Abstract
A quantitative, peripherally accessible biomarker for neuropathic pain has great potential to improve clinical outcomes. Based on the premise that peripheral and central immunity contribute to neuropathic pain mechanisms, we hypothesized that biomarkers could be identified from the whole blood of adult male rats, by integrating graded chronic constriction injury (CCI), ipsilateral lumbar dorsal quadrant (iLDQ) and whole blood transcriptomes, and pathway analysis with pain behavior. Correlational bioinformatics identified a range of putative biomarker genes for allodynia intensity, many encoding for proteins with a recognized role in immune/nociceptive mechanisms. A selection of these genes was validated in a separate replication study. Pathway analysis of the iLDQ transcriptome identified Fcγ and Fcε signaling pathways, among others. This study is the first to employ the whole blood transcriptome to identify pain biomarker panels. The novel correlational bioinformatics, developed here, selected such putative biomarkers based on a correlation with pain behavior and formation of signaling pathways with iLDQ genes. Future studies may demonstrate the predictive ability of these biomarker genes across other models and additional variables.
Keywords: allodynia, dorsal horn, Fc receptor, microarray, neuropathic pain, pathway analysis
Neuropathic pain may arise as a consequence of a lesion or disease affecting the somatosensory system (Jensen et al. 2011). Such pain is a major health problem that afflicts up to 8% of the general European population (Torrance et al. 2006; Bouhassira et al. 2008), substantially reduces quality of life for the afflicted individual and poses a significant economic burden to the health system and society. Symptoms may include hypersensitivity to noxious (hyperalgesia) and non-noxious (allodynia) stimuli, and spontaneous pain. Neuropathic pain presents challenges for diagnosis and treatment selection given its heterogeneity with respect to pain intensity, disease mechanisms, and treatment responses (Mogil et al. 1999a,b; Woolf and Decosterd 1999; Woolf and Mannion 1999; Finnerup and Jensen 2006; Costigan et al. 2009b; von Hehn et al. 2012), which are all incompletely understood.
The problems posed by disease heterogeneity are not unique to neuropathic pain. However, other fields facing such difficulties, like oncology, have addressed these clinical challenges through the development of biomarkers, defined by the NIH Biomarkers Working Group (2001) as ‘a characteristic that can be measured and evaluated as an indicator of normal biologic processes, pathologic processes or pharmacologic responses to therapeutic intervention’. Biomarkers should be distinguished from surrogate endpoints, for which stringent qualification is required to enable the surrogate to substitute for a clinical endpoint (NIH Biomarkers Working Group 2001). Biomarker development for neuropathic pain has had some potential leads in genetics (Foulkes and Wood 2008), quantification of primary afferent activity (Chizh and Hobson 2007), sampling the cerebrospinal fluid (Uceyler et al. 2007; Zin et al. 2010), and imaging of the pain matrix (Chizh and Hobson 2007; Tracey and Mantyh 2007), and more recently via pain phenotyping (von Hehn et al. 2012), but none are well accepted as objective biomarkers at present. An objective measure of pain intensity may prove a useful adjunct to the visual analogue scale and other subjective measures of pain reporting in a clinical scenario, to overcome the difficulties associated with inter-individual differences in reporting experiences or using rating scales, and when a subjective report cannot be solely relied upon (e.g. compensation claims, inability to communicate). An objective measure of pain intensity may be useful to assess analgesic efficacy during drug development, where a notoriously high placebo response and Type II error in clinical trials in pain is recognized as the single biggest hurdle in assessing drug efficacy (Dworkin 2011).
In fields such as oncology, the most recent advances in biomarker discovery and development have used transcriptome profiling of the blood as a window into disease processes that are not always easily accessible (Hanash et al. 2011). However, with regards to neuropathic pain, the RNA transcriptome has not been applied to biomarker screening in the blood or at any other site, but rather to mechanistic studies in rodent neuropathic pain models at the peripheral nerve injury site, dorsal root ganglia, and spinal cord (Lacroix-Fralish et al. 2011). Transcriptome analyses of the blood are likely to show promise for neuropathic pain, given the recognition that peripheral immune cells contribute to pain hypersensitivity at both peripheral and central sites in rodent models of nerve injury (Austin and Moalem-Taylor 2010; Grace et al. 2011a,b).
Therefore, we aimed to test whether the whole blood transcriptome could identify genes that would correlate with heterogeneity in mechanical allodynia intensity in the peripheral nerve-injured adult male rat, at a single time-point. Specifically, regarding the NIH biomarker definition (NIH, Biomarkers Working Group 2001), such an approach aimed to identify a potential measurement of the pathological pain process, namely mechanical allodynia intensity. If so, this approach may provide a useful strategy for future explorations of the broad applicability of such putative biomarkers for pre-clinical neuropathic pain assessment, neuropathic pain in general, and whether the whole blood transcriptome may prove useful for clinical evaluations. A key element in this novel approach is pain heterogeneity, as the shortcoming in virtually all the studies reviewed by Lacroix-Fralish et al. (2011), and in the field at large, is that means are compared between a largely homogeneous neuropathic pain group and a sham control. This simple pain versus no pain comparison of group means is not sufficient to discover biomarkers of pain intensity, as it is unable to differentiate whether degree of gene expression is related to the nature of the nerve injury and surrounding graded sequelae or the intensity of consequent pain behavior. Alternatively, by individually pairing the degree of allodynia with the respective gene expression from each rat, the variability in behavior can be powerfully harnessed to elucidate the gene expression strongly correlated with allodynia. Persson et al. (2009) correlated gene expression with the contrasting pain response phenotypes of different mouse strains, but this design is limited by other potential genetic differences between the strains. In contrast, our graded chronic constriction injury (CCI) model (Grace et al. 2010) presents an opportunity to intentionally generate a graded pain population that is not confounded by such genetic differences, allowing correlational investigation of transcriptional changes. The graded approach also has superior face validity, as it is consistent with the variable spectrum of clinical pain.
Therefore, we hypothesized that biomarkers of pain intensity, related to graded CCI, could be identified in peripheral blood, by correlating the whole blood transcriptome with pain behavior. Given the absence of prior correlational studies, new analyses were developed using Pathway Analysis to refine the gene list to a size suitable for testing in a validation group, so to enable future studies examining parameters beyond those tested here. To focus such analyses, the whole blood transcriptome was analyzed together with the transcriptome of the dorsal quadrant of the lumbar spinal cord, ipsilateral lumbar dorsal quadrant to sciatic injury (iLDQ). The inclusion of these conjoint whole blood and iLDQ analyses was based on the premise that signaling pathways of both tissues contributed to allodynia induced by sciatic nerve injury. Although it could be argued that the injured peripheral nerve or the dorsal root ganglia should also be included, given the prominent role of peripheral immune cells at these sites (Austin and Moalem-Taylor 2010), the iLDQ alone was selected at this time owing to the success of a recent transcriptome study in identifying peripheral immune contributions at this region (Costigan et al. 2009a). By analyzing the whole blood and iLDQ transcriptomes together, it was hypothesized that putative biomarkers identified in the whole blood would have a closer relationship with the mechanisms contributing to allodynia, than a simple correlation between the whole blood transcriptome and pain behavior. Thus, this study provides a new analytical approach that, based on the results to be reported here, bears future testing and investigation.
Materials and methods
Given the absence of prior correlational transcriptome studies, a new data pipeline was developed (summarized in Fig. 1). Development of the correlational data pipeline, detailed here, required the generation of data that were used to justify progression to the next step. Therefore, a significant proportion of the manuscript is dedicated to methods development to arrive at a list of putative blood-based biomarkers.
Fig. 1.
Transcriptome data pipeline developed and employed to identify putative biomarker panels. Red boxes represent putative biomarker panels; blue boxes represent experimental steps and results; green boxes represent bioinformatic steps; white boxes represent supplementary analyses. CCI, chronic constriction injury; iLDQ, ipsilateral lumbar dorsal quadrant; suppl, supplementary. *Genes common to both the iLDQ and whole blood gene lists were retained.
Subjects
Pathogen-free adult male Sprague-Dawley rats were used for the transcriptome study (300–350 g; Animal Resource Centre, Perth, WA, Australia; n = 10) and the prospective validation study (300–350 g; Harlan Laboratories, Madison, WI, USA; n = 12). Rats were housed in temperature- (18–21°C) and light-controlled (12 h light: dark cycle; lights on at 07:00 hours) rooms with standard rodent food and water available ad libitum and allowed to habituate to the holding facility for 1 week prior to experimentation. The transcriptome study was performed at the University of Adelaide, Australia, and all relevant procedures were approved by the University of Adelaide Animal Ethics Committee. The prospective validation study was performed at the University of Colorado at Boulder, USA, giving strength to translation of results across disparate research sites, and all relevant procedures were approved by the Institutional Animal Care and Use Committee of the University of Colorado at Boulder. Both studies were conducted in accordance with the ARRIVE guidelines and the NHMRC Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
Surgery
The CCI model of graded sciatic nerve injury was performed aseptically at the mid-thigh level of the left hindleg, as previously described, so to create a graded intensity of pain behavior (Grace et al. 2010). Briefly, animals were anesthetized with isoflurane, the skin of the hindquarters was shaved and the sciatic nerve gently elevated. Zero, 1, or 4 sterile chromic gut sutures (cuticular 4-0 chromic gut; Ethicon, Somerville, NJ, USA) were loosely tied around the isolated sciatic nerve (N; approximately 3–4 mm in length). Once the superficial muscle overlying the nerve was sutured with silk, and prior to surgical stapling of the skin incision, additional equal lengths of chromic gut were placed subcutaneously (S; approximately 3–4 mm in length), such that each animal was exposed to four equal lengths of chromic gut in total. Thus, the treatment groups (n = transcriptome, prospective validation studies), differing in pain behavior, were: sham (N0S0; n = 2, 3), low pain (N0S4; n = 2, 3); medium pain (N1S3; n = 3); and high pain (N4S0; n = 3).
Mechanical allodynia
Testing was conducted blind with respect to group assignment. Rats received at least three 60 min habituations to the test environment prior to behavioral testing. The von Frey test (Chaplan et al. 1994) was performed within the sciatic innervation region of the hindpaws as previously described in detail (Chacur et al. 2001; Milligan et al. 2001). Assessments were made prior to (baseline) and at postoperative days 3, 10, and 21. A logarithmic series of 10 calibrated Semmes-Weinstein monofilaments (von Frey hairs; Stoelting, Wood Dale, IL, USA) were applied randomly to the left and right hindpaws to define the threshold stimulus intensity required to elicit a paw withdrawal response. Log stiffness of the hairs was determined by log10 (milligrams × 10) and ranged from manufacturer designated 3.61 (0.407 g) to 5.18 (15.136 g) filaments. The behavioral responses were used to calculate absolute threshold (the 50% paw withdrawal threshold) by fitting a Gaussian integral psychometric function using a maximum-likelihood fitting method (Harvey 1986; Treutwein and Strasburger 1999), as described previously (Milligan et al. 2000, 2001). This fitting method allows parametric analyses that otherwise would not be statistically appropriate (Milligan et al. 2000, 2001). A reduction in von Frey threshold was interpreted as an increase in allodynia.
Tissue preparation, RNA extraction, chip hybridization, and PCR
Transcriptome study
Following final behavioral testing on day 21, the rats were anesthetized with sodium pentobarbital and circulating blood collected via cardiac puncture into RNAprotect Animal Blood Tubes (QIAGEN, Doncaster, VIC, Australia), and stored at −80°C. Following transcardial perfusion with isotonic saline, the L4–L6 segment of spinal cord (innervating the sciatic nerve) was carefully excised and the iLDQ was dissected, rapidly frozen in liquid nitrogen, and stored at −80°C.
