Abstract
Aims
This study aimed to determine the effect of a cell‐free supernatant of Lactobacillus paracasei ATCC 27092 with and without rice bran extract (RBE) on Salmonella Typhimurium 14028s growth, and to identify a metabolite profile with antimicrobial functions.
Methods and Results
Supernatant was collected from overnight cultures of L. paracasei incubated in the presence (LP+RBE) or absence (LP) of RBE and applied to S. Typhimurium. LP+RBE reduced 13·1% more S. Typhimurium growth than LP after 16 h (P < 0·05). Metabolite profiles of LP and LP+RBE were examined using nontargeted global metabolomics consisting of ultra‐high‐performance liquid chromatography coupled with tandem mass spectrometry. A comparison of LP and LP+RBE revealed 84 statistically significant metabolites (P < 0·05), where 20 were classified with antimicrobial functions.
Conclusions
LP+RBE reduced S. Typhimurium growth to a greater extent than LP, and the metabolite profile distinctions suggested that RBE favourably modulates the metabolism of L. paracasei. These findings warrant continued investigation of probiotic and RBE antimicrobial activities across microenvironments and matrices where S. Typhimurium exposure is problematic.
Significance and Impact of the Study
This study showed a novel metabolite profile of probiotic L. paracasei and prebiotic rice bran that increased antimicrobial activity against S. Typhimurium.
Keywords: L. paracasei subsp. paracasei, metabolomics, prebiotic, probiotic, rice bran, S. Typhimurium, synbiotic
Introduction
Globally, Salmonella enterica serovar Typhimurium accounts for the majority of human Salmonellosis cases each year. Salmonella Typhimurium causes widespread disease because of zoonotic transmission from cattle, poultry, swine, sheep, rodents and horses (Tsolis et al. 1999). An emerging strategy against S. Typhimurium involves the use of probiotics and prebiotics. As defined by the World Health Organization and Food and Drug Administration, probiotics are live micro‐organisms that, when administered in adequate amounts, confer a health benefit to the host (Anon 2006). Many widely investigated probiotic species belong to the bacterial genus Lactobacillus, which has a natural ability to reduce the growth of S. Typhimurium (Heredia‐Castro et al. 2015). In particular, Lactobacillus paracasei strains have been shown to effectively reduce the growth of enteric pathogens including S. Typhimurium, Listeria monocytogenes and Escherichia coli O157:H7 (Caridi 2002; Chiang and Pan 2012; Valerio et al. 2013).
Rice bran, the outer layer of the rice grain, is a natural and rich source of prebiotics that can be metabolized by the gut microbiome to modulate mucosal immune responses, reduce intestinal colonization of enteric pathogens (Kumar et al. 2012) and increase numbers of native probiotic lactobacilli (Henderson et al. 2012; Goodyear et al. 2015). Rice bran was also shown to support probiotic E. coli Nissle and Lactobacillus rhamnosus GG growth in gnotobiotic piglets (Yang et al. 2015).
Molecular mechanisms by which Lactobacillus sp. utilize and metabolize prebiotics are not well understood. Metabolomics has been minimally used to elucidate the functional significance of synbiotics via identification and quantification of small molecules (Ryan et al. 2011). Past investigations evaluated small‐molecule profiles produced by the microbiome in vitro and in vivo, including probiotics (Vitali et al. 2010; Mozzi et al. 2013; Weir et al. 2013). Metabolite profiles have been determined for an increasing number of foods fermented by lactic acid bacteria including rye, yogurt and wine (Settachaimongkon et al. 2014; Arbulu et al. 2015; Koistinen et al. 2016.).
It was hypothesized that a combination of L. paracasei and rice bran extract (RBE) would result in a unique profile of metabolites with antimicrobial activity that more effectively reduce the growth of S. Typhimurium compared to L. paracasei alone. This study aimed to compare the effectiveness of L. paracasei alone and L. paracasei with RBE supernatants at reducing S. Typhimurium growth and to evaluate their metabolomic profiles. The small molecule changes that occur in the presence and absence of RBE illustrate how L. paracasei and rice bran synergistically promote S. Typhimurium growth reduction.
Materials and methods
Bacterial strains and culture reagents
Lactobacillus paracasei ATCC 27092 was purchased from ATCC (Manassas, VA), and S. enterica subsp. enterica serovar Typhimurium 14028s Kanr (rPSM::GFP) was a generous gift from Dr Andres Vazquez‐Torres (University of Colorado). All bacterial cultures were stored at −80°C as 1‐ml aliquots supplemented with 20% glycerol in Luria–Bertani (LB) broth (MO BIO Laboratories, Inc. Carlsbad, CA) for S. Typhimurium, and deMan Rogosa Sharpe broth (MRS) (Becton, Dickinson, and Company Difco Laboratories, Franklin Lakes, NJ) for L. paracasei. MRS broth and agar, LB broth and MacConkey agar (Becton, Dickinson, and Company Difco Laboratories) were prepared and sterilized according to the manufacturer. To prepare MRS broth supplemented with RBE, 100 μg ml−1 RBE was added to the broth. The media was autoclaved with an 18‐min sterilization time, then stored at 4°C until use.
Rice bran extraction
RBE was prepared as described previously (Kumar et al. 2012). Briefly, 4 g of finely ground, heat‐stabilized Calrose rice bran (USDA‐ARS Rice Research Unit, Stuttgart, AK) was extracted in 42·6 ml of 80% methanol. The mixture was vortexed (232 Vortexer Fisher Scientific, Pittsburgh, PA, USA) on a high power setting for 5 min, incubated at −80°C overnight and centrifuged (Beckman Coulter Allegra X‐14R, Indianapolis, IN, USA) at 3724 g for 5 min. The supernatant was collected, and kept at −80°C until it could be dried in a speedvac concentrator (SPD1010; Thermo Scientific, Pittsburgh, PA, USA) at 45°C, with the heating time for 5 min, and a vacuum pressure of 7·5 torrs.
L. paracasei cell‐free supernatant preparation
The cell‐free supernatant (CFS) preparation was modified from a published procedure (Wang et al. 2012). Briefly, L. paracasei isolates were thawed from storage in −80°C, suspended in MRS broth and grown at 37°C until mid/late logarithmic phase. Approximately 1 × 107 cells were inoculated into 15 ml of MRS or MRS + 100 μg ml−1 RBE. The RBE concentration was determined based on dose–response experiments that observed 100 μg ml−1 of RBE, when added to fixed amounts of S. Typhimurium, reduced growth compared to a RBE‐free control culture of S. Typhimurium, as evidenced by differences in optical density at 600 nm (OD600) over 12 h (Fig. S1a). Each treatment was incubated at 37°C for 24 h. Lactobacillus paracasei supernatant (LP) and L. paracasei supernatant with RBE (LP+RBE) was collected by centrifuging two times at 3724 g for 10 min. The pH of the supernatant was adjusted using a pH meter (Corning Pinnacle 530, Cole‐Parmer, Vernon Hills, IL, USA) with 1 mol l−1 NaOH (Sigma Aldrich) until a pH of 4·5 was reached. CFS was filter‐sterilized through a 0·2‐μm pore (Pall Corporation LifeSciences Acrodisc syringe filters, Port Washington, NY, USA) into 1‐ml aliquots before being stored at −80°C. Sterility of all CFS was confirmed by plating the treatment on MRS agar and confirming the absence of any bacterial growth after 48 h. Three independent batches of CFS were used in this study.
S. Typhimurium growth reduction assay
Salmonella Typhimurium was thawed from storage in −80°C and suspended in sterile LB. Stocks were grown in a 24‐well plate at 37°C until early/mid logarithmic phase, and were assessed using the Cytation3 plate reader (BioTek Instruments Inc., Winooski, VT, USA). In a 96‐well plate, 20 μl of S. Typhimurium (approx. 2 × 106 cells) was diluted 10‐fold into 180 μl of sterile LB. To determine an appropriate treatment volume for the growth reduction assay, a dose curve analysis using LP was performed on S. Typhimurium (Fig. S1b). In brief, 100, 50 and 25 μl doses of 100% LP were added to approx. 2 × 105 S. Typhimurium cells, and growth was evaluated for 24 h at OD600. Fifty microlitres of treatment was the lowest dose at which a difference was observed in the inhibitory capacity of LP compared to a volume‐adjusted control.
To evaluate the effect of CFS on S. Typhimurium growth, a 50 μl of treatment with different percentages of CFS were added to each well: 0% vehicle control, 20, 40, 60, 80 and 100%. A negative control of S. Typhimurium inoculated with 50 μl LB broth was included on each well plate. To control for pH differences between treatments, all media was adjusted to a pH of 4·5 using 1N of NaOH or HCl before use. The plate was read on an automatic plate reader, incubated at 37°C for approx. 16 h, and individual wells were read at OD600 every 20 min. To evaluate differences between all treatments, OD600 was plotted over time. To further quantify growth reduction by LP and LP+RBE, per cent growth reduction was calculated from OD600 values using the following equation:
Experiments were repeated four times for all CFS concentrations, eight times for 100% LP and nine times for 100% LP+RBE using the same three batches of supernatant as described previously.
