Abstract
Despite influencing many aspects of T cell biology, the kinetics of T cell receptor (TCR) binding to peptide-major histocompatibility molecules (pMHC) remain infrequently determined in patient monitoring or for adoptive T cell therapy. Using specifically designed reversible fluorescent pMHC multimeric complexes, we performed a comprehensive study of TCR-pMHC off-rates combined with various functional assays on large libraries of self/tumor– and virus-specific CD8+ T cell clones from melanoma patients and healthy donors. We demonstrate that monomeric TCR-pMHC dissociation rates accurately predict the extent of cytotoxicity, cytokine production, polyfunctionality, cell proliferation, activating/inhibitory receptor expression, and in vivo antitumor potency of naturally occurring antigen-specific CD8+ T cells. Our data also confirm the superior binding avidities of virus-specific T cells as compared with self/tumor–specific T cell clonotypes (n > 300). Importantly, the TCR-pMHC off-rate is a more stable and robust biomarker of CD8+ T cell potency than the frequently used functional assays/metrics that depend on the T cell’s activation state, and therefore show major intra- and interexperimental variability. Taken together, our data show that the monomeric TCR-pMHC off-rate is highly useful for the ex vivo high-throughput functional assessment of antigen-specific CD8+ T cell responses and a strong candidate as a biomarker of T cell therapeutic efficacy.
Keywords: Immunology, Therapeutics
TCR-pMHC off-rate measurements are more reliable than cell-function assays and better suited to determine the functional potency of a patient’s T cells.
Introduction
Cytotoxic T lymphocytes mediate immune protection against a large number of infectious diseases, and recent developments in oncology confirmed their ability to eliminate cancers. To achieve successful immunity, T cells must be activated through specific interactions between T cell receptors (TCRs) and antigenic peptides presented by major histocompatibility molecules (pMHC) on antigen-presenting cells. This enables T cell expansion and differentiation into large numbers of effector cells with various functional capacities (i.e., killing, cytokine production, proliferation). Furthermore, T cells must migrate and localize to the infected or tumoral tissues, exerting their effector function and finally acquire memory properties, assuring long-lasting immunity.
Extensive research has been undertaken to determine which T cell properties are essential to generate protective and durable immune responses. T cell functional avidity, which measures in vitro T cell responses when exposed to increasing antigen concentrations, has been largely associated with the control of viral (1–3) or tumor (4, 5) load in animal models. In accordance with these observations, several findings in patients with HIV (6, 7) or hepatitis C (8, 9) infections further showed the key role of CD8+ T cells of high functional avidity in efficient viral control and clearance. Yet, others have challenged the functional superiority of such high-avidity cells, which may be prone to increased activation-induced cell death, senescence, or exhaustion (reviewed in ref. 10). In the context of antitumor responses, results obtained from melanoma patients also indicate that T cells of high functional avidities are required for efficient protection (11–13). Besides functional avidity, higher proportions of polyfunctional CD8+ or CD4+ T cells were also found in HIV (14, 15) and hepatitis C (16) controllers, when compared with individuals with progressive disease. Moreover, some reports proposed a direct link between functional avidity (i.e., antigen sensitivity) and polyfunctionality (i.e., T cell capacity to exert multiple effector functions) (17, 18). However, the ex vivo appraisal of T cell functionality/polyfunctionality is still often limited to assays of fixed stimulation doses and by the lack of universal standards of T cell assessment (reviewed in refs. 19, 20). It is therefore essential to improve our knowledge regarding the contribution of the different aspects of T cell function to clinical efficacy and to identify additional T cell–based parameters that may enable overcoming some of the limitations associated with functional assays.
The functional avidity of T cells is primarily controlled by the strength of TCR-pMHC interactions, a key parameter shown to impact on numerous aspects of T cell biology, including their thymic selection (21), activation and differentiation (22), autoimmune pathogenicity (23), and protection against infection and cancer (24). In fact, TCR-pMHC binding avidity may offer a key metric by which the quality of the T cell response can be directly evaluated, since it controls T cell activation, differentiation, and functional efficacy (25). Numerous studies indicate that, within the affinity range of physiological interactions (KD 100–1 μM), enhanced TCR-pMHC affinity or off-rate (koff) correlate with improved T cell functionality (26). However, most of these reports are based on artificial models (e.g., using affinity-optimized TCR variant panels or altered peptide ligand models), and thus only limited information is available on the overall impact and clinical relevance of TCR-pMHC binding avidity or kinetics (e.g., off-rates) in the context of naturally occurring antigen-specific CD8+ T cell responses. Moreover, identifying and selecting TCRs of higher avidity may be of particular importance in the tumoral setting, since most high avidity/affinity self/tumor antigen–reactive T cells are naturally eliminated or silenced by mechanisms of central and peripheral tolerance, emphasizing the need to select the remaining rare high-avidity cells for immunotherapy.
Reversible 2-color multimer-based approaches (i.e., Streptamers, NTAmers) have been developed to precisely quantify monomeric TCR-pMHC dissociation rates (i.e., off-rate or koff) directly on living T cells. Streptamers initially revealed that virus-specific CD8+ T cells with longer off-rates conferred better in vivo protection than T cells with shorter off-rates (27). However, owing to the faster decay of the multimeric complex onto monomeric pMHC when compared with Streptamers, NTAmers offer an increased sensitivity to detect T cells with low-avidity TCRs (26), such as those typically found in self/tumor–specific CD8+ T cell repertoires. Consequently, we recently showed that NTAmer-based koff strongly correlated with the killing capacity of TCR-engineered and natural tumor-specific human CD8+ T cells (28, 29).
With the aim to thoroughly evaluate possible correlations between T cell function and TCR-pMHC binding kinetics, we here undertook a large-scale analysis of combined multiple functions (i.e., killing, CD107a degranulation, cytokine production, proliferation, surface expression of activating/inhibitory receptors, and tumor control) and optimized off-rate measurements using NTAmers to characterize large libraries of tumor- and virus-specific CD8+ T cell clones isolated from melanoma patients and healthy donors. Our large data sets show that the TCR-pMHC off-rate is a major determinant controlling the functions of CD8+ T cells in vitro and in vivo. Our findings are also of practical importance, as we found that the TCR-ligand dissociation rate is a highly stable biomarker, more reliable and reproducible than the usual assessments based on multimer staining levels or functional T cell avidity, which may fluctuate depending on the T cell’s activation state.
Results
TCR-pMHC off-rate accurately correlates to overall T cell functional avidity.
To precisely address the relationship between the TCR-pMHC off-rate and the overall CD8+ T cell functional profile, we generated large libraries of HLA-A*0201–restricted CD8+ T cell clones, by direct ex vivo sorting and cloning of self/tumor–specific (i.e., Melan-A26–35 and NY-ESO-1157–165) and virus-specific (i.e., cytomegalovirus CMV/pp65495–504 and Epstein-Barr virus EBV/BMFL1259–267) effector memory (EM) T cells (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.92570DS1). We analyzed all clones for TCR-pMHC dissociation rates using NTAmers loaded with the native Melan-A, NY-ESO-1, EBV/BMFL1, or CMV/pp65 peptide because they provided a more physiological assessment of the TCR-pMHC recognition efficacy as opposed to the corresponding analog peptides (as detailed in Methods). Representative koff-based panels of self/tumor– and virus-specific CD8+ T cell clones were further characterized at the functional level, including assessment of cytotoxic activity, CD107a degranulation, and production of cytokines based on peptide titration assays, as well as proliferation (Supplemental Figure 2). Note that, for the same antigen specificity, most of the different functional readouts/measures were obtained during the same nonspecific restimulation cycle to make use of the antigen-specific CD8+ T cell clones in a similar resting state (>15 days after restimulation).
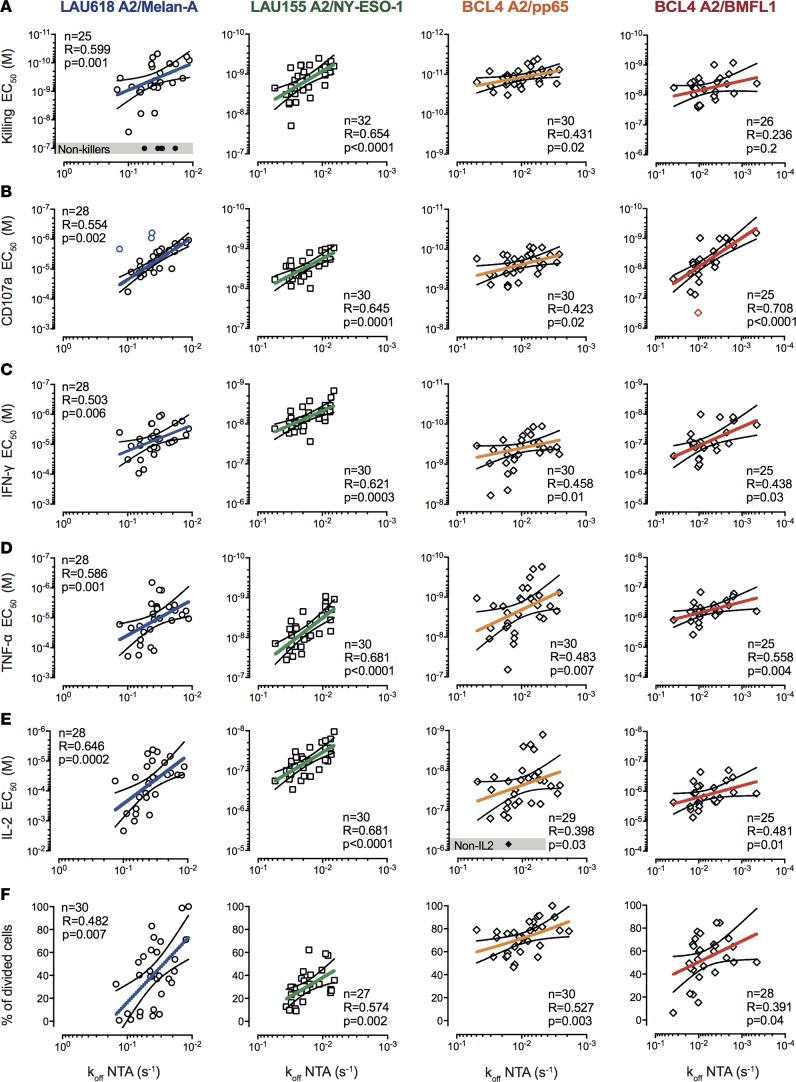
We observed, for all antigenic specificities, statistically significant correlations between TCR-pMHC off-rates and various functional avidity readouts (EC50, defined as the peptide concentration producing half-maximal response) or proliferative capacity (percentage of divided cells) (Figure 1 and Supplemental Figure 3, A and B). Yet, stronger correlations (P < 0.01–0.001, r > 0.5, and narrow confidence intervals) were generally found for self/tumor–specific (Melan-A and NY-ESO-1) than non–self/virus–specific (CMV/pp65 and EBV/BMFL1) T cells. By contrast, no positive correlations could be observed between TCR-pMHC off-rates and the maximally reached functions at saturating peptide doses (Bmax, maximal response) (Supplemental Figure 3C; data not shown). In turn, the maximal response depended on the in vivo differentiation status, with stronger Th2-related cytokine production by clones derived from the early-differentiated EM/CD28+ cells and greater granzyme B expression and killing by those from the late-differentiated EM/CD28– cells (Supplemental Figure 3D). Collectively, these results indicate that, within an antigen-specific repertoire, the kinetics of TCR-pMHC interactions represent a major determinant of the overall functional avidity of CD8+ T cells, regardless of their differentiation status (Supplemental Figure 3D) or function-specific activation thresholds (killing < CD107a < IFN-γ < TNF-α < IL-2) (Supplemental Figure 4A).