Total RNA (including microRNA) was purified from blood using a RNeasy Protect Animal Blood Kit (QIAGEN), as directed by the manufacturer. Spinal cord tissue was disrupted over dry ice in an RNase-free (DEPC treated) loose fitting glass homogenizer. Total RNA (including microRNA) was purified using a miRNeasy kit (QIAGEN), according to the manufacturer’s instructions. The RNA quality was assessed using the Agilent 2100 Bioanalyzer (Santa Clara, CA, USA) and RNA integrity numbers were > 7 for all samples.
cDNA was reverse transcribed from the total RNA and hybridized to the Affymetrix Rat Gene 1.0 ST Array (Santa Clara), according to the manufacturer’s instructions. Array intensity was quantified using the Affymetrix GeneChip 3000 7G scanner (Santa Clara) by the Adelaide Microarray Centre (SA, Australia).
Prospective validation study
Following final behavioral testing on day 21, the rats were anesthetized with sodium pentobarbital and 100 μL of circulating blood collected via cardiac puncture into tubes containing 1 mL of TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) for immediate processing. Total RNA was purified from whole blood using the standard phenol:chloroform extraction with TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions, and stored at −80°C.
Primer sequences were obtained from Genbank at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov; Table 1). cDNA was reverse transcribed from the total RNA and amplification performed using Quantitect SYBR Green PCR kit (QIAGEN, Valenica, CA, USA) in iCycler iQ 96-well PCR plates (Bio-Rad, Hercules, CA, USA) on a MyiQ single Color Real-Time PCR Detection System (Bio-Rad). The reaction mixture (26 μL) was composed of QuantiTect SYBR Green (containing fluorescent dye SYBR Green I, 2.5 mM MgCl2, dNTP mix, and Hotstart Taq Polymerase), 10 nM fluorescein, 500 nM of each forward and reverse primer (Invitrogen), nuclease-free water, and 1 μL of cDNA from each sample. Each sample was measured in duplicate. The reactions were initiated with a hotstart at 95°C for 25 min, followed by 40 cycles of 15 s at 94°C (denaturation), 30 s at 55–65°C (annealing), and 30 s at 72°C (extension). Melt curve analyses were conducted to assess uniformity of product formation, primer-dimer formation, and amplification of non-specific products. The PCR product was monitored in real-time, using the SYBR Green I fluorescence, using the MyiQ single Color Real-Time PCR Detection System (Bio-Rad). Threshold for detection of PCR product was set in the log-linear phase of amplification and the threshold cycle (CT) was determined for each reaction. The level of the target mRNA was quantified relative to the housekeeping gene (β-actin). The expression of β-actin was not significantly different between samples, suggesting that differences were likely caused by true changes in mRNA expression, rather than reflecting changes in cell number per sample.
Table 1.
PCR primer specifications
| Gene | Primer sequence (5′-3′) | GenBank Accession No. |
|---|---|---|
| Tnfrsf11b | F: GGAATAGATGTCACCCTGTG R: CGAGCTGTGTCTCCGTTTTA | NM_012870.2 |
| Thra | F: TTCCTGCCGGATGACATTGG R: AAGGCAGCTCGGAGAACATG | NM_001017960.1 |
| Avpr1a | F: TTGTGTCAGCAGCGTGAAGA R: GGTTTTCTGAATCGGTCCAG | NC_005106 |
| Csf3 | F: CAGCTGTGTGCCACCTACAA R: TCAGGCACTTTGTCTGCTGC | NC_005109.2 |
| Xylb | F: TAACATGGAGGTCTCGGCAT R: CTTAGGCATGATTCGGTAGC | NC_005107 |
| Olr952 | F: TCTACTGTCCACGTTGGCAT R: TCATGTGAGAGGAACAGGTG | NC_005106 |
| Hook1 | F: AGCAAAGGTTGAGCAAAGAG R: GCTGGAACTTTCACCTTCAG | NC_005104 |
| Trim30 | F: TGTCCTGAGAAGGAGTCAGA R: TCCATGAGCCCTTCAAATGC | NC_005100 |
| Cx3cl1 | F: TGTACTCTGCTGGCGGGTCA R: TCGTCTCCAGGATGATGGCG | NC_005118 |
| Irf2bp1 | F: CTGGTGACATCAAGGTCAAG R: GGGTGGGGTAGGGAATAATT | NC_005100 |
| LOC686781 | F: TACAGAGGGATCTGAGGCCA R: AGCGCATCTCCATGCTCTGA | NC_005100 |
| β-actin | F: TTCCTTCCTGGGTATGGAAT R: GAGGAGCAATGATCTTGATC | NM_031144.2 |
Data pipeline and analyses
Data pre-processing: correlational pathway analysis of whole blood and iLDQ genes using ‘Adelaide plots’
Array CEL files (Affymetrix) for the whole blood and iLDQ transcriptomes were normalized to a set of manufacturer designated control genes, that were unchanged between treatment groups, to ensure that differences were caused by true changes in mRNA expression, rather than reflecting changes in cell number per sample. Probes without signal significantly above background were eliminated. Following this, we departed from traditional preprocessing of microarray data that aims to identify significant differential gene expression between treatment group means (Ko et al. 2002; Yang et al. 2004; LaCroix-Fralish et al. 2006a; Lacroix-Fralish et al. 2006b; Griffin et al. 2007; Costigan et al. 2009a; Persson et al. 2009; Vega-Avelaira et al. 2009). Instead, independent Pearson correlations were performed between von Frey threshold and gene expression for the whole blood and iLDQ transcriptomes, which therefore accounted for the variability in behavior and were independent of treatment group (see Fig. 2c as example). The resulting gene list, consisting of 14 825 whole blood and 14 099 iLDQ genes, had correlation coefficients continuously ranging from |0.01| to |0.91| for the whole blood, and |0.01| to |0.94| for the iLDQ. Pathway Analysis programs are unable to deal with datasets of this size, and open source software are unable to weight genes, using the correlation coefficient, for example. Traditional data pre-processing addresses the problem of data reduction by selecting genes that are significantly differentially regulated between the treatment group means. In contrast, the continuous range of 14 825 or 14 099 correlations essentially posed a sorites paradox (Hyde 2011); that is, at what point along this continuous range of correlations does one define the breakpoint between meaningful and non-meaningful associations? The solution to this problem required that a threshold be determined to split the dataset according to a true association with allodynia. Such a threshold was identified by developing an analysis that was based on two premises: that the significance of a particular gene cannot be understood in isolation but only in the context of gene networks and signaling pathways (Kitano 2002), and that the genes most strongly correlated with allodynia would contain the signaling pathways relevant to allodynia.
Fig. 2.
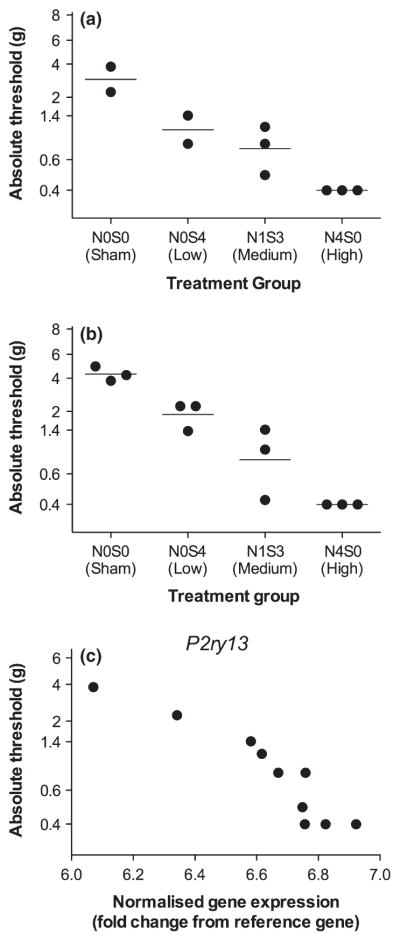
Graded chronic constriction injury of the sciatic nerve produces graded allodynia at day 21 post surgery. Graded neuropathy was induced by varying the distribution of four equivalent chromic gut pieces across the nerve (N) or subcutaneous (S) compartments. As such, treatment design was N0S0 (transcriptome study, n = 2 prospective validation study, n = 3), N0S4 (n = 2, 3), N1S3 (n = 3), and N4S0 (n = 3) to generate a single, heterogeneous group with respect to allodynia for correlational analysis in (a) the transcriptome study and; (b) the prospective validation study. (c) As an example, the correlation between ipsilateral lumbar dorsal quadrant P2ry13 gene expression and von Frey threshold is presented. Each point represents an individual rat.
Therefore, genes from the whole blood and iLDQ transcriptomes from each rat were independently ranked according to the absolute correlation coefficient (RAACC) between gene expression and von Frey threshold (hereafter referred to as the RAACC Gene list; Fig. 1). Beginning at the top (i.e. the highest correlation coefficient) of the RAACC Gene lists, bins were defined across the entire 14 825 whole blood gene and 14 099 iLDQ gene transcriptomes by selecting the top 1000 genes and then progressively frame-shifting downward by units of 100 genes (such that bin 1 contained genes 1–1000, bin 2 contained genes 101–1100, and so forth), so that each subsequent bin progressively excluded the prior 100 top ranked genes. This was done, as noted above, because pathway analysis programs are unable to handle such large datasets; hence, binning in this manner created a means for systematic analysis without losing data. Each 1000-gene bin was analyzed in DAVID 6.7 Pathway Analysis software (http://david.abcc.ncifcrf.gov) (Huang da et al. 2009a,b), which classified the binned genes into signaling pathways, based on the Kyoto Encyclopedia of Genes and Genomes database. To identify where the threshold between meaningful and non-meaningful correlations lay, it was necessary to determine the number of pathways that would be identified by chance using this approach. To do this, 1000 genes were randomly re-sampled from the iLDQ transcriptome and analyzed in DAVID 6.7 ten times. Subjecting these 10 bins of 1000 random genes to the DAVID 6.7 pathway analysis yielded an average of 1.8 pathways. Thus, as the bins created from the entire transcriptomes were progressively analyzed from bin 1 (highest correlations), the threshold was inferred at point at which the pathway number per bin fell below that expected by chance (≤ 1.8), that is the excluded genes (i.e. the preceding RAACC genes that were excluded from the bin) were the major contributors to signaling pathways related to allodynia.
The number of pathways classified from each bin by DAVID 6.7 was quantified for the binned iLDQ- and whole blood-RAACC Gene lists (Fig. 1). Within the iLDQ-RAACC Gene list (Fig. 1), bins 1 through 3 yielded between 12 and 21 pathways, but beyond these bins (i.e. beyond the top 300 genes), ≤ 2 pathways were identified per bin, suggesting that the top 300 genes (corresponding to r > |0.7|) were the major contributors to signaling pathways related to allodynia. Binned analysis of the whole blood-RAACC Gene list (Fig. 1) did not identify such an unequivocal threshold. Bins 1 through 5 yielded between 0 and 3 pathways, which inconsistently exceeded the number of pathways expected by chance. Beyond bin 5 (i.e. beyond the top 500 genes), ≤ 1 pathway per bin was identified, suggesting that the top 500 genes (corresponding to r > |0.7|) made only a meager contribution to the signaling pathways related to allodynia. Such a result is to be expected, given that the factors in the blood contributing to pain pathophysiology probably represent only a very small proportion of the total compartment.