Probiotic supernatant agar well diffusion assay
The agar well diffusion assay was modified from a published procedure (Aminnezhad et al. 2015). Briefly, Mueller–Hinton agar (Hardy Diagnostics, Santa Maria, CA) was inoculated with S. Typhimurium suspended in normal saline (0·89% g NaCl ml−1), equivalent to a 0·5 McFarland Standard (Hardy Diagnostics). Wells (8·0 mm) were punched into the agar and filled with 100 μl of the following treatments adjusted to a final pH of 4·5: vehicle control, vehicle control+RBE, LP and LP+RBE. Non‐pH‐adjusted normal saline was included as a negative control on each plate. Plates were incubated at 37°C overnight, and inhibition zone diameters were measured in millimetres. Measurements were collected from seven different plates and included supernatant collected from three independent cultures of LP and LP+RBE.
Supernatant extract preparation for metabolomics
Metabolomics was performed by Metabolon Inc © (Durham, NC). Briefly, 1 ml of supernatant samples were stored at −80°C in microcentrifuge tubes, and sent on dry ice to Metabolon in triplicate. The samples sent for analysis included vehicle control, vehicle control+RBE, LP and LP+RBE. Upon arrival, samples were stored at −80°C in liquid nitrogen until processing. To improve recovery of small molecules prior to detection, the protein fraction was removed by extracting the sample with a 5 : 1 methanol : water solution, using vigorous shaking at room temperature for 2 min followed by centrifugation at 680 g for 3 min. The extracted samples were split into four parts for analysis via ultra‐high‐performance liquid chromatography–tandem mass spectrometry (UPLC‐MS/MS) including two separate reverse phase UPLC‐MS/MS with positive ion mode electrospray ionization (ESI), reverse phase UPLC‐MS/MS with negative ion mode ESI and one sample for high liquid chromatography UPLC‐MS/MS‐negative ion mode ESI.
UPLC‐MS/MS analysis
Metabolite profiling was performed using a Waters ACQUITY UPLC, a Thermo Scientific (Waltham, MA, USA) Q‐Exactive heated electrospray ionization (HESI‐II) source, and an Orbitrap mass analyser operated at 35 000 mass resolution. For UPLC analysis, the sample extracts were dried and reconstituted in solvents appropriate for each of the four detection methods, and standards were included to ensure experimental consistency. Acidic positive ion conditions were optimized for either hydrophobic or hydrophilic compounds and were eluted from a C18 column (Waters UPLC BEH C18‐2·1 × 100 mm, 1·7 μm) using water and methanol (hydrophilic optimization) or methanol, acetonitrile and water (hydrophobic optimization) containing 0·05% perfluoropentanoic acid (PFPA) (hydrophilic optimization) or 0·5% PFPA (hydrophobic optimization) and 0·1% formic acid. Two aliquots were analysed using basic negative ion conditions; one was eluted on a separate C18 column using methanol and water with 6·5 mmol l−1 of ammonium bicarbonate at pH 8, and the other was eluted from a HILC column (Waters UPLC BEH Amide 2·1 × 150 mm, 1·7 μm) using a water–acetonitrile gradient with 10 mmol l−1 of ammonium formate, pH 10·8. The total scan range covered 70–1000 m/z.
Data extraction and compound identification
Raw data were extracted, peak‐identified and quality‐control processed as previously described (Brown et al. 2016) and compounds were then identified by comparison to library entities of purified standards or recurrent unknown entities, including over 3300 commercially available purified standard compounds. Identifications were made based on retention time/index with a narrow window of identification, mass to charge ratio (m/z) ±10 parts per million and chromatographic data including MS/MS forward and reverse scores between the experimental data and authentic standards. The raw counts of both supernatant profiles were converted into relative abundances and then median‐scaled to one. For each metabolite, fold difference (FD) was calculated by dividing the scaled relative abundance of LP+RBE by LP.
Metabolic pathway analysis
Pathway analysis was conducted as described previously (Brown et al. 2016). For selected lipids and amino acids/peptides, metabolite FD between LP+RBE and LP were visualized using Cytoscape 2.8.3 software. For each metabolite, node colour was determined by the direction of FD when comparing LP+RBE to LP, and node diameter was determined by the magnitude of the FD. The value on each node represents a pathway enrichment score, which was calculated by dividing the number of significant metabolites in pathway (k) by the total number of detected metabolites in pathway (m). This value was then divided by the fraction of the total number of significant metabolites in the data set (n) over the total number of detected metabolites in the complete data set (N):
Pathway enrichment scores greater than one indicated that a given pathway contained more metabolites with statistically significant FD between LP+RBE and LP compared to all pathways in the study.
Statistical analysis
All statistical analyses for the S. Typhimurium growth reduction and well diffusion assays were performed using GraphPad Prism 6.07 (San Diego, CA). For the S. Typhimurium growth reduction assays, treatments were analysed using a two‐way repeated measures anova with a Bonferroni post‐test to compare treatment means. For the agar well diffusion assay, treatments were compared using a one‐way anova with a Tukey post‐test to compare treatment means. Significance was determined at the level of P < 0·05. For metabolomic data, statistical analysis was performed by Metabolon Inc using ArrayStudio (Omnicsoft, Cary, NC) R ver. 2.142 and/pr SAS ver. 9.4. The relative abundance of each metabolite from LP and LP+RBE was scaled to that metabolite's median relative abundance and each scaled relative abundance was compared between LP and LP+RBE using a Welch's two‐sample t test. Statistical significance was determined at the level of P < 0·05.
Results
S. Typhimurium growth reduction by L. paracasei and rice bran extract
The dose‐dependent effects of LP and LP+RBE on S. Typhimurium growth were determined with increasing percentages of supernatant that were added and compared to a fixed volume of a vehicle control or vehicle control+RBE respectively. In both the LP and LP+RBE treatments, S. Typhimurium growth was reduced in a dose‐dependent manner (Fig. 1). By 5·0 h, all LP doses (20–100%) significantly reduced S. Typhimurium growth compared to the vehicle control. The average percent difference between LP and the vehicle control included: 22·2% for the 20% LP dose, 35·9% for 40% LP, 47·4% for 60% LP, 55·3% for 80% LP and 60·6% for the 100% LP (Fig. 1a). By the study endpoint of 16 h, two LP concentrations continued to show increased growth reduction compared to the vehicle control and included: 40% LP (41·7%) and 100% LP (68·4%) (Fig. 1a).
Figure 1.
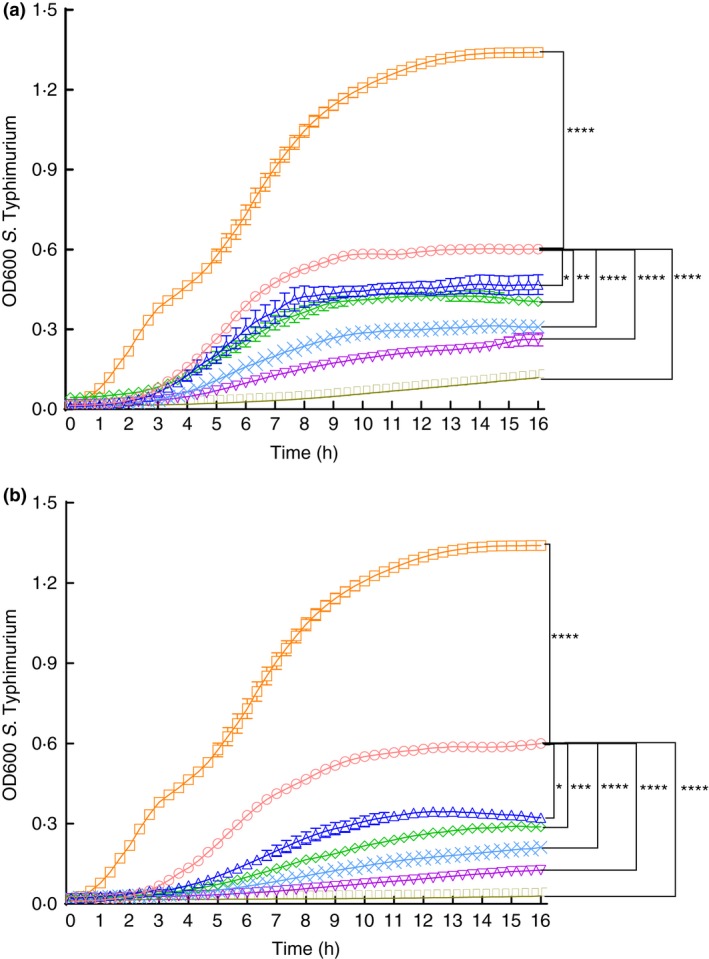
Lactobacillus paracasei supernatant in the presence and absence of rice bran extract reduces S. Typhimurium growth in a dose‐dependent manner. The dose‐dependent effects of (a) LP and (b) LP+RBE on S. Typhimurium growth were evaluated. Optical density at a wavelength of 600 nm was plotted on the y‐axis and time on the x‐axis. Each point represents the mean of four independent experiments and error bars indicate standard error. □ indicates negative control, ○ indicates vehicle control or vehicle control+RBE, ▵ indicates 20% LP or LP+RBE, ◊ indicates 40% LP or LP+RBE, X indicates 60% LP or LP+RBE, ▽ indicates 80% LP or LP+RBE, and ▯ indicates 100% LP or LP+RBE. Statistical significance between all treatments compared to the vehicle control occurred at (a) 5·0 h in LP and (b) 3·7 h in LP+RBE where *P < 0·05, **P < 0·01, ***P < 0·001 and ****P < 0·0001. In (a) and (b), results represent four independent experiments. [Colour figure can be viewed at wileyonlinelibrary.com]
LP+RBE treatments significantly reduced S. Typhimurium growth by 3·7 h. The average per cent difference for LP+RBE over RBE alone was 8·9% for the 20% LP+RBE dose, 16·1% for 40% LP+RBE, 21·6% for 60% LP+RBE, 25·5% for 80% LP+RBE and 37·0% for 100% LP+RBE (Fig. 1b). At 16 h, all LP+RBE doses showed increasing growth reduction: 38·8% for 20% LP+RBE, 43·9% for 40% LP+RBE, 55·2% for 60% LP+RBE, 67·6% for 80% LP+RBE and 82·0% for 100% LP+RBE (Fig. 1b). At 16 h, the 100% CFS dose showed highest S. Typhimurium growth reduction compared to the vehicle control: 68·4% for 100% LP and 82·0% for 100% LP+RBE (Fig. 1)
Next, 100% LP and 100% LP+RBE were compared to each other. At 9·0 h, LP+RBE was more effective than LP at reducing S. Typhimurium growth (P < 0·05) and became increasingly effective through 16 h (P < 0·0001) (Fig. 2a). The 100% LP+RBE reduced S. Typhimurium growth 13·1% more than 100% LP at 16 h (P < 0·05) (Fig. 2b). All LP+RBE doses (20, 40, 60, 80%) were more effective than LP at reducing S. Typhimurium growth by 16 h (data not shown).
