 Spiropyran-decorated zwitterionic hydrogel realizes reversible manipulation of mesenchymal stem cell fate through programmed light exposure.
Spiropyran-decorated zwitterionic hydrogel realizes reversible manipulation of mesenchymal stem cell fate through programmed light exposure.
Abstract
Specific protein-cell and drug-cell interactions have been widely used to manipulate stem cell fate. Despite extensive studies, most current platforms cannot realize reversible manipulation of stem cell differentiation. In this work, we report a photodynamic zwitterionic hydrogel capable of reversibly triggering and suspending the differentiation process via manipulating nonspecific interactions between cultured stem cells and the hydrogel. The differentiation state of stem cells can be altered by exposing the hydrogel to a selected light program, while differentiation can be immediately suspended when near-infrared exposure converts the hydrogel into a purely zwitterionic form. While many other studies apply specific interactions to control stem cell fate, this work provides a completely different approach—allowing reversible, real-time and localized manipulation of stem cell fate choice via nonspecific interactions.
Introduction
Hydrogels with advanced integrated functions have numerous potential applications in the biomedical field, especially for stem cell-based tissue engineering.1–3 Strategies to provide ‘smart’ capabilities to hydrogels primarily seek to achieve matrices that are instructive to neighboring stem cells, or that stimulate desired stem cell responses crucial to tissue regeneration processes.4,5 The ability to control the biophysical and biochemical properties of a hydrogel through an external stimulus is highly desirable and actively studied.5,6 Conventional smart hydrogels can be actuated via a change in pH, temperature or ionic strength as a bulk material.7,8 However, the most desired type of scaffold would realize dynamic heterogeneity like natural tissues, which is unrealistic to achieve using the bulk stimuli mentioned above.9 Unlike bulk stimuli-responsive hydrogel systems, light-induced hydrogels have numerous advantages—specific wavelengths can be delivered with high spatial and temporal precision, no chemical contamination is introduced, and closed systems such as hydrogel-cell constructs can be conveniently and easily actuated.9–13 The biophysical properties of photoresponsive hydrogels can be tuned through photolysis by utilizing photoscissile bonds11,14–16 or by means of bulk photodegradation.17–21 Moreover, the biochemical attributes of photoresponsive hydrogels can be manipulated by conjugating signaling ligands via site-specific photoconjugation chemistries such as thiol–ene addition,10,12,22 Michael-type chemistry23,24 and photo-induced click cycloaddition25 with the aim to upregulate specific stem cell activity pathways. These biophysical and biochemical manipulations can also be employed simultaneously, providing dynamic environments with the scope to answer fundamental questions about material regulation of live cell functions and advancing applications from drug delivery platforms to tissue engineering systems.11 Investigations of photoswitchable molecules including azobenzenes,26 stilbenes,27 spiropyrans,28,29 diarylethenes,30 fulgides,31 and others,32–34 have built a helpful toolbox employable for the construction of light-responsive systems and materials. However, none of these aforementioned light-induced materials enable both reversible photocleavage/conjugation reactions29 and restraint of stem cell differentiation. This makes it difficult to realize continuously programmed manipulation of stem cell fate and to provide a reference state from which biophysical and biochemical cues can be introduced and studied.
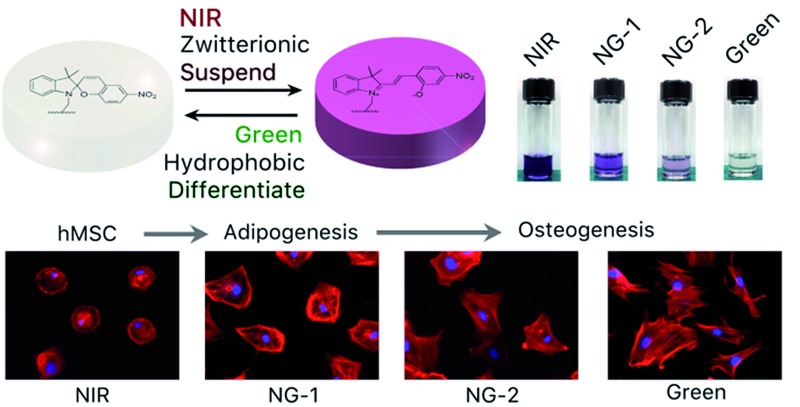
