Abstract
We describe here the synthesis of novel multivalent HIV V3 domain glycopeptides and their binding to broadly neutralizing antibodies PGT128 and 10-1074. Our binding data reveal a distinct mode of antigen recognition by the two antibodies and further suggest that multivalent glycopeptides could mimic the neutralizing epitopes more efficiently than the monomeric glycopeptide.
Graphical abstract
Synthetic bi- and trivalent HIV V3 glycopeptides show significantly enhanced affinity for HIV-neutralizing antibody 10-1074
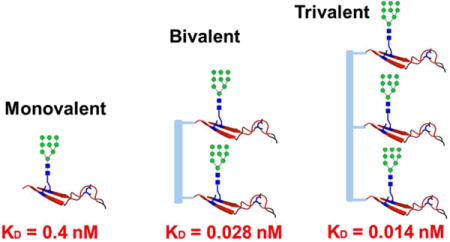
The HIV-1 envelope glycoprotein (Env) trimer is responsible for viral entry into host cells and is the primary target for vaccine design.1 The sequence variation and heavy glycosylation are two strong defense mechanisms for the virus to evade immune recognition, making it challenging to develop an effective vaccine.2, 3 Nevertheless, broadly neutralizing antibodies (bNAbs) do exist in sera from 10–30% of HIV-1 infected individuals, suggesting the possibility to raise neutralizing activities by vaccination.4 Recently, a series of glycan-reactive bNAbs have been isolated, including the PG9 and PG16 antibodies targeting the V1V2 domain,5, 6 and the PGT121-128 bNAb series targeting the V3 domain.7–10 Characterizing the epitopes of these bNAbs is a critical step for designing an effective vaccine against HIV-1.7, 11 However, the task has been hampered by the heavily heterogeneous glycosylation patterns of gp120 and the difficulty to obtain homogeneous immunogen glycoforms to study these epitopes. Toward this end, synthetic chemistry has showcased the power in defining the structural details of the glycan-associated epitopes of HIV-neutralizing antibodies.12–16 We have recently used a chemoenzymatic method to synthesize a library of homogeneous gp120 V1V2 glycopeptides for characterizing the glycan specificity and minimal epitopes of PG9 and PG16.17, 18. Danishefsky and co-workers identified a glycopeptide dimer as the preferred epitope of PG9 by applied chemical synthesis.19, 20 The PGT121-128 antibody series belongs to a new class of glycan dependent bNAbs that are more potent than PG9 and PG16.7 For example, PGT128 has shown great breadth in HIV neutralization,21 while 10-1074 has shown protection by passive immunization in animal models and HIV-1 infected individuals.22–25 Structural studies have revealed that these antibodies bind to the base of the gp120 V3 loop interaction with the N332 glycan that is highly conserved across the majority of HIV-1 isolates.9, 10 But so far, no synthetic glycopeptide antigens have been reported to mimic their epitopes. Since the HIV-1 gp120/gp41 Env subunits are present on the viral surface as a trimer,10 and since some of the V3-glycan specific antibodies have shown preference for native Env trimer over monomeric gp120,26 we hypothesized that multivalent V3 glycopeptides could be more efficient in recapitulating their epitopes.27 Herein, we report the synthesis of mono-, bi- and trivalent gp120 V3 glycopeptides as potential mimics of the V3 glycopeptide domains present in Env trimer. Our binding studies revealed a clearly different mode of antigen recognition of the glycopeptides by the PGT128 and 10-1074 antibodies. While PGT128 did not show preference among mono-, bi- and trivalent V3 glycopeptides, the 10-1074 showed dramatically enhanced binding to the bi- and trivalent V3 glycopeptide over the monomer, suggesting that the trivalent V3 glycopeptide could better mimic the actual epitope of bNAb 10-1074.
