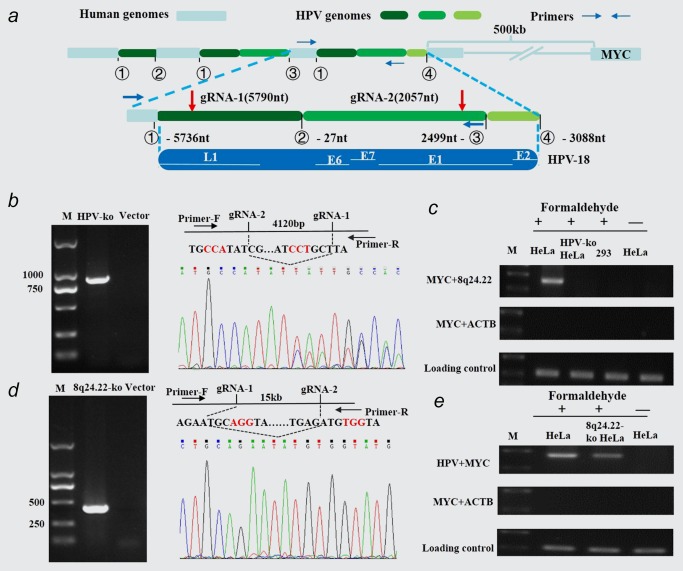
Long‐distance interaction among HPV, MYC and 8q24.22 via constituting a trimer in spatial location. (a) Proposed structure of the chromosome 8 locus containing HPV‐18 integration in HeLa cells. The top panel represented the rearranged haplotype with HPV‐18 insertion on chromosome 8q24.2 region. The bottom panel showed the partial HPV‐18 genome and corresponding genes. Red arrows referred to the location of two designed gRNAs in HPV genome, which were expected to cut about a 4,120 bp fragment. Blue arrows represented the primers used to detect the knockout HPV fragment. (b) HPV knock out in HeLa single clone strains detected by PCR assay (left panel) and DNA sequencing (right panel). (c) Effects of HPV knockout on the spatial co‐location of MYC locus and 8q24.22 region. The 3C assays were performed by using primers of MYC R and 8q24.22 F. (d) 8q24.22 double knockout in HeLa single clone strains detected by PCR assay (left panel) and DNA sequencing (right panel). (e) Effects of 8q24.22 double knockout on the spatial co‐location of integrated HPV and MYC locus. The 3C assays were performed by using primers of HPV F and MYC F. [Color figure can be viewed at wileyonlinelibrary.com]
We also generated stable 8q24.22 double knock‐out HeLa cell strains with a 15 kb DNA fragment removed from chromosome 8 (Fig. 3 d). To evaluate the potential influence of 8q24.22 region on the physical proximity of HPV, MYC and 8q24.22, we repeated 3C assays in 8q24.22 double‐ko HeLa cells. We found that the abundance of crosslinked HPV‐MYC fragments was noticeably decreased in 8q24.22 double knock‐out HeLa cells compared to wild type HeLa cells, but it still existed (Fig. 3 e), suggesting that the 8q24.22 region was important but not necessarily essential for the spatial interaction between the integrated HPV fragment and MYC locus. Taken together, the above results indicated that the spatial interaction among the three elements occurred at the same space and time by forming three‐dimensional structures of chromosomes in HeLa cells.
MYC expression was decreased when either integrated HPV or 8q24.22 region was knocked out
By single clone isolation, we generated a total of two HPV‐ko and nine 8q24.22‐ko HeLa cell strains. Southern blot assay showed that the abundance of integrated HPV fragment was significantly decreased in two HPV‐ko HeLa cell lines compared to wild type (Fig. 4 a). Further Real‐time PCR assay revealed that the gRNAs have achieved an 86 and 78% knock‐out in the HPV‐ko #1 and HPV‐ko #2 cell strains, respectively (Fig. 4 b). Among the 8q24.22‐ko HeLa cell strains, PCR assays identified one 8q24.22 region double deletion. The other eight strains are heterozygous 8q24.22‐ko in which the 8q24.22 region was removed randomly from one of the two chromosome 8.
Figure 4.
