Abstract
Our previous work showed myocellular differences in pediatric and adult dilated cardiomyopathy (DCM). However, a thorough characterization of the molecular pathways involved in pediatric DCM does not exist, limiting the development of age-specific therapies. To characterize this patient population, we investigated the transcriptome profile of pediatric patients. RNA-Seq from 7 DCM and 7 nonfailing (NF) explanted age-matched pediatric left ventricles (LV) was performed. Changes in gene expression were confirmed by real-time PCR (RT-PCR) in 36 DCM and 21 NF pediatric hearts and in 20 DCM and 10 NF adult hearts. The degree of myocyte hypertrophy was investigated in 4 DCM and 7 NF pediatric hearts and in 4 DCM and 9 NF adult hearts. Changes in gene expression in response to pluripotency-inducing factors were investigated in neonatal rat ventricular myocytes (NRVMs). Transcriptome analysis identified a gene expression profile in children compared with adults with DCM. Additionally, myocyte hypertrophy was not observed in pediatric hearts but was present in adult hearts. Furthermore, treatment of NRVMs with pluripotency-inducing factors recapitulated changes in gene expression observed in the pediatric DCM heart. Pediatric DCM is characterized by unique changes in gene expression that suggest maintenance of an undifferentiated state.
Keywords: Cardiology, Cell Biology
Pediatric DCM is characterized by unique changes in gene expression that suggest maintenance of an undifferentiated state.
Introduction
Heart failure (HF) is one of the leading causes of hospitalization in the US and is associated with substantial adverse effects on quality of life in adults and children. In children, HF is a heterogeneous disease process, with congenital heart disease being the most common indication for heart transplantation in infants and dilated cardiomyopathy (DCM) the most common indication in older children (1).
Treatment for adults with HF is based on results from multiple randomized controlled clinical trials that have demonstrated clear improvement in morbidity and mortality for guideline-recommended treatment modalities. Due to limitations in investigating pediatric populations, there has been only one randomized controlled trial in children with DCM (2). Importantly, the application of proven adult therapies to pediatric patients with DCM has resulted in no substantial improvement in survival over the past three decades, suggesting that pediatric DCM may not be biologically equivalent to its adult counterpart (3). In children with HF from DCM, five-year freedom from death or transplant remains low, ranging from 54%–63%. Therefore, there is a clear need for new approaches to better understand this disease process. Our work has shown differences in the molecular characteristics of pediatric and adult DCM hearts (4–8). These differences suggest that there are underlying myocardial cellular mechanisms uniquely regulated in children with HF.
In this study, we investigated global cardiac transcriptome in a subset of pediatric patients with DCM and in age-matched nonfailing (NF) controls. Significant changes in gene expression that affect cytokine signaling, signal transduction, and transcription were identified. Interestingly, these changes closely resemble maintenance of pluripotency in cardiac progenitor cell populations. Importantly, these changes were associated with patient age and were not observed in adult DCM patients. Furthermore, we identified age-specific morphological differences in hearts from pediatric and adult patients with DCM and found a significant increase in cell size in the adult DMC but no changes in pediatric DCM hearts. To our knowledge, these findings suggest a unique profile in the pathophysiology of pediatric DCM and provide several novel targets for therapeutic intervention and for future research on mechanisms involved in the pediatric failing heart.
Results
Patient characteristics.
Characteristics of patients are listed in Supplemental Table 1 and Supplemental Table 2 for pediatric and adult patients, respectively (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.94249DS1). The median age for pediatric NF donors was 14 years with an interquartile range (IQR) of 9.6 years and 52 years with an IQR of 27 years for adult NF donors. Median age for pediatric DCM patients was 3.9 years with an IQR of 11.3 years and 48 years with IQR of 32 years for adult DCM patients. As expected, angiotensin converting enzyme inhibitor, β blockers, and diuretic treatments were more commonly used in DCM patients compared with NF donors. There were no differences in the use of inotrope or vasopressin agents between NF controls and DCM patients in the pediatric or adult cohort.
Transcriptome profile of pediatric DCM hearts indicate enrichment of pluripotent markers.
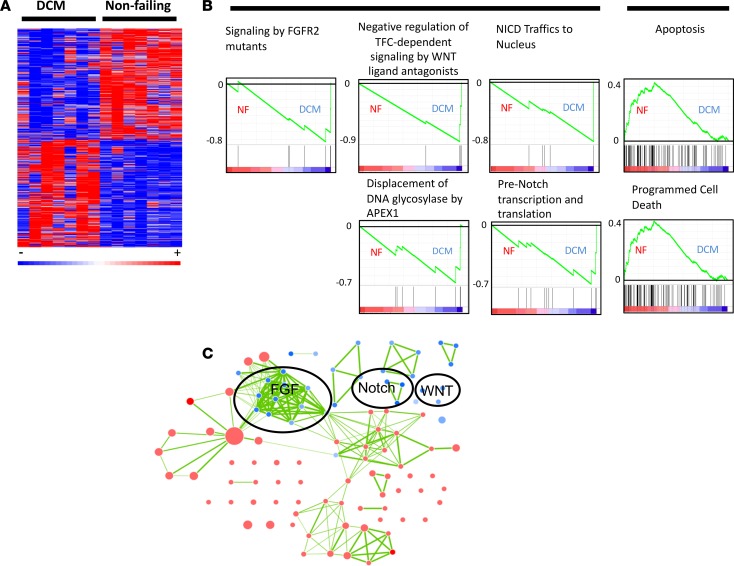
RNA-Seq of pediatric age-matched DCM patients and NF controls identified over 10,000 genes, 1,260 of which had significant differences (P < 0.05) in expression between NF controls and DCM patients (Supplemental Table 3 and Figure 1A). Pathway analysis performed using Gene Set Enrichment Analysis (GSEA) revealed multiple pathways associated with DCM patients (Table 1). Most prominently, FGF, WNT, and Notch signaling were significantly enriched, while the apoptotic pathway was significantly downregulated (Table 1). Enrichment plots of these pathways revealed that a small cohort of genes had a profound effect on significantly enriched pathways in DCM patients (Figure 1B and Supplemental Figure 1). As shown in Supplemental Figure 2, Ingenuity Pathway Analysis (IPA) produced similar analysis, with an expected decrease in apoptosis (Z score = –0.1, P = 0.0014). Network analysis of these pathways revealed a high degree of cross-talk, denoted by 112 nodes sharing 372 edges, suggesting a high degree of cooperation between mechanisms of signal transduction (Figure 1C). Interestingly, the combined activation of Notch, WNT, and FGF signaling is implicated in the induction of pluripotent stem cells (9–12).
Figure 1. Transcriptome profile of pediatric dilated cardiomyopathy (DCM) patients (n = 7 DCM and 7 nonfailing [NF] pediatric hearts).
(A) Heatmap of the 1,260 significantly changed genes (two-sided Wilcox rank-sum t test, P ≤ 0.05). (B) GSEA enrichment profiles in DCM patients and NF controls. These profiles highlight the influence of a small cohort of genes involved in FGF, WNT, and NOTCH signaling in DCM patients (heatmaps of enriched genes are provided in Supplemental Figure 1). Genes are represented by vertical bars, and enrichment is represented by a green line. (C) Cytoscape network of the significantly enriched pathways in both DCM and NF patients. Blue represents pathways associated with DCM patients, and red represents pathways associated with NF controls. Each node represents a single enriched pathway, and each link represents a gene common between pathways. The network accentuates the high degree of crosstalk between enriched pathways.