It was hypothesized that the meager organization of the top 500 genes from the whole blood-RAACC Gene list (Fig. 1) into signaling pathways could be improved, if analyzed with genes from the iLDQ, given that peripheral immunity has a recognized contribution to neuropathic pain pathophysiology within the CNS (Grace et al. 2011a,b). Therefore, the top 500 whole blood and 300 iLDQ genes from the respective RAACC Gene lists were merged to form an 800 gene combination list. To determine how many genes in the combination list might be false positives (Type I errors), the following analysis was undertaken: the whole blood- (14 825 genes) and iLDQ- (14 099 genes) RAACC Gene lists were combined and the binned analysis performed. As hypothesized, merging the whole blood- and iLDQ-RAACC gene lists resulted in an increased number of bins that yielded signaling pathways (exceeding the number expected by chance), beyond either the binned analysis of the whole blood or iLDQ in isolation. Bins 1 through 7 yielded between 2 and 23 pathways, but beyond these bins (i.e. beyond the top 700 genes), ≤ 1 pathways were identified per bin, suggesting that the top 700 genes were the major contributors to signaling pathways related to allodynia. This result indicated that when the top 500 whole blood and 300 iLDQ gene lists were merged to form an 800-gene combination list, ~100 genes were unlikely to be related to the severity of allodynia. Thus, the whole blood genes contributing to signaling pathways may be better defined when interpreted together with the iLDQ genes in this form of binned analysis. However, it was posited that the potential Type I errors present in the 800-gene list would be adequately eliminated through the identification of common pairs (Putative biomarker panel 1) or pathway derived gene list (Putative biomarker panel 2), thus the full combined 800-gene list was taken forward for further data refinement. It should be noted that 7 genes derived from both the iLDQ and whole blood transcriptomes are duplicated in the combination list (800 genes), and these are used to generate Putative biomarker panel 1: direct whole blood-iLDQ correlates (see results). As the combination list is eventually analyzed in DAVID 6.7, where duplicated genes are only counted once, any overlap is inconsequential.
The graphical representation of these binned analyses, depicted in Fig. 3, demonstrate that the most highly correlated RAACC genes are highly organized in functional signaling pathways, before abruptly diminishing. We are naming these new analyses ‘Adelaide plots’, akin to a ‘Manhattan plot’, because of the related function and etymological basis. Thus, the whole blood (500 genes), iLDQ (300 genes), and combination (800 genes) gene lists generated here are referred to as ‘Adelaide Gene lists’ (Fig. 1).
Fig. 3.
‘Adelaide plots’ define the gene list thresholds. Genes from the entire transcriptome were independently ranked according to absolute correlation coefficient (RAACC), for (a) the ipsilateral lumbar dorsal quadrant (iLDQ) transcriptome; (b) the whole blood transcriptome; (c) the combined iLDQ and whole blood transcriptomes. Beginning at the top of the RAACC Gene lists, bins were defined across the RAACC gene lists by selecting 1000 genes and then progressively frame-shifting by 100 genes (such that bin 1 contained genes 1–1000, bin 2 contained genes 101–1100 and so forth), so that each subsequent bin progressively excluded the prior 100 top ranked genes, and each bin analyzed in DAVID 6.7. The number of pathways classified from each bin was quantified and are displayed here as ‘Adelaide plots’, identifying the degree of organization into signaling pathways among the most highly correlated genes. As each bin was progressively analyzed in DAVID 6.7, the RAACC genes being major contributors to signaling pathways related to allodynia were identified as (a) the top 300 iLDQ genes, (b) the top 500 whole blood genes, (c) the top 700 combined iLDQ- and whole blood-RAACC Gene lists. These thresholds were inferred, as exclusion of these genes in Pathway Analyses (i.e. analyses beyond bins 4, 6, and 8, respectively) resulted in the pathway number per bin falling below that expected by chance (≤ 1.8, dotted line defined by random re-sampling of 1000 genes). Thus, Adelaide plots make it possible to identify the division between genes that contribute to signaling pathways and those that do not.
Data pre-processing: traditional differential expression of iLDQ genes between treatment groups
We next determined whether the new Adelaide plot analysis was better matched to our study design than a traditional analysis. As the iLDQ transcriptome would contain genes that were more closely related to allodynia than the whole blood, the iLDQ was selected to afford the differential analysis the best possible chance of success.
Microarray data processing was performed in the freely available statistical programming and graphics environment R (http://cran.r-project.org), with open source software available through Bioconductor project (http://www.bioconductor.org). Background subtraction was performed with RMA (Irizarry et al. 2003) and followed with quantile normalization, both implemented in affy library. Differential expression was determined with limma (Smyth 2005), using a linear model to compare all treatment groups with each other and isolate changes specific to iLDQ samples. Statistical significance of the results was low, as could be expected in an experiment with very few replicates in each treatment group.
In view of the large number of Type I errors that would be inevitably present in any selection of potentially regulated genes, functional analysis was undertaken to identify those with high likelihood of being associated with nerve injury. Forty-six named genes with p < 0.05, unadjusted for multiple testing, were identified. Gene networks were constructed in MetaCore (http://www.genego.com) and 22 of the initial 46 genes were found to have interconnecting relationships. The low number of significant iLDQ genes (i.e. significantly differentially regulated genes with interconnecting relationships) is unsurprising given the low number of replicates in each group, in contrast to the larger groups sizes that are typically required to overcome within-group variability. However, our approach suggests that when the variability is used as an endpoint, additional information can be gleaned with smaller group sizes.
Putative biomarker panel 1: direct whole blood-iLDQ correlates
The iLDQ- (300 genes) and whole blood- (500 genes) Adelaide Gene lists (Fig. 1) were compared. Genes common to both lists were identified (Common pairs identified, Fig. 1) and the magnitude of gene expression correlated between each whole blood-iLDQ pair, to quantify the reliability of gene expression in the blood to mirror that in the iLDQ.
Putative biomarker panel 2: contribution of whole blood genes to iLDQ signaling pathways
The iLDQ- (300 genes) and combination- (800 genes) Adelaide Gene lists were analyzed in DAVID 6.7. When the genes contributing to these pathways were identified, it was found that only 77 of the original 300 iLDQ genes and 106 of the original 800 combination genes contributed to the signaling pathways defined using DAVID 6.7. This resulted from the fact that 1 gene may contribute to multiple pathways. For example, in the iLDQ-Adelaide Gene list 77 of 300 genes generated 19 pathways, and the other 223 genes did not contribute. Therefore, the 77 and 106 genes noted above were selected for further analysis and referred to as ‘Pathway derived Gene lists’ (Fig. 1). The iLDQ genes (77 genes) that contributed to the combination Pathway derived gene list (106 genes) were then removed, leaving 29 whole blood genes that were both correlated with allodynia and also formed signaling pathways with iLDQ genes.
Prospective validation study
To validate the putative biomarkers identified via our novel data pipeline, a prospective validation study was performed in which a separate group of graded CCI rats were generated in different laboratory and whole blood collected (see ‘Prospective validation study’ above). Real-time PCR was performed on a selection of the top correlation coefficient ranked whole blood genes from each potential biomarker panel in Fig. 1: the whole blood-RAACC Gene list (top 2 genes); the 7 gene candidate biomarkers panel (top 2 genes); the 29 candidate biomarkers panel (top 4 genes); and 2 genes not correlated with von Frey threshold (r < 0.01) from the whole blood-RAACC Gene list. The derivation method of the 29 candidate biomarkers panel directly tested the study hypothesis, which if proved true, would increase the potential of the panel to yield suitable biomarkers. Therefore, a greater number of genes were selected from the 29 candidate biomarkers panel to subject the panel to more rigorous validation testing. In addition, Cx3cl1 [Entrez ID:89808] was also analyzed using real-time PCR because of the unique correlation with allodynia and its cognate receptor expression in the iLDQ (see Results). A Pearson correlation was performed between the magnitude of gene expression (relative to housekeeping gene) and von Frey threshold, recorded immediately prior to tissue collection at day 21 post surgery. The coefficient of variance (% CV) was determined between the pipeline and prospective correlation coefficients.
Distribution of published ‘pain genes’ within the iLDQ transcriptome dataset
The 39 genes that were identified as having ‘strong correlational’ or ‘causational’ evidential links with rodent neuropathic pain models in the recent meta-analysis by Lacroix-Fralish et al. (2011) were identified within the iLDQ transcriptome dataset. The relevant correlation coefficient was paired with each gene, and the ‘pain genes’ dataset was then split according to the concordance (23 genes; Table S2) or discordance (16 genes; Table S3) between the direction of correlation in this study and the published direction of gene regulation (Lacroix-Fralish et al. 2011). The data were binned into correlation coefficient tertiles and the distribution frequency plotted.
Results
The data pipeline used to identify the putative pain biomarkers is summarized in Fig. 1.
Mechanical allodynia
Behavior was quantified at 21 days post graded CCI surgery for both the transcriptome and prospective validation studies (Fig. 2). The degree of allodynia correlated with the graded CCI treatment group (transcriptome study: Pearson r = −0.99 [95% CI: −0.99, −0.50]; prospective validation study: Pearson r = −0.99 [95% CI: −1.00, −0.98]), similar to that found previously (Grace et al. 2010, 2011a). Whereas the experimental design employed different treatment groups, once graded allodynia had been achieved at day 21, all animals were treated as one heterogeneous group (transcriptome study: n = 10; prospective validation study: n = 12) for correlational analyses (e.g. Fig. 2c).
Putative biomarker panel 1: direct whole blood-iLDQ correlates
After derivation of Adelaide plots (Fig. 3), Adelaide Gene lists (Fig. 1) were determined for whole blood (500 genes) and iLDQ (300 genes). Following comparison of these lists, 7 genes were identified as common to both. These genes are ranked according to the correlation coefficient in Table 2, indicating the ability of gene expression in the blood to mirror that in the iLDQ, and are presented together with references to link the protein encoded by each gene with nociceptive hypersensitivity and immune signaling.
Putative biomarker panel 2: contribution of whole blood genes to iLDQ signaling pathways
Pathway derived Gene lists (Fig. 1) were generated by analyzing the iLDQ- (300 genes) and combination- (800 genes) Adelaide Gene lists (Fig. 1) in DAVID 6.7. The genes of the iLDQ-Pathway derived Gene list (77 genes) were removed from the combination-Pathway derived Gene list (106 genes). Thus, the 29 remaining whole blood genes were those that formed signaling pathways with iLDQ genes (Fig. 4), and were correlated with the severity of allodynia. These genes are ranked in Table 3 according to the correlation coefficient, and are presented together with references to link the protein encoded by each gene with nociceptive hypersensitivity and immune signaling.
Fig. 4.
Signaling pathways derived from (a) the ipsilateral lumbar dorsal quadrant (iLDQ), and (b) the combined whole blood and iLDQ gene lists. (a) The iLDQ-Adelaide gene list (300 genes; Figure 1) was classified into signaling pathways using DAVID 6.7. (b) The combined iLDQ and whole blood gene lists B (800 genes; Figure 1) were classified into signaling pathways using DAVID 6.7. Bars represent the −log(p value) for that pathway within the gene list (two-tailed Fisher’s exact test; threshold of 1.3 (p = 0.05) used).
Table 3.