In our previous study, ultra-low fouling zwitterionic poly(carboxybetaine)-based materials were found to be capable of restraining human mesenchymal stem cell (hMSC) differentiation, credited to the complete elimination of nonspecific interactions with cells.35–41 This finding encouraged us to develop a zwitterionic hydrogel platform responsive to external stimuli and utilize this platform and its unique reference state to explore the influence of nonspecific interactions on stem cell differentiation behavior. Spiropyran is a unique photoresponsive molecule we hypothesized would complement this platform, as its two isomers have vastly different properties. It can assume a zwitterionic hydrophilic form (merocyanine, MC), a closed-ring hydrophobic form (spiropyran, SP), or switch between these continuously and reversibly when exposed to invisible near-infrared (NIR) or visible green light.29 In a polar solution42 such as water, exposure to NIR light is able to convert SP moieties into MC moieties via two-photon excitation (TPE), while visible green light converts MC moieties back to SP moieties via single-photon excitation (SPE).43
In this work, a dynamic photoresponsive zwitterionic hydrogel containing spiropyran was developed, comprising a copolymer of static zwitterionic monomer carboxybetaine acrylamide (CBAA) and photoswitchable monomer spiropyran methacrylate (SPMA), crosslinked with zwitterionic carboxybetaine dimethacrylate (CBDMA). Prior to gel formation, CBAA was functionalized with cellular recognition peptide cyclic-RGD (cRGD) to mediate stem cell adhesion. As we have previously demonstrated, the cRGD sequence allows functionalized CB molecules to maintain a neutral charge and remain highly resistant to nonspecific protein binding.41 HMSCs were encapsulated within the described hydrogel during gel formation. The resulting cell-hydrogel constructs were then incubated in a bipotential (osteogenic and adipogenic) differentiation medium under photoirradiation by two constructively interacting NIR and green lights. Tuning the strength of each light enables the balance between hydrophobic SP and hydrophilic MC moieties in the construct to be deliberately modulated. Consequently, nonspecific interactions between hMSCs and their culture platform can be spatially and temporally regulated. The results show that differentiation restraint, lineage programming, and precise spatiotemporal control of differentiated hMSCs can be achieved with this photoactuated zwitterionic hydrogel platform.
Results and discussion
Hydrogel characterization
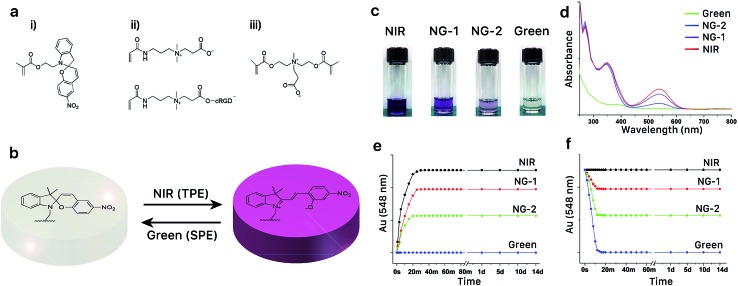
Synthesis of the photoswitchable SPMA monomer is described in the ESI and presented in ESI Fig. S1.† 44,45 A hydrogel was constructed with monomers, CBAA, cRGD-functionalized CBAA, SPMA and crosslinker CBDMA, as shown in Fig. 1a. The photochromic mechanism and properties of the synthesized hydrogel were evaluated and are presented in Fig. 1b–f. As illustrated in Fig. 1b, we speculated the hydrogel could be dynamically manipulated between two different states via exposure to different wavelengths of light. To test this, we exposed the hydrogel to infrared light (800 nm, 50 mW), green light (560 nm, 50 mW), or a combination of these wavelengths at different powers (50 mW infrared with 10 mW green (NG-1) or 50 mW infrared with 30 mW green (NG-2)) for 14 days. Photographs of hydrogels after exposure to different light programs are shown in Fig. 1c; the hydrogels appeared starkly different despite their chemically identical initial compositions. Hydrogels remained colorless under pure green light exposure, developed a dark purple color under pure NIR, and displayed intermediate hues under constructively interacting NIR and green light dependent on the intensity of each wavelength. The UV-Vis absorption spectra presented in Fig. 1d corroborated this MC ⇌ SP balance. NIR exposure converted SP to MC and maximized absorbance at the characteristic MC wavelength (548 nm), while green light reversed this conversion and diminished absorption at 548 nm. This dynamic photochromism process is summarized in Fig. 1e and f. As shown, the MC ⇌ SP conversion is stable (up to 14 days) and can be completed within 30 minutes. In addition, as presented in ESI Fig. S2,† this photoswitch is reversible through at least 10 NIR-green cycles. Notably, the mechanical and swelling properties of the hydrogels did not change significantly after different wavelength exposures, suggesting that this photochromism has little influence on the physical properties of the reported hydrogel (ESI Fig. S3†).
