Abstract
DNA double-strand breaks (DSBs) prevents cells from entering mitosis in the presence of genomic damage. Little is known about the response to DSBs once cells have already committed to mitosis. Here we review the genome-protective role of mitotic DNA damage response and evidence suggesting that its untimely activation induces chromosome segregation errors and paradoxically undermines genomic integrity. In contrast to normal cells, cancer cells coopt this pathway to propagate structural and numerical chromosomal instabilities. Cells derived from genomically unstable tumors exhibit evidence for a partially activated DNA damage response during mitosis, which leads to ongoing chromosome segregation errors. Thus, a thorough understanding of the consequences of mitotic DNA is key to our ability to devise novel anti-cancer therapeutic strategies.
Keywords: Mitosis, DNA damage response, DNA repair, Chromosomal instability (CIN), chromosome segregation, Cancer genomic instability, chromosome breaks, copy number variations
MITOTIC DNA DAMAGE RESPONSE: BEYOND CANONICAL DNA REPAIR
The primary function of mitosis is the faithful segregation of the genome into two copies that are equally shared among daughter cells. Mitotic cells are exquisitely sensitive to DNA double-stranded breaks (DSBs) [1–3] to which they can only mount a partial response that stops short of activating downstream DNA damage repair pathways [4] (Figure 1). While the compact architecture of mitotic chromatin is thought to confer a protective role, mitotic cells can still encounter DSBs as a result of persistent pre-mitotic errors [5,6] or due to the direct exposure to DNA-damaging agents [1]. The consequences of such damage have remained elusive. Mounting evidence suggests that untimely activation of the DNA damage response and DSB repair induces severe chromosome segregation defect [6,7]. These intricacies between the response to DNA damage in mitosis and interphase (Figure 1) have important implications not only for normal biology, but also for cancer cells which are frequently exposed to mitotic DNA damage either through unrepaired pre-mitotic errors or exogenous therapies. In this review, we first discuss how induction of the mitotic DNA damage response confers protective properties to mitotic chromosomes intended to maintain their structural integrity allowing for DNA repair once cells exit mitosis. This pathway is likely relevant in event of a handful of DNA breaks. We then examine the consequences of partially activating the mitotic DNA damage response and repair pathways on mitotic timing and chromosome segregation. Finally, we the debate putative roles and evolutionary origins of the mitotic DNA damage response focusing on how this pathway exists at a critical intersection between structural and numerical cancer chromosome instabilities. A better appreciation of this complex relationship is critical to our ability to understand the etiology of genomic instability and the means to target it for a therapeutic benefit.
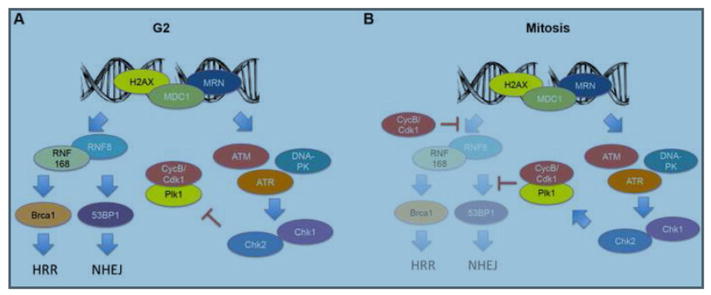
Figure 1. Differences in the DNA damage response between mitosis and the G2 phase of the cell cycle.