We first selected the mini-V3 glycopeptide derived from the JR-FL strain as the base glycopeptide with a deletion of the highly variable tip sequence, which allowed the high-resolution structure determination of the recombinant gp120 outer domain in complex with PGT128.21 A Man9GlcNAc2 glycan was placed at the conserved N332 site as previously structural study and our recent glycopeptide mapping data indicated that PGT128 and 10-1074 antibodies recognize high-mannose glycan at N332.8, 21, 28 A chemoenzymatic method was used to synthesize the cyclic V3 glycopeptide subunit carrying defined N-glycans, and the copper (I)-catalyzed alkyne-azide 3+2 cycloaddition (click chemistry) was employed to assemble multivalent glycopeptides on a peptide scaffold. Synthesis of the mini-V3 glycopeptide is depicted in Scheme 1. The precursor cyclic polypeptide 1 was synthesized by automated solid-phase peptide synthesis (SPPS), in which a GlcNAc moiety was placed at the pre-determined N332 site using Fmoc-Asn-(Ac3GlcNAc)-OH as a building block. An alkyne moiety was placed at the N-terminus of the peptide to allow site-specific ligation to a scaffold for multivalent presentation. In addition, a biotin tagged precursor 2 was synthesized to facilitate site-specific immobilization for binding analysis. The Man9GlcNAc glycan was prepared from Man9GlcNAc2Asn and converted to the activated glycan oxazoline 3 following previously reported procedure.29 The endoglycosidase mutant EndoA-N171A was used to transfer the high-mannose glycan to the precursor 1 and 2 to form natural glycosidic linkage.30 The corresponding alkyne-glycopeptide 4 and biotin-glycopeptide 5 were obtained in 87% and 89% yields after HPLC purification.
Scheme 1.
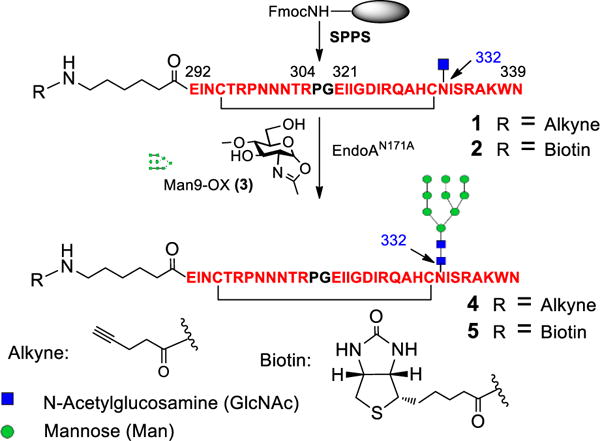
Chemoenzymatic synthesis of HIV-1 JR-FL mini-V3 glycopeptide carrying a high-mannose glycan at N332 site.
A linear bivalent scaffold 6 bearing two Lys(N3) residues and a biotin tag was synthesized by SPPS (Scheme 2). After HPLC purification, the azide group was ligated to the V3 glycopeptide 4 by click chemistry to afford the bivalent glycopeptide 7 in 68% yields. Similarly, a linear trivalent scaffold 8 bearing three Lys(N3) residues was also synthesized and ligated with glycopeptide 4 to afford the trivalent glycopeptide 9 in 43% yield. The purity and identity of the bi- and trivalent glycopeptides (7 and 9) were characterized by analytical HPLC and ESI-MS analysis (Supporting Information).
Scheme 2.
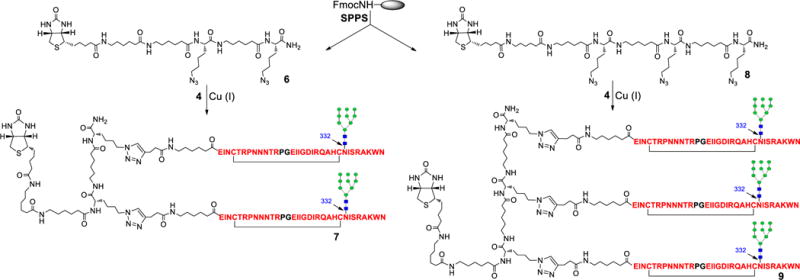
Synthesis of JR-FL mini-V3 bivalent glycopeptide antigen 7 and trivalent glycopeptide antigen 9.
The binding of the synthetic glycopeptides to the Fab domain of PGT128 and 10-1074 antibodies was first evaluated by SPR analysis. The biotin-tagged glycopeptide antigens (5, 7 and 9) were captured on Neutravidin coated CM5 chips to an identical response unit (RU), with almost identical amount of glycopeptides on the binding surface. The Fabs of PGT128 and10-1074 were run as analytes to evaluate the binding. Consistent with previous ELISA analysis,26 we did not observe significant binding preference for mono-, bi- and trivalent glycopeptides (5, 7 and 9) by the PGT128 Fab, which showed similar affinities with quick on and off rates (Figure 1a–c). The PGT128 IgG showed only slightly enhanced (2–3 fold) binding affinity for the bi- and trivalent glycopeptides 7 and 9 (KD = 1.5 and 1.1 μM, respectively) over the monovalent glycopeptide 5 (KD = 3.7 μM), probably due to a moderate avidity effect in the antibody-antigen interactions (Figure S1).
Figure 1.