Figure 2.
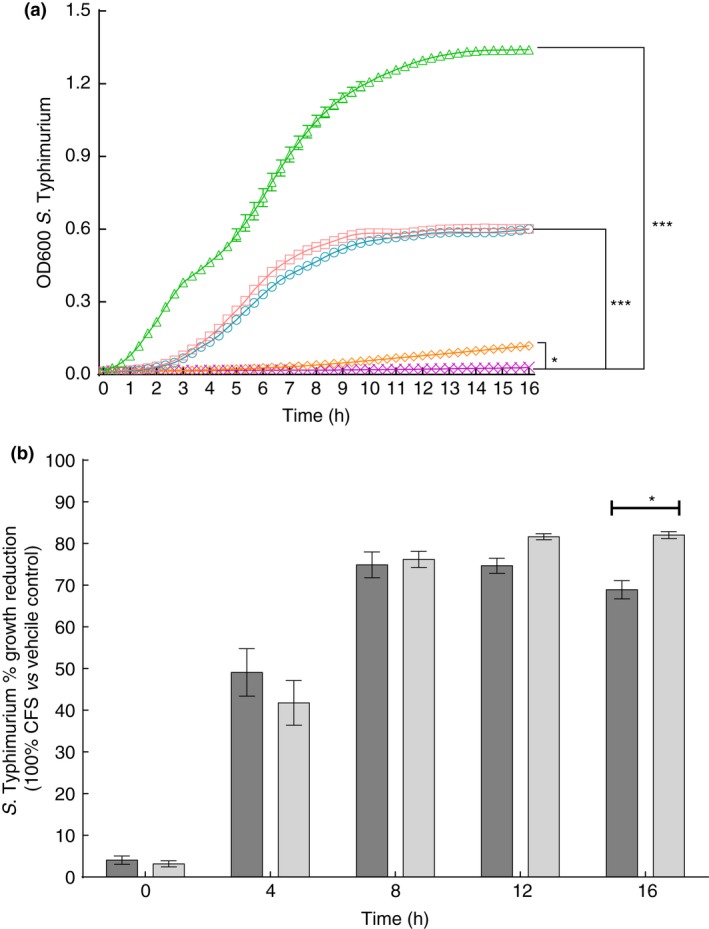
Rice bran extract enhances the ability of Lactobacillus paracasei supernatant to reduce S. Typhimurium growth. (a) The effectiveness of 100% LP and 100% LP+RBE at reducing S. Typhimurium growth were compared. The optical density at 600 nm is plotted on the y‐axis, and time on the x‐axis. Each point represents the mean of eight (LP) or nine independent experiments (LP+RBE) treatments and error bars indicate SE ▵ indicates negative control, □ indicates vehicle control, ○ indicates vehicle control+RBE, ◊ indicates LP and X indicates LP+RBE. By 9·0 h, LP+RBE reduced S. Typhimurium growth more than LP, vehicle control, vehicle control+RBE and the negative control, where*P < 0·05, ***P < 0·001 and ****P < 0·0001. (b) For the 100% CFS treatments, per cent S. Typhimurium growth inhibition of LP and LP+RBE treatments media was compared at 0, 4·0, 8·0, 12·0 and 16·0 h. At 16·0 h, LP+RBE reduced growth 13·1% more than LP *P < 0·05. Each bar represents the mean per cent difference of eight (LP) or nine (LP+RBE) independent experiments and error bars indicate standard error. Light grey bars represent LP and dark grey bars represent LP+RBE. [Colour figure can be viewed at wileyonlinelibrary.com]
Probiotic supernatant agar well diffusion assay against S. Typhimurium
To further assess the ability of CFS to reduce S. Typhimurium growth, the inhibitory zone diameters of LP, LP+RBE, vehicle control, vehicle control+RBE and normal saline (negative control) treatments were compared in an agar well diffusion assay where all treatments were pH adjusted to 4·5 prior to use. The mean inhibitory zone diameter (millimetres) and standard error are displayed for each treatment in Table 1 and represent seven independent experiments: normal saline (8·00 ± 0·00), vehicle control (8·93 ± 0·468), vehicle control+RBE (9·43 ± 0·517), LP (10·86 ± 0·261) and LP+RBE (12·07 ± 0·277). The diameters of the wells themselves were 8·0 mm, and normal saline did not create an inhibitory zone in any replicate. LP+RBE had a larger inhibitory zone diameter than all treatments: LP+RBE vs normal saline, vehicle control and vehicle control+RBE (P < 0·001), LP+RBE vs LP (P < 0·05).
Table 1.
Agar well diffusion of Lactobacillus paracasei supernatant against Salmonella Typhimurium in the presence and absence of rice bran extract
| Treatment | Zone of Inhibition (mm)a |
|---|---|
| Normal saline (negative control) | 8·00 ± 0·00 |
| Vehicle control | 8·93 ± 0·468 |
| Vehicle control+RBE | 9·43 ± 0·517 |
| LP | 10·86 ± 0·261 |
| LP+RBE | 12·07 ± 0·277 |
Inhibitory zone diameters (millimetres) against S. Typhimurium were determined for supernatants and control media. Results are reported as (mean ± standard error) and were collected from seven independently measured plates. Measurements were analysed using a one‐way anova, and pairs of treatments were compared using a Tukey post‐test, where statistical significance was determined as P < 0·05. LP+RBE had a significantly larger zone of inhibition compared to all other treatments: Normal saline (P < 0·0001), vehicle control (P < 0·0001), vehicle control+ RBE (P < 0·0001), LP (P < 0·05).
Metabolomics of probiotic L. paracasei supernatant and rice bran extract
Metabolome analysis of LP and LP+RBE led to identification of 362 metabolites that were organized by chemical class and summarized in Table 2. Of the 362 metabolites, 138 were classified as amino acids, 29 peptides, 29 carbohydrates, 11 TCA cycle, 54 lipids, 54 nucleotides, 20 cofactors and vitamins and 27 phytochemicals. There were a total of 84 metabolites with a FD that significantly differed between LP+RBE and LP (Table 3). Of these, 58 metabolites that had a higher and 26 had a lower relative abundance in LP+RBE compared to LP. Amino acid and lipid metabolite classes represented ~55% of metabolites differentially expressed between LP and LP+RBE. The relative abundance distributions for lipids and amino acids/small peptides across groups are depicted in Fig. 3.
Table 2.
Number of metabolites across classes that show a higher or lower fold change in Lactobacillus paracasei supernatant cultured in the presence or absence of rice bran extract
| Metabolite classification | Number of metabolites (P < 0·05) |
|---|---|
| Amino acids | 138 (19↑, 9↓) |
| Peptides | 29 (2↑) |
| Carbohydrates | 29 (7↑) |
| TCA cycle | 11 (2↑, 1↓) |
| Lipids | 54 (8↑, 9↓) |
| Nucleotides | 54 (12↑, 3↓) |
| Cofactor and vitamins | 20 (1↑, 1↓) |
| Other phytochemicals | 27 (7↑, 3↓) |
| Total number of identified metabolites | 362 (58↑, 26↓) |
Metabolite profiles of L. paracasei (LP) and L. paracasei + rice bran extract (LP+RBE) supernatant. For each metabolite, fold difference was calculated by dividing the scaled relative abundance in LP+RBE by LP. Fold differences were analysed using a Welch's two‐sample t‐test, and metabolites with a fold difference at statistically different (P < 0·05) levels between LP+RBE and LP are marked with ↑ or ↓ to denote the direction of change when comparing LP+RBE to LP.
Table 3.