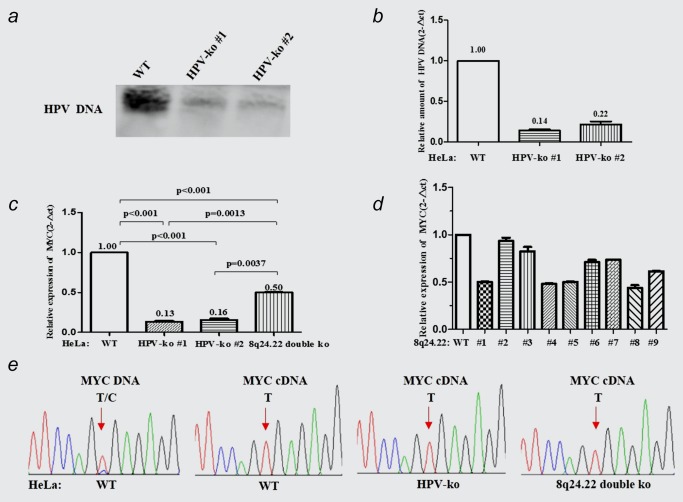
Effect of HPV and 8q24.22 knock‐out on MYC expression. (a, b) The efficiency of HPV DNA knockout in two HPV‐ko HeLa cell strains detected by southern blot analysis (a) and real‐time PCR (b). (c) Relative expression of MYC in HPV‐ko HeLa cell strains detected by Real‐time PCR. (d) Relative expression of MYC in 8q24.22 double (#1) or single (#2–9) knock‐out HeLa cell strains detected by Real‐time PCR. (e) The heterozygous SNP (rs4645948, T/C) in MYC DNA and cDNA were detected by PCR and sequencing in wild‐type, HPV‐ko and 8q24.22‐ko HeLa cells. [Color figure can be viewed at wileyonlinelibrary.com]
To assess the effect of the trimerized spatial interaction on MYC expression, we quantitatively measured the expression of MYC in these HeLa cell strains. The results of real‐time RT‐PCR assays showed that the relative expression of MYC gene in the two HPV‐ko HeLa cell lines was decreased to 13 and 16%, respectively (p < 0.001, p < 0.001). The expression of MYC was decreased to 50% in 8q24.22 double‐ko HeLa cells (#1) compared to that in wild type Hela cells(p < 0.001), but the effect of 8q24.22 double‐ko was significantly weaker than HPV‐ko cells (p = 0.0013, p = 0.0037; Fig. 4 c). As to the heterozygous 8q24.22‐ko HeLa cell strains, the expression of MYC was significantly decreased only in cell strains #4, #5 and #8, while showed no differences in #2, #3, #6, #7 and #9 cell strains when compared to that in the wild‐type HeLa cells (Fig. 4 d). Since MYC is highly expressed almost exclusively from the HPV‐18‐integrated haplotype in HeLa cells, the heterozygous 8q24.22 knock‐out which showed similar effect on MYC expression to that of 8q24.22 double knock‐out might be driven from the chromosome with HPV integration.
We also evaluated the haplotype‐imbalanced regulation on MYC expression. We detected a heterozygous SNP (rs4645948, T/C) in MYC gene in wild‐type HeLa cells through PCR amplification and sequencing (Fig. 4 e). However, sequencing analysis revealed that the MYC transcripts contained only the “T” base (mapped to the HPV‐integrated chromosome), no matter the transcripts were derived from the wild type HeLa cells, HPV‐ko HeLa cells or 8q24.22‐ko HeLa cells. Consistent with Andrew Adey's study,7 this result indicated that the contribution of “T” allele from the HPV‐integrated chromosome to the transcription of MYC gene was far greater than “C” allele.
HPV knockout induced senescence in HeLa cells
During the single clone selection, we noticed that it was very difficult to develop the stable HPV‐ko cells strains, its success rate was significantly lower than that of 8q24.22‐ko cells (2 vs. 9). It has been reported that MYC inactivation could induce the phenotype of cellular senescence in tumor cells.17, 18, 19 Since the expression of MYC was dramatically reduced when the HPV knock‐out, we postulated that HPV‐ko HeLa cell strains might develop phenotype of senescence. Indeed, the two HPV‐ko HeLa cell strains displayed obvious morphological changes soon in the following culture. To some extent, the shape of HeLa cells became huge and flat in the HPV‐ko HeLa cells, with a typical cell senescence morphology.20 We then performed β‐gal activity staining experiment to detect the senescence‐associated acidic β‐gal activity(SA‐β‐gal) in HPV‐ko and 8q24.22‐ko HeLa cells. The wild‐type and SOX6‐overexpressed HeLa cells were used as negative and positive controls, respectively.21 The results showed that the SA‐β‐gal positive cells in HPV‐ko HeLa cells was much more than that in Wild‐type HeLa cells (80.33 vs. 0.33%, p < 0.001). However, no obvious senescence was detected in 8q24.22 double–ko Hela cells, although the SA‐β‐gal positive cells in 8q24.22 double–ko HeLa cells was a little more than that in Wild‐type cells (3.67 vs. 0.33%; Fig. 5 a and 5B). These results indicated that significant decrease of MYC expression in HPV‐ko cells might contribute to the phenotype of senescence in HeLa cells.