Figure 1. Relationship between TCR dissociation rates and functional avidity of self/tumor– and virus-specific CD8+ T cell clones.
Correlations between EC50 values from (A) killing, (B) CD107a degranulation, (C) IFN-γ–, (D) TNF-α–, and (E) IL-2–production titration assays and NTAmer-derived TCR dissociation rates (koff). (F) Correlations between percentages of proliferating cells upon antigen-specific stimulation and NTAmer-derived TCR dissociation rates (koff). (A–F) Antigen-specific CD8+ T cell clones were generated upon direct ex vivo sorting from effector-memory (EM)/CD28+/– and/or EMRA/CD28–subsets. Each data point represents an A2/Melan-A26–35– (derived from patient LAU618, ○), A2/NY-ESO-1157–165– (patient LAU155, □), A2/pp65495–504–, or A2/BMFL1259–267– (healthy donor BCL4, ◊) specific individual T cell clone. Nonfunctional clones are represented in gray boxes. The number of clones displaying function n, as well as Spearman’s correlation (2 tailed, α = 0.05) coefficients R and P values are indicated. Color-coded and black lines are indicative of regression fitting and 95% confidence intervals, respectively. Of note, only very low numbers of outliers were identified when applying the ROUT method and are highlighted in color (71). The representative TCR-BV-CDR3 clonotype diversity of each antigenic specificity was LAU618/Melan-A, 77%; LAU155/NY-ESO-1, 43%; BCL4/pp65, 57%; and BCL4/BMFL1, 67%.
TCR-pMHC off-rate closely correlates to CD8+ T cell polyfunctionality.
Protective immunity against intracellular pathogens relies on the individual CD8+ T cell capacity to display multiple effector functions or polyfunctionality (10). We hypothesized that the kinetics of TCR-pMHC interactions could also affect their polyfunctionality. The coexpression levels of CD107a, IFN-γ, TNF-α, and IL-2 were characterized on a representative selection of self/tumor– and virus-specific CD8+ T cell clones with relative slow or fast TCR-pMHC off-rates (Figure 2). For all antigenic specificities and peptide titrations tested, the fraction of cells displaying more than 1 single function was always greater in CD8+ T cell clones with slower TCR-pMHC off-rates than with faster ones (Figure 2A). In line with these observations, we found that a significant proportion of antigen-specific CD8+ T cell clones with slow TCR-pMHC off-rates showed increased polyfunctional capacities (in terms of EC50 titration curves) when compared with the clones having fast TCR-pMHC off-rates (Figure 2, B–D). However, a strict correlation between off-rates and polyfunctionality was not always found, and limited differences were mostly observed in the EBV-specific CD8+ T cell responses. Taken together, these results show that the TCR-pMHC off-rate not only predicts single functional avidities of self/tumor– and virus-specific CD8+ T cells, but also their capacity to codevelop multiple effector functions.
Figure 2. Relationship between TCR dissociation rates and polyfunctionality of self/tumor– and virus-specific CD8+ T cell clones.
(A) CD107a, IFN-γ, TNF-α, and IL-2 coexpression titration assays of A2/Melan-A26–35– (derived from patient LAU618), A2/NY-ESO-1157–165– (patient LAU155), A2/pp65495–504–, or A2/BMFL1259–267– (healthy donor BCL4) specific clones with slow (n = 10) or fast (n = 10) TCR off-rates. Pie arcs depict the average fraction of cells displaying 0 to 4 functions. (B and C) Individual and (D) average ± SEM polyfunctional (coexpression of CD107a, IFN-γ, TNF-α, and IL-2) titration curves obtained for A2/Melan-A26–35– (derived from patient LAU618), A2/NY-ESO-1157–165– (patient LAU155), A2/pp65495–504–, or A2/BMFL1259–267– (healthy donor BCL4) specific clones with slow (n = 10, plain symbols and solid lines) or fast (n = 10, empty symbols and dotted lines) TCR off-rates. Vertical lines indicate EC50 values. The P values were determined by the extra sum-of-squares F test (α = 0.05). The representative TCR-BV-CDR3 clonotype diversity of each antigenic specificity was LAU618/Melan-A, 80%; LAU155/NY-ESO-1, 45%; BCL4/pp65, 65%; and BCL4/BMFL1, 80%.
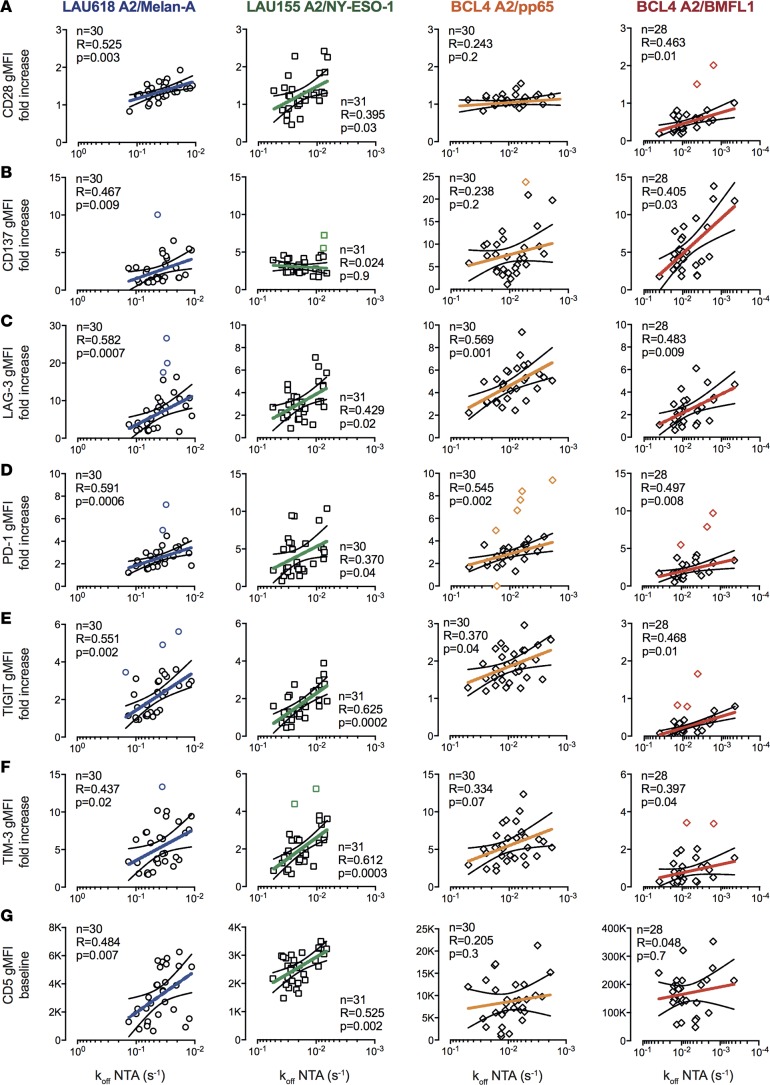
TCR-pMHC off-rate closely follows costimulatory/coinhibitory receptor expression in activated CD8+ T cells.
PD-1 surface expression on CD8+ T cells has been reported to positively correlate with TCR-pMHC binding avidity (30) or functional avidity (31). Here, we explored the relationship between NTAmer-derived off-rates and the expression of various costimulatory (CD28 and CD137) and coinhibitory (LAG-3, PD-1, TIGIT, and TIM-3) receptors (Figure 3). No consistent correlations were found when CD8+ T cell clones were assessed in a resting state (data not shown). In contrast, following 24 hours of stimulation with self/tumor or viral peptides, we observed substantial correlations between TCR-pMHC off-rates and the extent of increased expression of both costimulatory and coinhibitory receptors (Figure 3, A–F). These data indicate a direct impact of TCR-pMHC binding avidities on the susceptibility of CD8+ T cells to antigen-specific activation, and consequently on the upmodulation of both costimulatory and coinhibitory receptors upon stimulation.
Figure 3. Relationship between TCR dissociation rates and activating/inhibitory receptor expression of self/tumor– and virus-specific CD8+ T cell clones.
Correlations between fold increases in surface expression of (A) CD28, (B) CD137, (C) LAG-3, (D) PD-1, (E) TIGIT, and (F) TIM-3 upon antigen-specific stimulation and NTAmer-derived TCR dissociation rates (koff). (G) Correlations between baseline surface expression levels (geometric mean fluorescence intensity [gMFI]) of CD5 and NTAmer-derived TCR dissociation rates (koff). (A–G) Each data point represents an A2/Melan-A26–35– (derived from patient LAU618, ○), A2/NY-ESO-1157–165– (patient LAU155, □), A2/pp65495–504– or A2/BMFL1259–267– (healthy donor BCL4, ◊) specific individual T cell clone. The number of clones tested n, as well as Spearman’s correlation (2 tailed, α = 0.05) coefficients R and P values are indicated. Color-coded and black lines are indicative of regression fitting and 95% confidence intervals, respectively. Outliers were determined by the ROUT method and are highlighted in color (71). The representative TCR-BV-CDR3 clonotype diversity of each antigenic specificity was LAU618/Melan-A, 77%; LAU155/NY-ESO-1, 43%; BCL4/pp65, 57%; and BCL4/BMFL1, 67%.
We also investigated whether TCR-pMHC off-rates associated with CD5 expression, which is a measure of the strength for self-pMHC selecting ligands during thymocyte development (32). At baseline, most virus-specific CD8+ T cell clones displayed high expression levels of CD5, irrespective of their TCR-pMHC off-rates (Figure 3G). These data are in line with previous reports proposing that T cells with greater TCR sensitivity to self pMHC are most efficiently recruited in response to foreign antigens (33, 34). Positive correlations were only found in the context of self/tumor–specific CD8+ T cell clones, with slower off-rates associating with higher baseline levels of CD5 (Figure 3G and Supplemental Figure 5). This latter observation suggests that the expression levels of CD5 on self/tumor–specific T cells may also predict their capacity for increased homeostatic or antigen-specific response.