Putative biomarker panel 2: Twenty-nine contributors to iLDQ signaling pathways
| Gene name | Symbol | Entrez ID | Nociceptive hypersensitivity | Immune signaling | r | p |
|---|---|---|---|---|---|---|
| Tumor necrosis factor receptor superfamily, member 11b | Tnfrsf11b | [Entrez ID:25341] | (Doupis et al. 2009) | (Caidahl et al. 2010) | −0.900 | 0.001 |
| Thyroid hormone receptor alpha | ThrA | [Entrez ID:81812] | - | - | 0.898 | 0.001 |
| Arginine vasopressin receptor 1A | Avpr1a | [Entrez ID:25107] | (Mogil et al. 2011) | - | −0.805 | 0.005 |
| Colony stimulating factor 3 (granulocyte) | Csf3 | [Entrez ID:25610] | (Doupis et al. 2009; Ro et al. 2009) | (Ro et al. 2009) | −0.802 | 0.005 |
| Potassium large conductance calcium-activated channel, subfamily M, beta member 2 | Kcnmb2 | [Entrez ID:294961] | - | - | −0.794 | 0.006 |
| Galanin receptor 1 | Galr1 | [Entrez ID:50577] | (Alier et al. 2008) | −0.784 | 0.007 | |
| Calcitonin receptor-like (precursor to CGRP receptor) | Calcrl | [Entrez ID:25029] | (Ma et al. 2003; Lee and Kim 2007) | (De Corato et al. 2011) | 0.777 | 0.008 |
| Contactin 1 | Cntn1 | [Entrez ID:117258] | - | - | −0.776 | 0.008 |
| Protein phosphatase 3, regulatory subunit B, alpha isoform | Ppp3r2 | [Entrez ID:29749] | (Jung and Miller 2008; Miletic et al. 2011) | (Jung and Miller 2008) | −0.774 | 0.009 |
| Adenylate cyclase 8 | Adcy8 | [Entrez ID:29241] | - | - | −0.774 | 0.009 |
| Adenosine A2B receptor | Adora2b | [Entrez ID:29316] | (Abo-Salem et al. 2004) | (Ham and Rees 2008) | −0.768 | 0.009 |
| Protein kinase, cAMP dependent, catalytic, beta | Prkacb | [Entrez ID:293508] | (Gold and Gebhart 2010) | (Riether et al. 2011; Sugimoto et al. 2011) | 0.767 | 0.010 |
| Heat shock protein 1 | Hspb1 | [Entrez ID:24471] | (Kim et al. 2001) | (Kim et al. 2009) | −0.767 | 0.010 |
| Myosin, heavy polypeptide 2, skeletal muscle, adult | Myh2 | [Entrez ID:691644] | - | - | −0.761 | 0.011 |
| Deoxyribonuclease II beta | Dnase2b | [Entrez ID:59296] | - | (Okabe et al. 2008) | −0.759 | 0.011 |
| Purinergic receptor P2X, ligand-gated ion channel, 3 | P2rx3 | [Entrez ID:81739] | (Wirkner et al. 2007) | - | 0.757 | 0.011 |
| Chemokine (C-X3-C motif) ligand 1 | Cx3cl1 | [Entrez ID:89808] | (Milligan et al. 2008) | (Milligan et al. 2008) | −0.754 | 0.012 |
| Angiotensin II receptor, type 1b | Agtr1b | [Entrez ID:81638] | - | (Downie et al. 2009; Chon et al. 2011; Fuchtbauer et al. 2011) | −0.751 | 0.012 |
| Caspase recruitment domain family, member 6 | Card6 | [Entrez ID:294770] | - | (Dufner et al. 2006) | 0.747 | 0.013 |
| RT1 class II, locus Ba | Rt1-ba | [Entrez D:309621] | (Grace et al. 2011b) | (Grace et al. 2011b) | 0.739 | 0.015 |
| Coxsackie virus and adenovirus receptor | Cxadr | [Entrez ID:89843] | - | - | −0.738 | 0.015 |
| Adrenergic, beta-1-, receptor | Adrb1 | [Entrez ID:24925] | - | (Walker-Brown and Roberts 2009) | −0.730 | 0.016 |
| Inducible T-cell co-stimulator ligand | Icoslg | [Entrez ID:499415] | (Simpson et al. 2010) | 0.728 | 0.017 | |
| Tumor necrosis factor receptor superfamily, member 10b | Tnfrsf10b | [Entrez ID:364420] | - | (Hoffmann et al. 2009) | −0.728 | 0.017 |
| Purinergic receptor P2X, ligand-gated ion channel, 7 | P2rx7 | [Entrez ID:29665] | (Sperlagh et al. 2006; He et al. 2012) | (Sperlagh et al. 2006; Tsukimoto et al. 2009; He et al. 2012) | −0.725 | 0.018 |
| Histone cluster 3, H2ba | Hist3h2ba | [Entrez ID:303175] | - | - | −0.725 | 0.018 |
| Purinergic receptor P2Y, G-protein coupled 2 | P2ry2 | [Entrez ID:29597] | (Malin et al. 2008) | (Kronlage et al. 2010) | −0.723 | 0.018 |
| Prostaglandin E receptor 2, subtype EP2 | Ptger2 | [Entrez ID:81752] | (Kunori et al. 2011) | (Kunori et al. 2011) | −0.712 | 0.021 |
| Glutamate receptor, metabotropic 1 | Grm1 | [Entrez ID:24414] | (Kohara et al. 2007) | (Hermans and Challiss 2001; Chiocchetti et al. 2006) | −0.703 | 0.023 |
| Complement component 1, q subcomponent, alpha polypeptide | C1qa | [Entrez ID:298566] | (Twining et al. 2005; Griffin et al. 2007) | (Baruah et al. 2009; Schmid et al. 2009) | −0.702 | 0.024 |
Following a Pearson correlation between the whole blood and ipsilateral lumbar dorsal quadrant (iLDQ) transcriptomes with von Frey threshold, gene segments identified as making a major contribution to signaling pathways in Adelaide plots were selected. The whole blood- and iLDQ-Adelaide Gene lists were combined and classified into signaling pathways using DAVID 6.7 Pathway Analysis software. The iLDQ genes that contributed to signaling pathways were removed, leaving a list of whole blood genes that were correlated with the severity of allodynia, and were linked with iLDQ genes in signaling pathways. The table is ranked according to the correlation coefficient between the gene and von Frey threshold. A non-exhaustive list of references is cited for the link of the protein encoded by each gene to nociceptive hypersensitivity and immune signaling. Omitted references indicate that, to the best of our knowledge, the protein encoded by that gene currently has no known role.
Many of the 29 putative whole blood biomarker genes encode for proteins that form ligand-receptor pairings in the CNS. Of each potential pairing, the only ligand or receptor genes from the 29 putative whole blood biomarker genes to form cognate pairs with genes in the iLDQ-Pathway derived Gene list (77 genes; Fig. 1) were Csf3r [Entrez ID:298518] and Cx3cr1 [Entrez ID:171056]. When the cognate pairs were correlated, an insignificant inverse relationship was found between whole blood Csf3 [Entrez ID:25610] and iLDQ Csf3r (r = −0.47, p = 0.17; Fig. 5a), but a significant correlation was found between whole blood Cx3cl1 and iLDQ Cx3cr1 (r = 0.84, p = 0.003; Fig. 5b).
Fig. 5.
Relationship between blood ligand and ipsilateral lumbar dorsal quadrant (iLDQ) receptor gene expression in the graded chronic constriction injury model. Of the potential ligand/receptor pairing between the 29 whole blood genes contributing to iLDQ signaling pathways biomarker panel (Table 3) and the iLDQ-Pathway derived Gene list (Fig. 1), the only pairs to form with their cognate receptors in the iLDQ were Csf3 and Cx3cl1. A correlation between the ligands and their cognate receptors found (a) an insignificant inverse relationship between Csf3 and Csf3r; but (b) a significant relationship between Cx3cl1 and Cx3cr1.
Comparison between putative whole blood biomarkers from RAACC gene list and those derived from the Adelaide and pathway derived gene lists
Although it could be argued that whole blood biomarkers could be identified on the strength of the correlation between whole blood gene expression and von Frey threshold, we posited that genes more closely related to the mechanisms contributing to allodynia could be identified by taking the iLDQ signaling pathways into account. Therefore, the top 35 whole blood RAACC genes (equivalent to the number of non-repeated genes across both biomarker panels [Table 2, Table 3]) are presented in Table S1, together with references to link the protein encoded by each gene with nociceptive hypersensitivity, and immune signaling. Nineteen genes were found to contribute to nociceptive hypersensitivity and/or immune signaling in the Adelaide and Pathway derived biomarker panels (Table 2, Table 3), whereas only 7 such genes were identified in the whole blood-RAACC panel (Table S1).
Table 2.
Putative biomarker panel 1: Seven direct whole blood-iLDQ correlates
| Gene name | Symbol | Entrez ID | Nociceptive hypersensitivity | Immune signaling | Paired lumbar DH gene correlation | von Frey theshold correlation | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| r | p | r | p | |||||
| Hook homolog 1 (Drosophila) | Hook1 | [Entrez ID:313370] | - | - | 0.755 | 0.011 | −0.784 | 0.007 |
| Similar to Tripartite motif protein 30-like | Trim30 | [Entrez ID:499223] | - | (Shi et al. 2008) | −0.711 | 0.021 | 0.791 | 0.006 |
| Similar to Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | LOC295452 | [Entrez ID:295452] | - | - | 0.658 | 0.039 | −0.718 | 0.019 |
| Structural maintenance of chromosomes 2 | Smc2 | [Entrez ID:362519] | - | - | −0.633 | 0.050 | 0.743 | 0.014 |
| Complement component 1, q subcomponent, A chain | C1qa | [Entrez ID:298566] | (Twining et al. 2005; Griffin et al. 2007) | (Baruah et al. 2009; Schmid et al. 2009) | 0.560 | 0.092 | −0.702 | 0.024 |
| Transmembrane protein 176B | Tmem176b | [Entrez ID:171411] | - | (Louvet et al. 2005; Schmid et al. 2009; Condamine et al. 2010; Melchior et al. 2010) | 0.525 | 0.119 | −0.778 | 0.008 |
| SP100 nuclear antigen | Sp100 | [Entrez ID:363269] | - | (Negorev et al. 2009) | −0.518 | 0.125 | 0.706 | 0.022 |
Following a Pearson correlation between the whole blood and ipsilateral lumbar dorsal quadrant transcriptomes with von Frey threshold, gene segments identified as making a major contribution to signaling pathways in Adelaide plots were selected. The resulting whole blood- and ipsilateral lumbar dorsal quadrant-Adelaide Gene lists were compared and genes common to both were selected as putative biomarkers. The table is ranked according to the correlation coefficient between the gene expression and von Frey threshold. A non-exhaustive list of references is cited for the link of the protein encoded by each gene to nociceptive hypersensitivity and immune signaling. Omitted references indicate that, to the best of our knowledge, the protein encoded by that gene currently has no known role.
Prospective validation study
To validate the putative biomarkers identified via our novel data pipeline, a prospective validation study was performed in which a separate group of graded CCI rats were generated in a different laboratory and whole blood collected. Real-time PCR was performed on the top correlation coefficient ranked whole blood genes from the RAACC Gene list (top 2 genes; See Table S1), the 7 gene candidate biomarkers panel (top 2 genes; Table 2); the 29 candidate biomarkers panel (top 4 genes; Table 3); Cx3cl1, owing to its unique correlation with its cognate receptor in the iLDQ; and 2 genes not correlated with von Frey threshold (r < 0.01) from the whole blood-RAACC Gene list. Hence, a total of 9 putative biomarker genes and 2 negative control genes were analyzed. A Pearson correlation between gene expression (relative to the housekeeping gene, β-actin) and von Frey threshold revealed that 8 of 9 putative biomarker genes positively replicated, in addition to the replication of 2 negative controls. The results of the prospective validation study are presented in Table 4, and the genes correlated with von Frey threshold presented in Fig. 6, together with the regression line from the original transcriptome study. As the expression of β-actin was not significantly different between samples, this suggests that mRNA changes were likely caused by true changes in expression, rather than reflecting changes in cell number per sample.
Table 4.