Fig. 1. (a) Photoresponsive hydrogel components spiropyran methacrylate (i, SPMA), carboxybetaine acrylamide (ii, CBAA), cRGD-functionalized CBAA (iii) and crosslinker carboxybetaine dimethacrylate (CBDMA). (b) Reversible photochromism of spiropyran between its hydrophobic spiropyran (SP) and hydrophilic merocyanine (MC) forms. (c) Representative photograph of photodynamic hydrogels receiving NIR, NG-1, NG-2 and green light exposure. (d) UV-Vis absorption spectra of photodynamic hydrogels receiving NIR, NG-1, NG-2 and green light exposure. (e and f) Photodynamic process of the MC ⇌ SP conversion in hydrogels receiving different light exposures starting from either a green hydrogel (e) or NIR hydrogel (f).
In our previous studies, we have demonstrated the ultra-low fouling properties of polyCBAA, cRGD-functionalized polyCBAA and polyCBDMA.35,41 To examine the nonspecific interactions between SPMA and cells or culture media, we herein used surface plasmon resonance (SPR) to measure protein adsorption onto a polySPMA film from hMSC lysate and fetal bovine serum (FBS). The SPMA was grafted from a gold chip via surface-initiated atom transfer radical polymerization (ATRP) under previously reported conditions (ESI Fig. S4a†). We exposed the pSPMA films to NIR, NG-1, NG-2 and green light for one hour before evaluation, and found their resistance to nonspecific protein binding to differ starkly after each exposure condition. (ESI Fig. S4b and c†). While pSPMA films exhibited low protein fouling after exposure to pure NIR light, increased fouling was observed when the green wavelength was added and the highest fouling seen after pure green light exposure. This demonstrates that nonspecific interactions on SP-based substrates are highly dependent on the light system applied to them.
We proceeded to encapsulate hMSCs in the photodynamic hydrogel. The hydrogel-hMSC constructs were placed in bipotential media and cells were allowed to grow for 14 days under continuous exposure to pure NIR, NG-1, NG-2, or pure green light. As presented in ESI Fig. S5,† the encapsulated hMSCs exposed to each light condition retained nearly 100% viability over the 14 days. To explore their differentiation behavior, we then examined the expression of characteristic surface antigens, presence of histological markers, and characteristic mRNA production.
3D differentiation
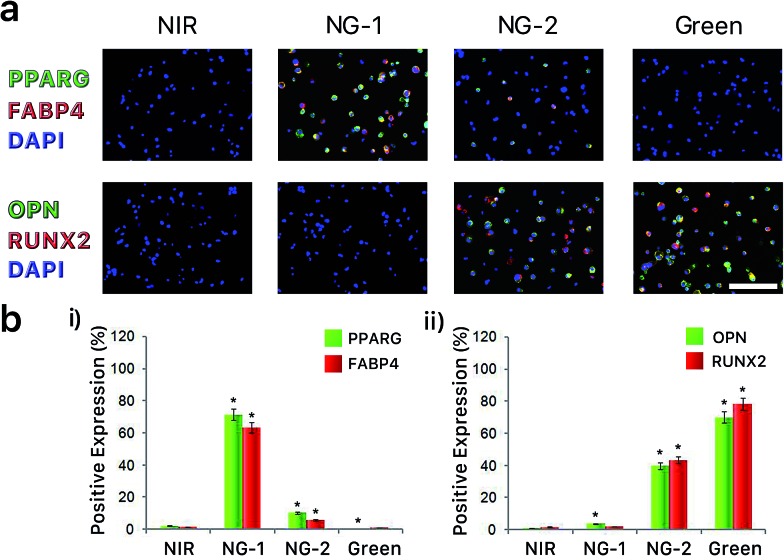
To examine the differentiation behavior of hMSCs encapsulated in photoactuated gels, we first assessed the expression of surface antigens characteristic to adipocyte (PPARG and FABP4) and osteocyte (OPN and RUNX2) lineages. When hMSC-hydrogel constructs were incubated under pure NIR exposure, neither adipogenic nor osteogenic surface antigens were observed (Fig. 2a). This suggests the differentiation potential of hMSCs is restrained when nonspecific interactions between the matrix and cells are eliminated, which is consistent with our previous finding.41 When nonspecific interactions were slightly enhanced under NIR-dominant mixed wavelength (NG-1) exposure, we observed clear expression of adipogenic surface antigens (PPARG and FABP4) by cells within the hydrogel. However, this adipogenic antigen expression was largely absent under green-dominant mixed (NG-2) and pure green photoirradiation, whereas osteogenic surface antigen (RUNX2 and OPN) expression was heightened when nonspecific interactions were further increased in these conditions (Fig. 2a). Quantitative results are summarized in Fig. 2b.