(A) Double strand DNA breaks (DSBs) elicit the phosphorylation of the modified histone H2AX (γ-H2AX) as well as the recruitment of mediator of the DNA damage checkpoint 1 (MDC1) and members of the MRN (Mre11-Rad50-Nbs1) complex. This, in turn leads to the activation of the transducers Ataxia-Telangiectasia mutated (ATM), ATR and DNA protein kinase (DNA-PK) and subsequent activation of Chk1 and Chk2 kinases. This leads to the inhibition of polo-like kinase 1 (Plk1) through the degradation of Bora and activation of the G2/M checkpoint preventing the onset of mitosis to allow time for DNA repair. Furthermore, RNF8 and RNF168 interact with MDC1, which allows their recruitment to DSB foci where they mediate the ubiquitination of γ-H2AX. This ubiquitination is key for the recruitment of Brca1 and 53BP1 to DSB foci and initiation of homologous recombination (HR)-mediated DNA repair and non-homologous end-joining (NHEJ), respectively. (B) During mitosis, induction of DSBs still leads to the immediate recruitment of γ-H2AX, MDC1, and members of the MRN complex. Mitotic cells also appear to exhibit activation of DNA damage transducers, however instead of inhibiting Plk1 evidence suggests that DNA damage response activation instead increases the levels of active phosphorylated Plk1. Importantly, mitotic cells inhibit DNA repair pathways though cyclin B/Cdk1-dependent, and mitosis specific, phosphorylation of Threonine-198 on RNF8, which prevents its interaction with MDC1 and recruitment to DSB foci. Furthermore, CDK1 and Plk1 phosphorylate 53BP1 on Threonine-1609 and Threonine-1618 preventing its binding to γ-H2AX. These two, mitosis specific phosphorylation events, effectively eliminate the recruitment of Brca1 and 53BP1 to DSBs and abrogate HR-mediated repair and NHEJ.
The genome protective role of mitotic DNA damage response
The fact that either the depletion of DNA damage response elements or their aberrant activation during mitosis perturbs the process of faithful chromosome segregation demonstrates that this pathway has evolved to play a central role in mitotic genome segregation beyond its canonical function during interphase of the cell cycle. Emerging evidence suggests that the DNA damage response may exist in a semi-activated state during mitosis even in the absence of DNA damage [8] and that activation of some of its components may be achieved through alternative means. Induction of DNA breaks during mitosis leads to rapid and efficient recruitment of early DNA damage response elements that mark the sites of DSBs for subsequent repair [9–11]. However it stops short of activating a full DNA repair program. It is postulated that recruitment of these elements, including the Mre11-Rad50-Nbs1 (MRN) complex, provides structural support during mitosis by tethering broken ends and marking DSB sites until the subsequent G1 phase [9,12,13], which represents the soonest opportunity for NHEJ-mediated repair. Indeed, depletion or inhibition of early components of the DNA damage response during mitosis significantly reduces clonogenic survival after exposure to ionizing radiation [4]. Thus, even if the response to DNA damage is blunted during mitosis, it appears that it exerts a genome-protective role by marking sites of damage for subsequent repair therefore preserving the structural integrity of chromosomes.
The exact point at which DSB repair is inactivated is not entirely clear. Evidence suggests repair can occur until early prophase. Replication stress, a common product of cellular oncogenic transformation, induces POLD3-dependent DNA synthesis in prophase, the earliest stage of mitosis [14]. This is supported by observation that TopBP1 marks sites of incompletely replicated DNA in early mitosis and promotes unscheduled DNA synthesis [13]. However, when TopBP1 foci persist beyond prophase, they transition into 53BP1 nuclear bodies, which are marked for repair in the subsequent G1 phase [13,14]. This might represent the point at which the role of the DNA damage response transitions from complete DNA repair to temporary marking of damaged DNA sites for the purpose of repair during the subsequent cell cycle. The genome protective role of the mitotic DNA damage response might explain why mitotic chromosomes are likely to remain physically in unity [9,15–17] when exposed to direct DSBs despite the inactivation of DSB repair pathways.
Beyond inactivating downstream DNA repair, there are examples suggesting that the DNA damage response may be ‘wired’ differently between mitosis and interphase of the cell cycle. For instance, during mitosis, Chk2 can be activated independently of ATM, likely through phosphorylation by DNA-PK, Plk1, or MPS1 kinases [8,18,19]. Furthermore, exposure to ionizing radiation leads to its phosphorylation at Serine-9 but not Threonine-68 [4,6]. Interestingly, the type and location of DNA damage may also have a role in the damage response signaling; deprotected telomeres during mitosis activate ATM but not its downstream target, Chk2 [20]. How various types of mitotic DNA damage engage DNA damage response and repair components is unknown and these intricacies warrant further in-depth investigation.