Figure 5.
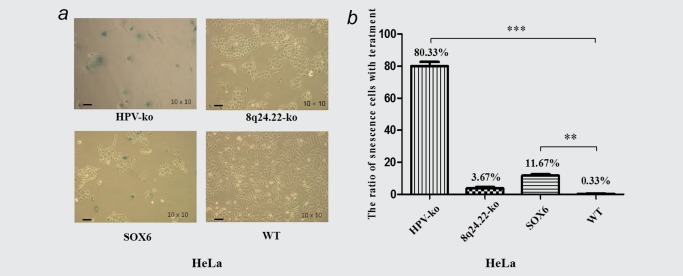
Effect of HPV and 8q24.22 knockout on senescence of HeLa cells. (a) Senescence‐associated acidic β‐gal activity detected by Cell Senescence Assay. Wild‐type HeLa cells and HeLa cells with overexpressed SOX6 were detected as negative and positive controls, respectively. The scale value is 400 μm. (b) The quantified analysis of senescence phenotype in four type of HeLa cells shown in 5a. [Color figure can be viewed at wileyonlinelibrary.com]
Discussion
In cervical cancer HeLa cells, the integrated HPV fragment has been found to activate the expression of MYC gene which located at downstream 500 kb specifically from the HPV‐integrated chromosome, indicating a long‐range interaction between the integrated viral genome DNA and MYC. Indeed, the interaction of such regulatory elements and their targeted genes have been demonstrated in three‐dimensional context, which implicated that distal elements can engage in gene regulation through long‐range looping interactions.22, 23 In this study, we confirmed for the first time the physical proximity between the integrated HPV fragment and MYC gene in HeLa cells.
By using 3C technique, we demonstrated the long‐distance interaction among the integrated HPV fragment, MYC gene and 8q24.22 region via forming a trimer. The 8q24.22 region is 3,300 kb distant from the integrated HPV fragment and 2,800 kb distant from MYC locus. We found that the physical proximity between the MYC locus and the 8q24.22 region was disappeared when the integrated HPV fragment was mainly removed, while the physical proximity between the MYC locus and the integrated HPV fragment was still existing, even though weakened when the 8q24.22 was removed. Importantly, the expression of MYC became much lower in HeLa cells with integrated HPV fragment removed by CRISPR/Cas9 genome editing system, as compared to that in HeLa cells with 8q24.22 region knockout. These results suggested that unlike the integrated HPV fragment which act as a dominant player, the 8q24.22 region was probably just a participant in regulating the expression of MYC (Fig. 6). In supporting our notion, several studies have showed that distal regulatory elements can interact with not only their target genes but also other elements, and the formation of intricate three‐dimensional patterns involves genes and their regulatory elements hundreds of kb distant from each other in linear relationship.24, 25, 26
Figure 6.
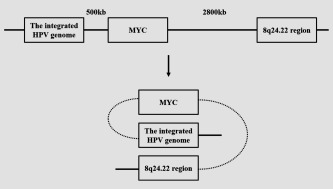
Schematic for the long distance regulation among the integrated HPV fragment, MYC and 8q24.22 region.
The insertion of HPV genome leads to complicated integration‐associated rearrangements and amplification in both viral genome and human chromosome 8q24.21 genomic DNA. Although we have successfully removed a 4,120 bp fragment of inserted viral DNA in two stable HeLa cell strains by CRISPR‐cas9 system, we could not demonstrate that all the copies of HPV have been knocked out because the gRNA achieved an 86 and 78% knock‐out in the two HPV‐ko HeLa cell strains, respectively. However, we indeed observed a significant downregulation of MYC expression in these cell strains, indicated that the deletion of the viral insertion induced by CRISPR‐cas9 system might be enough to disturb the long distance interaction between the inserted HPV fragment and the MYC alleles, and thereby decreasing the expression of MYC gene.