Prospective validation of select putative biomarker genes
| Gene name | Symbol | Entrez ID | Gene list | r | p | % CV |
|---|---|---|---|---|---|---|
| Tumor necrosis factor receptor superfamily, member 11b | Tnfrsf11b | [Entrez ID:25341] | 29 candidate biomarkers | −0.881 | 0.001 | 1.53 |
| Thyroid hormone receptor alpha | Thra | [Entrez ID:81812] | 29 candidate biomarkers | 0.639 | 0.01 | 23.88 |
| Arginine vasopressin receptor 1A | Avpr1a | [Entrez ID:25107] | 29 candidate biomarkers | −0.735 | 0.01 | 6.42 |
| Colony stimulating factor 3 (granulocyte) | Csf3 | [Entrez ID:25610] | 29 candidate biomarkers | 0.753 | 0.01 | 4.49 |
| Chemokine (C-X3-C motif) ligand 1 | Cx3cl1 | [Entrez ID:89808] | 29 candidate biomarkers | −0.713 | 0.01 | 3.96 |
| Xylulokinase homolog (H. influenzae) | Xylb | [Entrez ID:316067] | Whole blood RAACC Gene list | −0.664 | 0.05 | 21.75 |
| Olfactory receptor 952 | Olr952 | [Entrez ID:288880] | Whole blood RAACC Gene list | −0.635 | 0.05 | 24.58 |
| Hook homolog 1 (Drosophila) | Hook1 | [Entrez ID:313370] | 7 candidate biomarkers | −0.479 | 0.11 | 34.11 |
| Similar to Tripartite motif protein 30-like | Trim30 | [Entrez ID:499223] | 7 candidate biomarkers | 0.587 | 0.05 | 20.90 |
| Interferon regulatory factor 2 binding protein 1 | Irf2bp1 | [Entrez ID:308404] | Negative control | 0.148 | 0.67 | 40.83 |
| Similar to NFkB interacting protein 1 | LOC686781 | [Entrez ID:686781] | Negative control | 0.093 | 0.77 | 9.35 |
A prospective validation study was performed using a separate group of graded chronic constriction injury rats from different laboratory. Whole blood was collected and real-time PCR performed on select putative biomarker genes. The table presents the genes analyzed, the list from which they were derived, the Pearson correlation between gene expression and von Frey threshold, and the coefficient of variance (% CV) between the pipeline and prospective correlation coefficients.
Fig. 6.
Prospective validation of select putative biomarker genes. To validate the putative biomarkers identified via our novel data pipeline, a prospective validation study was performed using real-time PCR. A Pearson correlation was performed between gene expression (relative to housekeeping gene) and von Frey threshold. (a–d) The top 4 genes, from the 29 candidate biomarkers panel (Table 3) (Tnfrsf11b; Thra; Avpr1a; Csf3). (e, f) The top 2 genes, ranked by correlation coefficient, from the whole blood RAACC-Gene list were significantly correlated with von Frey threshold (Xylb; Olr952). (g) Of the top 2 genes from the 7 gene candidate biomarkers panel, only Trim30 was significantly correlated with von Frey threshold. (h) Cx3cl1 was also significantly correlated with von Frey threshold. The replication study regression line (black) and statistics, and the transcriptome study regression line (gray) are indicated for reference.
Distribution of published ‘pain genes’ within the iLDQ transcriptome dataset
The recent meta-analysis by Lacroix-Fralish et al. (2011) identified a list of spinal cord and dorsal root ganglion genes with ‘strong correlational’ or ‘causational’ evidential links with rodent neuropathic pain models. The distribution of these genes was identified within our iLDQ transcriptome, and the relationship between the direction of correlation and the published direction of gene regulation was classified as either concordant (23 genes; Table S2) or discordant (16 genes; Table S3). The distribution of concordant genes is more heavily weighted toward those that are negatively correlated with von Frey threshold (positively correlated with allodynia) than those that were discordant (Fig. 7). The discordant genes showed a normal distribution, clustered around r = 0, as did the distribution of the iLDQ transcriptome r frequency.
Fig. 7.
Distribution and concordance of published spinal cord and dorsal root ganglia ‘pain genes’ within the ipsilateral lumbar dorsal quadrant (iLDQ) transcriptome dataset. Genes that were identified as having ‘strong correlational’ or ‘causational’ evidential links with rodent neuropathic pain models in the recent meta-analysis by Lacroix-Fralish et al. (2011) were identified within the iLDQ transcriptome dataset (RAACC-Gene list; Figure 1). The ‘pain genes’ dataset was then split according to the concordance or discordance between the direction of correlation between gene expression and von Frey threshold, and the published direction of gene regulation (Lacroix-Fralish et al. 2011). (a) The data was binned in correlation coefficient tertiles and the distribution frequency plotted. (b) Individual points are plotted for concordant genes and overlaid with the correlation coefficient frequency of the entire iLDQ transcriptome. (c) Individual points are plotted for discordant genes and overlaid with the correlation coefficient frequency of the entire iLDQ transcriptome. Dashed lines indicate correlation coefficient tertiles; dotted lines indicate correlation coefficient thresholds defined here using Adelaide plots.
Discussion
We used our graded CCI model (Grace et al. 2010) to generate graded mechanical allodynia, based on the premise that elements of such behavior could be explained by graded pathology in the iLDQ, such as microglial activation. Tissues were collected from male rats at a single timepoint after full development of allodynia. Given that there is a recognized peripheral immune component to the CNS mechanisms of rodent neuropathic pain models (Grace et al. 2011a,b), we proposed the novel hypothesis that biomarkers of graded CCI could be identified in the blood. It was also hypothesized that whole blood genes correlated with von Frey threshold (RAACC-Gene list; Fig. 1) with a closer relationship to the mechanisms contributing to allodynia could be identified as putative biomarkers, based on their contribution to signaling pathways with iLDQ genes, leading to the development of new bioinformatic analyses. We successfully tested the first hypothesis by correlating the whole blood transcriptome with von Frey threshold and, after developing a new correlational bioinformatic pipeline, identified a range of putative biomarkers (Tables 2 and 3, Table S1). The majority of select putative biomarkers were successfully tested in an independent prospective validation study (Table 4). By comparing the number of genes that are linked with nociceptive hypersensitivity and/or immune signaling, according to current literature, across the RAACC and the bioinformatically derived biomarker panels, the second hypothesis was also confirmed.
Each approach yielded a panel of putative biomarkers accessible via the blood, which is in keeping with the call from Rolan et al. (2003) to use biomarker panels for complex diseases, such as neuropathic pain. This is especially relevant for the present study, where inflammatory processes, independent of neuropathic pain, may regulate and hence confound the interpretation of individual gene expression. Nevertheless, a few notable features and examples will be highlighted, regarding individual genes, to provide support for the potential of the biomarker panels, and the methods used to derive them. Firstly, many of the genes identified encode for proteins that have a known link to immune or nociceptive signaling (Table 2 and 3), thus providing significant face validity to our panels and validating previous results from disparate laboratories. For example, of the 9 genes that were tested in the prospective validation study, 4 have a recognized role in nociceptive signaling, 4 have a recognized role in immune signaling, and 4 have no recognized role in either process (Tables 2 and 3; Hook1 [Entrez ID:313370]; ThrA [Entrez ID:81812]; Xylb [Entrez ID:316067]; Olr952 [Entrez ID:288880]). Tnfrsf11b [Entrez ID:25341] encodes for a soluble cytokine receptor, also known as osteoprotegerin, which has been linked with many diseases, including painful diabetic neuropathy (Doupis et al. 2009). Avpr1a [Entrez ID:25107] encodes for an arginine vasopressin receptor, polymorphisms of which influence pain sensitivity to capsaicin across different mouse strains and in humans (Mogil et al. 2011). Csf3 encodes for an anti-inflammatory chemokine that is associated with painful diabetic neuropathy and has been proposed as a potential therapeutic for neuropathic pain (Doupis et al. 2009; Ro et al. 2009). Cx3cl1 encodes for the pro-inflammatory chemokine fractalkine, that is posited as a neuron-to-glial signal in pre-clinical neuropathic pain models (Milligan et al. 2008). Trim30 [Entrez ID:499223] encodes for a protein in the toll like receptor signaling cascade that functions as a negative regulator of NFκB activation (Shi et al. 2008). Secondly, some cross-validity is lent to the methods used to derive the putative biomarker panels, as C1qa [Entrez ID:298566] was identified in both panels. C1qa encodes for a subunit of the C1q complement protein, which may be causally linked to nociceptive hypersensitivity, if peripheral nerve injury induces activation of the classical complement pathway (Twining et al. 2005; Griffin et al. 2007). Finally, of the ligand/receptor cognates across the blood and iLDQ, a significant correlation was only found between Cx3cl1 in the blood and Cx3cr1 in the iLDQ (Fig. 5b). These genes encode for the chemokine fractalkine, and its cognate receptor, CX3CR1, which have a recognized mechanistic role in pre-clinical models of neuropathic pain. Fractalkine is posited to be a neuron-to-glia signal within the CNS (Milligan et al. 2008), whereas its cognate receptor is considered to be exclusively expressed by activated microglia in the dorsal horn of the spinal cord (Verge et al. 2004). Therefore, fractalkine gene expression in the blood may also serve as a biomarker for other diseases where microglial activation plays a role.
While the preceding discussion highlights the face validity of our biomarker panels, we have successfully demonstrated the strength of our approach by beginning the process of qualification (construct validation), in that 8 of 9 putative biomarker genes were positively replicated, in addition to the replication of 2 negative controls (Table 4, Fig. 6). The unsuccessful validation of 1 gene (Hook1) may be as a result of a Type I error in the transcriptome, which is potentially supported by Table 2, showing that the gene has no known function in either nociceptive or immune signaling. The replication study was also performed in a different laboratory, demonstrating the robustness of our findings. To build on our positive replication, it is necessary that other variables are tested, such as additional neuropathic pain models, additional pain modalities, sex, allodynia timecourse, and sensitivity to drug effect. Although beyond the scope of this study, the genes identified via our novel data pipeline may be useful to investigate additional variables, as they have a closer relationship with the mechanisms contributing to allodynia (Table 2, Table 3), compared with the whole blood-RAACC Gene list (Table S1). The targeted investigation of the putative biomarker genes (Tables 2 and 3) is more economical and has greater feasibility than conducting further transcriptome studies, and a potential causal relationship with allodynia suggests that the putative biomarker genes may be a more robust and reliable measure under different circumstances, compared with those simply correlated with allodynia (Table S1). Importantly, the pre-clinical development of the genes identified here, or their respective proteins, may lead to their translation as clinical neuropathic pain biomarkers that serve as tools for mechanism-based diagnoses; to target therapy by patient stratification; and/or to expedite drug development by enriching patient populations in clinical trials, quantify pain intensity and to detect treatment effect (Rolan 1997; Chizh et al. 2008; Lathia et al. 2009; Woodcock 2009).
The iLDQ transcriptome raises some additional points, regarding the mechanisms underlying nerve injury-induced nociceptive hypersensitivity. Firstly, despite the fact that the iLDQ is considered a neural tissue, many of the signaling pathways classified from the transcriptome of this tissue are in fact immune (Fig. 4a). These data further support the important role of central immunity in models of neuropathic pain (Austin and Moalem-Taylor 2010; Ren and Dubner 2010; Grace et al. 2011a,b), which may explain why neuronally targeted therapies for neuropathic pain are not disease modifying. Of these immune pathways, both the Fcγ and Fcε signaling pathways have potential, but as yet unrecognized, significance for neuropathic pain (see representative correlations in Figure S1). The antibody receptors, Fcγ and Fcε, are expressed by the innate immune cells of the CNS, microglia (Ulvestad et al. 1994; Inoue et al. 1999; Suemitsu et al. 2010), but also by neurons, leading to direct activation by IgG and IgE, respectively (Andoh and Kuraishi 2004a,b; Suemitsu et al. 2010; Qu et al. 2011), the former having been shown to induce mechanical allodynia when intrathecally administered to naïve mice (Cao et al. 2009). The consequences of Fc receptor activation on antigen presenting cells, such as microglia include cytokine secretion, and stimulation and modulation of T cells (Joller et al. 2011), which may have profound implications for pain pathophysiology (Grace et al. 2011b). Significant expression of spinal dorsal horn Fcγ receptor mRNAs have been identified by other transcriptome studies in pain models (Griffin et al. 2007; Costigan et al. 2009a). Therefore, the role of Fc receptors in neuropathic pain pathophysiology requires further investigation. As an aside, caution should be exercised when drawing conclusions from the signaling pathway classifications, as the Kyoto Encyclopedia of Genes and Genomes database contains only previously defined signaling pathways, and at present, the database is further limited by the absence of pathways that specifically encompass neuro-immune signaling.