CONSEQUENCES OF DNA DAMAGE DURING MITOSIS
The effect of DNA damage response activation on mitotic timing
Beyond its role in marking sites of DNA damage, the consequences of activating the DNA damage response during mitosis have remained elusive. Mounting evidence reveals that activation of the mitotic DNA damage response can paradoxically lead to deleterious effects on genome stability [6,7]. Partial induction of DNA damage response in mitosis perturbs several aspects of mitosis including mitotic timing and anaphase chromosome segregation (Figure 2). Work in multiple species has linked DNA damage response activation with a delay in mitotic progression (Figure 2B). DNA damage in Drosophila gastrula also delays anaphase onset [21]. In human cells, induction of DNA damage also leads to varying delays in anaphase onset [22–24]. The exact mechanism by which this mitotic delay occurs is not entirely understood, however it may be dependent on either the extent or the location of the damage incurred. Mitotic cells are exquisitely sensitive to telomere integrity, and damage to a single telomere is sufficient to significantly delay anaphase [24]. This effect is abrogated through the depletion of ATM, Chk1 or MPS1. Further, telomere fusion has been shown to induce an ATM-mediated mitotic arrest and death either during, or shortly after, mitosis [20,25,26]. This is supported by observations that damage at telomeres recruits additional response and repair factors compared to non-telomeric mitotic damage [24]. Along the same vein, cells exposed to as much as 12Gy – a dose of ionizing radiation that produces as many as ~420 DSBs per cell [27] but not specifically at the telomeres – are not the subject of an appreciable mitotic delay [6]. Thus, in the case of DNA damage-induced mitotic delay, location might be ascendant to the number of DSBs.
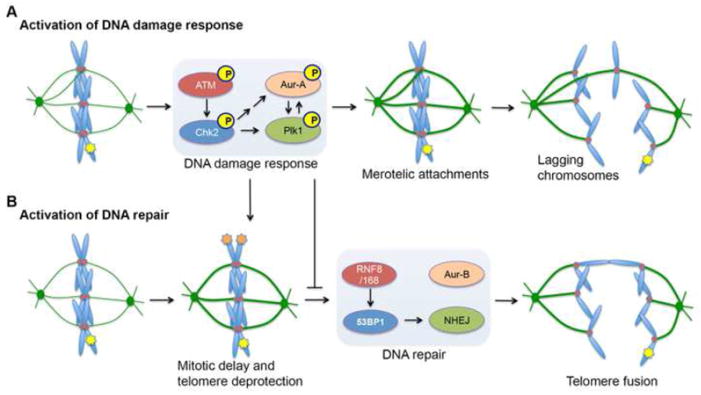
Figure 2. Consequences of activating the DNA damage response and DNA repair during mitosis.
(A) Induction of DNA double-strand breaks (DSBs, depicted by yellow star on chromosome arm) during mitosis elicits a partial DNA damage response. The ATM/Chk2 arm of this mitotic DNA damage response was shown to increase kinetochore-microtubule (kMT) attachment stability by activating the mitotic kinases Aurora-A and Plk1. Increased kMT attachment stability increases the probability that erroneous kMT attachments (termed merotelic attachments) persist until anaphase onset. In this scenario, individual chromatids are simultaneously attached to microtubules emanating from opposite spindle poles and when these attachments persist until anaphase, they produce lagging chromosomes, which are a hallmark of whole-chromosome mis-segregation and numerical chromosomal instability. (B) In addition, the mitotic DNA damage response also leads to delay in mitotic progression, which was shown to lead to the deprotection of telomeres (depicted by orange stars on chromosome ends). Interestingly, however, the mitotic DNA damage response inhibits NHEJ and it was recently shown that ectopic activation of NHEJ leads to Aurora-B-mediated fusion of deprotected telomeres. Consequently, this can lead to dicentric chromosomes during anaphase and structural chromosomal instability.
While the telomeric damage-induced arrest is related to ATM activation [24], subtle delays in anaphase onset in response to other forms of chromosome damage may be dependent on excessive stabilization of Cyclin A, whose degradation is required for mitotic progression [21]. Furthermore, mitotic cells with DNA damage accumulate the spindle assembly checkpoint proteins, Mad2, Bub1 and BubR1, at kinetochores, but this might reflect damage within centromeric DNA, which impacts on stable kinetochore attachment to microtubules leading to the activation of the SAC and a delay at the metaphase to anaphase transition [6,22].