It has been reported that MYC is highly expressed almost exclusively from the HPV‐18‐integrated haplotype in HeLa cells. Therefore, it would be of interest to determine whether the 3C sequences amplified with HPV, MYC and 8q24.22 were derived from the HPV‐integrated chromosome or from the other allele. We indeed identified the haplotype‐imbalanced regulation on MYC expression in HeLa cells by analyzing the contribution of each allele to the abundance of MYC expression. However, we could not determine the allele specificity of 8q24.22 by our current method because of the lack of the heterozygous SNPs in the crossing‐linked DNA fragments. Since some heterozygous single 8q24.22‐ko HeLa cell strains showed similar effect on MYC expression to that with 8q24.22 double knockout cells, it is reasonable to assume that these allele‐specific knockout of the 8q24.22 which decreased MYC expression were driven from the chromosome with HPV integration.
In addition to our findings of the long‐distance regulation of HPV on allele‐specific MYC expression, Ahmadiyeh et al. had discovered that the 8q24 risk loci which was located at the chromosomal region several hundred kb upstream of the MYC gene, could form a long‐range chromatin loop with MYC gene tissue‐specifically in prostate, breast and colon cancer cells.27 Furthermore, the distal Wnt/ß‐catenin responsive enhancers that localized to a region over 400 kb upstream from MYC gene also could interact with MYC promoter through large chromatin loops in colon cancer cells.28 These findings indicated that the MYC promoter could be regulated by distant chromatin elements such as enhancers through long‐range interaction system, and reflected the complexity of the transcriptional control of MYC in different tissues. We also analyzed the structure of 8q24.22 region based on the database of human genome, and found that there was no coded gene and no obvious enhancer sequence in this region. Future study should focus on the detailed mechanisms mediated the long distance interaction among the inserted HPV fragment, 8q24.22 and the MYC alleles. For example, CHIP experiment for enhancer markers such as histone modification elements may help to identify possible regulating element in 8q24.22 region.
Loss of HPV viral proteins E6 and E7 in cervical cancer cells could induce p53 or Rb expression, leading to cell cycle arrest and cell death.29 Knock down of E6 and E7 protein by RNA interference could induce senescence in HeLa Cells.30 Consistently, we observed that HeLa cells occurred senescence when the integrated HPV fragment was removed. Considering the phenomenon that the expression of MYC was dramatically reduced when the HPV knockout, we postulated that the cell senescence might be related to the significant downregulation of MYC expression, which had been proved to play a crucial role of integrated HPV in cervical cancer cell growth.17 But we also noticed that the double knockout of 8q24.22 region could not induce obvious senescence in HeLa cells, although it lead to about 50% decrease of MYC expression. The most possible reason for this is that in addition to the downregulation of MYC gene, other mechanisms including the expression of p53 or Rb induced by E6/E7 remove also played an important role in the development of cell senescence. Therefore, we cannot exclude the possibility that the low MYC expression might reflect the phenotype of senescent cells. However, based on the reasons mentioned above, we prefer to believe that the possibility of low MYC expression is related to the disturbed long distance interaction between HPV and MYC.
In summary, we demonstrated the long‐range interaction among the integrated HPV fragment, MYC locus and the 8q24.22 region in spatial location in HeLa cell line. This new regulation mechanism of virus integration on host cellular genes and cell malignant transformation will become a new study filed in tumors related to virus infection.
Supporting information
Supporting Information
Acknowledgement
The authors thank Z Zhao for providing this idea.
Brief description: A long‐range chromatin interaction between the integrated HPV fragment and MYC gene, as well as the 8q24.22 region was demonstrated by using Chromosome Conformation Capture (3C) method in HeLa cell line. The role of long‐range chromatin interaction on the expression of MYC was confirmed via knocking out the integrated HPV fragment or 8q24.22 region. This study provides an alternative explanation about the carcinogenic mechanism of HPV viral genome integration.
Conflict of interest: No potential conflict of interest was disclosed.
Conflict of Interest: The authors declare no conflict of interest.