Statistically significant metabolites from Lactobacillus paracasei supernatant prepared in the presence and absence of rice bran extract
| Metabolite | HMDB* | Fold differencea | P‐valueb |
|---|---|---|---|
| Amino acid | |||
| Methionine sulfone | – | ↑8·29 | 3·16 × 10−6 |
| 3‐sulfo‐L‐alanine | 02757 | ↑2·34 | 3·13 × 10−5 |
| Indole‐3‐carboxylic acid | 03320 | ↑1·94 | 0·00065 |
| Kynurenate | 00715 | ↑1·44 | 0·0086 |
| 1‐methylguanidine | 01522 | ↑1·34 | 0·0063 |
| Imidazole propionate | 02271 | ↑1·28 | 0·0047 |
| Cysteine s‐sulphate | 00731 | ↑1·24 | 0·013 |
| Succinimide | – | ↑1·22 | 0·0075 |
| Beta‐alanine | 00056 | ↑1·22 | 0·0089 |
| 4‐guanidinobutanoate | 03464 | ↑1·19 | 0·0024 |
| N‐formylphenylalanine | – | ↑1·18 | 0·048 |
| Asparagine | 00168 | ↑1·17 | 0·0015 |
| N‐acetylhistidine | 32055 | ↑1·13 | 0·020 |
| N6‐formyllysine | – | ↑1·13 | 0·013 |
| 4‐imidazoleacetate | 02024 | ↑1·12 | 0·024 |
| Phenethylamine | 02017 | ↑1·07 | 0·030 |
| Creatinine | 00562 | ↑1·08 | 0·037 |
| Carnitine | 00062 | ↑1·03 | 0·034 |
| Proline | 00162 | ↑1·03 | 0·040 |
| Leucine | 00687 | ↑1·03 | 0·042 |
| Histamine | 00870 | ↓0·94 | 0·042 |
| Acetylcarnitine | 00201 | ↓0·90 | 0·042 |
| N‐acetyltryptophan | 13713 | ↓0·88 | 0·030 |
| N‐acetylserine | 02931 | ↓0·87 | 0·037 |
| Spermine | 01256 | ↓0·70 | 0·044 |
| Indolelactate | 00671 | ↓0·68 | 0·0040 |
| Cystathionine | 00099 | ↓0·67 | 0·00019 |
| Methionine | 00696 | ↓0·64 | 0·023 |
| Hypotaurine | 00965 | ↓0·46 | 0·0037 |
| S‐adenosylhomocysteine (SAH) | 00939 | ↓0·39 | 0·0010 |
| Peptide | |||
| cGamma‐glutamylisoleucine | 11170 | ↑1·19 | 0·024 |
| Tyrosylglycine | – | ↑1·17 | 0·017 |
| Carbohydrate | |||
| Sucrose | 00258 | ↑7·39 | 5·76 × 10−7 |
| Arabonate/xylonate | – | ↑1·87 | 0·0059 |
| Ribonate | 00867 | ↑1·86 | 0·0015 |
| Erythrulose | 06293 | ↑1·46 | 0·029 |
| Erythronate | 00613 | ↑1·24 | 0·016 |
| N‐acetylglucosamine/N‐acetylgalactosamine | – | ↑1·12 | 0·018 |
| Glycerate | 00139 | ↑1·07 | 0·0026 |
| TCA cycle | |||
| Tricarballylate | 31193 | ↑1·79 | 6·83 × 10−5 |
| Citraconate/glutaconate | – | ↑1·16 | 0·017 |
| Alpha‐ketoglutarate | 00208 | ↓0·76 | 0·031 |
| Lipid | |||
| Linoleate (18:2n6) | 00673 | ↑3·18 | 0·0075 |
| 13‐HODE + 9‐HODE | – | ↑2·11 | 0·0082 |
| Eicosenoate (20:1) | 02231 | ↑1·65 | 0·0017 |
| 10‐hydroxystearate | – | ↑1·49 | 0·014 |
| Maleate | 00176 | ↑1·44 | 0·017 |
| Oleate/vaccenate (18:1) | – | ↑1·26 | 0·041 |
| Azelate (nonanedioate) | 00784 | ↑1·18 | 0·024 |
| Glycerol 3‐phosphate | 00126 | ↑1·05 | 0·031 |
| Alpha‐hydroxyisocaproate | 00746 | ↓0·86 | 0·016 |
| Alpha‐hydroxyisovalerate | 00407 | ↓0·85 | 0·025 |
| Trimethylamine N‐oxide | 00925 | ↓0·82 | 0·014 |
| 3‐hydroxyoctanoate | 01954 | ↓0·74 | 0·0015 |
| 3‐hydroxydecanoate | 02203 | ↓0·70 | 0·0085 |
| 3‐hydroxylaurate | 00387 | ↓0·65 | 0·0055 |
| Pinitol | 34219 | ↓0·61 | 0·015 |
| 5‐dodecenoate (12:1n7) | 00529 | ↓0·58 | 0·015 |
| Nucleotide | |||
| 2′‐deoxycytidine | 00014 | ↑1·60 | 0·0034 |
| N6‐succinyladenosine | 00912 | ↑1·51 | 0·0032 |
| 2′‐deoxyadenosine | 00101 | ↑1·31 | 0·0015 |
| Orotate | 00226 | ↑1·21 | 0·023 |
| Guanine | 00132 | ↑1·16 | 0·0026 |
| Guanosine | 00133 | ↑1·12 | 0·015 |
| Guanosine‐2′,3′‐cyclic monophosphate | 11629 | ↑1·11 | 0·046 |
| Hypoxanthine | 00157 | ↑1·08 | 0·046 |
| Cytidine | 00089 | ↑1·06 | 0·035 |
| cguanosine 2′‐monophosphate (2′‐GMP) | – | ↑1·06 | 0·036 |
| 2′‐deoxyuridine | 00012 | ↑1·05 | 0·043 |
| Cytidine 3′‐monophosphate (3′‐CMP) | – | ↓0·94 | 0·015 |
| Cytidine 5′‐monophosphate (5′‐CMP) | 00095 | ↓0·91 | 0·0071 |
| Adenosine | 00050 | ↓0·89 | 0·0015 |
| Cofactor and vitamin | |||
| Pyridoxate | 00017 | ↑1·19 | 0·020 |
| Pyridoxamine | 01431 | ↓0·80 | 0·026 |
| Phytochemicals/Other | |||
| Beta‐guanidinopropanoate | 13222 | ↑1·74 | 0·00042 |
| Harmane | – | ↑1·62 | 7·51 × 10−5 |
| Nicotianamine | – | ↑1·52 | 0·029 |
| 4‐hydroxybenzoate | 00500 | ↑1·40 | 0·00062 |
| Salicylate | 01895 | ↑1·25 | 0·019 |
| Pyrraline | – | ↑1·24 | 0·0057 |
| 2‐oxindole‐3‐acetate | – | ↑1·22 | 0·019 |
| Daidzein | 03312 | ↓0·86 | 0·040 |
| 2‐ketogluconate | – | ↓0·68 | 0·0066 |
| N‐glycolylneuraminate | 00833 | ↓0·61 | 0·011 |
*Human metabolome database (HMDB) numbers are given when available.
For each metabolite, fold difference is expressed as the scaled relative abundance in LP+RBE over LP. Arrows indicate the direction of change between treatments.
Each metabolite presented has a statistically significant (P < 0·05) fold difference, as determined by a Welch's two‐sample t‐test.
Indicates compounds that have not been officially confirmed based on a standard, but second‐order identity in Metabolon Inc. library.
Figure 3.
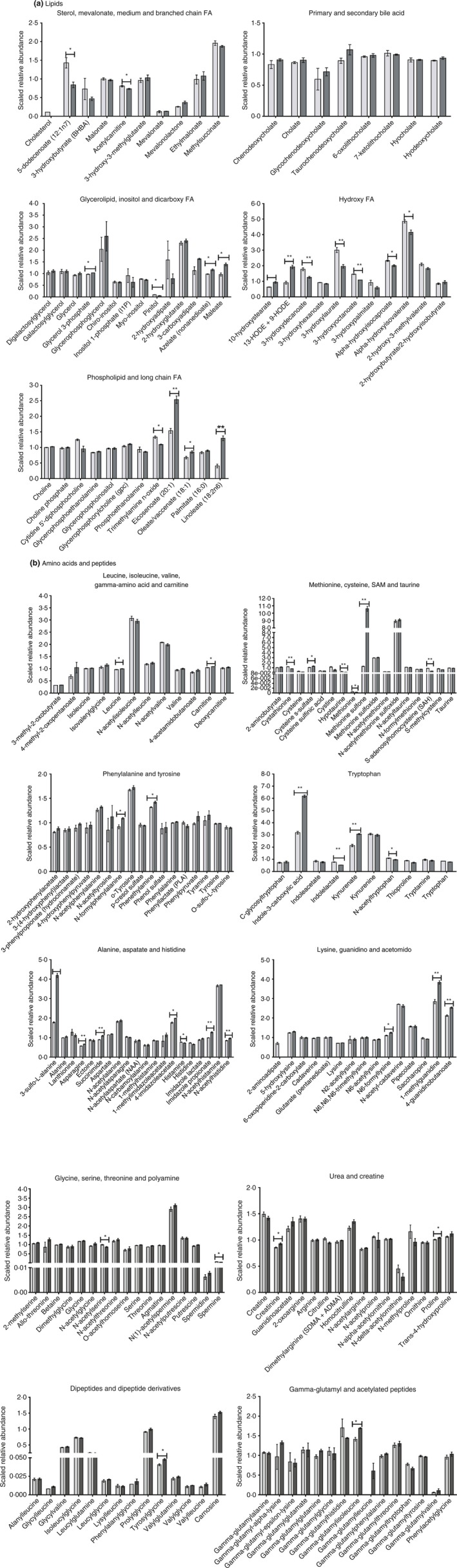
Rice bran extract alters the lipid, amino acid and peptide metabolite profiles of Lactobacillus paracasei supernatant. LP and LP+RBE were profiled using UPLC‐MS/MS. Each bar represents three independent samples and depicts the metabolite scaled relative abundance. Error bars depict standard error and *P < 0·05, and **P < 0·01. Metabolites were classified into metabolic pathways of (a) lipids and (b) amino acids/peptides based on their biochemical properties and/or physiological functions. Light grey bars represent LP and dark grey bars represent LP+RBE. FA indicates fatty acid and SAM indicates S‐adenosyl methione.