The benefits of delaying mitotic exit are unclear. One possibility would be enable time for the correction of erroneously attached kinetochore-microtubules when the cell senses that genomic integrity is at risk. However this proposition is at odds with observation discussed at a later point, showing how partial DNA damage response activation during mitosis also leads to indiscriminate increased kinetochore-microtubule stability undermining chromosome segregation fidelity [6]. On the other hand, it has long been known that cancer cells spend longer time in mitosis compared to their normal counterparts [28]. The basis of this delay is not fully understood, and persistent activation of mitotic DNA response pathways could represent a widespread mitosis-prolonging mechanism in cancer cells. After all, cancer cells experience mitotic DNA damage from unrepaired pre-mitotic errors caused by genomic instability. It was recently shown that cancer cells must balance their level of chromosome instability to maintain adequate adaptive potential while preventing genomic collapse from excessive instability [29,30]. One proposed mechanism for the ability of tumor cells to temper excessive instability is anaphase delay. Could cancer cells be coopting a well-intentioned pathway to modulate their genomic instability? The answer to this important question will emerge as we begin to tease out the precise molecular mechanisms that link the different types of DNA damage to mitotic progression.
DNA damage response activation during mitosis leads to whole chromosome missegregation
Induction of DNA damage during mitosis or independent pharmacologic activation of the DNA damage response in the absence of DSBs were shown to increase the formation of lagging chromosomes during anaphase leading to whole-chromosome missegregation and karyotype instability [6]. These segregation defects were a consequence of increased of kinetochore-microtubule attachment stability (Figure 2A). This was dependent on the activity of ATM and Chk2, as pharmacologic inhibition or genetic depletion of either transducer kinases during mitosis abolished the effect of the DNA damage response pathway on lagging chromosome rates after the induction of DNA damage. The regulation of kinetochore-microtubule attachments is a complex process and evidence demonstrates that the stability of these attachments must fall within a narrow permissible window to allow for faithful chromosome segregation [31–33]. Increasing attachment stability beyond this permissible range inappropriately stabilizes erroneous kinetochore-microtubule attachments and allows them to persist until anaphase onset, at which point they lead to lagging chromosomes and chromosome mis-segregation [34]. Interestingly, mitotic DNA damage also led to increased levels of phosphorylated Plk1 and Aurora A – two important mitotic kinases. Their enzymatic activity is necessary to mediate the effects of the DNA damage response on kinetochore-microtubule attachments and chromosome segregation [6,35]. This is in stark contrast to the DNA damage response during the G2 phase of the cell cycle, where DNA damage prevents mitotic entry through the inactivation of Plk1 and Aurora-A [36–38]. These observations, together with other evidence [4,6,7,24], suggest that DNA damage response signaling during mitosis may be wired differently compared to interphase of the cell cycle. A potential explanation for Plk1 activation during mitosis in response to DNA damage may lie in the requirement to suppress downstream DNA repair activity by phosphorylating evolutionarily conserved sites on RNF8 and 53BP1, both of which contain Plk1 consensus motifs [7,39]. Thus, the execution of the partial DNA damage response pathway in mitosis protects genome integrity from structural damage but this comes at the expense of increasing the likelihood of numerical chromosomal instability. The purpose of this paradoxical relationship is unknown; its putative roles in normal development and how it may impact genomic instability in cancer will be discussed below.