References
- 1. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2. Yugawa T, Kiyono T. Molecular mechanisms of cervical carcinogenesis by high‐risk human papillomaviruses: novel functions of E6 and E7 oncoproteins. Rev Med Virol 2009;19:97–113. [DOI] [PubMed] [Google Scholar]
- 3. Pan H, Griep AE. Altered cell cycle regulation in the lens of HPV‐16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev 1994;8:1285–99. [DOI] [PubMed] [Google Scholar]
- 4. Münger K, Basile JR, Duensing S, et al. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 2001;20:7888–98. [DOI] [PubMed] [Google Scholar]
- 5. Münger K, Baldwin A, Edwards KM, et al. Mechanisms of human papillomavirus‐induced oncogenesis. J Virol 2004;78:11451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang R, Shen C, Zhao L, et al. Dysregulation of host cellular genes targeted by human papillomavirus (HPV) integration contributes to HPV‐related cervical carcinogenesis. Int J Cancer 2016;138:1163–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Adey A, Burton JN, Kitzman JO, et al. The haplotype‐resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature 2013;500:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Macville M, Schrck E, Padilla‐Nash H, et al. Comprehensive and definitive molecular cytogenetic characterization of HeLa Cells by spectral karyotyping. Cancer Res 1999;59:141–50. [PubMed] [Google Scholar]
- 9. Peter M, Rosty C, Couturier J, et al. MYC activation associated with the integration of HPV DNA at the MYC locus in genital tumors. Oncogene 2006;25:5985–93. [DOI] [PubMed] [Google Scholar]
- 10. Gimenes F, Souza RP, de Abreu AL, et al. Simultaneous detection of human papillomavirus integration and c‐MYC gene amplification in cervical lesions: an emerging marker for the risk to progression. Arch Gynecol Obstet 2016;293:857–63. [DOI] [PubMed] [Google Scholar]
- 11. Veldman T, Liu X, Yuan H, et al. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc Natl Acad Sci USA 2003;100:8211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang YW, Chang HS, Lin CH, et al. HPV‐18 E7 conjugates to c‐Myc and mediates its transcriptional activity. Int J Biochem Cell Biol 2007;39:402–12. [DOI] [PubMed] [Google Scholar]
- 13. Lieberman‐Aiden E, Van Berkum Nynke L, Williams L, et al. Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science 2009;326:289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dekker J, Rippe K, Dekker M, et al. Capturing chromosome conformation. Science 2002;295:1306–11. [DOI] [PubMed] [Google Scholar]
- 15. Jiang S, Yang Z, Li W, et al. Re‐evaluation of the carcinogenic significance of hepatitis B virus integration in hepatocarcinogenesis. PLoS One 2012;7:e40363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J, Xu ZW, Liu S, et al. Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World J Gastroenterol 2015;21:9554–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu CH, van Riggelen J, Yetil A, et al. Cellular senescence is an important mechanism of tumor regression upon MYC inactivation. Proc Natl Acad Sci USA 2007;104:13028–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mannava S, Moparthy KC, Wheeler LJ, et al. Ribonucleotide reductase and thymidylate synthase or exogenous deoxyribonucleosides reduce DNA damage and senescence caused by C‐MYC depletion. Aging (Albany NY) 2012;4:917–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao FH, Hu XH, Li W, et al. Oridonin induces apoptosis and senescence in colorectal cancer cells by increasing histone hyperacetylation and regulation of p16, p21, p27 and c‐myc. BMC Cancer 2010;10:610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Munoz‐Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol 2014;15:482–96. [DOI] [PubMed] [Google Scholar]
- 21. Wang J, Ding S, Duan Z, et al. Role of p14ARF‐HDM2‐p53 axis in SOX6‐mediated tumor suppression. Oncogene 2016;35:1692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sanyal A, Lajoie BR, Jain G, et al. The long‐range interaction landscape of gene promoters. Nature 2012;489:109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gheldof N, Smith EM, Tabuchi TM, et al. Cell‐type‐specific long‐range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res 2010;38:4325–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Y, Zhang RP, McCord Y‐J, Ho et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 2012;148:908–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. BVS, Iyer G, Arya Lattice animal model of chromosome organization. Phys Rev 2012;86:011911. [DOI] [PubMed] [Google Scholar]
- 26. Jin F, Li Y, Dixon JR, et al. A high‐resolution map of the three‐dimensional chromatin interactome in human cells. Nature 2013;503:290–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahmadiyeh N, Pomerantz MM, Grisanzio C, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue ‐ specific long‐range interaction with MYC. Proc Natl Acad Sci USA 2010;107:9742–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yochum GS. Multiple Wnt/ß‐catenin responsive enhancers align with the MYC promoter through long‐range chromatin loops. PLoS One 2011;6:e18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. EM, Kennedy AVR, Kornepati M, Goldstein et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA‐guided endonuclease. J Virol 2014;88:11965–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hall Allison HS, Alexander Kenneth A. RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J Virol 2003;77:6066–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