Fig. 2. (a) After 14 days of culture in bipotential differentiation media, cell-hydrogel constructs exposed to NIR, NG-1, NG-2 and green light conditions were immunostained for adipogenic (PPARG, FABP4) and osteogenic (RUNX2, OPN) biomarkers. Scale bar, 50 μm. (b) Percentage of cells expressing either adipogenic (i) or osteogenic (ii) biomarkers when encapsulated in hydrogels after exposure to different light conditions during a 14 day incubation in bipotential differentiation media. Asterisks denote statistical significance compared with NIR hydrogels (**p < 0.001, t-test). Error bars represent standard error of the mean from 5 individual experiments.
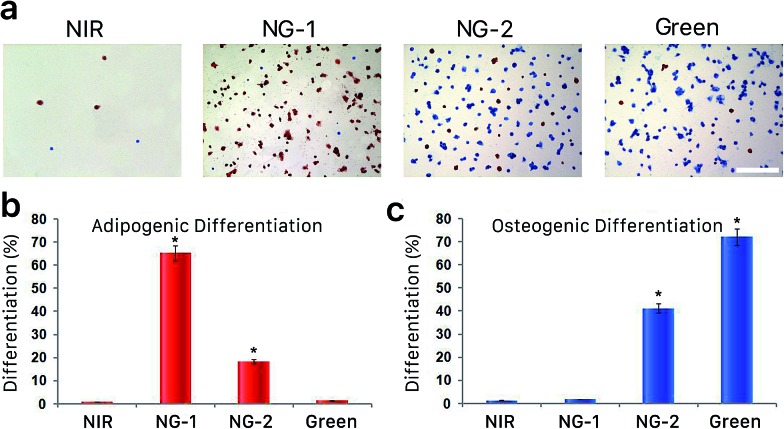
We further evaluated encapsulated hMSC differentiation via histological staining for neutral lipids (indicating adipogenesis) with Oil Red O, and for alkaline phosphatase (ALP, an osteogenic marker) with Fast Blue salt. These results, shown in Fig. 3a, validate our IHC analysis; neither marker was prominently found in the NIR-exposed constructs, while supplemental green light progressively increased nonspecific binding sites and initiated differentiation. The encapsulated hMSCs were prone to choosing an adipogenic lineage when fewer nonspecific interactions were available (NG-1) but increasingly committed to osteogenesis as more nonspecific binding was possible (NG-2 and green). Fig. 3b and c shows quantitative expression of the stained markers after 14 days of culture, and the dynamic differentiation rates observed under each illumination system are presented in ESI Fig. S6.†
Fig. 3. (a) Representative bright-field images of Oil Red O and Fast Blue salt-stained hMSC-hydrogel constructs following a 14 day incubation in bipotential differentiation media while exposed to different light conditions. Scale bar, 100 μm. Percentage of cells expressing neutral lipids (b) and alkaline phosphatase (ALP, c). Asterisks denote statistical significance compared with NIR hydrogels (**p < 0.05, t-test). Error bars represent standard error of the mean from 5 individual experiments.
In addition, we quantified the expression of characteristic mRNA via qRT-PCR (ESI Fig. S7†) and observed the same trend found by IHC analysis and histological staining. We also assessed the influence of cytoskeletal manipulation on differentiation trends under different light conditions, and these results are summarized in ESI Fig. S8 with relevant discussion in the ESI.†
2D differentiation
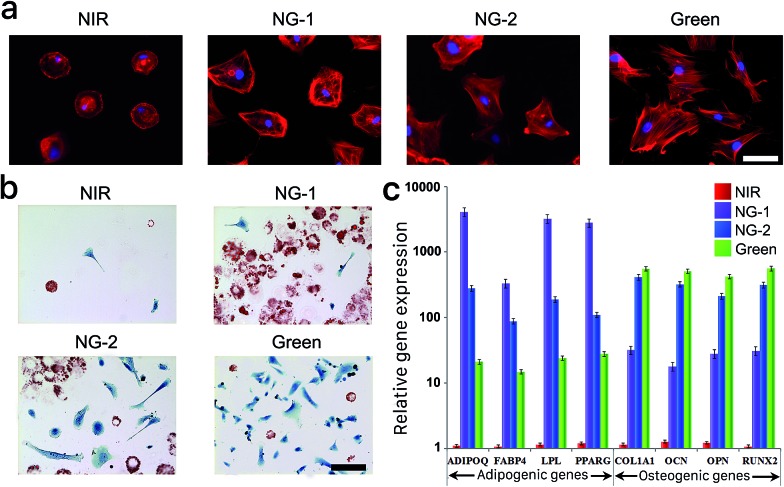
The differentiation behavior of hMSCs cultured in 3D photoactuated hydrogels demonstrates the critical role played by nonspecific binding in signal transfer and therefore the fate of hMSCs grown in these environments. Compared to 3D culture, seeding hMSCs on a 2D surface gives them more room to expand and grow; we therefore cultured hMSCs on the surface of photoswitchable hydrogels under the same light and media conditions described above to further explore their morphologies and lineage commitment. The fluorescent micrographs in Fig. 4a demonstrate the morphological differences we observed in 2D culture resulting from each light condition. The hMSCs retained their compact round shape under NIR exposure, as the nonfouling substrate greatly inhibited spreading. In contrast, the cells developed larger spread morphologies under the other light conditions, suggesting that hMSCs cultured on the surface of the reported hydrogel retained their stem cell phenotype under NIR exposure and committed to differentiated phenotypes under visible light. Based on the larger round adipocyte-like shape observed under NG-1 exposure and outstretched osteoblast morphology presented under NG-2 and green light systems (Fig. 4a), we speculated that the cells favored adipogenesis under weak visible light and osteogenesis under stronger green illumination as observed in 3D culture experiments.