Consequences of DSB repair activation during mitosis
During mitosis, DSB repair downstream of the DNA damage response is inactivated [4]. To understand the relevance of this inhibition, Orthwein et al. created unphosphorylatable mutants of RNF8 and 53BP1, which restored mitotic DSB repair [7]. Cells expressing these mutants were extremely sensitive to ionizing radiation and mitotic DNA damage. Similar to partial activation of the mitotic DNA damage response [6], activation of DNA repair also led to missegregation of whole chromosomes, as seen by an increased rate of kinetochore-positive micronucleus formation. In addition, however, these cells exhibited markedly increased rates of sister telomere fusions (Figure 2B). Telomeres can become deprotected during prolonged mitotic arrest [20,24], and activation of DNA damage repair during mitosis leads to telomere joining in an Aurora kinase B-dependent manner [7,26]. The consequences of this unscheduled repair are severe structural and numerical chromosomal aberrations that are triggered by telomere fusion. Therefore, normal cells have evolved to robustly silence DNA repair mechanisms downstream of DNA damage during mitosis to prevent aberrant NHEJ of broken DNA ends and deprotected telomeres, which would lead to mitotic catastrophe. Activation of DNA repair during mitosis risks propagating structural chromosomal instability as the dicentric chromosomes that are produced after telomere fusions are subject to further structural damage when cells undergo anaphase (Figure 2B).
PUTATIVE FUNCTIONS OF THE MITOTIC DNA DAMAGE RESPONSE IN NORMAL CELLS
The evolutionary relevance of maintaining the ability to mount a partial DNA damage response during mitosis, at a time when the cell is chiefly preoccupied with the process of chromosome segregation, is not entirely clear. One possibility is that cells may need to address DNA damage during mitosis that arises from unrepaired DNA damage upon release from a G2/M checkpoint arrest. The G2/M checkpoint has an estimated threshold of 10–20 DSBs below which cells can enter mitosis [26]. A second possibility is that cells need to address DNA damage that arises from DNA replication defects. For instance, incomplete replication of chromosomes would lead to chromatin bridges that must be resolved to allow for the physical segregation of chromosomes. This may require a DNA damage response-mediated activity to resolve chromatin bridges during mitosis in order to allow for the completion of cytokinesis. Recent observations reveal that lagging chromosomes at the spindle mid-zone is vulnerable to breakage [40], yet it is unclear whether these breaks are the direct consequence of physical interference by the cytokinetic furrow or if they arise from a enzymatically-induced break. Recent work on telomere fusion showed that induction of DNA damage responsive pathways underlies nuclease-dependent resolution of chromatin bridges during anaphase [41]. In a similar vein, Duda et al. showed that another nuclease, MUS81, resolves persistent replication intermediates at mitotic onset, in a DNA-damage response-dependent manner, to prevent errors in chromosome segregation [42]. Thus the mitotic DNA damage response would protect the structural integrity of mitotic chromosomes during segregation by facilitating the resolution of missegregating DNA at the spindle mid-zone, albeit at the risk of inducing numerical karyotype aberrations.
A third putative function of DNA damage response activation during mitosis would be to trigger cellular senescence or death in response to unrepaired damage in order to clear cells with a defective genome from otherwise normal tissues. This is particularly critical to the cycling stem cell population where genomic damage can be inherited and propagated for many generations. While a small number of DSBs during mitosis may be marked for repair during the subsequent G1 phase [9–11], a more significant amount of damage would have to be repaired through error-prone, NHEJ pathway [43]. In these cases, cell death or senescence may be a preferable choice to error-prone repair. By directly causing whole chromosome missegregation and subsequent aneuploidy, the DNA damage response could induce a p53/p21-mediated cellular senescence in the subsequent G1 phase in response to aneuploidy [44]. Unlike p53 activation from DNA damage, aneuploidy is an irreparable defect and as such a p53-mediated arrest would therefore be irreversible or at least long lasting. Interestingly, work on cleavage-stage human embryos reveals that segmental deletions, duplications, and amplifications from breakage-fusion-bridge cycles occur at measurable rates [45]. Instead of attempting to repair damage at these early stages of embryogenesis, a putative role for the mitotic DNA damage response during development could be to clear these cells through irreversible induction of aneuploidy and p53-mediated arrest.