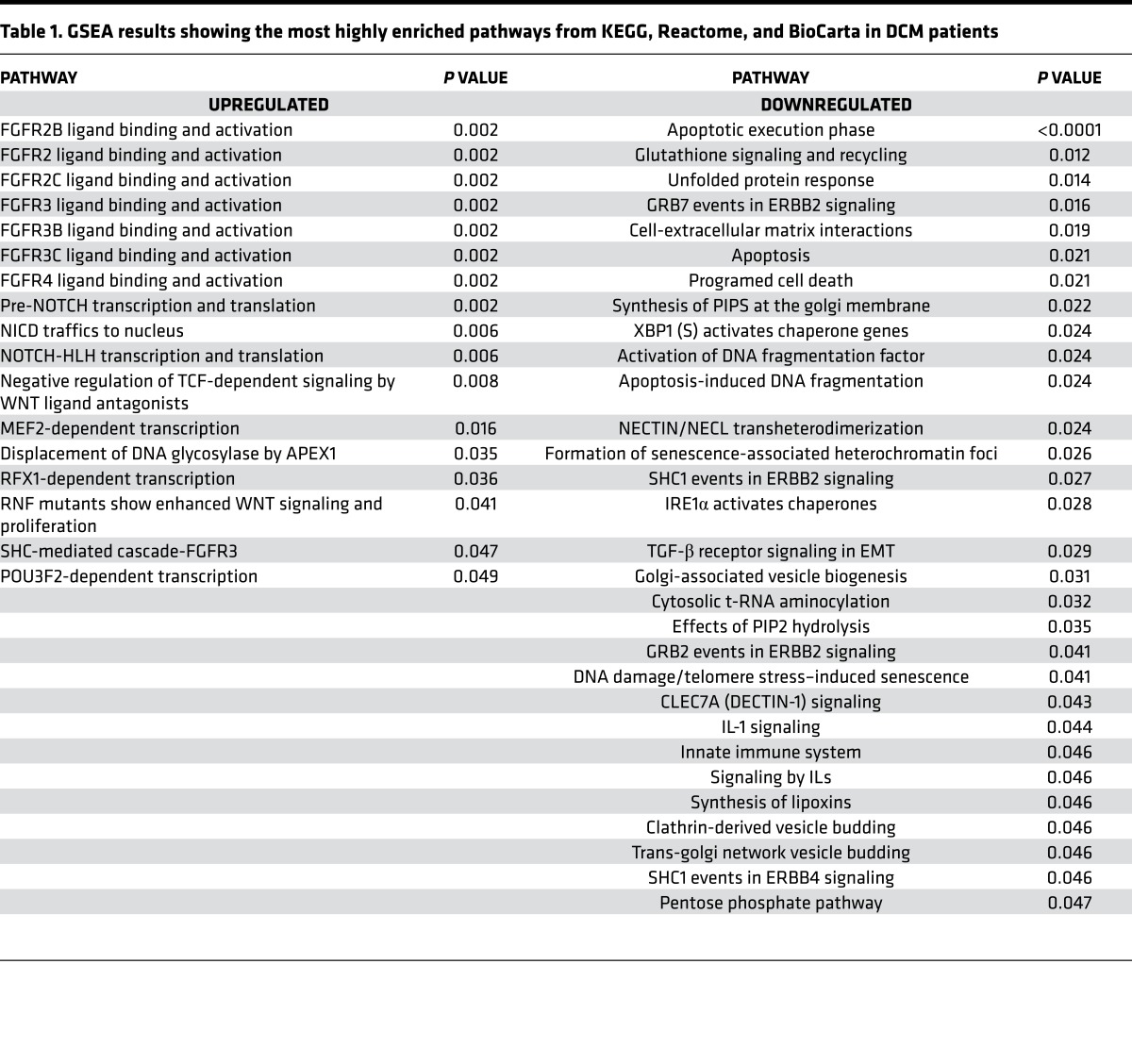
Table 1. GSEA results showing the most highly enriched pathways from KEGG, Reactome, and BioCarta in DCM patients.
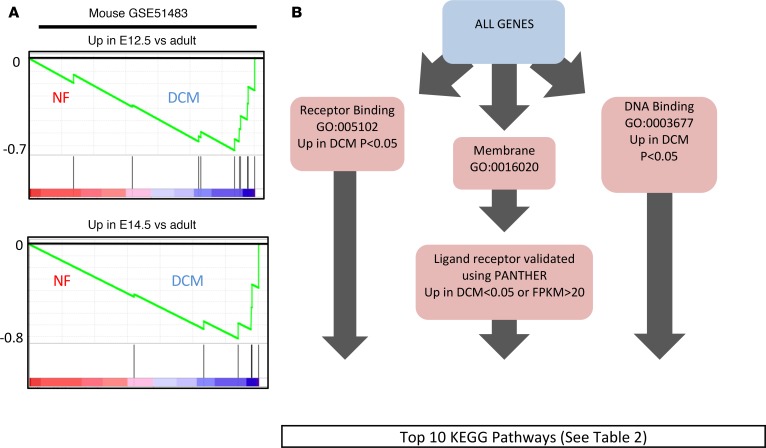
To identify the potential function of these pathways in cardiac tissue, genes enriched in pediatric DCM patients were compared with genes enriched in mouse cardiac development (GSE51483) (Figure 2A and Supplemental Figure 3). Pediatric DCM patients had a significant enrichment of genes unique to embryonic day 12–14 in mouse hearts (day 12.5, P = 0.014; day 14.5, P = 0.02), suggesting that the gene expression profile in cardiac tissue of pediatric DCM patients is comparable with dedifferentiation to an embryonic-like state. In support of these findings, IPA predicted a decrease in development of cardiovascular tissue (Z score = –0.664, P = 0.0054) (Supplemental Figure 4).
Figure 2. Enrichment of pluripotent-associated genes in pediatric dilated cardiomyopathy (DCM).
(A) GSEA enrichment plots showing an enrichment of mouse embryonic day 12.5 (E12.5) and E14.5 genes in pediatric DCM patients (heatmaps of enriched genes are provided in Supplemental Figure 3). These plots demonstrate the coexistence of embryonic-expressed genes in the context of pediatric DCM. (B) Schematic of the workflow used to unbiasedly identify all genes involved in signal transduction. KEGG database enrichment of pathways identified from the results of isolating all genes involved in signal transduction. See Table 2 for a list of the top 10 KEGG pathways identified.
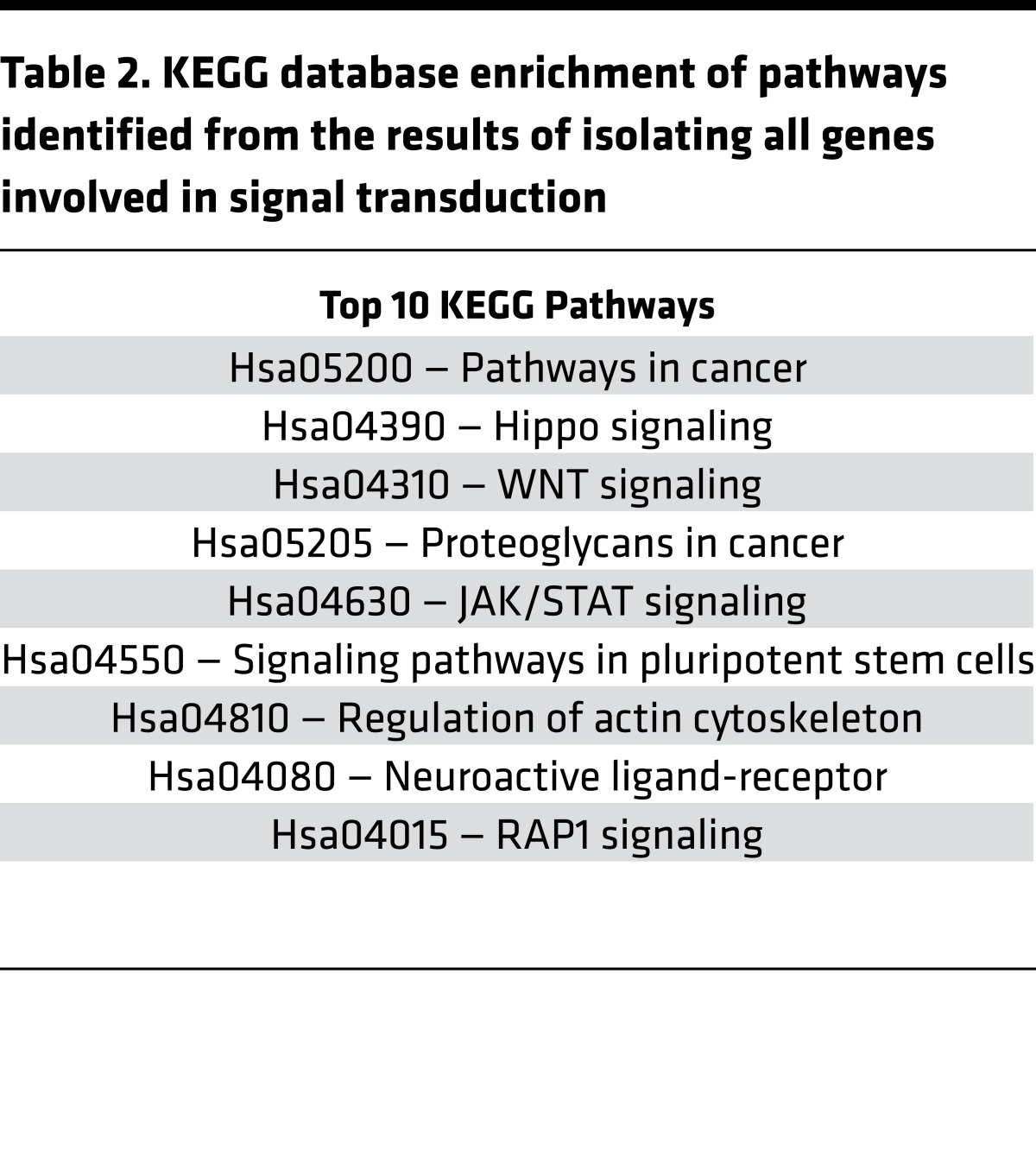
To identify dysregulated signaling pathways, RNA-Seq data was filtered to detect all genes involved in signal transduction, including receptor ligands, membrane receptors, and genes with DNA-binding elements (Figure 2B). Analysis of the resulting cohort of genes using the KEGG database revealed enrichment of signaling pathways involved in pluripotent stem cell signaling (Table 2).