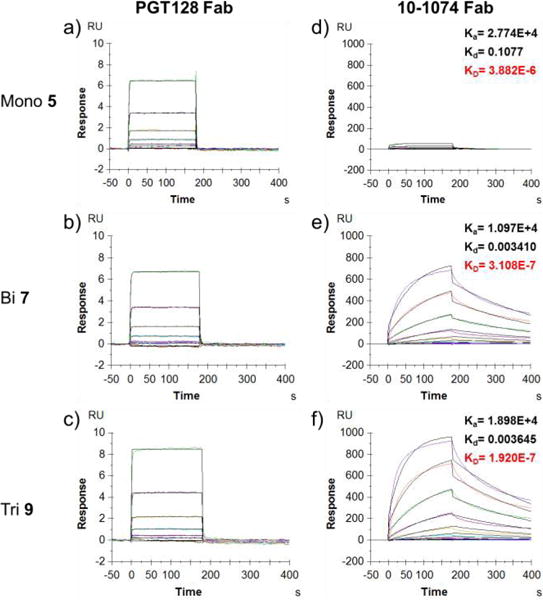
SPR sensorgrams of binding between mono-, bi- and trivalent glycopeptides (5, 7 and 9) and antibody Fab fragments. a–c: with PGT128 Fab; d-f: with 10-1074 Fab. The Fab fragments were run from 1000 nM with 1:2 serial dilutions. Data were fit with a 1:1 Langmuir binding model (fitting was shown in black). Ka was given in M−1S−1; Kd in S−1 and KD in M.
The 10-1074 Fab displayed weak binding to the monovalent glycopeptide 5 (KD = 3.88 μM), but showed enhanced binding affinity to the bi- and trivalent glycopeptide antigens (7 and 9, KD = 0.31 and 0.19 μM, respectively) (Figure 1d–f). This corresponded to a 13 and 20-fold enhancement of affinity over the monovalent glycopeptide 5. Interestingly, there was no significant difference in the binding affinities of 10-1074 Fab between the bi- and trivalent glycopeptides (7 and 9). Since the Fab binding could eliminate the avidity effect, these results suggest that two V3 glycopeptide units in the bi- or trivalent glycopeptides were most likely involved in the interaction with the 10-1074 Fab. The binding of the 10-1074 IgG was also consistent with that of the Fab binding (see Figure S2). For both 10-1074 Fab and IgG binding, the enhancement of the overall affinities for the bi- and trivalent glycopeptides (7 and 9) were contributed mainly by a decrease in the off rates. The slow dissociation for the bi- and trivalent glycopeptides further suggested that 10-1074 Fab is interacting with two distinct binding sites to accommodate the glycopeptides, indicating an apparent multivalent effect in antigen recognition.31, 32
The SPR binding results were verified by ELISA analysis (Figure 2). The PGT128 antibody showed only a moderate enhancement in the binding to bi- and trivalent glycopeptides (7 and 9) over the monovalent glycopeptide 5 (Figure 2a). However, the binding of 10-1074 IgG was greatly enhanced with the bi- and trivalent glycopeptides. The EC50 values for the bi- and trivalent glycopeptides (7 and 9) were more than 25-fold lower than that of the monovalent glycopeptide 5. Significant enhancement of binding by bi- and trivalent antigens was also observed in ELISA analysis using the 10-1074 Fab (Figure 2c). Taken together, both the SPR and ELISA analyses suggested that the bi- and trivalent glycopeptides recapitulated the 10-1074 epitope better than the monovalent glycopeptide.
Figure 2.
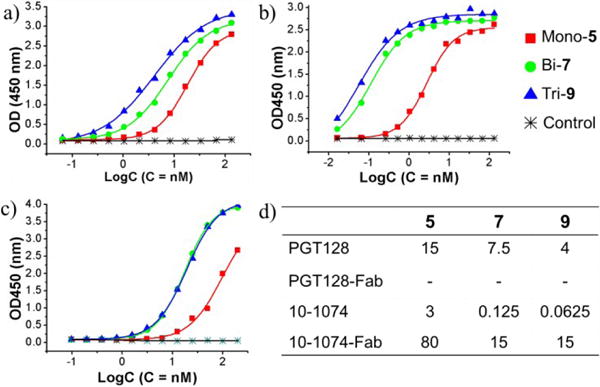
ELISA analysis of the binding between mono-, bi- and trivalent glycopeptides (5, 7 and 9) and antibody IgGs. a) with PGT128 IgG; b) with 10-1074 IgG; c) with 10-1074 Fab; d) EC50 values (nM). Dash denoted no binding.