Non-transformed mitotic cells may also undergo cell death, or crisis, in response to chromosomal damage in the absence of p53. The mechanism underlying this has long been elusive. Recently Hayashi et al. found that telomere deprotection and fusion during mitosis elicits a DNA damage response leading to p53-independent cell death [25]. Normal cells in which p53 has been inactivated are prone to enter mitosis with persistent DNA damage and they often contain deprotected and fused telomeres during mitosis. The authors found that ATM underlies both telomere fusion through DSB repair as well as a robust mitotic arrest [24]. This prolonged arrest in turn leads to an Aurora B-mediated telomere deprotection and mitotic catastrophe and cell death during, or shortly after, mitosis. Others have also found that in cases where the G2/M checkpoint is overridden cells trigger an Aurora-B and Survivin-mediated mitotic arrest [46]. Interestingly, cells that lack the ability for mitotic arrest escape crisis and go on to develop genomic instability [25], demonstrating that mitotic DNA damage response can also play a role in safeguarding genome stability in the absence of p53-mediated senescence.
MITOTIC DNA DAMAGE IN CANCER CELLS: LINKING STRUCTURAL AND NUMERICAL CHROMOSOMAL INSTABILITIES
Cancer cells can experience mitotic DNA damage as a result of accumulated pre-mitotic damage from replication stress or a defective G2/M checkpoint [5,6,47]. Alternatively, they can also experience de novo damage after cells have already committed to mitosis when exposed to DNA damaging therapies or when lagging chromosomes or a chromatin bridges becomes trapped in the cleavage furrow during cytokinesis [40,41]. Another source of mitotic DNA damage could arise from errors during chromosome segregation that had occurred during the preceding mitosis. Lagging chromosomes can become encapsulated into micronuclei that have defective micronuclear envelopes and DNA replication machinery [48]. This in turn leads to micronuclear envelope collapse and chromosome pulverization, a process that leads to complex structural rearrangement, a process otherwise known as chromothripsis [49,50] (Figure 3). If left uncorrected, pulverized chromosomes can persist into the subsequent mitosis inducing a mitotic DNA damage response and closing the loop between structural and numerical chromosome instabilities. Of note, telomere fusion can also lead to a similar process [41]. How mitotic DNA damage and the ensuing response pathways interplay with cancer genomic instability is only beginning to emerge. In a number of chromosomally unstable cancer-derived cell lines, pharmacologic inhibition of Chk2 during mitosis reduced inherent chromosome segregation errors [6]. In these cells, increased basal levels of γ-H2AX and phosphorylated Chk2 were observed, suggesting baseline activation of the DNA damage response during mitosis. Further, the extent of reduction in segregation errors upon Chk2 inhibition was directly proportional the amount of γ-H2AX foci seen, supporting the notion that activation of the DNA damage response during mitosis may underlie frequent chromosome segregation errors in cancer cells [6]. In colorectal cancer-derived chromosomally unstable cell lines, cells with pre-mitotic replication stress were more likely to manifest γ-H2AX foci in early mitosis and these cells exhibited errors in anaphase chromosome segregation [5]. Thus, activation of the mitotic DNA damage response may be a widespread phenomenon in chromosomally unstable cancer cells and it arises from an inherent and persistent level of DNA damage during mitosis. More importantly, these studies point toward the possibility that cancer cells have coopted an otherwise genome protective role of the mitotic DNA damage response in order to propagate genomic instability by linking pre-mitotic, or mitotic, DNA damage to numerical and structural chromosomal instabilities that results from chromosome segregation errors during anaphase.
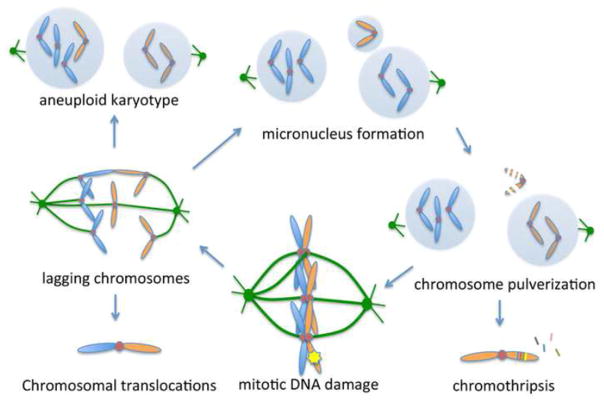
Figure 3. The mitotic DNA damage response links structural and numerical cancer chromosome instabilities.