Secondly, examination of the distribution of published ‘pain genes’ (Lacroix-Fralish et al. 2011) within our iLDQ transcriptome identified genes that were either concordant or discordant with the direction of correlation and the published direction of gene regulation (Fig. 7; Table S2, Table S3). Provided that these data are not confounded by artifacts of the transcriptome, they lead to a number of potential conclusions. The concordant genes are skewed toward up-regulation as compared to discordant genes in our model (r < −0.16), with ~60% of concordant ‘pain genes’ falling within the lower tertile and therefore negatively correlated with von Frey threshold (positively correlated with allodynia). These data suggest that severity of allodynia is generally associated with activation and recruitment of new genes and processes, rather than down-regulation of existing ones. Of the genes in the lower tertile (r < −0.16), the Adelaide plot (Fig. 3a) suggests that the top 300 genes (corresponding to r < −0.7) are probably the most important predictors of pain severity (Gfap [Entrez ID:24387]; Ctss [Entrez ID:50654]; C1qc [Entrez ID:362634]; C1qb [Entrez ID:29687]; Aif1 [Entrez ID:29427]; C3 [Entrez ID:24232]). Of the discordant genes, those that fall in the outer tertiles (Rgs4 [Entrez ID:29480]; Scn11a [Entrez ID:29701]; Chrna3 [Entrez ID:25101]; C1s [Entrez ID:192262]; Gfra1 [Entrez ID:25454]; Calca [Entrez ID:24241]; S100a4 [Entrez ID:24615]; Gadd45a [Entrez ID:25112]) are especially noteworthy, as they have an opposite relationship with pain severity to that previously reported (Lacroix-Fralish et al. 2011). However, the meaning and behavioral significance of this is as yet unknown, and requires further study and validation.
A final note concerns the conclusions drawn from studies of single factors in models of neuropathic pain, for which a number of recent studies serve as useful, but not isolated, examples (Ren and Neugebauer 2010; Gao et al. 2011; He et al. 2012; Wilson et al. 2011). These studies use exclusive terms, such as ‘mediate’, to describe the effect of a single factor on pain behavior. The Adelaide plots generated from this study suggest that at least 300 genes contribute to allodynia at the spinal level (Fig. 3a). Therefore, it is inaccurate to use such exclusive terms to describe single factor pain studies, and we suggest that such conclusions are avoided in the future. Moreover, these data suggest that pharmacologically targeting a single site well down the mechanistic pathway to treat pain may be a futile approach, owing to the apparent widespread adaptations in multiple systems and possible redundancy of nociception amplification systems.
Our novel, proof-of-concept, graded CCI transcriptome study design departed from previous pain transcriptome studies in two main ways. Firstly, heterogeneity was intentionally created in a single population for analysis, thus avoiding the difficulties associated with few replicates in each treatment group. Gene selection was based on relationship with the endpoint of interest, allodynia, rather than on the treatment group. Thus, we were able to differentiate, at least in part, between gene expression specific to the model (such as changes in whole blood related to the chromic gut sutures placement in each surgery group) and that specific to allodynia. Furthermore, we did not attempt to limit variability within treatment groups by pooling RNA prior to chip hybridization, for example, as has been done previously (Griffin et al. 2007; Costigan et al. 2009a). Secondly, we used a novel data pipeline to analyze the graded transcriptome. Data pre-processing for a binary study design is relatively straightforward, in that only the genes significantly up- or down-regulated compared to controls are carried through to the next stage of analysis (Ko et al. 2002; Yang et al. 2004; LaCroix-Fralish et al. 2006a; Lacroix-Fralish et al. 2006b; Griffin et al. 2007; Costigan et al. 2009a; Vega-Avelaira et al. 2009). Even some correlational studies have employed a similar pre-processing step by excluding non-significant expression of normalized fold changes compared to a baseline (Cameron et al. 2007; Persson et al. 2009). However, it is possible that selecting genes for further analysis based on overabundance may overlook subtle changes in gene expression. This is especially relevant for highly regulated and controlled genes, or those whose product is highly potent. Therefore, our approach differed, in that we developed the Adelaide plot to identify genes of interest. The use of pathway analysis in the data preprocessing is likely to minimize and eliminate Type I errors, which is supported by the fact that we identified genes encoding for proteins that have established or potential links with the mechanisms underlying neuropathic pain (Table 2, Table 3). Adelaide plots are a powerful and versatile tool to identify the level of organization in signaling pathways of large datasets and would be valuable to employ in future studies. A potential weakness in using Pathway Analysis software, such as DAVID 6.7, is that genes are selected on the basis of a known function and contribution to known signaling pathways. Hence, potential biomarker candidates may have been overlooked among the genes that have not yet been characterized. Tools using next-generation sequence data that identify novel expressed sequence tags or gene products may be a valuable alternative to partially circumvent this limitation. Our study design is based on the premise that the rats with graded CCI differ only in respect to the mechanisms underlying graded allodynia, as discussed previously (Grace et al. 2010). However, there may be other mechanisms that could confound pre-clinical validation and clinical translation, such as graded Wallerian degeneration (Lin et al. 2001), which may lead to a graded immune response (Griffin et al. 1992). Therefore, future studies should consider examining additional neuropathic pain models that have the capacity to produce a heterogeneous response.
We are the first to use the transcriptome to identify putative blood-based biomarker panels in a rodent model of nerve injury. The criteria for such biomarkers were that they were correlated with von Frey threshold and that they contributed to signaling pathways with iLDQ genes. To achieve this aim, we developed a novel data pipeline to integrate graded CCI, pain behavior, whole blood and iLDQ transcriptomes, using the power of Pathway Analysis. The panels identified from this approach include a direct correlation between gene expression common to both the whole blood and iLDQ, and identification of whole blood genes that directly contribute to the iLDQ signaling pathways. Future qualification studies may demonstrate the predictive ability of these biomarker genes across further models and additional variables.
Acknowledgments
Supported by the Pain and Anaesthesia Research Clinic. Dr. Mark Hutchinson was a NHMRC CJ Martin Fellow (ID 465423; 2007–2010) and is an Australian Research Council Research Fellow (DP110100297; 20110297; 2012). Dr. Peter Grace was the recipient of a Faculty of Health Sciences Divisional PhD Scholarship (2007–2010). Dr. Linda Watkins is supported, in part, by a career award from NIDA/NIH (DA024044). All authors declare that there are no financial or commercial conflicts of interest.
Abbreviations used
- CCI
chronic constriction injury
- CNS
central nervous system
- iLDQ
ipsilateral lumbar dorsal quadrant
- N
neuronal
- S
Subcutaneous
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
Figure S1. Representative correlations from Fcγ and Fcε signaling pathways.
Table S1. The top 35 whole blood genes, ranked according to absolute correlation coefficient between gene expression and von Frey threshold.
Table S2. Concordant gene list.
Table S3. Discordant gene list.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copyedited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Abo-Salem OM, Hayallah AM, Bilkei-Gorzo A, Filipek B, Zimmer A, Muller CE. Antinociceptive effects of novel A2B adenosine receptor antagonists. J Pharmacol Exp Ther. 2004;308:358–366. doi: 10.1124/jpet.103.056036. [DOI] [PubMed] [Google Scholar]
- Alier KA, Chen Y, Sollenberg UE, Langel U, Smith PA. Selective stimulation of GalR1 and GalR2 in rat substantia gelatinosa reveals a cellular basis for the anti- and pro-nociceptive actions of galanin. Pain. 2008;137:138–146. doi: 10.1016/j.pain.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Andoh T, Kuraishi Y. Direct action of immunoglobulin G on primary sensory neurons through Fc gamma receptor I. FASEB J. 2004a;18:182–184. doi: 10.1096/fj.02-1169fje. [DOI] [PubMed] [Google Scholar]
- Andoh T, Kuraishi Y. Expression of Fc epsilon receptor I on primary sensory neurons in mice. Neuro Report. 2004b;15:2029–2031. doi: 10.1097/00001756-200409150-00007. [DOI] [PubMed] [Google Scholar]
- Austin PJ, Moalem-Taylor G. The neuro-immune balance in neuropathic pain: involvement of inflammatory immune cells, immune-like glial cells and cytokines. J Neuroimmunol. 2010;229:26–50. doi: 10.1016/j.jneuroim.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Baruah P, Dumitriu IE, Malik TH, Cook HT, Dyson J, Scott D, Simpson E, Botto M. C1q enhances IFN-gamma production by antigen-specific T cells via the CD40 costimulatory pathway on dendritic cells. Blood. 2009;113:3485–3493. doi: 10.1182/blood-2008-06-164392. [DOI] [PubMed] [Google Scholar]
- Bouhassira D, Lanteri-Minet M, Attal N, Laurent B, Touboul C. Prevalence of chronic pain with neuropathic characteristics in the general population. Pain. 2008;136:380–387. doi: 10.1016/j.pain.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Caidahl K, Ueland T, Aukrust P. Osteoprotegerin: a Biomarker With Many Faces. Arterioscler Thromb Vasc Biol. 2010;30:1684–1686. doi: 10.1161/ATVBAHA.110.208843. [DOI] [PubMed] [Google Scholar]
- Cameron B, Galbraith S, Zhang Y, et al. Gene expression correlates of postinfective fatigue syndrome after infectious mononucleosis. J Infect Dis. 2007;196:56–66. doi: 10.1086/518614. [DOI] [PubMed] [Google Scholar]
- Cao L, Tanga FY, Deleo JA. The contributing role of CD14 in toll-like receptor 4 dependent neuropathic pain. Neuroscience. 2009;158:896–903. doi: 10.1016/j.neuroscience.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacur M, Milligan ED, Gazda LS, Armstrong C, Wang H, Tracey KJ, Maier SF, Watkins LR. A new model of sciatic inflammatory neuritis (SIN): induction of unilateral and bilateral mechanical allodynia following acute unilateral peri-sciatic immune activation in rats. Pain. 2001;94:231–244. doi: 10.1016/S0304-3959(01)00354-2. [DOI] [PubMed] [Google Scholar]
- Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- Chiocchetti A, Miglio G, Mesturini R, et al. Group I mGlu receptor stimulation inhibits activation-induced cell death of human T lymphocytes. Br J Pharmacol. 2006;148:760–768. doi: 10.1038/sj.bjp.0706746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizh BA, Hobson AR. Using objective markers and imaging in the development of novel treatments of chronic pain. Expert Rev Neurother. 2007;7:443–447. doi: 10.1586/14737175.7.5.443. [DOI] [PubMed] [Google Scholar]