Table 2. KEGG database enrichment of pathways identified from the results of isolating all genes involved in signal transduction.
Expression of genes involved in pluripotent stem cell signaling and cardiac proliferation is unique to pediatric DCM patients.
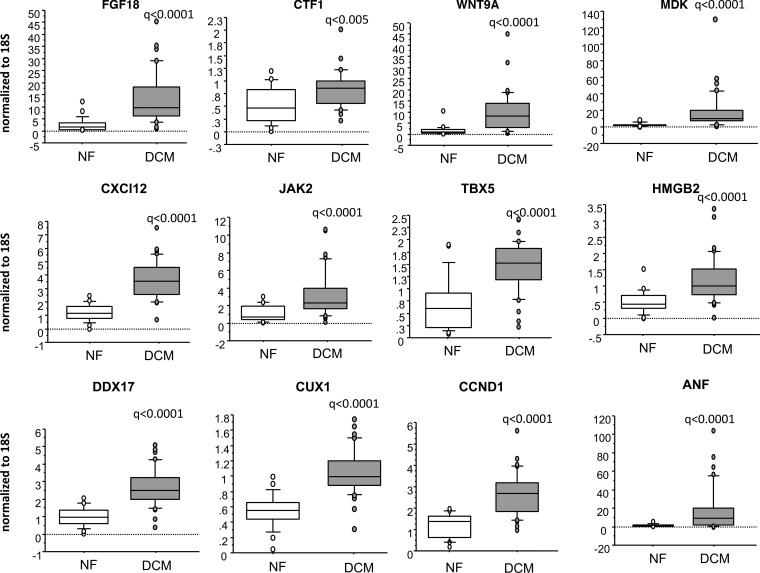
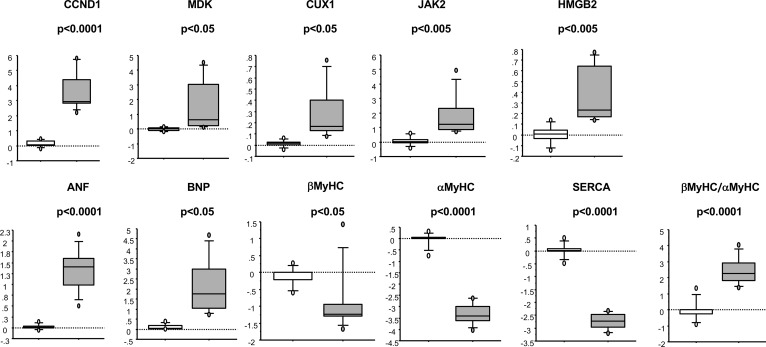
A subset of genes involved in pluripotent stem cell signaling, cardiomyocyte differentiation, and cell cycle progression was selected for validation by real-time (RT-PCR). As shown in Figure 3, the observed increase in expression of cytokines FGF18, CTF1, WNT9A, MDK, and CXCL12; signal transduction gene JAK2; and chromatin/transcriptional regulators TBX5, HMGB2, DDX17, CUX1, and CCND1 were confirmed by RT-PCR. To determine if gene expression correlated with patient age, regression analysis was performed. As shown in Supplemental Table 4 and Supplemental Figure 5, expression of FGF18, CTF1, WNT9A, CXCL12, MDK, and CUX1 correlated with patient age in the pediatric DCM population (P < 0.1).
Figure 3. Genes involved in pluripotency are uniquely dysregulated in pediatric DCM patients.
RT-PCR confirmation of a subset of genes dysregulated in pediatric DCM patients (n = 36 DCM and 21 NF pediatric hearts). Gene expression was normalized to 18S. q values (two-sided Wilcox rank-sum t test) are shown.
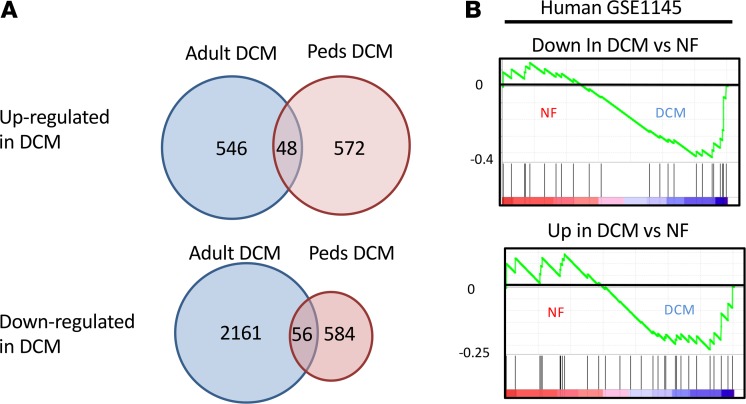
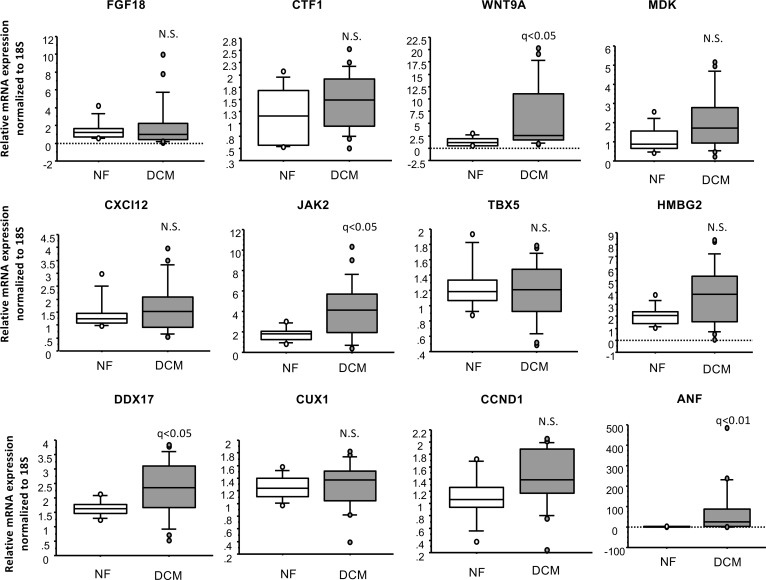
To determine if transcriptome changes in pediatric DCM patients were enriched with adult DCM–related genes, gene expression profiles from human adult DCM and NF controls from GSE1145 were compared with our pediatric DCM dataset (Figure 4A). Only 8.25% of dysregulated pediatric DCM genes were found to be dysregulated in adult DCM patients. Furthermore, GSEA of the two datasets was not significant (Figure 4B and Supplemental Figure 6). When expression of genes related to pluripotency and proliferation was analyzed in adult DCM patients, WNT9A, DDX17, and JAK2 were the only genes dysregulated in the adult population (Figure 5). Atrial natriuretic factor (ANF) was included as a control, since it has been previously shown to be upregulated in pediatric and adult DCM patients (4).