The lipid metabolite changes included, but were not limited to medium‐chain, branched chain, long‐chain and other fatty acids (Fig. 4a). FD between lipids, when comparing LP+RBE to LP included: linoleate (3·18 FD), 13‐HODE + 9‐HODE (2·11 FD), eicosenoate (1·65 FD), 10‐hydroxystearate (1·49 FD), maleate (1·44 FD), oleate/vaccenate (1·26 FD), azelate (1·18 FD), glycerol 3‐phosphate (1·05 FD), alpha‐hydroxyisocaproate (0·86 FD), alpha‐hydroxyisovalerate (0·85 FD), trimethylamine N‐oxide (0·82 FD), 3‐hydroxyoctanoate (0·74 FD), 3‐hydroxydecanoate (0·70 FD), 3‐hydroxylaurate (0·65 FD), pinitol (0·61 FD) and 5‐dodecenoate (0·58 FD).
Figure 4.
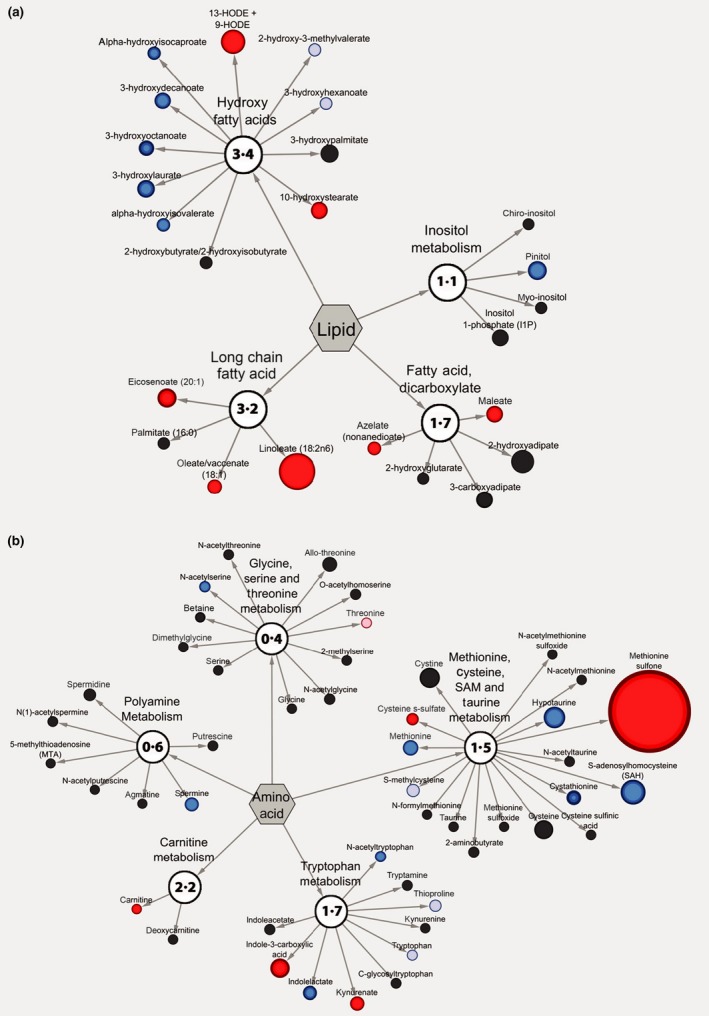
Cytoscape visualization illustrates the metabolic pathways most differentially regulated between treatments. Cytoscape visualization of (a) lipid and (b) amino acid metabolic pathways that differ between LP and LP+RBE. For each metabolite, node diameter is proportional to the fold difference in LP+RBE compared to LP. Node colours indicate the direction of a metabolite's fold difference, where red indicates metabolites with a higher scaled abundance in LP+RBE (P < 0·05), blue indicates lower abundance in LP+RBE (P < 0·05), pink indicates trending higher in LP+RBE (0·05 < P < 0·10), and light blue indicates trending lower in LP+RBE (0·05 < P < 0·10). Black nodes indicate metabolites with fold differences that were not significantly altered between treatments. The pathway enrichment score is the number in the circles for each subclassification.
Fold differences between amino acids/peptides when comparing LP+RBE to LP included: methionine sulfone (8·29 FD), 3‐sulfo‐l‐alanine (2·34 FD), indole‐3‐carboxylic acid (1·94 FD), kynurenate (1·44 FD), 1‐methylguanidine (1·34 FD), imidazole propionate (1·28 FD), cysteine s‐sulphate (1·24 FD), succinimide (1·22 FD), 4‐guanidinobutanoate (1·19 FD), N‐formylphenylalanine (1·18 FD), asparagine (1·17 FD), N‐acetylhistidine (1·13 FD), N6‐formyllysine (1·13 FD), 4‐imidazoleacetate (1·12 FD), phenethylamine (1·07 FD), creatinine (1·08 FD), carnitine (1·03 FD), proline (1·03 FD), leucine (1·03 FD), histamine (0·94 FD), acetylcarnitine (0·90 FD), N‐acetyltryptophan (0·88 FD), N‐acetylserine (0·87 FD), spermine (0·70 FD), indolelactate (0·68 FD), cystathionine (0·67 FD), methionine (0·64 FD), hypotaurine (0·46 FD) and S‐adenosylhomocysteine (0·39 FD) (Fig. 4b). A literature search of the significant metabolites in the comparison of LP+RBE to LP revealed metabolites with established antimicrobial activities, many of which included the above lipids and amino acids/peptides (Table 4).
Table 4.
Metabolites from Lactobacillus paracasei supernatant prepared in the presence and absence of rice bran extract with reported antimicrobial activities
| Fold differencea | P valueb | Functions | References | |
|---|---|---|---|---|
| Amino acids/peptides | ||||
| Methionine sulfone | ↑8·29 | 3·16 × 10−6 | Inhibits S. Typhimurium growth by blocking glutamate synthesis | Hentchel and Escalante‐Semerena (2015) |
| Indole‐3‐carboxylic acid | ↑1·94 | 0·00065 | Inhibited the growth of Staphylococcus aureus, Pseudomonas aeruginosa, Candida albicans, Escherichia coli, Klebsiella pneumoniae and multidrug resistant strains of these species | Hinton and Ingram (2000) |
| Carnitine | ↑1·03 | 0·034 | Inhibited the growth of Streptococcus agalactiae in vitro. | Atroshi et al. (1998) |
| N‐acetylserine | ↓ 0·87 | 0·037 | May increase the susceptibility of S. Typhimurium to methicillin in vitro by modifying cysteine metabolism | Oppezzo and Anton (1995) |
| Spermine | ↓0·70 | 0·044 | May increase susceptibility of S. Typhimurium and other bacteria to Beta‐lactam antibiotics | Kwon and Lu (2007) |
| S‐adenosylhomocysteine (SAH) | ↓0·39 | 0·001 | In high enough concentrations it can inhibit bacterial S‐adenosyl methionine metabolism, which is important for quorum sensing and polyamine synthesis | Simms and Subbaramaiah (1991), Parveen and Cornell (2011) |
| TCA cycle | ||||
| Tricarballylate | ↑1·79 | 6·83 × 10−5 | Toxic to S. enterica if accumulated to high levels. | Boyd et al. (2012) |
| Lipids | ||||
| 3‐hydroxyoctanoate | ↓0·74 | 0·0015 | It and its derivatives inhibited the growth of S. Typhimurium, E. coli, S. aureus, L. monocytogenes, P. aeruginosa, C. albicans and Microsporum gypseum. | Radivojevic et al. (2016) |
| Linoleate (18:2n6) | ↑3·18 | 0·0075 | Bactericidal and bacteriostatic against S. aureus, Streptococcus pyogenes and E. coli by inhibiting fatty acid synthesis | Zheng et al. (2005) |
| 13‐Hydroxyoctadecadienoate and 9‐Hydroxy‐10,12‐octadecadienoate | ↑2·11 | 0·0082 | Detected in an oxylipin extract that inhibited growth of Penicillium funiculosum and Enterococcus hirae in vitro | Martin‐Arjol et al. (2010) |
| Maleate | ↑1·44 | 0·017 | Reduced the growth of S. Typhimurium on refrigerated turkey frankfurters | Gadang et al. (2008) |
| coleate/vaccenate (18:1) | ↑1·26 | 0·041 | Reduced growth of pathogens on poultry skin, including S. Typhimurium and also prevented fatty acid elongation in a variety of pathogens | Zheng et al. (2005), Hinton and Ingram (2000) |
| Azelate (nonanedioate) | ↑1·18 | 0·024 | Reduced growth of S. aureus, Staphylococcus epidermidis, and Propionibacterium acnes in vitro | Charnock et al. (2004) |
| Alpha‐hydroxyisocaproate | ↓0·86 | 0·016 | Bactericidal against several strains of both Gram‐negative and Gram‐positive bacteria in vitro | Sakko et al. (2012) |
| Pinitol | ↓0·61 | 0·015 | Synergistically enhances the potency of beta‐lactam antimicrobials by lowering the effective dose eightfold. As part of an Ademsia aegiceras extract, it had bacteriostatic and/or bactericidal effects against S. aureus, E. coli, Proteus mirabilis and Pseudomonas aeruginosa | Ahmad et al. (2016) |
| Nucleotides | ||||
| cHypoxanthine | ↑1·08 | 0·046 | Inhibits Salmonella enteriditis and Escherichia coli growth when added to breast milk | Al‐Shehri et al. (2015) |
| Other phytochemicals | ||||
| Harmane | ↑1·62 | 7·51 × 10−5 | Inhibits the growth of and functions as a bactericidal agent against Salmonella spp. and other pathogens in vitro by intercalating with DNA | Cowan (1999), Arshad et al. (2008) |
| 4‐hydroxybenzoate | ↑1·40 | 0·00062 | Inhibited the growth of S. aureus, E. coli, Saccharomyces cerevisiae and Fusarium culmorum in vitro | Kosová et al. (2015) |
| Daidzein | ↓0·86 | 0·040 | Extracted from soy milk, where it inhibited the growth of S. Typhimurium, and a variety of enteric pathogens | Chin et al. (2012) |
| 2‐ketogluconate | ↓0·68 | 0·0066 | Reduced the growth of a variety of micro‐organisms in studies evaluating secondary metabolites produced by Pseudomonas fluorescens | Cheng et al. (2015) |
For each metabolite, fold difference is expressed as the scaled relative abundance in LP+RBE over LP. Arrows indicate the direction of change between treatments.