TCR-pMHC off-rate predicts the in vivo functional potency of self/tumor–specific CD8+ T cells.
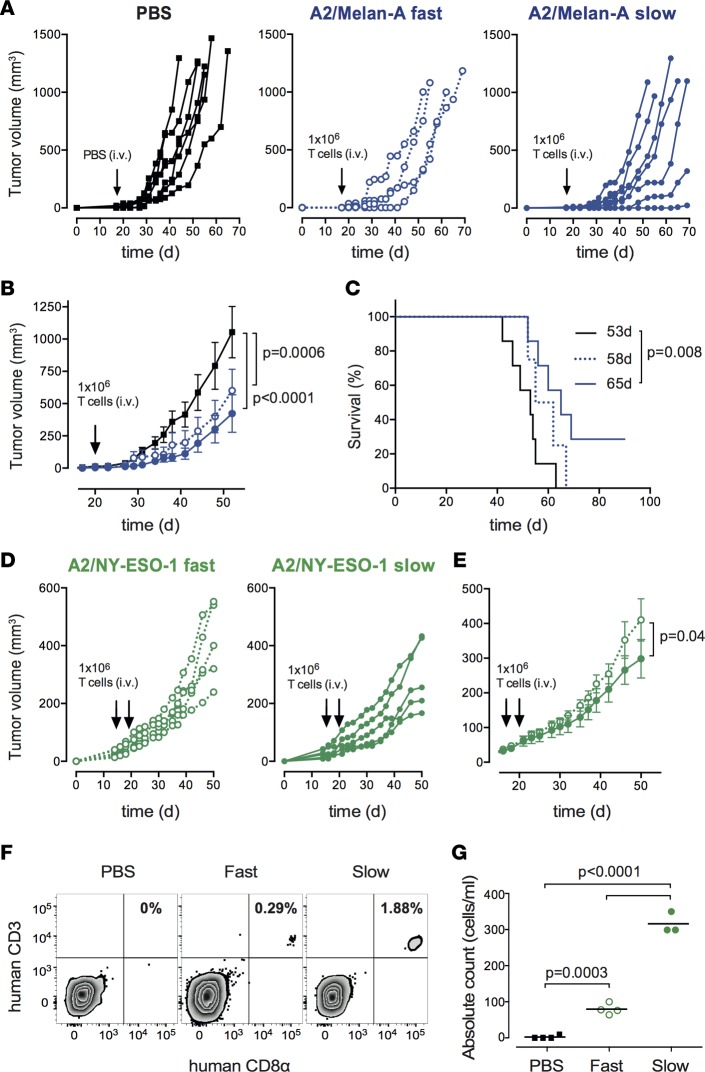
To further substantiate the relevance of our in vitro observations, we evaluated the impact of TCR-pMHC off-rates on the ability of self/tumor–specific CD8+ T cells to control tumor growth in vivo. We first adoptively transferred A2/Melan-A26–35–specific CD8+ T cell clones of slow versus fast TCR-pMHC off-rates into immunodeficient NSG mice bearing human melanoma Me275 tumors (Figure 4A). The transfer of fast off-rate T cell clones showed intermediate tumor growth control. In contrast, T cell clones with slow off-rates mediated a more significant delay in tumor growth when compared with the untreated (PBS) group (Figure 4B). Furthermore, a significantly prolonged survival was only observed for mice treated with A2/Melan-A26–35–specific clones of slow TCR-pMHC off-rates (Figure 4C). To confirm those observations, we then performed similar experiments using the A2/NY-ESO-1 antigenic model, but this time, all mice received s.c. injections of human recombinant IL-2 to enhance the T cell antitumor efficacy (Figure 4D). In line with the observations made on Melan-A26–35–specific T cells, NY-ESO-1157–165–specific CD8+ T cell clones of slow TCR-pMHC off-rates provided a significant delay in tumor growth in comparison to the clones with fast off-rates (Figure 4E). Finally, we monitored the peripheral persistence of NY-ESO-1157–165–specific T cells at days 2 and 14 following adoptive transfer. Analysis of tail bleeds taken at day 2 revealed that there was a significantly improved engraftment of slow off-rate T cell clones compared with fast off-rate T cell clones (Figure 4F). Yet, tumor-specific T cells did not persist beyond 14 days after T cell transfer (data not shown), in line with a previous report (35). In summary, these data provide further evidence that the TCR-pMHC off-rate represents an excellent biomarker to predict the immunotherapeutic potential of tumor-specific CD8+ T cells, and could therefore be selectively used to enhance the efficacy of adoptive T cell therapy (27).
Figure 4. Relationship between TCR dissociation rates and tumor control in immunodeficient mice upon adoptive T cell transfer.
(A) Individual or (B) average ± SEM tumor growth and (C) Kaplan–Meier survival curves of tumor-bearing NSG mice adoptively transferred with PBS (control, n = 7; black solid lines) or 1 × 106 A2/Melan-A26–35–specific T cell clones with fast (n = 4; blue dotted lines) or slow (n = 7; blue solid lines) TCR off-rates. (D) Individual or (E) average ± SEM tumor growth curves of tumor-bearing NSG mice adoptively transferred twice with 1 × 106 A2/NY-ESO-1157–165–specific T cell clones with fast (n = 5; green dotted lines) or slow (n = 5; green solid lines) TCR off-rates. Tumor volume and survival curve P values were determined by 2-way ANOVA and log-rank tests, respectively. (F) Representative staining and (G) absolute counts of human CD8+ T cells from blood taken from tail veins at day 2 following adoptive transfer of 4 × 106 A2/NY-ESO-1157–165–specific CD8+ T cell clones with fast (n = 4; green empty circles) or slow (n = 3; green full circles) TCR off-rates. As control, 3 mice received PBS (n = 4; black squares). P values were determined by 1-way ANOVA multiple comparison tests.
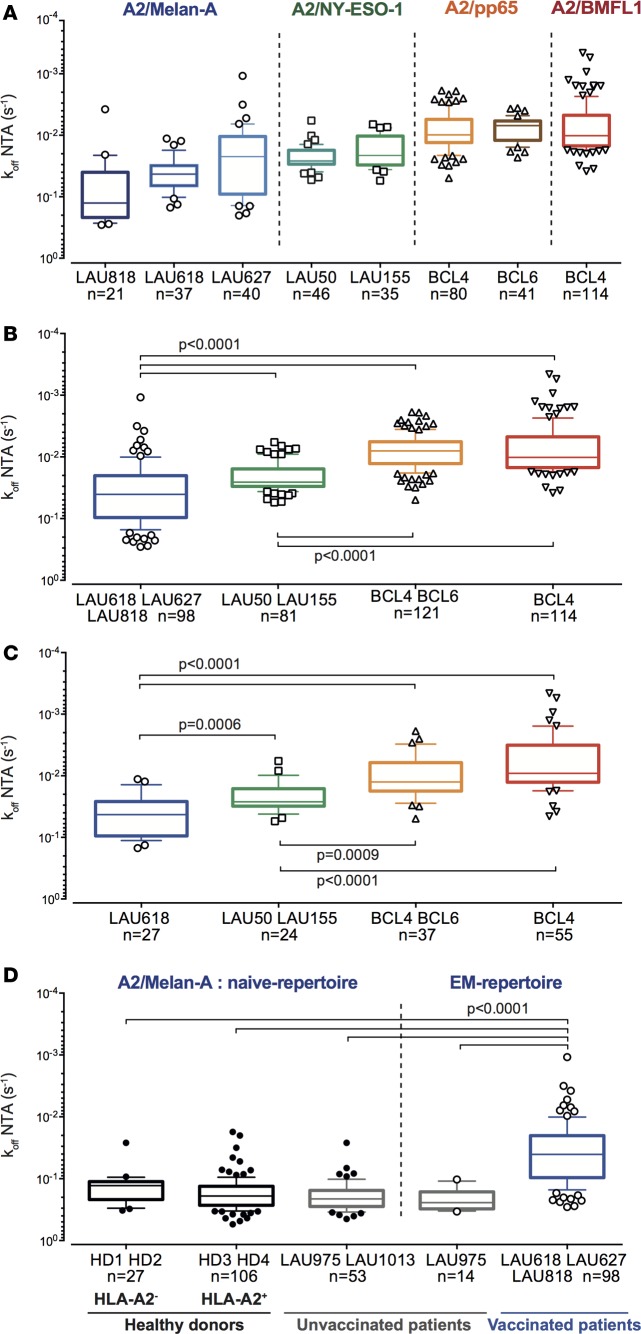
TCR-pMHC off-rates vary according to the antigenic specificity of CD8+ T cells.
Only limited information is available on the overall quality of TCR-pMHC binding avidity of self/tumor–specific versus non–self/pathogen–specific CD8+ T cell repertoires (36, 37). To address this point, we performed a comprehensive analysis of TCR-pMHC off-rates on 414 EM CD8+ T cell clones specific for (a) the differentiation antigen A2/Melan-A26–35, (b) the cancer testis antigen A2/NY-ESO-1157–165, (c) the viral CMV/pp65495–504 antigen, and (d) the viral EBV/BMFL1259–267 antigen isolated from 5 melanoma patients and 2 healthy donors (Figure 5, A and B, and Supplemental Figure 6, A and B). TCR-pMHC off-rate repertoires varied according to the T cell antigenic specificity. As such, A2/Melan-A26–35–specific CD8+ T cells displayed significantly faster TCR-pMHC off-rates than the A2/NY-ESO-1157–165–specific ones. Moreover, both tumor-specific TCR repertoires exhibited significantly faster TCR-pMHC off-rates than repertoires specific for herpes virus antigens (A2/pp65495–504 and A2/BMFL1259–267). Due to the presence of highly frequent TCR clonotypes potentially biasing the NY-ESO-1– and CMV-specific and to a lesser extent the EBV- and Melan-A–specific CD8+ T cell repertoires (38–40), we performed an extensive TCR-BV-CDR3 clonotyping of 353 EM CD8+ T cell clones (Figure 5C and Supplemental Table 1). We identified 143 individual clonotypes (specific for A2/Melan-A– and A2/NY-ESO-1–tumor antigens, and A2/pp65– and A2/BMLF1–viral epitopes), representing approximately 40% of the clonotype diversity, and depending on the antigenic specificity (Melan-A26–35 > EBV/BMFL1259–267 > NY-ESO-1157–165 and CMV/pp65495–504). The same TCR-pMHC off-rate hierarchy (virus-specific > self/tumor–specific CD8+ T cells) was observed when considering all CD8+ T cell clones (Figure 5B) or only the individual TCR clonotypes (Figure 5C). Finally, similar differences were obtained when the CD8+ T cell clones were subdivided according to their ex vivo differentiation status (early-differentiated EM/CD28+ or late-differentiated EM/EMRA/CD28–; Supplemental Figure 6C).
Figure 5. TCR dissociation rates according to the antigenic specificity, clonotype repertoire, and ex vivo differentiation status of CD8+ T cell clones.