- Chizh BA, Greenspan JD, Casey KL, Nemenov MI, Treede RD. Identifying biological markers of activity in human nociceptive pathways to facilitate analgesic drug development. Pain. 2008;140:249–253. doi: 10.1016/j.pain.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H, Neumann J, Boer P, Joles JA, Braam B. Enhanced Angiotensin II type 1 receptor expression in leukocytes of patients with chronic kidney disease. Eur J Pharmacol. 2011;666:205–210. doi: 10.1016/j.ejphar.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Condamine T, Le Texier L, Howie D, et al. Tmem176B and Tmem176A are associated with the immature state of dendritic cells. J Leukoc Biol. 2010;88:507–515. doi: 10.1189/jlb.1109738. [DOI] [PubMed] [Google Scholar]
- Costigan M, Moss A, Latremoliere A, et al. T-cell infiltration and signaling in the adult dorsal spinal cord is a major contributor to neuropathic pain-like hypersensitivity. J Neurosci. 2009a;29:14415–14422. doi: 10.1523/JNEUROSCI.4569-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan M, Scholz J, Woolf CJ. Neuropathic pain: a maladaptive response of the nervous system to damage. Annu Rev Neurosci. 2009b;32:1–32. doi: 10.1146/annurev.neuro.051508.135531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Corato A, Lisi L, Capuano A, Tringali G, Tramutola A, Navarra P, Dello Russo C. Trigeminal satellite cells express functional calcitonin gene-related peptide receptors, whose activation enhances interleukin-1beta pro-inflammatory effects. J Neuroimmunol. 2011;237:39–46. doi: 10.1016/j.jneuroim.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Doupis J, Lyons TE, Wu S, Gnardellis C, Dinh T, Veves A. Microvascular reactivity and inflammatory cytokines in painful and painless peripheral diabetic neuropathy. J Clin Endocrinol Metab. 2009;94:2157–2163. doi: 10.1210/jc.2008-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie LE, Vessey K, Miller A, Ward MM, Pianta MJ, Vingrys AJ, Wilkinson-Berka JL, Fletcher EL. Neuronal and glial cell expression of angiotensin II type 1 (AT1) and type 2 (AT2) receptors in the rat retina. Neuroscience. 2009;161:195–213. doi: 10.1016/j.neuroscience.2009.02.084. [DOI] [PubMed] [Google Scholar]
- Dufner A, Pownall S, Mak TW. Caspase recruitment domain protein 6 is a microtubule-interacting protein that positively modulates NF-kappaB activation. Proc Natl Acad Sci USA. 2006;103:988–993. doi: 10.1073/pnas.0510380103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin R. An audience with Robert Dworkin. Nat Rev Drug Discov. 2011;10:570. doi: 10.1038/nrd3530. [DOI] [PubMed] [Google Scholar]
- Finnerup NB, Jensen TS. Mechanisms of disease: mechanism-based classification of neuropathic pain-a critical analysis. Nat Clin Pract Neurol. 2006;2:107–115. doi: 10.1038/ncpneuro0118. [DOI] [PubMed] [Google Scholar]
- Foulkes T, Wood JN. Pain genes. PLoS Genet. 2008;4:e1000086. doi: 10.1371/journal.pgen.1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchtbauer L, Groth-Rasmussen M, Holm TH, Lobner M, Toft-Hansen H, Khorooshi R, Owens T. Angiotensin II Type 1 receptor (AT1) signaling in astrocytes regulates synaptic degeneration-induced leukocyte entry to the central nervous system. Brain Behav Immun. 2011;25:897–904. doi: 10.1016/j.bbi.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Gao HM, Zhou H, Zhang F, Wilson BC, Kam W, Hong JS. HMGB1 acts on microglia Mac1 to mediate chronic neuroinflammation that drives progressive neurodegeneration. J Neurosci. 2011;31:1081–1092. doi: 10.1523/JNEUROSCI.3732-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MS, Gebhart GF. Nociceptor sensitization in pain pathogenesis. Nat Med. 2010;16:1248–1257. doi: 10.1038/nm.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace PM, Hutchinson MR, Manavis J, Somogyi AA, Rolan PE. A novel animal model of graded neuropathic pain: utility to investigate mechanisms of population heterogeneity. J Neurosci Methods. 2010;193:47–53. doi: 10.1016/j.jneumeth.2010.08.025. [DOI] [PubMed] [Google Scholar]
- Grace PM, Hutchinson MR, Bishop A, Somogyi AA, Mayrhofer G, Rolan PE. Adoptive transfer of peripheral immune cells potentiates allodynia in a graded chronic constriction injury model of neuropathic pain. Brain Behav Immun. 2011a;25:503–513. doi: 10.1016/j.bbi.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Grace PM, Rolan PE, Hutchinson MR. Peripheral immune contributions to the maintenance of central glial activation underlying neuropathic pain. Brain Behav Immun. 2011b;25:1322–1332. doi: 10.1016/j.bbi.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Griffin JW, George R, Lobato C, Tyor WR, Yan LC, Glass JD. Macrophage responses and myelin clearance during Wallerian degeneration: relevance to immune-mediated demyelination. J Neuroimmunol. 1992;40:153–165. doi: 10.1016/0165-5728(92)90129-9. [DOI] [PubMed] [Google Scholar]
- Griffin RS, Costigan M, Brenner GJ, Ma CH, Scholz J, Moss A, Allchorne AJ, Stahl GL, Woolf CJ. Complement induction in spinal cord microglia results in anaphylatoxin C5a-mediated pain hypersensitivity. J Neurosci. 2007;27:8699–8708. doi: 10.1523/JNEUROSCI.2018-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham J, Rees DA. The adenosine a2b receptor: its role in inflammation. Endocr Metab Immune Disord Drug Targets. 2008;8:244–254. doi: 10.2174/187153008786848303. [DOI] [PubMed] [Google Scholar]
- Hanash SM, Baik CS, Kallioniemi O. Emerging molecular biomarkers–blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol. 2011;8:142–150. doi: 10.1038/nrclinonc.2010.220. [DOI] [PubMed] [Google Scholar]
- Harvey LO. Efficient estimation of sensory thresholds. Behav Res Methods Istrum Comput. 1986;18:623–632. [Google Scholar]
- He WJ, Cui J, Du L, Zhao YD, Burnstock G, Zhou HD, Ruan HZ. Spinal P2X(7) receptor mediates microglia activation-induced neuropathic pain in the sciatic nerve injury rat model. Behav Brain Res. 2012;226:163–170. doi: 10.1016/j.bbr.2011.09.015. [DOI] [PubMed] [Google Scholar]
- von Hehn CA, Baron R, Woolf CJ. Deconstructing the neuropathic pain phenotype to reveal neural mechanisms. Neuron. 2012;73:638–652. doi: 10.1016/j.neuron.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans E, Challiss RA. Structural, signalling and regulatory properties of the group I metabotropic glutamate receptors: prototypic family C G-protein-coupled receptors. Biochem J. 2001;359:465–484. doi: 10.1042/0264-6021:3590465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann O, Zipp F, Weber JR. Tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) in central nervous system inflammation. J Mol Med (Berlin) 2009;87:753–763. doi: 10.1007/s00109-009-0484-x. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hyde D. Sorites Paradox. Zalta E, editor. [Accessed: 6 March 2012];The Stanford Encyclopedia of Philosphy. 2011 Online: http://plato.stanford.edu/archives/win2011/entries/sorites-paradox/
- Inoue H, Sawada M, Ryo A, et al. Serial analysis of gene expression in a microglial cell line. Glia. 1999;28:265–271. doi: 10.1002/(sici)1098-1136(199912)28:3<265::aid-glia10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TS, Baron R, Haanpaa M, Kalso E, Loeser JD, Rice AS, Treede RD. A new definition of neuropathic pain. Pain. 2011;152:2204–2205. doi: 10.1016/j.pain.2011.06.017. [DOI] [PubMed] [Google Scholar]
- Joller N, Weber SS, Oxenius A. Antibody-Fc receptor interactions in protection against intracellular pathogens. Eur J Immunol. 2011;41:889–897. doi: 10.1002/eji.201041340. [DOI] [PubMed] [Google Scholar]
- Jung H, Miller RJ. Activation of the nuclear factor of activated T-cells (NFAT) mediates upregulation of CCR2 chemokine receptors in dorsal root ganglion (DRG) neurons: a possible mechanism for activity-dependent transcription in DRG neurons in association with neuropathic pain. Mol Cell Neurosci. 2008;37:170–177. doi: 10.1016/j.mcn.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Lee SJ, Park SY, Yoo HJ, Kim SH, Kim KJ, Cho HJ. Differentially expressed genes in rat dorsal root ganglia following peripheral nerve injury. Neuro Report. 2001;12:3401–3405. doi: 10.1097/00001756-200110290-00050. [DOI] [PubMed] [Google Scholar]
- Kim H, Moon C, Ahn M, Byun J, Lee Y, Kim MD, Matsumoto Y, Koh CS, Shin T. Heat shock protein 27 upregulation and phosphorylation in rat experimental autoimmune encephalomyelitis. Brain Res. 2009;1304:155–163. doi: 10.1016/j.brainres.2009.09.060. [DOI] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Ko J, Na DS, Lee YH, Shin SY, Kim JH, Hwang BG, Min BI, Park DS. cDNA microarray analysis of the differential gene expression in the neuropathic pain and electro-acupuncture treatment models. J Biochem Mol Biol. 2002;35:420–427. doi: 10.5483/bmbrep.2002.35.4.420. [DOI] [PubMed] [Google Scholar]
- Kohara A, Nagakura Y, Kiso T, Toya T, Watabiki T, Tamura S, Shitaka Y, Itahana H, Okada M. Antinociceptive profile of a selective metabotropic glutamate receptor 1 antagonist YM-230888 in chronic pain rodent models. Eur J Pharmacol. 2007;571:8–16. doi: 10.1016/j.ejphar.2007.05.030. [DOI] [PubMed] [Google Scholar]
- Kronlage M, Song J, Sorokin L, et al. Autocrine purinergic receptor signaling is essential for macrophage chemotaxis. Sci Signal. 2010;3:ra55. doi: 10.1126/scisignal.2000588. [DOI] [PubMed] [Google Scholar]
- Kunori S, Matsumura S, Okuda-Ashitaka E, Katano T, Audoly LP, Urade Y, Ito S. A novel role of prostaglandin E2 in neuropathic pain: blockade of microglial migration in the spinal cord. Glia. 2011;59:208–218. doi: 10.1002/glia.21090. [DOI] [PubMed] [Google Scholar]
- LaCroix-Fralish ML, Tawfik VL, Spratt KF, DeLeo JA. Sex differences in lumbar spinal cord gene expression following experimental lumbar radiculopathy. J Mol Neurosci. 2006a;30:283–295. doi: 10.1385/JMN:30:3:283. [DOI] [PubMed] [Google Scholar]
- Lacroix-Fralish ML, Tawfik VL, Tanga FY, Spratt KF, DeLeo JA. Differential spinal cord gene expression in rodent models of radicular and neuropathic pain. Anesthesiology. 2006b;104:1283–1292. doi: 10.1097/00000542-200606000-00025. [DOI] [PubMed] [Google Scholar]
- Lacroix-Fralish ML, Austin JS, Zheng FY, Levitin DJ, Mogil JS. Patterns of pain: meta-analysis of microarray studies of pain. Pain. 2011;152:1888–1898. doi: 10.1016/j.pain.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Lathia CD, Amakye D, Dai W, et al. The value, qualification, and regulatory use of surrogate end points in drug development. Clin Pharmacol Ther. 2009;86:32–43. doi: 10.1038/clpt.2009.69. [DOI] [PubMed] [Google Scholar]
- Lee SE, Kim JH. Involvement of substance P and calcitonin gene-related peptide in development and maintenance of neuropathic pain from spinal nerve injury model of rat. Neurosci Res. 2007;58:245–249. doi: 10.1016/j.neures.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Lin YW, Tseng TJ, Lin WM, Hsieh ST. Cutaneous nerve terminal degeneration in painful mononeuropathy. Exp Neurol. 2001;170:290–296. doi: 10.1006/exnr.2001.7704. [DOI] [PubMed] [Google Scholar]
- Louvet C, Chiffoleau E, Heslan M, et al. Identification of a new member of the CD20/FcepsilonRIbeta family overexpressed in tolerated allografts. Am J Transplant. 2005;5:2143–2153. doi: 10.1111/j.1600-6143.2005.01007.x. [DOI] [PubMed] [Google Scholar]
- Ma W, Chabot JG, Powell KJ, Jhamandas K, Dickerson IM, Quirion R. Localization and modulation of calcitonin gene-related peptide-receptor component protein-immunoreactive cells in the rat central and peripheral nervous systems. Neuroscience. 2003;120:677–694. doi: 10.1016/s0306-4522(03)00159-3. [DOI] [PubMed] [Google Scholar]