For comparative analysis, we also synthesized the corresponding non-glycosylated trivalent peptide 10 (Scheme S1) and the trivalent high-mannose glycan 11 (Scheme S2), respectively. Consistent with previous array results,8 both 10-1074 Fab and IgG did not recognize the nonglycosylated trivalent peptide, neither did 10-1074 bind with the trivalent high-mannose glycan structures without the V3 peptide portions (Figure. 3). We also synthesized monovalent and trivalent JR-FL V3 glycopeptides carrying a sialylated complex N-glycan at the N332 site, and performed its binding for the 10-1074 antibody. Under the same conditions, no binding was observed for both monomeric and trimeric forms of the complex type glycopeptides (data not shown). This result implied that both the high-mannose N-glycan and the V3 peptide elements were critical for the recognition by antibody 10-1074. For PGT128, the Fab did not show any binding to the trivalent peptide 10 and the trivalent high-mannose glycan 11 under the same binding conditions (Figure 3). But weak recognition to the trivalent high-mannose glycan was detected by the PGT128 IgG (Figure S3). To assess whether the observed distinct binding mode of PGT128 and 10-1074 is generalizable to other HIV-1 strains, we synthesized mono-, bi- and trivalent V3 glycopeptides derived from the CN54 strain, carrying a high-mannose N-glycan at the conserved N332 site of gp120 (Scheme S3–5). Similar to the binding with the JR-FL glycopeptides, the 10-1074 Fab displayed significantly enhanced binding affinity for the bi- and trivalent glycopeptides (13 and 14, KD = 0.32 and 0.21 μM, respectively) over the monovalent glycopeptide 12 (KD = 4.96 μM), indicating a significant binding preference of 10-1074 for the bi- and trivalent glycopeptide (Figure S4). For PGT128, again no significant binding preference among mono-, bi- and trivalent glycopeptides was observed. Taken together, the binding data from both the JR-FL and CN54 V3 glycopeptides indicate that the multivalent presentation could dramatically enhance the affinity of 10-1074, however, the PGT128 does not show preference to multivalent antigen presentation. For PGT128, previous structural studies showed that mostly the heavy chain interacts with the V3 loop.21 On the other hand, a recent X-ray structural analysis of 10-1074 Fab in complex with an Env trimer (BG505 SOSIP.664) indicates that both the heavy and light chains are involved in the primary interaction with the N332 high-mannose N-glycan and the “GDIR” peptide motif in the gp120 V3 loop, together with secondary or minimal interactions with the N301 and/or N156 glycans.10 In addition, the X-ray crystal structure study of PGT124 antibody, a homologous antibody of 10-1074, showed similar mode of recognition with both the chains of the antibody.33 Our binding analysis with the well-defined synthetic glycopeptides appear to be consistent with these structural studies in that a high-affinity binding of 10-1074 requires both the high-mannose N-glycan at N332 in the context of the V3 polypeptide. We also performed an ELISA binding of the JR-FL Env trimer and gp120 with 10-1074 Fab. Our preliminary binding analysis indicated that 10-1074 showed comparable binding affinity to the Env trimer and the gp120 (Figure S5). This result implies that the synthetic multivalent glycopeptides (7 and 9) may position glycans within the context of the multivalent scaffold at proximity, so that the 10-1074 might be able to accommodate an additional N-glycan from another glycopeptide domain to enhance the affinity. Antibody 10-1074 is a highly potent anti-HIV-1 antibody with broadly neutralizing activities. Recent clinical trials have demonstrated that 10-1074 could efficiently suppress viremia in HIV-1-infected individuals.25 On the other hand, a recent report has shown that immunization with a synthetic mini-V3 glycopeptide is able to induce glycan-dependent antibodies in macaques.34 Thus, the identification of a high-affinity antigenic assembly that also offers the desired glycan homogeneity as demonstrated by the synthetic bi- and trivalent V3 glycopeptides constitutes a critical step for a successful vaccine design.
Figure 3.
SPR sensorgrams of the binding between antibody Fab fragments and nonglycosylated trivalent V3 peptide or trivalent glycans. a) with trivalent V3 peptide (10); b) with trivalent Man9GlcNAc2 glycan (11). Fabs were run from 1000 nM with 1:2 serial dilutions. Data were fit with a 1:1 Langmuir binding model (fitting was shown in black). Ka was given in M−1S−1; Kd in S−1 and KD in M.S1)
In conclusion, the chemoenzymatic synthesis of the homogeneous V3 glycopeptide and its bi- and trivalent forms has enabled the characterization of the glycan specificity and subunit requirement in antigen recognition by two broadly HIV-neutralizing antibodies, PGT128 and 10-1074. These studies have revealed that both antibodies recognize an essential V3 glycopeptide subunit with a high-mannose N-glycan installed at N332 site. In contrast to PGT128, the remarkably enhanced affinity of 10-1074 to the bi- and trivalent V3 glycopeptides suggest that the synthetic multivalent V3 glycopeptides may efficiently mimic the neutralizing epitope of the broadly neutralizing antibody on viral surface, which provide a valuable component for vaccine design. Immunization of a synthetic glycoconjugate vaccine based on the trivalent glycopeptide structure is in progress and the results will be reported in due course.