(A and B) NTAmer-derived TCR dissociation rates (koff) of EM/EMRA CD28+/– clones (n = 414) specific for the differentiation antigen A2/Melan-A26–35 (derived from melanoma patients LAU618, LAU627, and LAU818 following vaccination with Melan-A/peptide, incomplete Freund’s adjuvant, and CpG), the cancer testis A2/NY-ESO-1157–165 (from patients LAU50 and LAU155 with naturally occurring T cell responses), or the persistent herpes viruses A2/pp65495–504 or A2/BMFL1259–267 (from healthy donors BCL4 and BCL6), categorized according to (A) the respective patients and donors or (B) antigenic specificity. (C) NTAmer-derived TCR dissociation rates (koff) of individual TCR-BV-CDR3 clonotypes specific for the tumor epitopes A2/Melan-A26–35 (n = 27) and A2/NY-ESO-1157–165 (n = 24), and the persistent herpes virus epitopes A2/pp65495–504 (n = 37) and A2/BMFL1259–267 (n = 55). (D) NTAmer-derived TCR dissociation rates (koff) of A2/Melan-A26–35–specific clones derived from HLA-A2–negative (HD1 and HD2), HLA-A2–positive (HD3 and HD4) healthy donors, HLA-A2–positive unvaccinated (LAU975 and LAU1013) and A2/Melan-A26–35–vaccinated (LAU618, LAU627, and LAU818) melanoma patients, categorized according to the patient/donor groups and the differentiation status of T cell clones. (A–D) Data are depicted as box (25th to 75th percentiles) and whisker (10th to 90th percentiles) with the middle line representing the median. Numbers of clones n, as well as Kruskal-Wallis test (α = 0.05) derived P values are indicated. Significant differences between the A2/Melan-A26–35– and the A2/NY-ESO-1157–165–specific groups were obtained by Mann-Whitney test (2 tailed).
The differences found between A2/Melan-A26–35–specific and A2/NY-ESO-1157–165–specific repertoires may result from the fact that the A2/Melan-A26–35–specific clones were derived following peptide vaccination in combination with CpG and incomplete Freund’s adjuvant (IFA) adjuvant (41), when compared with the NY-ESO-1 repertoire obtained from patients with naturally occurring T cell responses. Thus, we investigated the quality of the natural A2/Melan-A26–35–specific CD8+ T cell repertoires found in unvaccinated melanoma patients (n = 2), as well as in A2-positive and A2-negative individuals without melanoma (n = 4), known to express an unusually large peripheral repertoire of naive (CD45RA+CCR7+) A2/Melan-A26–35–reactive CD8+ T cells (42). Unvaccinated patients exhibited differentiated A2/Melan-A26–35–specific T cell repertoires of significantly faster off-rates when compared with the ones derived from vaccinated melanoma patients (Figure 5D). Strikingly, similar rapid off-rates were observed for the CD45RA+CCR7+ naive-specific T cell repertoires derived from unvaccinated patients as well as from A2-positive and A2-negative healthy individuals. These observations reveal the overall inferior quality of the TCR-pMHC binding repertoires specific for the self–A2/Melan-A26–35 epitope, when compared with the ones specific for the cancer testis A2/NY-ESO-1 or viral antigens. Yet, several clones with slower off-rates could still be detected, indicating the presence of rare self/Melan-A–specific T cells of high binding avidity within the endogenous unvaccinated repertoire. Finally, our data show that higher-avidity T cells can be selected following therapeutic vaccination, emphasizing the relevance of therapeutic vaccination approaches in enhancing the quality of a tumor-specific repertoire.
TCR-pMHC off-rate is a stable and robust biomarker independent of the activation state of the T cell.
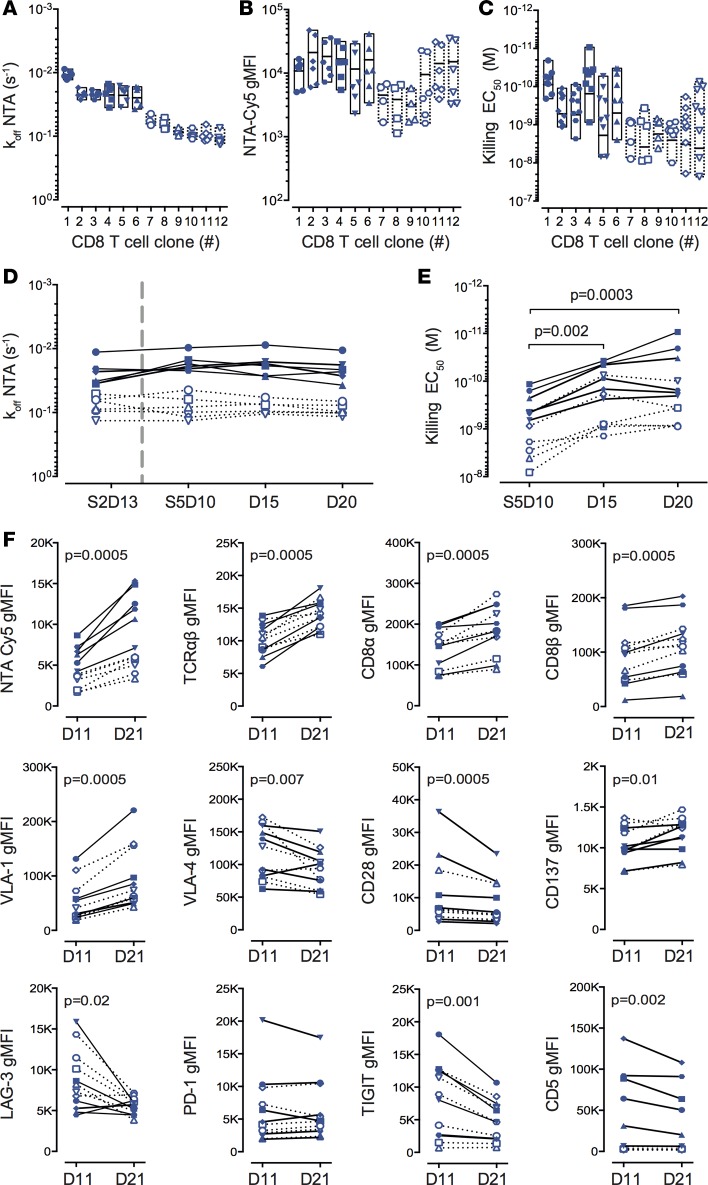
CD8+ T cell functional avidity represents a biological readout that is potentially influenced by multiple factors, such as TCR-pMHC binding avidity, TCR and CD8 surface expression, as well as various molecules regulating TCR signaling and T cell function (10). In that regard, the TCR-pMHC off-rate may provide a more reliable biophysical parameter than the widely used functional-related methods to assess T cell potency. To investigate this question, we first compared the variations obtained following separate experimental measurements (n = 4 to 9) of TCR-based dissociation rates, multimer staining intensity levels, and EC50 killing avidity of 12 representative Melan-A–specific CD8+ T cell clones. For each individual clone, the interexperimental off-rate values nicely clustered together, in sharp contrast to the repeat multimer staining and functional avidity experiments showing large disparities (Figure 6, A–C). Furthermore, the average dissociation rates of these clones strongly correlated with average EC50 killing avidity, but not with average multimer staining intensity (Supplemental Figure 7A). Finally, no correlation was found between functional avidity and multimer staining levels, in agreement with previous reports (reviewed in ref. 26). We next performed longitudinal measurements of TCR-pMHC off-rates and EC50 killing avidity on a representative panel of A2/Melan-A26–35–specific CD8+ T cell clones following nonspecific in vitro stimulation with phytohemagglutinin (PHA) and feeder cells (Supplemental Figure 7, B and C). We observed a remarkable stability of TCR-pMHC off-rate measurements upon stimulation, even when tested at a 6-month interval on T cell clones that underwent several additional rounds of PHA/feeder expansion (Figure 6D). In contrast and as previously described (43), for a given T cell clone, the killing avidity greatly varied and was augmented up to 10-fold, according to the time elapsed since the last stimulation (Figure 6E). These data indicate that the functional avidity reflects the in vitro activation status of CD8+ T cells, in line with the upregulation of cell-surface expression of TCRαβ, CD8αβ, and VLA-1 integrin, and conversely the downregulation of VLA-4 integrin and several coinhibitory receptors such as CD5, LAG-3, and TIGIT or the costimulatory receptor CD28 (Figure 6F). Importantly, the TCR-pMHC binding off-rate measurement is independent of TCRαβ levels, and stands out as a more stable and reliable biomarker than the usually performed assessments of multimer staining levels (i.e., mean fluorescence intensity) or EC50 functional avidity.
Figure 6. Interexperimental and over-time variations of TCR dissociation rates, pMHC multimer staining, and functional avidity assays.
(A) NTAmer-derived TCR dissociation rates (koff), (B) NTAmer surface staining levels (geometric mean fluorescence intensity [gMFI]) and (C) killing avidity values (EC50) obtained in independent assays (n > 4) for A2/Melan-A26–35–specific CD8+ T cell clones with slow (n = 6, plain symbols and solid lines) or fast (n = 6, empty symbols and dotted lines) TCR off-rates. (A–C) Data are depicted as individual values and boxes (minimum to maximum, with the middle line representing the mean). (D) NTAmer-derived TCR dissociation rates (koff), (E) killing avidity values (EC50), and (F) surface staining levels (gMFI) obtained over time (D10/11, D15, and D20/21; D = day) following nonspecific stimulation (by PHA and irradiated feeder cells) for A2/Melan-A26–35–specific T cell clones with slow (n = 6, plain symbols and solid lines) or fast (n = 6, empty symbols and dotted lines) TCR off-rates. S2 represents the off-rate measurements of the same clones 6 months before the fifth round of stimulation (S5). The P values were determined by the Friedman (α = 0.05) and Wilcoxon matched-pair signed-rank (2-tailed) tests.
Discussion
Several observations support the importance of considering both quantitative (i.e., magnitude of response) and qualitative (i.e., functional avidity, polyfunctionality) determinants of the T cell response, in order to predict in vivo efficacy (reviewed in ref. 10). However, ex vivo functional avidity or EC50 (using titrated functional assays) and polyfunctionality assessments remain laborious and time consuming, and are often not possible because relatively large cell numbers must be withdrawn from patients. Importantly, and as shown in the current study, EC50 values largely depend on the T cell’s activation state, and are thus influenced by intraexperimental (i.e., over-time experimental measurements following T cell stimulation) and interexperimental (i.e., separate experimental measurements) variability/fluctuations (Figure 6). Moreover, functional avidity varies greatly depending on the functional readouts (e.g., cytotoxicity versus cytokine production), which mostly reflects modulation of the function-specific activation thresholds (cytotoxicity < cytokine production) (Figure 1 and Supplemental Figure 4A). Taken together, these observations show that there is a strong need to identify a T cell–based biomarker that overcomes the major limitations associated with functional assays and provides a reliable, simple-to-use, amenable-to-standardization immune metric for immunotherapy of cancer or chronic microbial infections.