Figure 4. Comparison between pediatric and adult DCM dysregulated genes.
(A) Venn diagram of genes differentially regulated in pediatric and adult DCM patients compared with NF controls. Adult DCM dataset is from GSE1145. (B) GSEA profiles of the enrichment plots showing no enrichment of adult DCM genes in the pediatric dataset (heatmaps of enriched genes are provided in Supplemental Figure 6).
Figure 5. Gene expression analysis in adult DCM patients.
Gene expression analysis was performed by RT-PCR (n = 20 DCM and 10 NF adult hearts). Gene expression was normalized to 18S. q values (two-sided Wilcox rank-sum t test) are shown.
Identification of transcriptional regulators.
In spite of the significant enrichment of cell cycle and stem cell–associated genes, expression of the essential stem cell factors octamer-binding transcription factor 4 (OCT4), sex determining region Y-box 2 (SOX2), kruppel-like factor 4 (KLF4), and NANOG was not observed. However, promoter analysis of the 1,260 differentially regulated genes, in comparison with background DNA fragments, revealed a significant enrichment of KLF4 (P < 0.001, 310 genes), OCT4 (P < 0.001, 104 genes), NANOG (P < 0.001, 190 genes), and SOX2 (P < 0.001, 185 genes) binding sites (Supplemental Table 5).
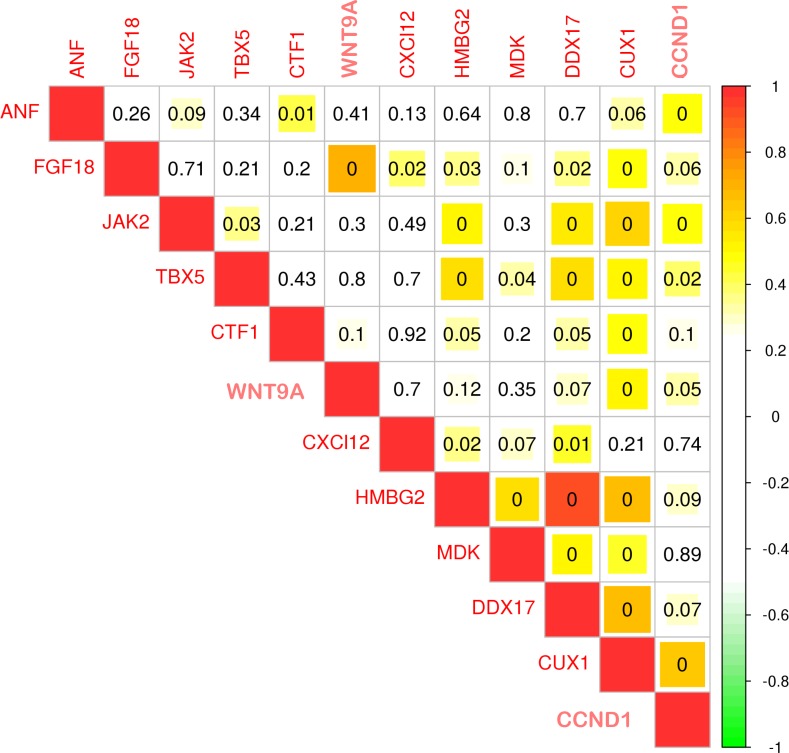
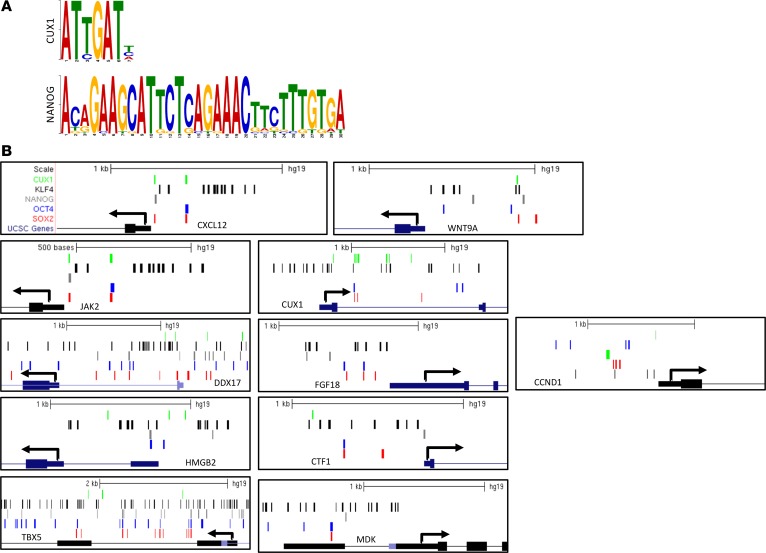
Additionally, to determine if there is a relationship between the validated genes, regression analysis was performed for all 11 RT-PCR–validated genes. Expression of the transcription factor CUX1 positively correlated with all other genes (Figure 6). Moreover, the presence of binding motifs for KLF4, OCT4, SOX2 (motifs from the JASPAR database), NANOG (created from GSE61475), and CUX1 (created from GSM1003622) was investigated (Figure 7, A and B) in all 11 genes. The promoter region of these genes was defined as 500 bp upstream of the transcription start site. This analysis revealed the presence of OCT4 and KLF4 in all validated genes. The SOX2 binding site was not identified in the HMGB2 promoter, the NANOG binding site was not detected in the CUX1 promoter, and the CUX1 binding site was not detected in the promoter of FGF18 and MDK (Figure 7B).
Figure 6. Expression of the transcription factor CUT-like homeobox 1 (CUX1) correlates with other genes tested by RT-PCR.
Spearman rank correlation plot using the RT-PCR data shown in Figure 3. CUX1 has a positive correlation with most genes in the cohort, implicating CUX1 as a potential transcriptional regulator in DCM patients. Numbers in boxes represent correlation P values (≤0.0001). Colors represent R values show on the right of the graph (bar/numbers are representative of R values).
Figure 7. Putative binding site for transcription factors involved in stem cell signaling.
(A) Motifs generated for CUX1 and NANOG using the MEME suit. These motifs were then used for transcription factor binding site analysis for selected gene promoters. (B) Location of CUX1, OCT4, NONAG, KLF4, and SOX2 binding sites in the promoters of the validated stem cell genes.
Cardiomyocyte hypertrophy is not present in pediatric DCM hearts.