All metabolites presented have a statistically significant (P < 0·05) fold difference as determined by a Welch's two‐sample t‐test.
Indicates compounds that have not been officially confirmed based on a standard, but second‐order identity in Metabolon Inc. library.
Visualization of metabolic pathways between LP+RBE and LP supernatant treatments
The Cytoscape software visualization tool in MetaboLync © was used to highlight lipids (Fig. 4a) and amino acids/peptides (Fig. 4b) in LP+RBE and LP that contained metabolites with reported antimicrobial functions from Table 4. For a given metabolite, node colour indicated the direction and statistical significance, and node diameter indicated the magnitude of the FD. A pathway enrichment score was calculated for each metabolic pathway as described in the methods. Lipid metabolic pathways visualized in descending order of pathway enrichment score were: hydroxy fatty acids (3·4), long‐chain fatty acids (3·2), dicarboxylate fatty acid (1·7) and inositol (1·1) (Fig. 4a). Amino acid/peptide metabolic pathways visualized, in descending order of pathway enrichment score were: carnitine (2·2), tryptophan (1·7), methionine, cysteine, S‐adenosyl methionine and taurine (1·5), polyamine (0·6), and glycine, serine and threonine (0·4) (Fig. 4b).
Discussion
This study demonstrated that a synbiotic of L. paracasei and RBE contains small molecules with the capacity to reduce S. Typhimurium growth, and appeared to function through pH‐independent mechanisms (Figs 1 and 2). The metabolome of LP and LP+RBE identified 84 small molecules that could explain how LP+RBE reduced S. Typhimurium growth more effectively than LP (Table 3). The presence of MRS components did not contribute to treatment differences between LP and LP+RBE because each treatment contained identical amounts of MRS media. The MRS and MRS+RBE media had similar compounds identified from both LP and LP+RBE treatments, respectively, and therefore the differential effects of LP and LP+RBE were distinguished by FD in the compounds measured between the two treatment groups. The metabolite differences were measured by the relative abundances for each metabolite, and were attributed to L. paracasei metabolism of RBE. Given that these metabolites identified had antimicrobial activities against a variety of pathogens (Table 4), there is promising potential for these supernatants to be used with a broad spectrum of pathogen protection applications relevant to food safety as well as human and animal health.
RBE‐mediated alterations to L. paracasei lipid metabolism, primarily fatty acid production, may explain how LP+RBE can reduce S. Typhimurium growth more than L. paracasei alone. Bacterial production of fatty acids may occur through fermentation of complex carbohydrates, catabolism of complex lipids with fatty acid moieties, de‐novo fatty acid synthesis or via the bioconversion of amino acids into fatty acids (Buccioni et al., 2012). Maleate, azelate, alpha‐hydroxyisocaproate, lineolate, oleate/vaccenate and 13‐HODE were abundant in LP+RBE compared to LP and had reported antimicrobial activity. Maleate is commonly used as a bacteriostatic agent against S. Typhimurium on frankfurters (Gadang et al. 2008). Azelate is used as a topical dermatosis agent to reduce the growth of Staphylococcus aureus, Staphylococcus epidermidis and Propionibacterium acnes (Charnock et al. 2004). Both maleate and azelate are present in the rice plant (Kim et al., 2014; Agarrwal et al., 2016), and L. paracasei metabolism increases their bioavailability. Linoleate is also detected at high levels in rice bran (Kaur et al., 2012). Multiple species of lactobacilli have the capability to produce linoleic acid, as well as catabolize linoleic acid into oleate/vaccenate and 13‐HODE (Black et al. 2013; Druart et al. 2014; Pessione, 2012). Collectively, these fatty acids in LP+RBE demonstrated to have bacteriostatic functions against multiple pathogenic micro‐organisms, including S. Typhimurium, and this may be mediated in part due to interference with bacterial fatty acid synthesis (Hinton and Ingram 2000; Zheng et al. 2005; Martin‐Arjol et al. 2010).
In addition to lipids, RBE amino acids and small peptides serve as another metabolite source that L. paracasei can bioconvert into different compounds or catabolize into free amino acids with antimicrobial functions (Neis et al. 2015). Alterations to methionine and tryptophan metabolism may serve as major mechanisms by which RBE enhances the antimicrobial activity of L. paracasei against S. Typhimurium. Methionine sulfone (8·29‐fold increase in LP+RBE), which forms during methionine oxidation, has been shown to inhibit S. Typhimurium growth by blocking glutamate synthesis (Hentchel and Escalante‐Semerena 2015). Lactobacilli modulate the oxidative state of their environment by producing hydrogen peroxide (Pessione 2012). The supernatant environment created by LP+RBE, coupled with normal hydrogen peroxide production by L. paracasei may facilitate methionine oxidation into methionine sulfone. Although methionine sulfone is typically considered a marker of oxidative damage (Hoshi and Heinemann 2001), its presence in controlled amounts may be beneficial as an antimicrobial defence. Multiple studies have investigated the roles of Lactobacillus sp. in tryptophan metabolism, which have linked the production of tryptophan‐derived metabolites to modulations of the mucosal immune and enteric nervous systems (Zelante et al. 2013; Clarke et al. 2014). Indole‐3‐carboxylic acid was increased and is synthesized from tryptophan by both plant and microbial species (Kavitha et al. 2010; Böttcher et al. 2014). As an antimicrobial agent, indole‐3‐carboxylic acid has been shown to inhibit the growth of a variety of bacterial and fungal pathogens, including multidrug‐resistant bacteria (Zutz et al. 2016). In the presence of RBE, L. paracasei may increase its production or the bioavailability of indole‐3‐carboxylic acid during tryptophan metabolism.
Metabolism of RBE by L. paracasei may have increased the bioavailability of other rice bran phytochemicals, including harmane and 4‐hydroxybenzoate (Table 3). Harmane is a beta‐carboline alkaloid present in plants, and has been shown to have growth inhibitory and bactericidal effects against some Salmonella sp. (Arshad et al. 2008), and works mechanistically to intercalate DNA (Cowan 1999). 4‐hydroxybenzoate, a derivative of benzoic acid produced by plants and some species of bacteria, has been demonstrated to reduce the growth of a variety of Gram‐positive and ‐negative bacteria, yeasts and moulds (Peng et al. 2006; Kosová et al. 2015). Increased expression of both hypoxanthine and tricarballylate in LP+RBE suggested that RBE modulations to L. paracasei nucleotide and TCA cycle metabolism are important to its S. Typhimurium growth‐inhibitory properties (Table 3). Tricarballylate was reported to inhibit citric acid cycle enzymes and was reported to have toxic effects on S. enterica when it was accumulated intracellularly at high levels (Boyd et al. 2012). Past investigations have associated tricarballylate production with the metabolic activity of healthy intestinal and rumen flora (McDevitt and Goldman 1991; Cook et al. 1994), suggesting that in the presence of RBE, L. paracasei may increase the production of tricarballylate to levels that S. Typhimurium cannot tolerate so that it has a decreased ability to replicate and/or invade host cells.
Four of the lipid and three of the amino acid groups had pathway enrichment scores greater than one, indicating these pathways contribute to LP+RBE effects on growth inhibition. For example, spermine and pinitol decreased 0·70‐fold and 0·61‐fold in LP+RBE, yet in past investigations they increased the function of methicillin and beta‐lactam antimicrobials respectively (Kwon and Lu 2007; Lievin‐Le Moal et al., 2011; Ahmad et al. 2016). These studies suggest that regardless of absolute or relative metabolite abundance, the mere presence or absence of metabolites within the supernatant influence the functions of other metabolites present in the supernatant. The level of inhibition, antagonism, additive or synergistic functions for a given combination of metabolites in the supernatant is complex to investigate as a bioactive mixture, and the reductionist approaches needed may yield conflicting results. Thus, the relationships between metabolites, for inhibition of pathogen growth, remains an area of ongoing investigation that can be further explored with targeted metabolite profile analyses and merits consideration of bioactivity‐guided fractionation methods. The metabolomics investigation conducted herein compared the mechanisms of LP+RBE and LP alone by looking at levels of metabolite abundance and metabolic pathway activity. In order to distinguish the unique mechanisms by which RBE modulates L. paracasei metabolism, the metabolome could be compared with gene expression levels obtained from metatranscriptomics and genomics analyses of LP+RBE and LP. This approach would seek to determine direct changes to L. paracasei gene expression of metabolic enzymes in the presence of RBE. Evaluating the small‐molecule profiles of a symbiotic should integrate information from metabolites, overlapping pathways and metabolic networks to provide a comprehensive understanding of overall function.