Here, using an extensive and representative panel of antigen-specific CD8+ T cells generated in the context of natural or postvaccination immune responses, we show that the TCR-ligand dissociation rate globally correlated to all aspects of CD8+ T cell functions tested (i.e., cytotoxic activity, CD107a degranulation, cytokine production, proliferation and coreceptor modulation; Figures 1 and 3), including polyfunctionality (Figure 2) of both self/tumor– and virus-specific CD8+ T cells. Nonetheless, virus-specific T cells displayed weaker, although statistically significant correlations, than tumor-specific T cells, which may in part be the consequence of their overall slower TCR off-rates. These data nicely fit with the model proposing that enhanced TCR affinity or off-rate correlates with improved T cell responsiveness, but that this correlation is no longer linear above a certain TCR binding avidity threshold (reviewed in ref. 26). Specifically, using artificial affinity-enhanced TCRs, several reports (30, 44, 45) have shown that maximal T cell responsiveness occurs within an optimal window of TCR-pMHC binding interactions, usually lying in the upper physiological affinity range (KD between 10 and 1 μM), and encompassing naturally occurring non–self/virus–specific TCR repertoires (36, 37). Moreover, the monomeric TCR-pMHC off-rate also predicted the relative tumor control activity in vivo (Figure 4). Importantly, as a biophysical readout, the TCR-pMHC off-rate represents a more stable and robust parameter of T cell potency, compared with the fluctuating biological metrics, such as T cell functional avidity or multimer-staining levels, which instead depend on the activation status of the cell (Figure 6). Our observations are in agreement with other studies showing that functional avidity is not a constant parameter in individual T cell clones, but gradually increases with time after in vitro restimulation (43, 46) or during the early course of acute viral infection in vivo (47). Enhanced antigen sensitivity is notably influenced by the differential expression of TCRαβ and accessory molecules (i.e., increased CD8αβ and VLA-1 versus reduced CD28, LAG-3, and TIGIT expression) (Figure 6). Altogether, our data show that the TCR-pMHC off-rate stands out as a major and stable determinant of CD8+ T cell function, allowing the accurate monitoring of the quality of naturally occurring or vaccination-induced self/tumor–specific T cell responses, but also for identifying the most potent CD8+ T cells for adoptive transfer therapy.
Thus far, a debate remains regarding which parameter(s) of the TCR-pMHC interactions (e.g., KD, koff, kon) could better predict T cell activation and subsequent response potency. Several studies reported that the dissociation rate (koff) was the most significant factor (27, 45), whereas others proposed that the dissociation constant KD was the preeminent correlate of T cell responsiveness (44, 48). However, the association rate parameter, kon, may also contribute to the response potency (49, 50). In that regard, Aleksic et al. (51) and Govern et al. (52) proposed that these apparently contradictory observations might in fact reflect the impact of fast versus slow association rates on the TCR-pMHC binding duration. Indeed, at the cell interface, fast kon rates would allow rapid rebinding of the same TCR-ligand complex after dissociation, resulting in enhanced effective dissociation half-lives. Molecular TCR-pMHC binding interactions are usually assessed by surface plasmon resonance (SPR) measurements in solution (3D binding), which fail to take into account the kon-associated rapid rebinding effect of the TCR to the same pMHC. The NTAmer-based approach deviates in that regard from SPR measurements. Using a panel of CD8+ T cells engineered to express TCR variants of increasing affinities for pMHC, we previously observed that TCRs with fast kon had prolonged NTAmer-based dissociation half-lives compared with those with slow kon (28). Thus, NTAmers may somehow reflect additional membrane-associated kinetic aspects (i.e., impact of rebinding and CD8 coreceptor), which are typically integrated by the 2D surface–based kinetic analyses (reviewed in ref. 53). Despite its current limitations (T cell cloning requirement, no direct kon readouts), the NTAmer technology allows for rapid and accurate real-time off-rate measurements of large panels of naturally occurring antigen-specific CD8+ T cells that may display a broad range of TCR-pMHC affinities, including weak interactions (refs. 28, 29, and current study). Finally, a tight correlation between TCR off-rates and T cell antigenic sensitivity was not always observed, and notably depended on the antigenic specificity of the cells, but also on the T cell functional readout (Figures 1 and 3). However, robust statistical evaluation did not identify consistent outlier clones (i.e., the same clone that behaved as an outlier in one functional assay was not an outlier in the other functional assays). Thus, the few outlier data that we observed might best be explained by the variability/fluctuations related to biological measures (Figure 6), yet we cannot entirely exclude an impact of the kon parameter, possibly influencing T cell responsiveness (49, 50). In-depth kon evaluation of such exceptions would be highly useful, although only feasible once novel technologies that can interrogate all TCR-pMHC binding parameters directly on living T cells become available.
Extending previous studies showing a positive correlation between PD-1 expression and TCR-pMHC avidity (30) or functional avidity (31), NTAmer-based off-rates nicely predicted the upmodulation of both costimulatory (CD28, 4-1BB) and coinhibitory (PD-1, LAG-3, TIM-3, TIGIT) receptors upon antigen-specific stimulation (Figure 3). Thus, our results indicate that T cells of higher binding avidity are more susceptible to activation and subsequent upregulation of activating/inhibitory receptors than lower-avidity ones. Expression of inhibitory receptors such as PD-1 is usually considered a hallmark of T cell exhaustion in chronic infection and cancer, and consequently high-avidity T cells may be more prone to functional impairment. However, Odorizzi et al. (54) recently found that genetic absence of PD-1 on CD8+ T cells does not prevent exhaustion during chronic LCMV infection. Instead, PD-1 also plays a critical role in protecting T cells from overstimulation, excessive proliferation, and terminal differentiation (54), and identifies highly reactive antitumor T lymphocytes (55). Moreover, T cell differentiation and activation are major drivers of inhibitory receptor expression (56). In line with these observations, the extent of coreceptor upmodulation observed following stimulation (Figure 3) likely reveals the overall antigen sensitivity of the T cells, which is mostly driven by TCR-pMHC binding avidity.
Another major finding is that the TCR-pMHC dissociation rate parameter allows the direct comparison across various antigen-specific T cell repertoires, in contrast to functional assays. The latter ones rely on the stability of the pMHC complexes, which is not the case for monomeric TCR-pMHC dissociation experiments. Indeed, the stability of peptide binding to MHC may highly vary between different antigens even when presented by the same HLA-A*0201 molecule. This may help explaining why direct comparisons of in vitro functional avidities (i.e., EC50) between tumor- and virus-specific T cell clones, or between Melan-A26–35 and NY-ESO-1157–165 or CMV/pp65495–504 and EBV/BMFL1259–267 specificities show such divergent differences (Figure 1 and Supplemental Figure 4B). For instance, Melan-A– and EBV-specific T cell clones generally exhibit the lowest EC50 functional avidities, whereas NY-ESO-1– and CMV-specific T cell clones share the highest ones. In contrast, this is no longer an issue for the off-rate measurements, which rely by definition on the dissociation rate between the TCR and a given pMHC complex at the monomeric level. Consequently, we were able to directly compare large T cell clonotype repertoires (n > 300) across 4 different antigenic specificities and confirm strong binding differences between self/tumor and virus-specific CD8+ T cells (Figure 5 and refs. 36, 37). Specifically, virus-specific CD8+ T cell repertoires were endowed with longer TCR-pMHC dissociation rates than self/tumor–specific ones. These data nicely support the concept that many tumor antigens are in fact self-antigens, and consequently mechanisms of central and peripheral tolerance shape the self/antigen-specific repertoires towards lower TCR avidities by removing high-avidity self-reactive T cells (23, 57).
Fluorochrome-conjugated pMHC reagents are widely used for the detection and analysis of antigen-specific CD8+ T cells. Various reports have previously shown that certain functional antigen-specific CD8+ T cells fail to bind tetrameric MHC ligands, which could represent up to several percent of the CD8+ T cell subset (58–60). Moreover, this is of particular importance when staining tumor-specific CD8+ T cells, known to express lower TCR-pMHC affinity/avidity repertoires than virus-specific cells (Figure 5 and refs. 36, 37). We therefore used pMHC multimer and NTAmer molecules to detect tumor-specific CD8+ T cells, which consistently displayed higher sensitivity than Streptamers or pentamers (Supplemental Figure 1A) or pMHC tetramer molecules (data not shown). However, we cannot entirely exclude that a sizeable fraction of antigen-specific T cells may not be stained by these higher-sensitivity tools and may therefore be ignored in our experimental setting.
The Melan-A/MART-126–35 antigenic peptide is among the best-studied human tumor–associated antigens. We have previously documented that the frequency of naive A2/Melan-A26–35–specific CD8+ T cells is unusually high, because of the large numbers selected in the thymus (42). A recent study reported that medullary thymic epithelial cells express a truncated Melan-A transcript, which precludes clonal deletion (central tolerance) to this antigen due to the lack of the expression of the immunodominant 26–35 epitope (61). Another interesting explanation might lay in the impact of certain germ line TCR gene segments, notably the TRAV12-2 gene dominant in the Melan-A antigen–specific T cell repertoire, on contributing substantial binding affinity for the HLA-A2/Melan-A26–35 complex (62). One additional plausible cause of the presence of this large Melan-A26–35–reactive T cell repertoire is that it could be positively selected through the recognition of unknown Melan-A cross-reactive peptides expressed in the thymus (63, 64). Here, we found that naive Melan-A26–35–reactive repertoires isolated from either healthy individuals or unvaccinated melanoma patients depicted an overall poor TCR binding avidity, when compared with the primed repertoires from vaccinated patients (Figure 5). Thus, our observations are compatible with central tolerance mechanisms, possibly involving other cross-reactive self-antigens, and restricting the Melan-A26–35–reactive T cell repertoire to the lower-avidity range. Yet, although rare, our large-scale study could identify few self/Melan-A26–35–specific naive CD8+ T cells of higher binding avidities within healthy individual’s and patient’s repertoires, extending and refining prior studies performed using conventional pMHC class-I fluorescent multimers (65). Therefore, it is possible that therapeutic vaccination allows for the selection and expansion of a wide Melan-A–reactive TCR avidity repertoire, which includes highly specific T cells sharing similar binding avidities to those present in the cancer testis A2/NY-ESO-1–specific repertoire.
Finally, our results highlight the importance of optimizing the choice of tumor antigens for the development of cancer-based immunotherapies. Notably, it remains to be determined whether T cell repertoires targeting tumor-derived neoantigens can display greater TCR-pMHC binding avidities than self/tumor–antigen ones, since neoantigen-specific T cells are more likely to escape thymic negative selection (66). It is tempting to speculate that potent neoantigen-specific CD8+ T cells would display TCR off-rates of a magnitude closer to the kinetics of virus-specific CD8+ T cells shown in this study.