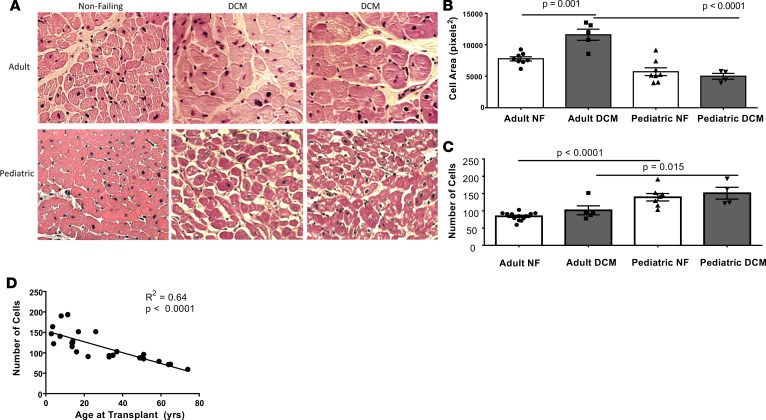
To determine morphologic characteristics of pediatric and adult DCM patients, cell area and number were quantified in pediatric and adult NF controls and DCM patients. In pediatric DCM patients, no changes in cardiomyocyte cell area were observed when compared with pediatric NF controls, whereas in adult patients with DCM, the cardiomyocytes were hypertrophied compared with NF controls (Figure 8, A and B). Moreover, IPA suggests decreased hypertrophy in pediatric DCM patients (Supplemental Figure 7A). Further, there are significantly more cardiomyocytes in left ventricles (LV) from pediatric DCM patients when compared with adult DCM patients or from pediatric NF controls when compared with adult NF controls (Figure 8C). Interestingly, the number of cardiomyocytes correlates with the age of the patient at the time of transplant or donation (Figure 8D). Although hypertrophy was not observed in pediatric DCM myocytes, IPA predicted a decrease in contractility (Z score = –1.578, P = 0.002) and function of the cardiovascular system (Z score = –2.07, P = 0.00022) (Supplemental Figure 6, B and C)
Figure 8. Pediatric and adult DCM cell area and number from 4 dilated cardiomyopathy (DCM) and 7 nonfailing (NF) pediatric hearts and from 4 DCM and 9 NF adult hearts.
(A) Representative images from adult and pediatric NF and DCM left ventricle (LV). 40× magnification. (B) Cardiomyocyte area quantified from adult and pediatric NF and DCM LV. Cardiomyocytes from adult DCM LV have significantly increased area when compared with cardiomyocytes from adult NF LV and pediatric DCM LV (adult DCM vs. adult NF, P = 0.001; adult DCM vs. pediatric DCM, P < 0.0001 [ANOVA with Sidak’s correction]). (C) Quantification of the number of myocytes in adult and pediatric NF and DCM LV. There is a significantly greater number of myocytes in pediatric NF LV compared with adult NF LV. P < 0.0001 (ANOVA with Sidak’s correction). (D) The number of cardiomyocytes correlates with the age of the patient at the time of transplant (or donation) as determined by Spearman rank correlation.
Pluripotent factors activate the fetal gene program (FGP) and induce expression of genes dysregulated in pediatric DCM.
To test the hypothesis that the pattern of gene expression observed in pediatric DCM was consistent with pluripotent signals, NRVMs were treated with WNT activator and leukemia inhibitory factor (LIF), factors involved in maintenance of a proliferative/reprogrammed state in cardiac progenitor cells (13). In addition, cells were treated with FGF2, which induces cardiomyocyte differentiation (14). As shown in Figure 6, treatment with these factors results in upregulation of CCND1, MDK, CUX1, JAK2, and HMGB2 and no changes in expression of FGF18, WNT9A, CXCL12, TBX5, and DDX17 (data not shown). In addition, treatment with these factors resulted in a marked activation of the FGP with upregulation of ANF and BNP, no changes in β myosin heavy chain (βMyHC), an increase in the βMyHC/αMyHC ratio, and downregulation of αMyHC and SR-Ca2+ ATPase (SERCA).
Discussion
Current treatment of pediatric DCM patients is based on clinical trials in adult patients and is without a single Level A evidence treatment recommendation (15). Basing pediatric HF treatment guidelines on data from adults with HF does not take into consideration the possibility of age-related intrinsic differences in biologic factors driving the disease process. We have previously shown that myocellular characteristics of pediatric DCM patients are different than their adult counterparts, including pediatric-specific downregulation of the β2-adrenergic receptor (4), an age-specific miRNA expression profile (5), lack of interstitial fibrosis (8), and a unique molecular response to phosphodiesterase 3 inhibitor therapy (6). These results suggest an age- or development-dependent molecular profile in pediatric DCM patients, warranting an investigation of changes in gene expression specific to the pediatric DCM population. Here, we show that pediatric DCM patients display a unique cardiac gene expression profile that may be linked to incomplete differentiation.
Pluripotent stem cell signaling in pediatric DCM.
Transcriptome analysis shows dysregulation of genes involved in various cellular processes, including pluripotent stem cell signaling, cell growth, and dedifferentiation. Interestingly, previous studies have shown an increase in cardiac progenitor cells in nondiseased infant hearts that are in an incomplete state of cardiac differentiation (16–18). As reported by Amir et al., these cells coexpress cardiac (SERCA) and stem cell markers and have the ability to differentiate into cardiomyocytes (16). Although there is an age-dependent decline in the relative density of these cells in hearts with normal cardiac function, work by Wehman et al. (19) showed an increase in cardiac stem cell numbers in end-stage pediatric HF patients when compared with congenital heart disease controls with normal cardiac function. The number of cardiac stem cells did not decline with age in children with HF, in contrast with the decline seen in age-matched controls without HF. In addition, the authors show that HF-associated cells may have a diminished proliferative ability and reduced functionality, associated with shorter telomeres. The gene expression results presented here are consistent with these previous reports.
As shown in Figure 1, combined database analysis of differentially expressed genes suggests enrichment of FGF (signaling by FGFR2 mutants), WNT (negative regulation of T cell factor–dependent [TFC-dependent] signaling by WNT ligand antagonists and displacement of DNA glycosylase by apurinic/apyrimidinic endodeoxyribonuclease 1 [APEX1]), and Notch (Notch intracellular domain [NICD] traffic to nucleus and pre-Notch transcription and translation) signaling pathways. Combined activation of Notch, WNT, and FGF signaling is implicated in the induction of pluripotent stem cells (9–12). Moreover, when expression of dysregulated genes was filtered to detect genes specifically involved in signal transduction, enrichment of genes involved in cell cycle progression and stem cell pluripotency signaling was observed (Table 2). A detailed description of these genes is provided in the Supplemental Discussion.
Changes in gene expression are enriched in embryonic development.
Importantly, the observed changes in gene expression are enriched in mouse cardiac embryonic development, suggesting a pattern concordant with dedifferentiation (Figure 2 and Supplemental Figure 4). Although this dedifferentiation process is known to be involved in adult HF, it accompanies myocyte hypertrophy. The lack of myocyte hypertrophy (Figure 8 and Supplemental Figure 7), a predicted decrease in apoptosis (Table 1, Figure 1B, and Supplemental Figure 2), and a profile concordant with dedifferentiation suggest a unique myocellular phenotype in children with DCM. Previous studies showed that transgenic overexpression of the transcription factor NKX2–5 resulted in increased expression of ANF and BNP without cardiac hypertrophic remodeling (20), whereas cardiac-specific deletion of GATA4 resulted in cardiac dysfunction and an increase in βMyHC gene expression, but no cellular hypertrophy in response to pressure overload (21). Although changes in gene expression of GATA4 or NKX2–5 were not observed in pediatric DCM, these studies support the possibility of gene expression remodeling and cardiac dysfunction without myocyte hypertrophy. Alternatively, it is possible that these cells never fully differentiated and simply maintain expression of genes present during embryonic development. Since hyperplasia, rather than hypertrophy, is the common mechanism of cell growth during development, these cells may lack the ability to become hypertrophic. Importantly, independent of the presence of cellular hypertrophy, the transcriptome profile of pediatric DCM patients is predicted to promote cardiac dysfunction (Supplemental Figure 7).
Changes in expression of a subset of genes are associated with patient age, suggesting that age of the patient at the time of disease presentation may result in different myocellular adaptations. Furthermore, there is a significant correlation between age at transplant and number of cardiomyocytes. It is unclear if these results indicate myocyte loss with aging or if the increase in cell number in children is due to smaller myocyte size, resulting in more cells per field. Regardless, a thorough characterization of DCM patients based on age at time of disease presentation may be necessary to optimize identification of appropriate therapeutic targets.
Changes in gene expression indicate a high degree of correlation between selected genes.