Supplementary Material
Acknowledgments
We thank Prof. Pamela Bjorkman for discussions during the work and her lab members for kindly providing PGT128, 10-1074 and the Fab proteins. This work was supported by the National Institutes of Health (NIH grant R01AI113896).
Footnotes
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Julien JP, et al. Science. 2013;342:1477. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pancera M, et al. Nature. 2014;514:455. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart-Jones GB, et al. Cell. 2016;165:813. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X, et al. Science. 2010;329:856. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker LM, et al. Science. 2009;326:285. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLellan JS, et al. Nature. 2011;480:336. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker LM, et al. Nature. 2011;477:466. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mouquet H, et al. Proc Natl Acad Sci, USA. 2012;109:E3268. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong L, et al. Nat Struct Mol Biol. 2013;20:796. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gristick HB, et al. Nat Struct Mol Biol. 2016;23:906. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulp DW, Schief WR. Curr Opin Virol. 2013;3:322. doi: 10.1016/j.coviro.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unverzagt C, Kajihara Y. Chem Soc Rev. 2013;42:4408. doi: 10.1039/c3cs35485g. [DOI] [PubMed] [Google Scholar]
- 13.Murakami M, Kiuchi T, Nishihara M, Tezuka K, Okamoto R, Izumi M, Kajihara Y. Sci Adv. 2016;2:e1500678. doi: 10.1126/sciadv.1500678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shivatare SS, et al. Nat Chem. 2016;8:338. doi: 10.1038/nchem.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang LX. Curr Opin Chem Biol. 2013;17:997. doi: 10.1016/j.cbpa.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horiya S, MacPherson IS, Krauss IJ. Nat Chem Biol. 2014;10:990. doi: 10.1038/nchembio.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amin MN, McLellan JS, Huang W, Orwenyo J, Burton DR, Koff WC, Kwong PD, Wang LX. Nat Chem Biol. 2013;9:521. doi: 10.1038/nchembio.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toonstra C, Amin MN, Wang LX. J Org Chem. 2016;81:6176. doi: 10.1021/acs.joc.6b01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aussedat B, et al. J Am Chem Soc. 2013;135:13113. doi: 10.1021/ja405990z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alam SM, et al. Proc Natl Acad Sci USA. 2013;110:18214. [Google Scholar]
- 21.Pejchal R, et al. Science. 2011;334:1097. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautam R, et al. Nature. 2016;533:105. doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein F, et al. Nature. 2012;492:118. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shingai M, et al. Nature. 2013;503:277. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caskey M, et al. Nat Med. 2017;23:185. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longo NS, et al. J Virol. 2016;90:10574. doi: 10.1128/JVI.01012-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fasting C, Schalley CA, Weber M, Seitz O, Hecht S, Koksch B, Dernedde J, Graf C, Knapp EW, Haag R. Angew Chem Int Ed. 2012;51:10472. doi: 10.1002/anie.201201114. [DOI] [PubMed] [Google Scholar]
- 28.Orwenyo J, Cai H, Giddens J, Amin MN, Toonstra C, Wang LX. ACS Chem Biol. 2017 doi: 10.1021/acschembio.7b00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umekawa M, Huang W, Li B, Fujita K, Ashida H, Wang LX, Yamamoto K. J Biol Chem. 2008;283:4469. doi: 10.1074/jbc.M707137200. [DOI] [PubMed] [Google Scholar]
- 30.Huang W, Li C, Li B, Umekawa M, Yamamoto K, Zhang X, Wang LX. J Am Chem Soc. 2009;131:2214. doi: 10.1021/ja8074677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaufman EN, Jain RK. Cancer Res. 1992;52:4157. [PubMed] [Google Scholar]
- 32.Mammen M, Choi SK, Whitesides GM. Angew Chem Int Ed Engl. 1998;37:2754. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 33.Garces F, et al. Cell. 2014;159:69. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alam SM, et al. Sci Transl Med. 2017;9:eaai7521. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