Large-scale ex vivo assessment of TCR-pMHC binding kinetics was until recently technically challenging, underestimating the overall impact and clinical relevance of this biophysical parameter in the context of antigen-specific CD8+ T cell repertoires. Based on monomeric TCR-pMHC off-rate measurements (i.e., NTAmers), we here demonstrated that the koff parameter represents a powerful biomarker to characterize in vitro and in vivo CD8+ T cell potency within antigen-specific CD8+ T cell responses. Yet, robust techniques allowing for the rapid identification and isolation of CD8+ T cells of highest avidity and functions directly ex vivo from tissues or blood samples and at the single-cell level are still required. In that regard, Nauerth and colleagues (67) proposed that small polyclonal virus-specific CD8+ T cell populations could be analyzed directly ex vivo without the need of previous TCR cloning or T cell sorting. The recent implementation of an ex vivo platform allowing for the single-cell serial determination of 2D TCR-pMHC affinity (based on micropipette adhesion frequency) and TCR clonotyping is also highly promising (68). In conclusion, recent technological breakthroughs now enable the rapid development of TCR-pMHC binding kinetics–based simple assays as sensitive and reliable biomarkers of CD8+ T cell activity and clinical efficacy.
Methods
Patients, healthy donors, and ethics statement.
Peripheral blood samples were collected from HLA-A*0201–negative (HD1 and HD2), HLA-A*0201–positive (HD3 and HD4), HLA-A*0201–positive and CMV/EBV chronically infected (BCL4 and BCL6) healthy donors (HDs) (39) and from HLA-A*0201–positive stage III/IV metastatic melanoma patients included in immunotherapy studies (patient LAU50, NCT00112242; patient LAU155, NCT00002669; and patients LAU975, LAU1013, LAU618, LAU627, and LAU818, NCT00112229; www.clinicaltrials.gov) (38, 41, 69). Patients LAU618, LAU627, and LAU818 received 8 to 12 monthly low-dose vaccinations injected s.c. with 100 μg high-affinity Melan-A26–35 (A27L) analog peptide mixed with 0.5 mg CpG 7909/PF-3512676 (Pfizer and Coley Pharmaceutical Group) and emulsified in IFA (Montanide ISA-51, Seppic). Peripheral blood mononuclear cells (PBMCs) centrifuged in Ficoll-Hypaque (Pharmacia) were cryopreserved in 10% DMSO and stored in liquid nitrogen until further use.
Generation of antigen-specific CD8+ T cell clones.
Thawed PBMCs were positively enriched using anti-CD8-coated magnetic microbeads (Miltenyi Biotec), stained in PBS, 0.2% BSA, and 5 mM EDTA with PE-labeled HLA-A*0201 multimers (loaded with analog Melan-A26–35 (A27L), NY-ESO-1157–165 (C165A), and EBV/BMFL1259–267 (C260A), or native CMV/pp65495–504 peptide) (TCMetrix Sàrl) at 4°C for 45 minutes, followed by cell surface markers (APC anti-CD28, FITC anti-CD45RA [BD Pharmingen], PE-Cy7 anti-CCR7 [BioLegend], and APC-A750 anti-CD8 [Beckman Coulter], Supplemental Table 2) at 4°C for 30 minutes. Cells were then sorted into defined differentiated subpopulations (naive, CD45RA+CCR7+CD28+; EM, CD45RA–CCR7–CD28+/–; or EMRA, CD45RA+CCR7–CD28–) of antigen-specific CD8+ T cells on a FACSAria (BD Biosciences) or Astrios (Beckman Coulter) flow cytometer. Sorted cells were cloned by limiting dilution in Terasaki plates and expanded in RPMI 1640 medium supplemented with 8% human serum, 150 U/ml human recombinant IL-2 (gift of GlaxoSmithKline), 1 μg/ml PHA (Sodiag), and 1 × 106/ml 30-Gy-irradiated allogeneic PBMCs. The antigenic specificity of CD8+ T cell clones was controlled by HLA-A*0201/peptide multimer staining (TCMetrix Sàrl). Extensive TCR-BV-CDR3 clonotyping was performed on the T cells from patients LAU618, LAU155, and LAU50 and from healthy donors BCL4 and BCL6, as previously described (39), allowing selecting representative sets of dominant (with frequency > 5%) and nondominant TCR-BV-CDR3 clonotypes. Clonotype diversity varied from 43% to 80%, depending on the antigenic specificity (Melan-A26–35 > EBV/BMFL1259–267 > NY-ESO-1157–165 and CMV/pp65495–504) and is indicated throughout the manuscript.
NTAmer staining and dissociation kinetics measurements.
The pMHC multimer and NTAmer molecules used in this study carry 8 to 12 pMHC monomers per conjugate, similarly to Dextramer molecules. Importantly, multimers and NTAmers provided a superior ex vivo detection of A2/Melan-A–specific CD8+ T cells from PBMCs of 2 melanoma patients, when compared with pentamers (5 pMHC monomers) or Streptamers (5–7 pMHC monomers) (Supplemental Figure 1A). NTAmers are dually labeled pMHC multimers built on NTA-Ni2+-His-tag interactions (70) and were used for dissociation kinetic measurements as described previously (28, 29). Briefly, individual antigen-specific CD8+ T cell clones were stained for 45 minutes at 4°C in PBS, 0.2% BSA, and 5 mM EDTA with antigen-specific NTAmers, in which the HLA-A*0201 molecules were loaded with the native Melan-A26–35, NY-ESO-1157–165, EBV/BMFL1259–267, or CMV/pp65495–504 peptide. Of note, Melan-A– and NY-ESO-1–specific T cells isolated from melanoma patients as well as EBV-specific T cells from healthy donor BCL4 were initially sorted with the analog-peptide multimers. Yet, all Melan-A–, NY-ESO-1–, and EBV-derived T cell clones presented a high degree of cross-reactivity, since native-peptide NTAmers showed a comparable capacity to stably label each generated specific clone and thus should not have introduced a significant bias in the analysis. NTAmer staining was assessed at 4°C on a SORP-LSR II flow cytometer (BD Biosciences). Following 1 minute of baseline acquisition, imidazole (100 mM) was added and Cy5 fluorescence measured during the following 10 minutes. Data were analyzed using the kinetic module of FlowJo software (v.9.7.6, Tree Star) and modeled (1-phase exponential decay) using Prism software (v.6, GraphPad).
Chromium release cytolytic assay.
Chromium release cytolytic assays were performed as previously described (13). Briefly, 51Cr-labeled HLA-A*0201–positive TAP-deficient T2 cells were pulsed with serial dilutions of native Melan-A26–35, NY-ESO-1157–165, EBV/BMFL1259–267, or CMV/pp65495–504 peptides, and incubated with antigen-specific CD8+ T cell clones at an E/T ratio of 10:1 for 4 hours. NY-ESO-1157–165 and EBV BMFL1259–267 peptides were preincubated for 1 hour at room temperature with 2 mM disulfide-reducing agent tris(2-carboxyethyl)phosphine (TCEP, Pierce Biotechnology). Percentages of specific lysis were calculated as 100 × (experimental – spontaneous release)/(total – spontaneous release). EC50 and Bmax values were derived by dose-response curve analysis (log[agonist] versus response) using Prism software. Non–killer clones were defined as displaying a maximal lysis less than 25% and/or for which an EC50 value could not be accurately determined. These non–killer clones were excluded from the statistical analyses.
CD107a degranulation and intracellular cytokine staining.
HLA-A*0201–positive TAP-deficient T2 cells were pulsed 1 hour at 37°C with serial dilutions of the native Melan-A26–35, NY-ESO-1157–165, EBV/BMFL1259–267, or CMV/pp65495–504 peptides, washed, and incubated with antigen-specific CD8+ T cell clones at an E/T ratio of 1:2 for 6 hours in the presence of FITC anti-CD107a (BD Pharmingen; Supplemental Table 2) and brefeldin A (10 μg/ml, Sigma-Aldrich). NY-ESO-1157–165 and EBV BMFL1259–267 peptides were preincubated for 1 hour at room temperature with the disulfide-reducing agent TCEP (2 mM). Cells were then stained in PBS, 0.2% BSA, 5 mM EDTA, and 0.2% NaN3 with Pacific-Blue anti-CD8α (Beckman Coulter) at 4°C for 30 minutes, fixed in PBS 1% formaldehyde, 2% glucose, and 5 mM NaN3 for 20 minutes at room temperature, and finally stained in PBS, 0.2% BSA, 5 mM EDTA, 0.2% NaN3, and 0.1% saponin (Sigma-Aldrich) with PerCPCy5.5 anti–IL-2, APC anti–IL-13, PE-Cy7 anti–IFN-γ, A700 anti–TNF-α (BD Pharmingen; Supplemental Table 2), and PE anti–IL-4 (Biolegend) for 30 minutes at 4°C before acquisition on a Gallios (Beckman Coulter) flow cytometer. Percentages of CD107a/cytokine–positive T cells were analyzed using FlowJo software (v.10.0.7, Tree Star). EC50 and Bmax values were derived by dose-response curve analysis (log[agonist] versus response) using Prism software. Non-cytokine clones were defined as displaying a maximal response less than 25% and for which an EC50 value could not be determined accurately. These non-cytokine clones were not included in the statistical analyses. CD107a, IL-2, IFN-γ, and TNF-α coexpression were analyzed using SPICE software (v.5.35, National Institute of Allergy & Infectious Diseases).
Proliferation assay.
30-Gy-irradiated HLA-A*0201–positive PBMCs were pulsed 1 hour at 37°C with native Melan-A26–35 (10 μM), NY-ESO-1157–165 (1 μM), EBV/BMFL1259–267 (1 μM), or CMV/pp65495–504 (0.01 μM) peptides, washed, and incubated with CellTraceViolet-stained antigen-specific CD8+ T cell clones (Thermo Fisher Scientific) at an E/T ratio of 1:2 in RPMI 1640 medium supplemented with 8% human serum and 50 U/ml human recombinant IL-2. NY-ESO-1157–165 and EBV/BMFL1259–267 peptides were preincubated for 1 hour at room temperature with the disulfide-reducing agent TCEP (2 mM). After 7 days, antigen-specific CD8+ T cell clones were acquired on the Gallios flow cytometer. Percentages of divided cells were analyzed using the proliferation module of FlowJo software (v.9.7.6).
Surface marker expression/modulation assay.