Expression of a subset of genes dysregulated in pediatric DCM patients was validated by RT-PCR and resulted in 100% concordance between RNA-Seq and RT-PCR; all genes selected for validation were dysregulated by RT-PCR, even when P values approached P = 0.05 (HMGB2 and DDX17) (Figure 3). Although only a subset of genes was validated, this high concordance supports the findings extrapolated for all dysregulated genes. Moreover, RT-PCR analysis showed a significant age-dependent dysregulation in expression of several genes (Supplemental Table 4), indicating that age of disease onset may affect DCM phenotype in pediatric patients. Expression of dysregulated genes was filtered to detect genes specifically involved in signal transduction; enrichment of genes involved in cell cycle progression and pluripotency stem cell signalling was observed (Table 2) and include CCND1 (P = 0.0023) (22), CCND2 (P = 0.00025) (22), CUX1 (P = 0.00468) (23), kinesin-like protein 22 (KIF22) (P = 0.006) (24), chromobox protein homologue 7 (CBX7) (P = 0.0057) (25), KLF2 (P = 0.003) (26), HMGB2 (P = 0.043) (27), and DDX17 (P = 0.05) (28). Additionally, TBX5 (P = 0.015) and GATA6 (P = 0.0038) were upregulated, both of which have been implicated in reprogramming of pluripotent stem cells (13, 29). For a thorough description of validated genes, please see Supplementary Discussion.
As shown in Figure 6, there is a putative relationship between several dysregulated genes. Moreover, expression of the transcription factor CUX1 positively correlates with all genes. In addition, in spite of the significant enrichment of cell cycle and stem cell associated genes, expression of the essential stem cell factors OCT4, SOX2, and NANOG was not observed. However, promoter analysis of dysregulated genes show putative binding sites for CUX1, OCT4, KLF4, SOX2, and NANOG, suggesting that these factors are important for the observed changes in gene expression.
Pluripotent factors induce pathologic remodeling.
Treatment of cardiac progenitor cells with the pluripotency-inducing factors (WNT activator and LIF; ref. 13) and a factor involved in cardiomyocyte differentiation (FGF2; ref. 14) results in stably reprogrammed cells with multipotency properties toward cardiovascular lineages (13). As shown in Figure 9, treatment of NRVMs with these factors resulted in an increased expression of a subset of genes dysregulated in pediatric DCM, as well as in activation of the FGP, a hallmark of pathologic remodeling. These results indicate that induction of pluripotency may negatively affect proper myocyte maturation that, if maintained, can be detrimental to cardiomyocyte function.
Figure 9. Treatment of NRVMs with pluripotent factors activate the fetal gene program (FGP) and induce expression of genes dysregulated in pediatric DCM.
Cells were treated with leukemia inhibitory factor (LIF), WNT activator, and FGF2 for 24 hours and with LIF and FGF2 for another 24 hours. Gene expression was determined by RT-PCR and normalized to 18S. n = 6 independent experiments. CCND1, cyclin D1; MDK, midkine; Cux1, CUT-like homeobox 1; JAK2, janus kinase 2; HMGB2, high-mobility group binding 2; ANF, atrial natriuretic factor; BNP, B-type natriuretic factor; MyHC, myosin heavy chain; SERCA, SR-Ca2+ ATPase. P values were determined by two-sided Wilcox rank-sum t test.
Conclusions.
The results presented here suggest a specific phenotype associated with pediatric DCM that is characterized by lack of myocyte hypertrophy and an pediatric-specific gene expression profile. These results underscore the importance of pediatric-focused investigations and justify attempts to develop targeted therapies for pediatric HF.
Limitations.
There are several limitations to the study. First, since the observed differences in pediatric DCM hearts are based on tissue bank studies, these investigations are cross-sectional, and although in vitro mechanistic studies were performed, in vivo studies are needed to determine if these genes are detrimental to heart function. Furthermore, it is possible that some of these changes are compensatory and do not reflect a pathological response. Second, transcriptome analysis was performed in whole heart tissue, which contains a combination of cardiomyocytes, endothelial cells, and fibroblasts. Therefore, the observed changes in gene expression may not be myocyte specific. In addition, NF control hearts are from brain-dead donors, and the observed changes in gene expression could be influenced by underlying physiologic and metabolic alterations related to their cause of death. Third, although care was taken to properly age-match DCM patients and NF controls when choosing samples subjected to RNA-Seq, RT-PCR confirmation included younger DCM patients. The most common age of presentation in pediatric DCM is in infancy, but NF control infant hearts are not available due to the rarity of unused organs in this population. Therefore, it was not feasible to age-match hearts for the purposes of the RT-PCR confirmation studies. Thus, to the best of our ability, patient age was taken into consideration, and regression analysis of patient age and gene expression/cell morphology was performed. Finally, the vast majority of patients in this study had idiopathic DCM (IDC), allowing us to make comparisons to an analogous adult cohort. However, while IDC remains the most common cause of pediatric DCM, it is difficult to know how applicable our findings are to children with other etiologies of DCM.
Methods
Human samples.
NF hearts were from adult or pediatric organ donors, whose hearts could not be placed for technical reasons (size or blood type mismatch; Children’s Hospital Colorado and University of Colorado Hospital). All patients with DCM had nonischemic cardiomyopathy without any definitive contributing comorbidity. Children with DCM were eligible for inclusion in this study if they had a fractional shortening < 26% or a left ventricular end-diastolic dimension (LVIDd) Z score of > 2 as measured by M-mode at the time of heart transplantation. All pediatric patients were evaluated for structural congenital heart disease, primary arrhythmias, inborn errors of metabolism, neuromuscular disorders, genetic syndromes, and familial disease. Patients with suspicion of myocarditis underwent endomyocardial biopsy at the discretion of the attending cardiologist. The diagnosis of viral myocarditis in this study is reserved for those patients with evidence of lymphocytic infiltrate associated with myocyte destruction on myocardial biopsy or on histologic evaluation of the explanted heart. During the timecourse of this study, cardiac MRI was not readily available for the evaluation of myocarditis and was not performed for any included patient. Patients with congenital heart disease, neuromuscular disorders, or inborn errors of metabolism were excluded from this study. At the time of cardiac transplantation or donation, cardiac tissue was rapidly dissected, flash frozen in liquid nitrogen, and stored long-term at –800C until further use.
RNA extraction and RT-PCR.