The experiments described herein illustrated the in vitro efficacy of LP+RBE over LP alone in reducing S. Typhimurium growth, and warrant investigation for the efficacy of LP+RBE against other enteric pathogens. These data, demonstrating enhanced antimicrobial activity of LP+RBE, support results from various animal models (Kumar et al. 2012; Yang et al. 2015). In mice, rice bran simultaneously reduced S. Typhimurium shedding and promoted Lactobacillus growth (Kumar et al. 2012), and in pigs, there was less diarrhoea from Rotavirus (Yang et al. 2015), suggesting that LP+RBE may benefit the host for prevention of pathogen colonization, and for the host immune system to mount a protective response before the onset of extensive disease. The dose of RBE used in these studies has influenced the observable levels of pathogen inhibition, and future investigations merit attention to RBE dose with an intact and varied microbiome composition and structure for demonstrating antimicrobial activities with L. paracasei. Given the growing concern for antimicrobial resistance in S. Typhimurium, alternative treatments and preventive measures are needed. Understanding the metabolite profile of a synbiotic can lead to a comprehensive understanding for how food components enhance the natural antimicrobial activity of L. paracasei. In an era of multidrug resistance, synbiotics of Lactobacillus and rice bran have strong potential to provide broad‐spectrum protection against many pathogens by serving as viable, natural alternatives or by applications in combination with standard antimicrobial drugs to reduce the dosages needed to be efficacious.
Conflict of Interest
No conflict of interest declared.
Supporting information
Figure S1 Dose–response studies were performed for selection of (a) RBE concentration and (b) vehicle control volume to use in the S. Typhimurium growth reduction assay.
Acknowledgements
This work was funded by the Bill and Melinda Gates Foundation (OPP1015267). Furthermore, we thank Dr Anna McClung from the USDA‐ARS Dale Bumpers Rice Research Unit (Stuttgart, AK) for supplying rice bran, and Dr Irfan A. Ghazi, from the University of Hyderabad, for assistance with the preliminary RBE dose curve analyses.
References
- Agarrwal, R. , Padmakumari, A.P. , Bentur, J.S. and Nair, S. (2016) Metabolic and transcriptomic changes induced in host during hypersensitive response mediated resistance in rice against the Asian rice gall midge. Rice 9, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, F. , Misra, L. , Gupta, V.K. , Darokar, M.P. , Prakash, O. , Khan, F. and Shukla, R. (2016) Synergistic effect of (+)‐pinitol from Saraca asoca with beta‐lactam antibiotics and studies on the in silico possible mechanism. J Asian Nat Prod Res 18, 172–183. [DOI] [PubMed] [Google Scholar]
- Al‐Shehri, S.S. , Knox, C.L. , Liley, H.G. , Cowley, D.M. , Wright, J.R. , Henman, M.G. , Hewavitharana, A.K. , Charles, B.G. et al (2015) Breastmilk‐saliva interactions boost innate immunity by regulating the oral microbiome in early infancy. PLoS ONE 10, e0135047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminnezhad, S. , Kermanshahi, R.K. and Ranjbar, R. (2015) Evaluation of synergistic interactions between cell‐free supernatant of lactobacillus strains and amikacin and genetamicin against Pseudomonas aeruginosa . Jundishapur J Microbiol 8, e16592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anon (2006) Probiotics in food: health and nutritional properties and guidelines for evaluation. FAO Food Nutr Pap 85, 2. [Google Scholar]
- Arbulu, M. , Sampedro, M.C. , Gómez‐Caballero, A. , Goicolea, M.A. and Barrio, R.J. (2015) Untargeted metabolomic analysis using liquid chromatography quadrupole time‐of‐flight mass spectrometry for non‐volatile profiling of wines. Anal Chim Acta 858, 32–41. [DOI] [PubMed] [Google Scholar]
- Arshad, N. , Zitterl‐Eglseer, K. , Hasnain, S. and Hess, M. (2008) Effect of Peganum harmala or its beta‐carboline alkaloids on certain antibiotic resistant strains of bacteria and protozoa from poultry. Phytother Res 22, 1533–1538. [DOI] [PubMed] [Google Scholar]
- Atroshi, F. , Rizzo, A. , Westermarck, T. and Ali‐vehmas, T. (1998) Effects of tamoxifen, melatonin, coenzyme Q10, and L‐carnitine supplementation on bacterial growth in the presence of mycotoxins. Pharmacol Res 38, 289–295. [DOI] [PubMed] [Google Scholar]
- Black, B.A. , Zannini, E. , Curtis, J.M. and Gänzle, M.G. (2013) Antifungal hydroxy fatty acids produced during sourdough fermentation: microbial and enzymatic pathways, and antifungal activity in bread. Appl Environ Microbiol 79, 1866–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher, C. , Chapman, A. , Fellermeier, F. , Choudhary, M. , Scheel, D. and Glawischnig, E. (2014) The biosynthetic pathway of indole‐3‐carbaldehyde and indole‐3‐carboxylic acid derivatives in Arabidopsis . Plant Physiol 165, 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, J.M. , Teoh, W.P. and Downs, D.M. (2012) Decreased transport restores growth of a Salmonella enterica apbC mutant on tricarballylate. J Bacteriol 194, 576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.G. , Rao, S. , Weir, T.L. , O'malia, J. , Bazan, M. , Brown, R.J. and Ryan, E.P. (2016) Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab 4, eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccioni, A. , Decandia, M. , Minieri, S. , Molle, G. and Cabiddu, A. (2012) Lipid metabolism in the rumen: new insights on lipolysis and biohydrogenation with an emphasis on the role of endogenous plant factors. Anim Feed Sci Technol 174, 1–25. [Google Scholar]
- Caridi, A. (2002) Selection of Escherichia coli‐inhibiting strains of Lactobacillus paracasei subsp. paracasei . J Ind Microbiol Biotechnol 29, 303–308. [DOI] [PubMed] [Google Scholar]
- Charnock, C. , Brudeli, B. and Klaveness, J. (2004) Evaluation of the antibacterial efficacy of diesters of azelaic acid. Eur J Pharm Sci 21, 589–596. [DOI] [PubMed] [Google Scholar]
- Cheng, X. and van deer Vort, M. and Raaijmakers, J.M. , (2015) Gac‐mediated changes in pyrroquinoline quinone biosynthesis enhance the antimicrobial activity of Pseudomonas fluorescens SBW25. Environ Microbiol Rep 7, 139–147. [DOI] [PubMed] [Google Scholar]
- Chiang, S.S. and Pan, T.M. (2012) Beneficial effects of Lactobacillus paracasei subsp. paracasei NTU 101 and its fermented products. J Appl Microbiol and Biotechnol 93, 903–916. [DOI] [PubMed] [Google Scholar]
- Chin, Y.P. , Tsui, K.C. , Chen, M.C. , Wang, C.Y. , Yang, C.Y. and Lin, Y.L. (2012) Bactericidal activity of soymilk fermentation broth by in vitro and animal models. J Med Food 15, 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, G. , Stilling, R.M. , Kennedy, P.J. , Stanton, C. , Cryan, J.F. and Dinan, T.G. (2014) Minireview: Gut microbiota: the neglected endocrine organ. Mol Endocrinol 28, 1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, G.M. , Wells, J.E. and Russell, J.B. (1994) Ability of Acidaminococcus fermentans to oxidize trans‐aconitate and decrease the accumulation of tricarballylate, a toxic end product of ruminal fermentation. Appl Environ Microbiol 60, 2533–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan, M.M. (1999) Plant products as antimicrobial agents. Clin Microbiol Rev 12, 564–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druart, C. , Alligier, M. , Salazar, N. , Neyrinck, A.M. and Delzenne, N.M. (2014) Modulation of the gut microbiota by nutrients with prebiotic and probiotic properties. Adv Nutr 5, 624S–633S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadang, V.P. , Hettiarachchy, N.S. , Johnson, M.G. and Owens, C. (2008) Evaluation of antibacterial activity of whey protein isolate coating incorporated with nisin, grape seed extract, malic acid, and EDTA on a turkey frankfurter system. J Food Sci 73, M389–M394. [DOI] [PubMed] [Google Scholar]
- Goodyear, A. , Kumar, A. , Ehrhart, E.J. , Swanson, K.S. , Grusak, M.A. , Leach, J.E. , Dow, S.W. , Mcclung, A. et al (2015) Dietary rice bran supplementation prevents Salmonella colonization differentially across varieties and by priming intestinal immunity. J Funct Foods 18, Part A, 653–664. [Google Scholar]