For coreceptor modulation assays, antigen-specific CD8+ T cell clones were incubated for 24 hours in the absence or presence of HLA-A*0201 unlabeled tetramers loaded with native Melan-A26–35 (1 μg/ml), NY-ESO-1157–165 (1 μg/ml), EBV BMFL1259–267 (0.1 μg/ml), or CMV pp65495–504 (0.01 μg/ml) peptides. Cells were then stained in PBS, 0.2% BSA, 5 mM EDTA, and 0.2% NaN3 with (a) A488 anti-PD1 (Serotec), PE-Cy7 anti-CD5 (BD Pharmingen), APC anti-TIGIT (eBioscience), and BrV421 anti-CD28 (Biolegend), or with (b) FITC anti–LAG-3 (Enzo), PE anti–TIM-3 (R&D Systems), and APC anti-CD137 (BD Pharmingen) at 4°C for 30 minutes and acquired on the Gallios flow cytometer. Marker expression (geometric mean fluorescence intensity [gMFI]) was analyzed using FlowJo software (v.10.0.7) and their modulation was calculated as (gMFI of stimulated cells)/(gMFI of un-stimulated cells).
For over-time expression assays, tumor-specific CD8+ T cell clones were stimulated and expanded upon PHA and irradiated feeder cells, and stained over time (at day 10, 15, and 20) in PBS, 0.2% BSA, 5 mM EDTA, and 0.2% NaN3 with FITC anti-CD8β, PE-Cy7 anti-CD8α, PE anti–pan-TCRαβ (Beckman Coulter), PE anti–VLA-1, PE-Cy7 anti-CD5, APC anti–VLA-4, APC anti-CD137, BrV421 anti-PD1 (BD Pharmingen), APC anti-TIGIT (eBioscience), BrV421 anti-CD28 (Biolegend), or FITC anti–LAG-3 (Enzo) at 4°C for 30 minutes, and acquired, using identical settings, on the Gallios flow cytometer. Supplemental Table 2 contains a detailed list and information of all antibodies used in this study.
Adoptive T cell transfer in immunodeficient mice.
NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice (Jackson Laboratory, stock number 005557) were bred in a conventional animal facility at the University of Lausanne under specific pathogen–free status. Six- to nine-week-old female mice were anesthetized with isoflurane and subcutaneously injected with 1 × 106 A2/Melan-A26–35–positive and A2/NY-ESO-1157–165–positive human melanoma Me275 tumor cells (grown in DMEM medium supplemented with 10% FCS, and previously passed in NSG mice for A2/NY-ESO-1157–165–specific experiments). Once the tumors became palpable (around day 14 to 20), 1 × 106 human tumor-specific CD8 T cell clones were injected intravenously in the tail vein. For A2/NY-ESO-1157–165–specific experiments, 1 × 106 T cell clones were administrated twice at day 14 and day 21, followed by 3 daily subcutaneously injections of human recombinant IL-2 (3 × 104 U), starting at the day of T cell transfer. Tumor volumes were measured by caliper twice per week and calculated as follows: volume = length × width × width/2. Mice were sacrificed by CO2 inhalation before the tumor volume exceeded 1,000 mm3 or when necrotic skin lesions were observed at the tumor site. In separate experiments, we collected blood from tail veins at day 2 and 14 after infusion of 4 × 106 A2/NY-ESO-1157–165–specific T cell clones and analyzed the frequency of persisting human CD8+ T cells by flow cytometry. This study was approved by the Veterinary Authority of the Canton de Vaud (Permit number VD1850.5) and performed in accordance with Swiss ethical guidelines.
Statistics.
Data were analyzed using Prism software (v.6, GraphPad) by nonparametric Spearman correlation, nonlinear regression (95% confidence intervals and 10% ROUT coefficient Q; see ref. 71), extra sum-of-squares F, Kruskal-Wallis, Mann-Whitney, Friedman, Wilcoxon-paired, 2-way ANOVA and log-rank tests. The associated P values (2-tailed and α = 0.05 when applicable), as well as numbers of experiments and sample sizes are indicated throughout.
Study approval.
Study protocols were designed, approved, and conducted according to the relevant regulatory standards from (a) the ethical commission of the University of Lausanne (Lausanne, Switzerland), (b) the Protocol Review Committee of the Ludwig Institute for Cancer Research (New-York), and (c) Swissmedic (Bern, Switzerland). Healthy donors and patient recruitment, study procedures, and blood withdrawal were done upon written informed consent.
Author contributions
MA, JS, MH, and NR conceived and designed the study. MA, BC, LCI, MND, JS, GCM, PR, DES, and MH acquired data (provided animals, acquired and managed patients, provided facilities, etc.). MA, MH, and NR analyzed and interpreted data. MA, PR, DES, MH, and NR wrote and/or revised the manuscript. NR supervised the study.
Supplementary Material
Acknowledgments
The authors thank the patients and the healthy donors for their dedicated collaboration to this study. We acknowledge Nicole Montandon and Patricia Werffeli for excellent technical and secretarial help, and Petra Baumgaertner, Alena Donda, Philippe Gannon, and Kalliopi Ioannidou for collaboration and advice. This study was sponsored and supported by the ISREC Foundation (Switzerland), the MEDIC Foundation (Switzerland), the Promedica Foundation (Switzerland), the Swiss National Science Foundation (310030-159417, 31003A-156469 and Sinergia CRSII3-160708), and the Wilhelm Sander-Foundation (Germany).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(14):e92570. https://doi.org/10.1172/jci.insight.92570.
Contributor Information
Mathilde Allard, Email: Mathilde.Allard@chuv.ch.
Barbara Couturaud, Email: couturaud.barbara@gmail.com.
Laura Carretero-Iglesia, Email: laura.carreteroi@gmail.com.
Minh Ngoc Duong, Email: mn.duong@hotmail.com.
Julien Schmidt, Email: Julien.Schmidt@unil.ch.
Pedro Romero, Email: pedro.romero@unil.ch.
Michael Hebeisen, Email: michael.hebeisen@unil.ch.
Nathalie Rufer, Email: nathalie.rufer@unil.ch.
References
- 1.Speiser DE, Kyburz D, Stübi U, Hengartner H, Zinkernagel RM. Discrepancy between in vitro measurable and in vivo virus neutralizing cytotoxic T cell reactivities. Low T cell receptor specificity and avidity sufficient for in vitro proliferation or cytotoxicity to peptide-coated target cells but not for in vivo protection. J Immunol. 1992;149(3):972–980. [PubMed] [Google Scholar]
- 2.Alexander-Miller MA, Leggatt GR, Berzofsky JA. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc Natl Acad Sci U S A. 1996;93(9):4102–4107. doi: 10.1073/pnas.93.9.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallimore A, Dumrese T, Hengartner H, Zinkernagel RM, Rammensee HG. Protective immunity does not correlate with the hierarchy of virus-specific cytotoxic T cell responses to naturally processed peptides. J Exp Med. 1998;187(10):1647–1657. doi: 10.1084/jem.187.10.1647-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeh HJ, Perry-Lalley D, Dudley ME, Rosenberg SA, Yang JC. High avidity CTLs for two self-antigens demonstrate superior in vitro and in vivo antitumor efficacy. J Immunol. 1999;162(2):989–994. [PubMed] [Google Scholar]
- 5.Bullock TN, Mullins DW, Colella TA, Engelhard VH. Manipulation of avidity to improve effectiveness of adoptively transferred CD8(+) T cells for melanoma immunotherapy in human MHC class I-transgenic mice. J Immunol. 2001;167(10):5824–5831. doi: 10.4049/jimmunol.167.10.5824. [DOI] [PubMed] [Google Scholar]
- 6.Almeida JR, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204(10):2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger CT, et al. High-functional-avidity cytotoxic T lymphocyte responses to HLA-B-restricted Gag-derived epitopes associated with relative HIV control. J Virol. 2011;85(18):9334–9345. doi: 10.1128/JVI.00460-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yerly D, et al. Increased cytotoxic T-lymphocyte epitope variant cross-recognition and functional avidity are associated with hepatitis C virus clearance. J Virol. 2008;82(6):3147–3153. doi: 10.1128/JVI.02252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neveu B, et al. Selection of high-avidity CD8 T cells correlates with control of hepatitis C virus infection. Hepatology. 2008;48(3):713–722. doi: 10.1002/hep.22379. [DOI] [PubMed] [Google Scholar]
- 10.Viganò S, Utzschneider DT, Perreau M, Pantaleo G, Zehn D, Harari A. Functional avidity: a measure to predict the efficacy of effector T cells? Clin Dev Immunol. 2012;2012:153863. doi: 10.1155/2012/153863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudley ME, Nishimura MI, Holt AK, Rosenberg SA. Antitumor immunization with a minimal peptide epitope (G9-209-2M) leads to a functionally heterogeneous CTL response. J Immunother. 1999;22(4):288–298. doi: 10.1097/00002371-199907000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Dutoit V, et al. Heterogeneous T-cell response to MAGE-A10(254-262): high avidity-specific cytolytic T lymphocytes show superior antitumor activity. Cancer Res. 2001;61(15):5850–5856. [PubMed] [Google Scholar]
- 13.Speiser DE, et al. A novel approach to characterize clonality and differentiation of human melanoma-specific T cell responses: spontaneous priming and efficient boosting by vaccination. J Immunol. 2006;177(2):1338–1348. doi: 10.4049/jimmunol.177.2.1338. [DOI] [PubMed] [Google Scholar]
- 14.Betts MR, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harari A, Petitpierre S, Vallelian F, Pantaleo G. Skewed representation of functionally distinct populations of virus-specific CD4 T cells in HIV-1-infected subjects with progressive disease: changes after antiretroviral therapy. Blood. 2004;103(3):966–972. doi: 10.1182/blood-2003-04-1203. [DOI] [PubMed] [Google Scholar]
- 16.Ciuffreda D, et al. Polyfunctional HCV-specific T-cell responses are associated with effective control of HCV replication. Eur J Immunol. 2008;38(10):2665–2677. doi: 10.1002/eji.200838336. [DOI] [PubMed] [Google Scholar]
- 17.Wilde S, et al. Human antitumor CD8+ T cells producing Th1 polycytokines show superior antigen sensitivity and tumor recognition. J Immunol. 2012;189(2):598–605. doi: 10.4049/jimmunol.1102165. [DOI] [PubMed] [Google Scholar]
- 18.Almeida JR, et al. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113(25):6351–6360. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clay TM, Hobeika AC, Mosca PJ, Lyerly HK, Morse MA. Assays for monitoring cellular immune responses to active immunotherapy of cancer. Clin Cancer Res. 2001;7(5):1127–1135. [PubMed] [Google Scholar]
- 20.Keilholz U, Martus P, Scheibenbogen C. Immune monitoring of T-cell responses in cancer vaccine development. Clin Cancer Res. 2006;12(7 Pt 2):2346s–2352s. doi: 10.1158/1078-0432.CCR-05-2540. [DOI] [PubMed] [Google Scholar]