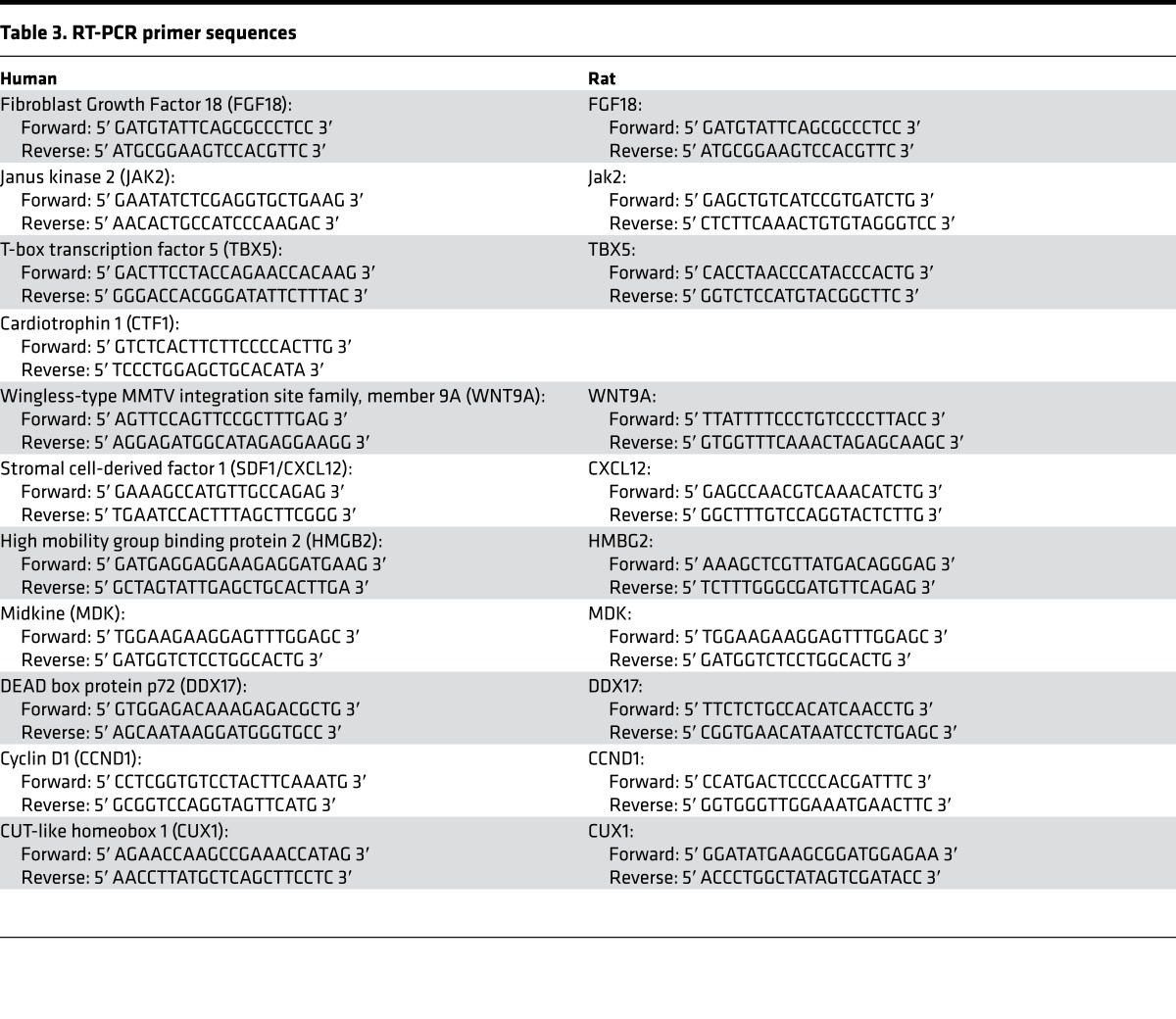
Total RNA was extracted from homogenized LV free wall using the mirVana kit (Ambion) and reverse transcribed into complementary DNA using the iScript cDNA Synthesis Kit (Bio-Rad) as previously described (4). Candidate gene expression was measured by RT-PCR, performed with Power Syber Green PCR Master Mix (Applied Biosystems, Invitrogen) as described (4) (n = 36 DCM and 21 NF pediatric hearts, and n = 20 DCM and 10 NF adult hearts). Primer sequences are listed below.
Human.
Fibroblast Growth Factor 18 (FGF18) (Table 3).
Table 3. RT-PCR primer sequences.
RNA-Seq.
Sequencing (1 × 50) was performed on an Illumina hiSEQ. Samples were demultiplexed and aligned to Hg19 using TopHat2 (30). Aligned reads were normalized and annotated using the cufflinks pipeline (n = 7 DCM and 7 NF pediatric hearts). The patients chosen for RNA-Seq analysis included 7 children with DCM who were age-matched with the NF controls (GSE99321).
Histologic analysis.
Images of H&E stained slides from 9 adult NF LV, 4 adult DCM LV, 7 pediatric NF LV, and 4 pediatric DCM LV were obtained at 40× magnification on a Zeiss Axiovert 200 microscope using an Axiocam MRC camera. A minimum of 5 images from each patient were analyzed for cell area and number of cells using Image J software. The total cell area and number of cells per field for each patient were averaged to determine cell area and number of cells for that patient. Two independent observers (KCW and CCS) conducted the cell measurements in a blinded fashion, and results were averaged.
Neonatal rat ventricular myocyte (NRVM) preparation and treatment.
NRVMs were isolated from the ventricle of 1- to 3-day-old Sprague Dawley rats (Charles River Laboratories) by enzymatic digestions as described previously (31). Cells were treated with 40 ng/ml FGF2, 1,000 U/ml LIF, and 8 μM Chir (WNT/β-catenin activator) (13) for 24 hours and FGF2 and LIF for another 24 hours.
Data analysis.
Significant changes in gene expression were calculated using a two-sided Wilcox rank-sum t test with a p-value threshold of ≤ 0.05. Raw counts of ≥ 10 in any one sample were necessary for inclusion. Differentially expressed genes were then centered about their mean value and clustered hierarchically using an uncentered correlation and average linkage in Cluster 3.0, and they were displayed as a heatmap using Java TreeView. IPA was used to investigate biological processes associated with pediatric DCM.
Several databases were queried to ascertain changes in cell signaling and transcript abundance using GSEA (32). GSEA is a statistical tool that allows global gene expression investigations, such as RNA-Seq and microarray, to be assessed for enrichment of sets of genes based on biological variables using publicly available data. Enrichment is defined based on all the gene-specific scores for a given gene set. All detected genes (independent of significance) were analyzed by GSEA, and significant enrichment as calculated by GSEA is presented. A full description of analysis and data interpretation can be found in http://software.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html
Gmt files supplied by the MSig database maintained by the Broad Institute were used for KEGG and BioCarta data (33), and the 2015 version of the Reactome database (34) was directly downloaded from http://www.reactome.org/pages/download-data/ Transcription factor profiles were also assayed using gmt files supplied by the Broad Institute. To examine cardiac-specific gene expression profiles, representative developmental gene profiles were created from GSE51483, and disease profiles were created from GSE1145. These gene profiles were saved as a gmt file and assayed using GSEA. The results of these studies were displayed visually in CytoScape 3.0 using the expression profile application (35). To attain a higher resolution of the expression profile associated with DCM, gene ontology (GO) terms (36) were used to identify potential receptor ligands, transmembrane receptors, and all genes with a DNA binding motif.
Statistics.
Pearson’s χ2 test was performed to determine if inotrope use was similar between the NF controls and DCM patients. RT-PCR results, cell size, and cell number were analyzed using ANOVA with Sidak’s correction to compare adult and pediatric NF and DCM patients. Regression analysis were based on age and gene expression.
Study approval.
The study complies with the Declaration of Helsinki. Written informed consent, approved by the University of Colorado IRB, for cardiac tissue research use was obtained from DCM subjects or family members of organ donors whose hearts were not suitable for transplantation. All animal work was approved by the IACUC of the University of Colorado Anschutz Medical Campus.
Author contributions
PDT performed RNA-Seq analysis, data interpretation, and manuscript writing; KCW performed histology analysis, data interpretation, and manuscript writing; AKF performed statistical analysis and manuscript review; DAJ performed RT-PCR and data analysis; KN performed RT-PCR and data analysis; MRGT performed experimental planning and manuscript review; JJ and JCC performed patient selection and tissue collection. SDM performed experimental planning, patient selection, and manuscript writing; BLS performed study conception, experimental planning, and manuscript writing; and CCS performed study conception, experimental planning, data analysis, and manuscript writing.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the Children’s Hospital Colorado (CHC) and University of Colorado Hospital (UCH) Cardiothoracic surgical teams including David Campbell and Max Mitchell (CHC), and David Fullerton, and Bret Reece (UCH) for the procurement of heart tissue. The authors would also like to acknowledge Peter Buttrick, Amrut Ambardekar, and Michael Bristow for maintaining the adult tissue bank, and Alix Michael and Scott Kirby from Children’s Hospital Colorado for assistance with collection of historical clinical data on the pediatric patients. Finally, the authors would like to acknowledge Michael Bristow for critically evaluating the manuscript. This work was supported by the Jack Cooper Millisor Chair in Pediatric Heart Disease; the Boedecker Foundation; a gift from the Nair Family; American Heart Association (AHA) Grant-in-aid (grant 13GRNT16950045); and NIH (grant HL126928 and HL107715). Postdoctoral fellow salary was supported by AHA (grant 16POST29970010 to KCW). Also supported by NIH Colorado Clinical and Translational Science Award (grant UL1 TR001082). Contents are the authors’ sole responsibility and do not necessarily represent official NIH views.