- Henderson, A.J. , Kumar, A. , Barnett, B. , Dow, S.W. and Ryan, E.P. (2012) Consumption of rice bran increases mucosal immunoglobulin A concentrations and numbers of intestinal Lactobacillus spp. J Med Food 15, 469–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentchel, K.L. and Escalante‐Semerena, J.C. (2015) In Salmonella enterica, the Gcn5‐related acetyltransferase MddA (formerly YncA) acetylates methionine sulfoximine and methionine sulfone, blocking their toxic effects. J Bacteriol 197, 314–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia‐Castro, P.Y. , Mendez‐Romero, J.I. , Hernandez‐Mendoza, A. , Acedo‐Felix, E. , Gonzalez‐Cordova, A.F. and Vallejo‐Cordoba, B. (2015) Antimicrobial activity and partial characterization of bacteriocin‐like inhibitory substances produced by Lactobacillus spp. isolated from artisanal mexican cheese. J Dairy Sci 98, 8285–8293. [DOI] [PubMed] [Google Scholar]
- Hinton, A. Jr and Ingram, K.D. (2000) Use of oleic acid to reduce the population of the bacterial flora of poultry skin. J Food Prot 63, 1282–1286. [DOI] [PubMed] [Google Scholar]
- Hoshi, T. and Heinemann, S.H. (2001) Regulation of cell function by methionine oxidation and reduction. J Physiol (Lond) 531, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, A. , Jassal, V. , Thind, S.S. and Aggarwal, P. (2012) Rice bran oil an alternate bakery shortening. J Food Sci Technol 49, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavitha, A. , Prabhakar, P. , Vijayalakshmi, M. and Venkateswarlu, Y. (2010) Purification and biological evaluation of the metabolites produced by Streptomyces sp. TK‐VL_333. Res Microbiol 161, 335–345. [DOI] [PubMed] [Google Scholar]
- Kim, G.R. , Jung, E.S. , Lee, S. , Lim, S.H. , Ha, S.H. and Lee, C.H. (2014) Combined mass spectrometry‐based metabolite profiling of different pigmented rice (Oryza sativa L.) seeds and correlation with antioxidant activities. Molecules 19, 15673–15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koistinen, V.M. , Katina, K. , Nordlund, E. , Poutanen, K. and Hanhineva, K. (2016) Changes in the phytochemical profile of rye bran induced by enzymatic bioprocessing and sourdough fermentation. Food Res Int 89, 1106–1115. [Google Scholar]
- Kosová, M. , Hrádková, I. , Mátlová, V. , Kadlec, D. , Šmidrkal, J. and Filip, V. (2015) Antimicrobial effect of 4‐hydroxybenzoic acid ester with glycerol. J Clin Pharm Ther 40, 436–440. [DOI] [PubMed] [Google Scholar]
- Kumar, A. , Henderson, A. , Forster, G.M. , Goodyear, A.W. , Weir, T.L. , Leach, J.E. , Dow, S.W. and Ryan, E.P. (2012) Dietary rice bran promotes resistance to Salmonella enterica serovar Typhimurium colonization in mice. BMC Microbiol 12, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, D.H. and Lu, C.D. (2007) Polyamine effects on antibiotic susceptibility in bacteria. Antimicrob Agents Chemother 51, 2070–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievin‐Le Moal, V. , Amsellem, R. and Servin, A.L. (2011) Impairment of swimming motility by antidiarrheic Lactobacillus acidophilus strain LB retards internalization of Salmonella enterica serovar Typhimurium within human enterocyte‐like cells. Antimicrob Agents Chemother 55, 4810–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Arjol, I. , Bassas‐Galia, M. , Bermudo, E. , Garcia, F. and Manresa, A. (2010) Identification of oxylipins with antifungal activity by LC‐MS/MS from the supernatant of Pseudomonas 42A2. Chem Phys Lipids 163, 341–346. [DOI] [PubMed] [Google Scholar]
- McDevitt, J. and Goldman, P. (1991) Effect of the intestinal flora on the urinary organic acid profile of rats ingesting a chemically simplified diet. Food Chem Toxicol 29, 107–113. [DOI] [PubMed] [Google Scholar]
- Mozzi, F. , Ortiz, M.E. , Bleckwedel, J. , De Vuyst, L. and Pescuma, M. (2013) Metabolomics as a tool for the comprehensive understanding of fermented and functional foods with lactic acid bacteria. Food Res Int 54, 1152–1161. [Google Scholar]
- Neis, E.P.J.G. , Dejong, C.H.C. and Rensen, S.S. (2015) The role of microbial amino acid metabolism in host metabolism. Nutrients 7, 2930–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppezzo, O.J. and Anton, D.N. (1995) Involvement of cysB and cysE genes in the sensitivity of Salmonella Typhimurium to mecillinam. J Bacteriol 177, 4524–4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveen, N. and Cornell, K.A. (2011) Methylthioadenosine/S‐adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol Microbiol 79, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, X. , Adachi, K. , Chen, C. , Kasai, H. , Kanoh, K. , Shizuri, Y. and Misawa, N. (2006) Discovery of a marine bacterium producing 4‐Hydroxybenzoate and its alkyl esters, parabens. Appl Environ Microbiol 72, 5556–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessione, E. (2012) Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front Cell Infect Microbiol 2, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojevic, J. , Skaro, S. , Senerovic, L. , Vasiljevic, B. , Guzik, M. , Kenny, S.T. , Maslak, V. , Nikodinovic‐Runic, J. et al (2016) Polyhydroxyalkanoate‐based 3‐hydroxyoctanoic acid and its derivatives as a platform of bioactive compounds. Appl Microbiol Biotechnol 100, 161–172. [DOI] [PubMed] [Google Scholar]
- Ryan, E.P. , Heuberger, A.L. , Weir, T.L. , Barnett, B. , Broeckling, C.D. and Prenni, J.E. (2011) Rice bran fermented with Saccharomyces boulardii generates novel metabolite profiles with bioactivity. J Agric Food Chem 59, 1862–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakko, M. , Tjäderhane, L. , Sorsa, T. , Hietala, P. , Järvinen, A. , Bowyer, P. and Rautemaa, R. (2012) 2‐Hydroxyisocaproic acid (HICA): a new potential topical antibacterial agent. Int J Antimicrob Agents 39, 539–540. [DOI] [PubMed] [Google Scholar]
- Settachaimongkon, S. , Nout, M.J.R. , Antunes Fernandes, E.C. , Hettinga, K.A. , Vervoort, J.M. , Van Hooijdonk, T.C.M. , Zwietering, M.H. , Smid, E.J. et al (2014) Influence of different proteolytic strains of Streptococcus thermophilus in co‐culture with Lactobacillus delbrueckii subsp. bulgaricus on the metabolite profile of set‐yoghurt. Int J Food Microbiol 177, 29–36. [DOI] [PubMed] [Google Scholar]
- Simms, S.A. and Subbaramaiah, K. (1991) The kinetic mechanism of S‐adenosyl‐L‐methionine: glutamylmethyltransferase from Salmonella Typhimurium. J Biol Chem 266, 12741–12746. [PubMed] [Google Scholar]
- Tsolis, R.M. , Townsend, S.M. , Miao, E.A. , Miller, S.I. , Ficht, T.A. , Adams, L.G. and Baumler, A.J. (1999) Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature‐tagged mutagenesis. Infect Immun 67, 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio, F. , Lonigro, S.L. , Di Biase, M. , De Candia, S. , Callegari, M.L. and Lavermicocca, P. (2013) Bioprotection of ready‐to‐eat probiotic artichokes processed with Lactobacillus paracasei LMGP22043 against foodborne pathogens. J Food Sci 78, M1757–M1763. [DOI] [PubMed] [Google Scholar]
- Vitali, B. , Ndagijimana, M. , Cruciani, F. , Carnevali, P. , Candela, M. , Guerzoni, M.E. and Brigidi, P. (2010) Impact of a synbiotic food on the gut microbial ecology and metabolic profiles. BMC Microbiol 10. doi: 10.1186/1471‐2180‐10‐4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Liu, Y. , Sidhu, A. , Ma, Z. , Mcclain, C. and Feng, W. (2012) Lactobacillus rhamnosus GG culture supernatant ameliorates acute alcohol‐induced intestinal permeability and liver injury. Am J Physiol‐Gastrointes Liver Physiol 303, G32–G41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, T.L. , Manter, D.K. , Sheflin, A.M. , Barnett, B.A. , Heuberger, A.L. and Ryan, E.P. (2013) Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE 8, e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Twitchell, E. , Li, G. , Wen, K. , Weiss, M. , Kocher, J. , Lei, S. , Ramesh, A. et al (2015) High protective efficacy of rice bran against human rotavirus diarrhea via enhancing probiotic growth, gut barrier function, and innate immunity. Sci Rep 5, 15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante, T. , Iannitti, R.G. , Cunha, C. , De Luca, A. , Giovannini, G. , Pieraccini, G. , Zecchi, R. , D'angelo, C. et al (2013) Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin‐22. Immunity 39, 37, 2–85. [DOI] [PubMed] [Google Scholar]
- Zheng, C.J. , Yoo, J.S. , Lee, T.G. , Cho, H.Y. , Kim, Y.H. and Kim, W.G. (2005) Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS Lett 579, 5157–5162. [DOI] [PubMed] [Google Scholar]
- Zutz, C. , Bacher, M. , Parich, A. , Kluger, B. , Gacek‐Matthews, A. , Schuhmacher, R. , Wagner, M. , Rychli, K. et al (2016) Valproic acid induces antimicrobial compound production in doratomyces microspores. Front Microbiol 7, eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Dose–response studies were performed for selection of (a) RBE concentration and (b) vehicle control volume to use in the S. Typhimurium growth reduction assay.