- 21.Moran AE, Hogquist KA. T-cell receptor affinity in thymic development. Immunology. 2012;135(4):261–267. doi: 10.1111/j.1365-2567.2011.03547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozga AJ, et al. pMHC affinity controls duration of CD8+ T cell-DC interactions and imprints timing of effector differentiation versus expansion. J Exp Med. 2016;213(12):2811–2829. doi: 10.1084/jem.20160206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zehn D, Bevan MJ. T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity. 2006;25(2):261–270. doi: 10.1016/j.immuni.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hebeisen M, Oberle SG, Presotto D, Speiser DE, Zehn D, Rufer N. Molecular insights for optimizing T cell receptor specificity against cancer. Front Immunol. 2013;4:154. doi: 10.3389/fimmu.2013.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone JD, Chervin AS, Kranz DM. T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity. Immunology. 2009;126(2):165–176. doi: 10.1111/j.1365-2567.2008.03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hebeisen M, Allard M, Gannon PO, Schmidt J, Speiser DE, Rufer N. Identifying individual T cell receptors of optimal avidity for tumor antigens. Front Immunol. 2015;6:582. doi: 10.3389/fimmu.2015.00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nauerth M, et al. TCR-ligand koff rate correlates with the protective capacity of antigen-specific CD8+ T cells for adoptive transfer. Sci Transl Med. 2013;5(192):192ra87. doi: 10.1126/scitranslmed.3005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hebeisen M, et al. Identification of rare high-avidity, tumor-reactive CD8+ T cells by monomeric TCR-ligand off-rates measurements on living cells. Cancer Res. 2015;75(10):1983–1991. doi: 10.1158/0008-5472.CAN-14-3516. [DOI] [PubMed] [Google Scholar]
- 29.Gannon PO, et al. Quantitative TCR:pMHC dissociation rate assessment by NTAmers reveals antimelanoma T cell repertoires enriched for high functional competence. J Immunol. 2015;195(1):356–366. doi: 10.4049/jimmunol.1403145. [DOI] [PubMed] [Google Scholar]
- 30.Hebeisen M, et al. SHP-1 phosphatase activity counteracts increased T cell receptor affinity. J Clin Invest. 2013;123(3):1044–1056. doi: 10.1172/JCI65325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon S, et al. PD-1 expression conditions T cell avidity within an antigen-specific repertoire. Oncoimmunology. 2016;5(1):e1104448. doi: 10.1080/2162402X.2015.1104448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med. 1998;188(12):2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandl JN, Monteiro JP, Vrisekoop N, Germain RN. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity. 2013;38(2):263–274. doi: 10.1016/j.immuni.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fulton RB, et al. The TCR’s sensitivity to self peptide-MHC dictates the ability of naive CD8(+) T cells to respond to foreign antigens. Nat Immunol. 2015;16(1):107–117. doi: 10.1038/ni.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Straetemans T, et al. T-cell receptor gene therapy in human melanoma-bearing immune-deficient mice: human but not mouse T cells recapitulate outcome of clinical studies. Hum Gene Ther. 2012;23(2):187–201. doi: 10.1089/hum.2010.126. [DOI] [PubMed] [Google Scholar]
- 36.Aleksic M, et al. Different affinity windows for virus and cancer-specific T-cell receptors: implications for therapeutic strategies. Eur J Immunol. 2012;42(12):3174–3179. doi: 10.1002/eji.201242606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole DK, et al. Human TCR-binding affinity is governed by MHC class restriction. J Immunol. 2007;178(9):5727–5734. doi: 10.4049/jimmunol.178.9.5727. [DOI] [PubMed] [Google Scholar]
- 38.Derré L, et al. Distinct sets of alphabeta TCRs confer similar recognition of tumor antigen NY-ESO-1157-165 by interacting with its central Met/Trp residues. Proc Natl Acad Sci U S A. 2008;105(39):15010–15015. doi: 10.1073/pnas.0807954105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iancu EM, et al. Clonotype selection and composition of human CD8 T cells specific for persistent herpes viruses varies with differentiation but is stable over time. J Immunol. 2009;183(1):319–331. doi: 10.4049/jimmunol.0803647. [DOI] [PubMed] [Google Scholar]
- 40.Speiser DE, et al. Single cell analysis reveals similar functional competence of dominant and nondominant CD8 T-cell clonotypes. Proc Natl Acad Sci U S A. 2011;108(37):15318–15323. doi: 10.1073/pnas.1105419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Speiser DE, et al. Unmodified self antigen triggers human CD8 T cells with stronger tumor reactivity than altered antigen. Proc Natl Acad Sci U S A. 2008;105(10):3849–3854. doi: 10.1073/pnas.0800080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romero P, Speiser DE, Rufer N. Deciphering the unusual HLA-A2/Melan-A/MART-1-specific TCR repertoire in humans. Eur J Immunol. 2014;44(9):2567–2570. doi: 10.1002/eji.201445004. [DOI] [PubMed] [Google Scholar]
- 43.Hesse MD, Karulin AY, Boehm BO, Lehmann PV, Tary-Lehmann M. A T cell clone’s avidity is a function of its activation state. J Immunol. 2001;167(3):1353–1361. doi: 10.4049/jimmunol.167.3.1353. [DOI] [PubMed] [Google Scholar]
- 44.Tan MP, et al. T cell receptor binding affinity governs the functional profile of cancer-specific CD8+ T cells. Clin Exp Immunol. 2015;180(2):255–270. doi: 10.1111/cei.12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalergis AM, et al. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat Immunol. 2001;2(3):229–234. doi: 10.1038/85286. [DOI] [PubMed] [Google Scholar]
- 46.Falcioni F, et al. Influence of CD26 and integrins on the antigen sensitivity of human memory T cells. Hum Immunol. 1996;50(2):79–90. doi: 10.1016/0198-8859(96)00121-8. [DOI] [PubMed] [Google Scholar]
- 47.Slifka MK, Whitton JL. Functional avidity maturation of CD8(+) T cells without selection of higher affinity TCR. Nat Immunol. 2001;2(8):711–717. doi: 10.1038/90650. [DOI] [PubMed] [Google Scholar]
- 48.Tian S, Maile R, Collins EJ, Frelinger JA. CD8+ T cell activation is governed by TCR-peptide/MHC affinity, not dissociation rate. J Immunol. 2007;179(5):2952–2960. doi: 10.4049/jimmunol.179.5.2952. [DOI] [PubMed] [Google Scholar]
- 49.Huang J, et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature. 2010;464(7290):932–936. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huppa JB, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463(7283):963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aleksic M, et al. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity. 2010;32(2):163–174. doi: 10.1016/j.immuni.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Govern CC, Paczosa MK, Chakraborty AK, Huseby ES. Fast on-rates allow short dwell time ligands to activate T cells. Proc Natl Acad Sci U S A. 2010;107(19):8724–8729. doi: 10.1073/pnas.1000966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu C, Jiang N, Huang J, Zarnitsyna VI, Evavold BD. Insights from in situ analysis of TCR-pMHC recognition: response of an interaction network. Immunol Rev. 2013;251(1):49–64. doi: 10.1111/imr.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Odorizzi PM, Pauken KE, Paley MA, Sharpe A, Wherry EJ. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J Exp Med. 2015;212(7):1125–1137. doi: 10.1084/jem.20142237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gros A, et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124(5):2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Legat A, Speiser DE, Pircher H, Zehn D, Fuertes Marraco SA. Inhibitory receptor expression depends more dominantly on differentiation and activation than “exhaustion” of human CD8 T cells. Front Immunol. 2013;4:455. doi: 10.3389/fimmu.2013.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bouneaud C, Kourilsky P, Bousso P. Impact of negative selection on the T cell repertoire reactive to a self-peptide: a large fraction of T cell clones escapes clonal deletion. Immunity. 2000;13(6):829–840. doi: 10.1016/S1074-7613(00)00080-7. [DOI] [PubMed] [Google Scholar]
- 58.Tungatt K, et al. Antibody stabilization of peptide-MHC multimers reveals functional T cells bearing extremely low-affinity TCRs. J Immunol. 2015;194(1):463–474. doi: 10.4049/jimmunol.1401785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khan N, Cobbold M, Cummerson J, Moss PA. Persistent viral infection in humans can drive high frequency low-affinity T-cell expansions. Immunology. 2010;131(4):537–548. doi: 10.1111/j.1365-2567.2010.03326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang J, et al. Detection, phenotyping, and quantification of antigen-specific T cells using a peptide-MHC dodecamer. Proc Natl Acad Sci U S A. 2016;113(13):E1890–E1897. doi: 10.1073/pnas.1602488113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinto S, et al. Misinitiation of intrathymic MART-1 transcription and biased TCR usage explain the high frequency of MART-1-specific T cells. Eur J Immunol. 2014;44(9):2811–2821. doi: 10.1002/eji.201444499. [DOI] [PubMed] [Google Scholar]
- 62.Cole DK, et al. Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. J Biol Chem. 2009;284(40):27281–27289. doi: 10.1074/jbc.M109.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loftus DJ, et al. Identification of epitope mimics recognized by CTL reactive to the melanoma/melanocyte-derived peptide MART-1(27-35) J Exp Med. 1996;184(2):647–657. doi: 10.1084/jem.184.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dutoit V, et al. Degeneracy of antigen recognition as the molecular basis for the high frequency of naive A2/Melan-a peptide multimer(+) CD8(+) T cells in humans. J Exp Med. 2002;196(2):207–216. doi: 10.1084/jem.20020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pittet MJ, et al. Alpha 3 domain mutants of peptide/MHC class I multimers allow the selective isolation of high avidity tumor-reactive CD8 T cells. J Immunol. 2003;171(4):1844–1849. doi: 10.4049/jimmunol.171.4.1844. [DOI] [PubMed] [Google Scholar]
- 66.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 67.Nauerth M, et al. Flow cytometry-based TCR-ligand Koff -rate assay for fast avidity screening of even very small antigen-specific T cell populations ex vivo. Cytometry A. 2016;89(9):816–825. doi: 10.1002/cyto.a.22933. [DOI] [PubMed] [Google Scholar]
- 68.Zhang SQ, et al. Direct measurement of T cell receptor affinity and sequence from naïve antiviral T cells. Sci Transl Med. 2016;8(341):341ra77. doi: 10.1126/scitranslmed.aaf1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Speiser DE, et al. Rapid and strong human CD8+ T cell responses to vaccination with peptide, IFA, and CpG oligodeoxynucleotide 7909. J Clin Invest. 2005;115(3):739–746. doi: 10.1172/JCI23373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmidt J, Guillaume P, Irving M, Baumgaertner P, Speiser D, Luescher IF. Reversible major histocompatibility complex I-peptide multimers containing Ni(2+)-nitrilotriacetic acid peptides and histidine tags improve analysis and sorting of CD8(+) T cells. J Biol Chem. 2011;286(48):41723–41735. doi: 10.1074/jbc.M111.283127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Motulsky HJ, Brown RE. Detecting outliers when fitting data with nonlinear regression - a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics. 2006;7:123. doi: 10.1186/1471-2105-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.