Fig. 4. (a) Representative fluorescent micrographs of hMSCs stained with TRITC-phalloidin (red) and DAPI (blue) grown under different light conditions. Scale bars, 15 μm. (b) Representative bright-field images of Oil Red O- and Fast Blue salt-stained hMSCs grown under the same set of light conditions as in (a). Scale bars, 100 μm. (c) Adipogenic and osteogenic gene activities of hMSCs cultured under each light condition. The expression of all osteogenic and adipogenic genes was significantly different between hydrogels exposed to each light condition (*p < 0.05, t-test). All cells presented in this figure were grown on the surface of the photoresponsive gel for 14 days in bipotential differentiation media. Error bars represent standard error of the mean from 5 individual experiments.
We repeated histological staining of the surface-cultured cells grown under each light condition, with representative images presented in Fig. 4b. HMSCs cultured under visible light differentiated predictably according to the degree of nonspecific interaction possible, committing to adipogenesis under weaker green light intensity (NG-1) and increasingly osteogenesis under higher intensity (NG-2) or pure green light. Furthermore, we isolated mRNAs from the surface-cultured hMSCs and utilized qRT-PCR to examine genes characteristic to each lineage. Congruent with our other analyses of 2D- and 3D-cultured cells, adipogenic gene expression was highest under NG-1 exposure and osteogenic gene expression dominated under NG-2 and green conditions (Fig. 4c). None of the lineage-specific genes were expressed at significant levels by hMSCs cultured on the nonfouling NIR-exposed gel, again indicating their multipotency was maintained.
Programmed differentiation
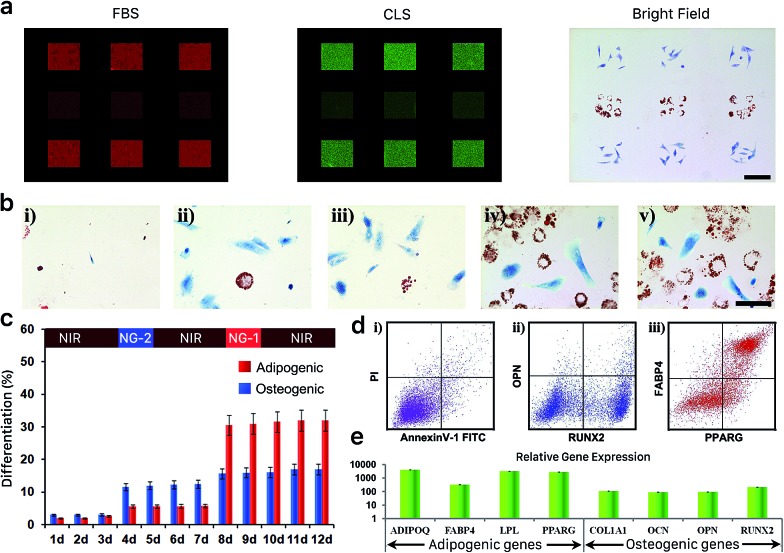
Achieving robust spatiotemporal control of cell-scaffold constructs is a key challenge in the regeneration of complex tissues or organs. We hypothesized the photoactuated platform presented here would allow hMSC differentiation lineages to be precisely programmed in real time and with respect to their location in the gel. A patterned and constructively interacting irradiation method was employed first to evaluate spatial control. In brief, the entire hydrogel sample was exposed to NIR light from the bottom, and green light of varying intensities was applied through a patterned chrome photomask from the top. In the example shown in Fig. 5, 30 mW green light is applied through the 1st and 3rd patterned rows of the photomask and 10 mW green light is applied through the 2nd patterned row. Green light sources of different powers were isolated to avoid crosstalk. During the patterned light exposure, we added protein solutions from either culture medium (FBS) or hMSC lysate (CLS) onto the gel and incubated them for 24 hours. The resulting protein adsorption was evaluated with fluorescent tagging by Rhodamine (FBS) and FITC (CLS), as described in the ESI.† As presented in Fig. 5a, the 1st and the 3rd rows presented strong binding affinity to both FBS and CLS proteins while the 2nd row presented weaker binding to these solutions. Notably, the remainder of the hydrogel exposed to pure NIR resisted fouling by both FBS and CLS proteins, in sharp contrast to the patterned area exposed to visible light. After confirming this spatial control of protein fouling, we seeded hMSCs on the hydrogel surface to pursue patterned differentiation. Using the same masking pattern, we cultured the cell-hydrogel construct in bipotential media for 14 days and assessed their differentiation behavior with histological staining. As shown in Fig. 5a, while most hMSCs did not differentiate under pure NIR exposure, they differentiated actively in the patterned areas exposed to green light. It was apparent that the patterned exposures enabled delicate spatial control of differentiation behavior on the same scaffold. HMSCs grown in the 1st and 3rd rows conclusively chose osteogenic differentiation, while those grown in the 2nd row preferred adipogenesis.