Footnotes
Conflict of interest: CCS is a scientific founder and shareholder at miRagen Inc. BLS received research support from Forest Laboratories Inc. CCS, SDM, and BLS are scientific founders and shareholders at CoramiR Inc.
Reference information: JCI Insight. 2017;2(14):e94249. https://doi.org/10.1172/jci.insight.94249.
Contributor Information
Anis Karimpour-Fard, Email: Anis.Karimpour-Fard@ucdenver.edu.
James Jaggers, Email: james.jaggers@childrenscolorado.org.
Karin Nunley, Email: karin.nunley@ucdenver.edu.
References
- 1.Towbin JA, et al. Incidence, causes, and outcomes of dilated cardiomyopathy in children. JAMA. 2006;296(15):1867–1876. doi: 10.1001/jama.296.15.1867. [DOI] [PubMed] [Google Scholar]
- 2.Shaddy RE, et al. Carvedilol for children and adolescents with heart failure: a randomized controlled trial. JAMA. 2007;298(10):1171–1179. doi: 10.1001/jama.298.10.1171. [DOI] [PubMed] [Google Scholar]
- 3.Kantor PF, Abraham JR, Dipchand AI, Benson LN, Redington AN. The impact of changing medical therapy on transplantation-free survival in pediatric dilated cardiomyopathy. J Am Coll Cardiol. 2010;55(13):1377–1384. doi: 10.1016/j.jacc.2009.11.059. [DOI] [PubMed] [Google Scholar]
- 4.Miyamoto SD, et al. Beta-adrenergic adaptation in paediatric idiopathic dilated cardiomyopathy. Eur Heart J. 2014;35(1):33–41. doi: 10.1093/eurheartj/ehs229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stauffer BL, Russell G, Nunley K, Miyamoto SD, Sucharov CC. miRNA expression in pediatric failing human heart. J Mol Cell Cardiol. 2013;57:43–46. doi: 10.1016/j.yjmcc.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakano SJ, Miyamoto SD, Movsesian M, Nelson P, Stauffer BL, Sucharov CC. Age-related differences in phosphodiesterase activity and effects of chronic phosphodiesterase inhibition in idiopathic dilated cardiomyopathy. Circ Heart Fail. 2015;8(1):57–63. doi: 10.1161/CIRCHEARTFAILURE.114.001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano SJ, et al. Cardiac Adenylyl Cyclase and Phosphodiesterase Expression Profiles Vary by Age, Disease, and Chronic Phosphodiesterase Inhibitor Treatment. J Card Fail. 2017;23(1):72–80. doi: 10.1016/j.cardfail.2016.07.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woulfe KC, et al. Fibrosis and Fibrotic Gene Expression in Pediatric and Adult Patients With Idiopathic Dilated Cardiomyopathy. J Card Fail. 2017;23(4):314–324. doi: 10.1016/j.cardfail.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CY, et al. Inhibition of Notch signaling facilitates the differentiation of human-induced pluripotent stem cells into neural stem cells. Mol Cell Biochem. 2014;395(1-2):291–298. doi: 10.1007/s11010-014-2130-3. [DOI] [PubMed] [Google Scholar]
- 10.Huang G, Ye S, Zhou X, Liu D, Ying QL. Molecular basis of embryonic stem cell self-renewal: from signaling pathways to pluripotency network. Cell Mol Life Sci. 2015;72(9):1741–1757. doi: 10.1007/s00018-015-1833-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkins K, Joy S, McKay T. Cell signalling pathways underlying induced pluripotent stem cell reprogramming. World J Stem Cells. 2014;6(5):620–628. doi: 10.4252/wjsc.v6.i5.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romorini L, et al. AKT/GSK3β signaling pathway is critically involved in human pluripotent stem cell survival. Sci Rep. 2016;6:35660. doi: 10.1038/srep35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lalit PA, et al. Lineage Reprogramming of Fibroblasts into Proliferative Induced Cardiac Progenitor Cells by Defined Factors. Cell Stem Cell. 2016;18(3):354–367. doi: 10.1016/j.stem.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ Res. 2012;111(9):1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirk R, et al. The International Society for Heart and Lung Transplantation Guidelines for the management of pediatric heart failure: Executive summary. [Corrected] J Heart Lung Transplant. 2014;33(9):888–909. doi: 10.1016/j.healun.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Amir G, et al. Dynamics of human myocardial progenitor cell populations in the neonatal period. Ann Thorac Surg. 2008;86(4):1311–1319. doi: 10.1016/j.athoracsur.2008.06.058. [DOI] [PubMed] [Google Scholar]
- 17.Mishra R, et al. Characterization and functionality of cardiac progenitor cells in congenital heart patients. Circulation. 2011;123(4):364–373. doi: 10.1161/CIRCULATIONAHA.110.971622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson DL, Mishra R, Sharma S, Goh SK, Deshmukh S, Kaushal S. A strong regenerative ability of cardiac stem cells derived from neonatal hearts. Circulation. 2012;126(11 Suppl 1):S46–S53. doi: 10.1161/CIRCULATIONAHA.111.084699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wehman B, et al. Pediatric End-Stage Failing Hearts Demonstrate Increased Cardiac Stem Cells. Ann Thorac Surg. 2015;100(2):615–622. doi: 10.1016/j.athoracsur.2015.04.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takimoto E, et al. Up-regulation of natriuretic peptides in the ventricle of Csx/Nkx2-5 transgenic mice. Biochem Biophys Res Commun. 2000;270(3):1074–1079. doi: 10.1006/bbrc.2000.2561. [DOI] [PubMed] [Google Scholar]
- 21.Chen HW, et al. Dynamic changes of gene expression profiles during postnatal development of the heart in mice. Heart. 2004;90(8):927–934. doi: 10.1136/hrt.2002.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hotchkiss A, Robinson J, MacLean J, Feridooni T, Wafa K, Pasumarthi KB. Role of D-type cyclins in heart development and disease. Can J Physiol Pharmacol. 2012;90(9):1197–1207. doi: 10.1139/y2012-037. [DOI] [PubMed] [Google Scholar]
- 23.Truscott M, Harada R, Vadnais C, Robert F, Nepveu A. p110 CUX1 cooperates with E2F transcription factors in the transcriptional activation of cell cycle-regulated genes. Mol Cell Biol. 2008;28(10):3127–3138. doi: 10.1128/MCB.02089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y, et al. Inhibition of KIF22 suppresses cancer cell proliferation by delaying mitotic exit through upregulating CDC25C expression. Carcinogenesis. 2014;35(6):1416–1425. doi: 10.1093/carcin/bgu065. [DOI] [PubMed] [Google Scholar]
- 25.Klauke K, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol. 2013;15(4):353–362. doi: 10.1038/ncb2701. [DOI] [PubMed] [Google Scholar]
- 26.Yeo JC, et al. Klf2 is an essential factor that sustains ground state pluripotency. Cell Stem Cell. 2014;14(6):864–872. doi: 10.1016/j.stem.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Taniguchi N, Caramés B, Hsu E, Cherqui S, Kawakami Y, Lotz M. Expression patterns and function of chromatin protein HMGB2 during mesenchymal stem cell differentiation. J Biol Chem. 2011;286(48):41489–41498. doi: 10.1074/jbc.M111.236984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alqahtani H, et al. DDX17 (P72), a Sox2 binding partner, promotes stem-like features conferred by Sox2 in a small cell population in estrogen receptor-positive breast cancer. Cell Signal. 2016;28(2):42–50. doi: 10.1016/j.cellsig.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Wamaitha SE, et al. Gata6 potently initiates reprograming of pluripotent and differentiated cells to extraembryonic endoderm stem cells. Genes Dev. 2015;29(12):1239–1255. doi: 10.1101/gad.257071.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sucharov CC, Mariner PD, Nunley KR, Long C, Leinwand L, Bristow MR. A beta1-adrenergic receptor CaM kinase II-dependent pathway mediates cardiac myocyte fetal gene induction. Am J Physiol Heart Circ Physiol. 2006;291(3):H1299–H1308. doi: 10.1152/ajpheart.00017.2006. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabregat A, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44(D1):D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.