- Malin SA, Davis BM, Koerber HR, Reynolds IJ, Albers KM, Molliver DC. Thermal nociception and TRPV1 function are attenuated in mice lacking the nucleotide receptor P2Y2. Pain. 2008;138:448–496. doi: 10.1016/j.pain.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior B, Garcia AE, Hsiung BK, et al. Dual induction of TREM2 and tolerance-related transcript, Tmem176b, in amyloid transgenic mice: implications for vaccine-based therapies for Alzheimer’s disease. ASN Neuro. 2010;2:e00037. doi: 10.1042/AN20100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miletic G, Sullivan KM, Dodson AM, Lippitt JA, Schneider JA, Miletic V. Changes in calcineurin message, enzyme activity and protein content in the spinal dorsal horn are associated with chronic constriction injury of the rat sciatic nerve. Neuroscience. 2011;188:142–147. doi: 10.1016/j.neuroscience.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan ED, Mehmert KK, Hinde JL, Harvey LO, Martin D, Tracey KJ, Maier SF, Watkins LR. Thermal hyperalgesia and mechanical allodynia produced by intrathecal administration of the human immunodeficiency virus-1 (HIV-1) envelope glycoprotein, gp120. Brain Res. 2000;861:105–116. doi: 10.1016/s0006-8993(00)02050-3. [DOI] [PubMed] [Google Scholar]
- Milligan ED, O’Connor KA, Nguyen KT, et al. Intrathecal HIV-1 envelope glycoprotein gp120 induces enhanced pain states mediated by spinal cord proinflammatory cytokines. J Neurosci. 2001;21:2808–2819. doi: 10.1523/JNEUROSCI.21-08-02808.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan ED, Sloane EM, Watkins LR. Glia in pathological pain: a role for fractalkine. J Neuroimmunol. 2008;198:113–120. doi: 10.1016/j.jneuroim.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogil JS, Wilson SG, Bon K, et al. Heritability of nociception I: responses of 11 inbred mouse strains on 12 measures of nociception. Pain. 1999a;80:67–82. doi: 10.1016/s0304-3959(98)00197-3. [DOI] [PubMed] [Google Scholar]
- Mogil JS, Wilson SG, Bon K, et al. Heritability of nociception II ‘Types’ of nociception revealed by genetic correlation analysis. Pain. 1999b;80:83–93. doi: 10.1016/s0304-3959(98)00196-1. [DOI] [PubMed] [Google Scholar]
- Mogil JS, Sorge RE, LaCroix-Fralish ML, et al. Pain sensitivity and vasopressin analgesia are mediated by a gene-sex-environment interaction. Nat Neurosci. 2011;14:1569–1573. doi: 10.1038/nn.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negorev DG, Vladimirova OV, Maul GG. Differential functions of interferon-upregulated Sp100 isoforms: herpes simplex virus type 1 promoter-based immediate-early gene suppression and PML protection from ICP0-mediated degradation. J Virol. 2009;83:5168–5180. doi: 10.1128/JVI.02083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH Biomarkers Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- Okabe Y, Kawane K, Nagata S. IFN regulatory factor (IRF) 3/7-dependent and -independent gene induction by mammalian DNA that escapes degradation. Eur J Immunol. 2008;38:3150–3158. doi: 10.1002/eji.200838559. [DOI] [PubMed] [Google Scholar]
- Persson AK, Gebauer M, Jordan S, et al. Correlational analysis for identifying genes whose regulation contributes to chronic neuropathic pain. Mol Pain. 2009;5:7. doi: 10.1186/1744-8069-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L, Zhang P, Lamotte RH, Ma C. Neuronal Fc-gamma receptor I mediated excitatory effects of IgG immune complex on rat dorsal root ganglion neurons. Brain Behav Immun. 2011;25:1399–1407. doi: 10.1016/j.bbi.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren K, Dubner R. Interactions between the immune and nervous systems in pain. Nat Med. 2010;16:1267–1276. doi: 10.1038/nm.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W, Neugebauer V. Pain-related increase of excitatory transmission and decrease of inhibitory transmission in the central nucleus of the amygdala are mediated by mGluR1. Mol Pain. 2010;6:93. doi: 10.1186/1744-8069-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riether C, Kavelaars A, Wirth T, Pacheco-Lopez G, Doenlen R, Willemen H, Heijnen CJ, Schedlowski M, Engler H. Stimulation of beta-adrenergic receptors inhibits calcineurin activity in CD4(+) T cells via PKA-AKAP interaction. Brain Behav Immun. 2011;25:59–66. doi: 10.1016/j.bbi.2010.07.248. [DOI] [PubMed] [Google Scholar]
- Ro LS, Chen S, Chao P, Lee Y, Kwok-Tong T. The potential application of granulocyte colony stimulating factor therapy on neuropathic pain. Chang Gung Med J. 2009;23:235–246. [PubMed] [Google Scholar]
- Rolan P. The contribution of clinical pharmacology surrogates and models to drug development–a critical appraisal. Br J Clin Pharmacol. 1997;44:219–225. doi: 10.1046/j.1365-2125.1997.t01-1-00583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolan P, Atkinson AJ, Jr, Lesko LJ. Use of biomarkers from drug discovery through clinical practice: report of the Ninth European Federation of Pharmaceutical Sciences Conference on Optimizing Drug Development. Clin Pharmacol Ther. 2003;73:284–291. doi: 10.1016/s0009-9236(02)17625-9. [DOI] [PubMed] [Google Scholar]
- Schmid CD, Melchior B, Masek K, Puntambekar SS, Danielson PE, Lo DD, Sutcliffe JG, Carson MJ. Differential gene expression in LPS/IFNgamma activated microglia and macrophages: in vitro versus in vivo. J Neurochem. 2009;109(Suppl 1):117–125. doi: 10.1111/j.1471-4159.2009.05984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Deng W, Bi E, et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB 2 and TAB 3 for degradation. Nat Immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- Simpson TR, Quezada SA, Allison JP. Regulation of CD4 T cell activation and effector function by inducible costimulator (ICOS) Curr Opin Immunol. 2010;22:326–332. doi: 10.1016/j.coi.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Smyth G. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry RA, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- Sperlagh B, Vizi ES, Wirkner K, Illes P. P2X7 receptors in the nervous system. Prog Neurobiol. 2006;78:327–346. doi: 10.1016/j.pneurobio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Suemitsu S, Watanabe M, Yokobayashi E, Usui S, Ishikawa T, Matsumoto Y, Yamada N, Okamoto M, Kuroda S. Fcgamma receptors contribute to pyramidal cell death in the mouse hippocampus following local kainic acid injection. Neuroscience. 2010;166:819–831. doi: 10.1016/j.neuroscience.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Sugimoto T, Morioka N, Sato K, Hisaoka K, Nakata Y. Noradrenergic regulation of period1 expression in spinal astrocytes is involved in protein kinase A, c-Jun N-terminal kinase and extracellular signal-regulated kinase activation mediated by alpha1- and beta2-adrenoceptors. Neuroscience. 2011;185:1–13. doi: 10.1016/j.neuroscience.2011.04.024. [DOI] [PubMed] [Google Scholar]
- Torrance N, Smith BH, Bennett MI, Lee AJ. The epidemiology of chronic pain of predominantly neuropathic origin Results from a general population survey. J Pain. 2006;7:281–289. doi: 10.1016/j.jpain.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Tracey I, Mantyh PW. The cerebral signature for pain perception and its modulation. Neuron. 2007;55:377–391. doi: 10.1016/j.neuron.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Treutwein B, Strasburger H. Fitting the psychometric function. Percept Psychophys. 1999;61:87–106. doi: 10.3758/bf03211951. [DOI] [PubMed] [Google Scholar]
- Tsukimoto M, Tokunaga A, Harada H, Kojima S. Blockade of murine T cell activation by antagonists of P2Y6 and P2X7 receptors. Biochem Biophys Res Commun. 2009;384:512–518. doi: 10.1016/j.bbrc.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Twining CM, Sloane EM, Schoeniger DK, Milligan ED, Martin D, Marsh H, Maier SF, Watkins LR. Activation of the spinal cord complement cascade might contribute to mechanical allodynia induced by three animal models of spinal sensitization. J Pain. 2005;6:174–183. doi: 10.1016/j.jpain.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Uceyler N, Rogausch JP, Toyka KV, Sommer C. Differential expression of cytokines in painful and painless neuropathies. Neurology. 2007;69:42–49. doi: 10.1212/01.wnl.0000265062.92340.a5. [DOI] [PubMed] [Google Scholar]
- Ulvestad E, Williams K, Vedeler C, Antel J, Nyland H, Mork S, Matre R. Reactive microglia in multiple sclerosis lesions have an increased expression of receptors for the Fc part of IgG. J Neurol Sci. 1994;121:125–131. doi: 10.1016/0022-510x(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Vega-Avelaira D, Geranton SM, Fitzgerald M. Differential regulation of immune responses and macrophage/neuron interactions in the dorsal root ganglion in young and adult rats following nerve injury. Mol Pain. 2009;5:70. doi: 10.1186/1744-8069-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verge GM, Milligan ED, Maier SF, Watkins LR, Naeve GS, Foster AC. Fractalkine (CX3CL1) and fractalkine receptor (CX3CR1) distribution in spinal cord and dorsal root ganglia under basal and neuropathic pain conditions. Eur J Neurosci. 2004;20:1150–1160. doi: 10.1111/j.1460-9568.2004.03593.x. [DOI] [PubMed] [Google Scholar]
- Walker-Brown J, Roberts MR. Differential contribution of beta-adrenergic receptors expressed on radiosensitive versus radioresistant cells to protection against inflammation and mortality in murine endotoxemia. Shock. 2009;32:541–547. doi: 10.1097/SHK.0b013e3181a6eda2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NM, Jung H, Ripsch MS, Miller RJ, White FA. CXCR4 signaling mediates morphine-induced tactile hyperalgesia. Brain Behav Immun. 2011;25:565–573. doi: 10.1016/j.bbi.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirkner K, Sperlagh B, Illes P. P2X3 receptor involvement in pain states. Mol Neurobiol. 2007;36:165–183. doi: 10.1007/s12035-007-0033-y. [DOI] [PubMed] [Google Scholar]
- Woodcock J. Chutes and ladders on the critical path: comparative effectiveness, product value, and the use of biomarkers in drug development. Clin Pharmacol Ther. 2009;86:12–14. doi: 10.1038/clpt.2009.33. [DOI] [PubMed] [Google Scholar]
- Woolf CJ, Decosterd I. Implications of recent advances in the understanding of pain pathophysiology for the assessment of pain in patients. Pain. 1999;(Suppl 6):S141–147. doi: 10.1016/S0304-3959(99)00148-7. [DOI] [PubMed] [Google Scholar]
- Woolf CJ, Mannion RJ. Neuropathic pain: aetiology, symptoms, mechanisms, and management. Lancet. 1999;353:1959–1964. doi: 10.1016/S0140-6736(99)01307-0. [DOI] [PubMed] [Google Scholar]
- Yang L, Zhang FX, Huang F, Lu YJ, Li GD, Bao L, Xiao HS, Zhang X. Peripheral nerve injury induces trans-synaptic modification of channels, receptors and signal pathways in rat dorsal spinal cord. Eur J Neurosci. 2004;19:871–883. doi: 10.1111/j.0953-816x.2004.03121.x. [DOI] [PubMed] [Google Scholar]
- Zin CS, Nissen LM, O’Callaghan JP, Moore BJ, Smith MT. Preliminary study of the plasma and cerebrospinal fluid concentrations of IL-6 and IL-10 in patients with chronic pain receiving intrathecal opioid infusions by chronically implanted pump for pain management. Pain Med. 2010;11:550–561. doi: 10.1111/j.1526-4637.2010.00821.x. [DOI] [PubMed] [Google Scholar]