Fig. 5. (a) FBS and CLS protein adsorption and hMSC differentiation on the hydrogel receiving patterned exposure. (b) Representative bright-field images of Oil Red O and Fast Blue salt-stained hMSCs following 12 day bipotential differentiation media incubation within hydrogels exposed to programmed light conditions. Scale bar, 30 μm. (c) Percentage of cells differentiating to adipocytes (lipid was stained red by Oil Red O) or osteoblasts (ALP was stained blue by Fast Blue salt) in bipotential differentiation media for 12 days when exposed to a light program. (d) Representative flow cytometry profile of cell apoptosis/necrosis (i) and the expression of the osteogenic biomarkers (ii) and the adipogenic biomarkers (iii) for hMSCs grown on the hydrogel surface receiving the 12 day light program. (e) Representative gene expression profile of hMSCs grown on the hydrogel surface receiving the 12 day light program.
We further endeavored to manipulate the fate choice of cultured hMSCs in real time in order to precisely control the temporal differentiation composition on a single scaffold. We exposed the surface-cultured cells in bipotential differentiation media to a continuous wavelength-sweep program—NIR, 3 days → NG-2, 1 day → NIR, 3 days → NG-1, 1 day → NIR, 3 days. NIR was applied through the full timeline, while a chosen visible light program (i.e. NG-1 or NG-2) was applied for 24 hours every three days. As presented in Fig. 5b(i), during the first three days of culture under NIR exposure, hMSC differentiation potential was largely restrained. When we applied NG-2 for one day, we began to observe significant osteogenesis on the hydrogel while their adipogenic potential was still largely inhibited. Notably, when we switched from NG-2 back to NIR exposure, differentiation was suspended due to the elimination of nonspecific interactions. As shown in Fig. 5b(iii), we did not observe further significant differentiation during the 2nd three-day exposure to pure NIR. During the one-day NG-1 application that followed, we observed resumed lineage commitment—as seen in Fig. 5b(iv), significant adipogenesis was noted on the platform while enhanced osteogenesis was not seen. This adipogenesis was again suspended by reverting the construct to NIR illumination only. As seen by comparing Fig. 5b(v) to (iv), we did not observe significantly enhanced differentiation during the final three-day NIR period. A quantitative summary of the differentiation assessed by histological staining during this continuous wavelength-sweep program is summarized in Fig. 5c. Notably, as presented in ESI Fig. S9,† the sequence of applied illumination conditions did not affect the overall differentiated composition.
We finally analyzed the cultured cells via flow cytometry after this 12 day programmed culture, and found the cells experienced neither significant death nor apoptosis on the reported platform (Fig. 5d). This assessment indicated the high biocompatibility of the photoactuated platform and differentiation programming strategy. Examination of characteristic surface antigen and mRNA expression by cells on the hydrogel surface further confirmed the positive differentiation of hMSCs after this 12 day culture program. The quantitative differentiation profiles as assayed through each method are summarized in Fig. 5c–e.
Conclusions
In this work, we demonstrate a photoswitchable zwitterionic hydrogel platform that enables dynamic control of hMSC differentiation spatially and temporally. This hydrogel can be continuously and reversibly shifted between a zwitterionic hydrophilic state and a hydrophobic state when exposed to a combination of green and NIR lights, all without causing cell damage. The combined capabilities of this photodynamic platform—control of the nonspecific interactions possible without changing the hydrogel composition, and restraint of stem cell differentiation in a zwitterionic hydrogel free of nonspecific interactions—make precise direction of stem cell self-renewal and differentiation possible. As a result, the composition, location, and differentiation status of stem cells can be easily manipulated and adjusted at will on a single hydrogel scaffold by simply controlling lights and avoiding chemical contamination. This strategy will advance the fundamental understanding of stem cell differentiation mechanisms, as well as provide a platform for hMSC-based regenerative medicine.
Acknowledgments
This work was funded by the National Science Foundation (DMR 1307375 and CBET1264477), the Office of Naval Research (N00014-15-1-2277) and the National Natural Science Foundation of China (Grants 51173129, 51325305). We thank Dr Greg Martin, director of the Keck Microscopy Center at the University of Washington, for helping with microscopic imaging. We thank Dr Michele Black, director of the Flow and Imaging Cytometry Core Lab at the University of Washington, for providing help with flow cytometry. Finally, we thank Dr Raftery and Dr Haiwei Gu of the Northwest Metabolomics Research Center (NW-MRC) for assisting with LC-MS assessment.
Footnotes
References
- Roy I., Gupta M. N. Chem. Biol. 2003;10:1161–1171. doi: 10.1016/j.chembiol.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Kopeček J. Biomaterials. 2007;28:5185–5192. doi: 10.1016/j.biomaterials.2007.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Barbero A., Suárez I. J., Sierra-Martín B., Fernández-Nieves A., de las Nieves F. J., Marquez M., Rubio-Retama J., López-Cabarcos E. Adv. Colloid Interface Sci. 2009;147:88–108. doi: 10.1016/j.cis.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Hoffman A. S. Adv. Drug Delivery Rev. 2013;65:10–16. doi: 10.1016/j.addr.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Park K. Adv. Drug Delivery Rev. 2012;64:49–60. [Google Scholar]
- Lutolf M. P. Nat. Mater. 2009;8:451–453. doi: 10.1038/nmat2458. [DOI] [PubMed] [Google Scholar]
- Theato P., Sumerlin B. S., O'Reilly R. K., Epps III T. H. Chem. Soc. Rev. 2013;42:7055–7056. doi: 10.1039/c3cs90057f. [DOI] [PubMed] [Google Scholar]
- Oliveira M. B. and Mano J. F., Polymers in Regenerative Medicine: Biomedical Applications from Nano- to Macro-Structures, 2015, pp. 49–90. [Google Scholar]
- Tomatsu I., Peng K., Kros A. Adv. Drug Delivery Rev. 2011;63:1257–1266. doi: 10.1016/j.addr.2011.06.009. [DOI] [PubMed] [Google Scholar]
- DeForest C. A., Anseth K. S. Nat. Chem. 2011;3:925–931. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloxin A. M., Kasko A. M., Salinas C. N., Anseth K. S. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForest C. A., Polizzotti B. D., Anseth K. S. Nat. Mater. 2009;8:659–664. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercole F., Davis T. P., Evans R. A. Polym. Chem. 2010;1:37–54. [Google Scholar]
- Wirkner M., Alonso J. M., Maus V., Salierno M., Lee T. T., García A. J., del Campo A. Adv. Mater. 2011;23:3907–3910. doi: 10.1002/adma.201100925. [DOI] [PubMed] [Google Scholar]
- Griffin D. R., Schlosser J. L., Lam S. F., Nguyen T. H., Maynard H. D., Kasko A. M. Biomacromolecules. 2013;14:1199–1207. doi: 10.1021/bm400169d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForest C. A., Tirrell D. A. Nat. Mater. 2015;14:523–531. doi: 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- Kloxin A. M., Tibbitt M. W., Anseth K. S. Nat. Protoc. 2010;5:1867–1887. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbanks B. D., Singh S. P., Bowman C. N., Anseth K. S. Macromolecules. 2011;44:2444–2450. doi: 10.1021/ma200202w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D. R., Kasko A. M. J. Am. Chem. Soc. 2012;134:13103–13107. doi: 10.1021/ja305280w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azagarsamy M. A., Alge D. L., Radhakrishnan S. J., Tibbitt M. W., Anseth K. S. Biomacromolecules. 2012;13:2219–2224. doi: 10.1021/bm300646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Li J., Tan S., Wang R., Zhang Y. J. Am. Chem. Soc. 2013;135:18718–18721. doi: 10.1021/ja409000b. [DOI] [PubMed] [Google Scholar]
- DeForest C. A., Sims E. A., Anseth K. S. Chem. Mater. 2010;22:4783–4790. doi: 10.1021/cm101391y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Shoichet M. S. Nat. Mater. 2004;3:249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- Wylie R. G., Ahsan S., Aizawa Y., Maxwell K. L., Morshead C. M., Shoichet M. S. Nat. Mater. 2011;10:799–806. doi: 10.1038/nmat3101. [DOI] [PubMed] [Google Scholar]
- Adzima B. J., Tao Y., Kloxin C. J., DeForest C. A., Anseth K. S., Bowman C. N. Nat. Chem. 2011;3:256–259. doi: 10.1038/nchem.980. [DOI] [PubMed] [Google Scholar]
- Bandara H. D., Burdette S. C. Chem. Soc. Rev. 2012;41:1809–1825. doi: 10.1039/c1cs15179g. [DOI] [PubMed] [Google Scholar]
- Fuß W., Kosmidis C., Schmid W. E., Trushin S. A. Angew. Chem., Int. Ed. 2004;43:4178–4182. doi: 10.1002/anie.200454221. [DOI] [PubMed] [Google Scholar]
- Minkin V. I. Chem. Rev. 2004;104:2751–2776. doi: 10.1021/cr020088u. [DOI] [PubMed] [Google Scholar]
- Klajn R. Chem. Soc. Rev. 2013;43:148–184. doi: 10.1039/c3cs60181a. [DOI] [PubMed] [Google Scholar]
- der Molen S. J., ávan Wees B. J. Chem. Commun. 2006:3597–3599. [Google Scholar]
- Heinz B., Malkmus S., Laimgruber S., Dietrich S., Schulz C., Rück-Braun K., Braun M., Zinth W., Gilch P. J. Am. Chem. Soc. 2007;129:8577–8584. doi: 10.1021/ja071396i. [DOI] [PubMed] [Google Scholar]
- Koumura N., Zijlstra R. W., van Delden R. A., Harada N., Feringa B. L. Nature. 1999;401:152–155. doi: 10.1038/43646. [DOI] [PubMed] [Google Scholar]
- Zhao P., Fang C., Xia C., Wang Y., Liu D., Xie S. Appl. Phys. Lett. 2008;93:013113. [Google Scholar]
- di Nunzio M. R., Gentili P. L., Romani A., Favaro G. ChemPhysChem. 2008;9:768–775. doi: 10.1002/cphc.200700789. [DOI] [PubMed] [Google Scholar]
- Jiang S., Cao Z. Adv. Mater. 2010;22:920–932. doi: 10.1002/adma.200901407. [DOI] [PubMed] [Google Scholar]
- Zhang L., Cao Z., Bai T., Carr L., Ella-Menye J.-R., Irvin C., Ratner B. D., Jiang S. Nat. Biotechnol. 2013;31:553–556. doi: 10.1038/nbt.2580. [DOI] [PubMed] [Google Scholar]
- Keefe A. J., Jiang S. Nat. Chem. 2012;4:60–64. [Google Scholar]
- Bai T., Liu S., Sun F., Sinclair A., Zhang L., Shao Q., Jiang S. Biomaterials. 2014;35:3926–3933. doi: 10.1016/j.biomaterials.2014.01.077. [DOI] [PubMed] [Google Scholar]
- Chien H.-W., Tsai W.-B., Jiang S. Biomaterials. 2012;33:5706–5712. doi: 10.1016/j.biomaterials.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Carr L. R., Zhou Y., Krause J. E., Xue H., Jiang S. Biomaterials. 2011;32:6893–6899. doi: 10.1016/j.biomaterials.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Bai T., Sun F., Zhang L., Sinclair A., Liu S., Ella-Menye J. R., Zheng Y., Jiang S. Angew. Chem., Int. Ed. 2014;126:12943–12948. doi: 10.1002/anie.201405930. [DOI] [PubMed] [Google Scholar]
- Wojtyk J. T., Wasey A., Kazmaier P. M., Hoz S., Buncel E. J. Phys. Chem. A. 2000;104:9046–9055. doi: 10.1021/jp068575r. [DOI] [PubMed] [Google Scholar]
- Zhu M.-Q., Zhang G.-F., Li C., Aldred M. P., Chang E., Drezek R. A., Li A. D. J. Am. Chem. Soc. 2010;133:365–372. doi: 10.1021/ja106895k. [DOI] [PubMed] [Google Scholar]
- Elizalde L. E., Ledezma R., López R. G. Synth. Commun. 2005;35:603–610. [Google Scholar]
- Friedle S., Thomas S. W. Angew. Chem., Int. Ed. 2010;122:8140–8143. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.