By inducing chromosome segregation defects, mitotic DNA damage can lead to telomere fusions and structural aberrations leading to chromosome translocations. Lagging chromosomes can missegregate leading to aneuploidy or alternatively can be sequestered into micronuclei which are defective in DNA replication and lead to pulverization of their enclosed chromosome. This widespread structural chromosomal damage can persist into the subsequent mitosis leading to continued cycle of genomic instability. Alternatively, pulverized chromosomes can be repaired in a random fashion leading to focal complex rearrangements and deletions, a process known as chromothripsis.
The role of the mitotic DNA damage response in propagating structural and numerical chromosomal aberrations in cancer cells has important therapeutic implications. First, it suggests that widely used DNA-damaging therapies, such as ionizing radiation, cisplatin, doxorubicin, among others, may impart their cytotoxic effect at least in part, through their interference with the process of anaphase chromosome segregation [51]. While chromosomally unstable cancer cells exhibit elevated rates of chromosome missegregation, they need to optimize these rates in order to thrive [29]. Markedly low chromosome missegregation rates cannot support genomic heterogeneity required for tumor evolution whereas excessively elevated rates are likely deleterious to cellular viability [30,52]. This implies the presence of an evolutionary pressure on chromosome missegregation rates that balance survival with cellular viability and allows clonal populations to rapidly acquire abnormal karyotypes supporting increased fitness. Therefore the exposure of an already chromosomally unstable tumor to DNA-damaging therapies is likely to drive tumor cells from an optimal fitness state that supports tumor evolution into a state so overwhelmingly unstable that it is no longer compatible with viability. This hypothesis is supported by a large number of clinical observations where moderate rates of chromosomal instability are most likely to confer therapeutic resistance and poor prognosis whereas either extremely high or extremely low chromosome missegregation rates were associated with a favorable prognosis and higher odds of treatment response [53–57]. This hypothesis was also experimentally tested in an orthotopic mouse model of human glioblastoma where chromosome missegregation rates were genetically suppressed during exposure to radiation treatment and the resultant tumors exhibited striking radiation resistance [51]. These findings are relevant to concerted efforts aiming to target components of the DNA damage response [58–61]; inhibition of the DNA damage response would be expected to yield different outcomes depending on the phase of the cell cycle during which therapy is delivered and the effect of these therapies on the mitotic tumor cell population should be carefully considered. In support of this, Lee et al. developed a genetic model based on a human artificial chromosome to directly measure chromosome loss in response to treatment with widely used anti-cancer drugs [62]. They found that drugs, which target the DNA damage response, were among the most likely to induce numerical chromosomal aberrations. Thus, cancer cells have distorted the use of a pathway intended to safeguard the genome in order to propagate further genomic instability. Understanding of this fact however offers new ways of targeting tumors based on their inability to cope with excessive genomic damage.
CONCLUDING REMARKS
The role of the DNA damage response during mitosis is only beginning to emerge, though it is clear that it does not simply mirror what is known about the DNA damage response during interphase of the cell cycle (see outstanding questions section). It is reasonable to postulate that the machinery responsible for safeguarding the genome during G1, S, and G2 phases of the cell cycle would also be intimately involved in the intricate process of mitotic chromosome segregation. However, the purpose and hallmarks of this relationship remain to be fully understood. By inducing chromosome missegregation, the DNA damage response has the potential to indirectly clear cells with damaged genomes by inducing irreversible senescence through a p53-dependent mechanism in response to aneuploidy or by mediating crisis in a p53-independent manner particularly in response to telomeric damage. Nonetheless, in cancer cells, which often lack a functional p53-signaling pathway and are more proficient in averting crisis, the mitotic DNA damage response appears to have been coopted to propagate genomic diversity by turning structural chromosomal damage into numerical instability. A proper appreciation of this process is both essential to our understanding of how DNA-damaging therapies influence genomic integrity as well as the development of novel therapeutic strategies that incorporate chromosome missegregation and its effect on the viability of tumor cell populations.
OUTSTANDING QUESTIONS.
What are the mechanisms by which DNA damage response proteins act during normal mitosis to safeguard faithful chromosome segregation
On a global scale how different is the mitotic DNA damage response from that of interphase of the cell cycle? Is the mitotic DNA damage response – albeit truncated – wired differently than the canonical response?
How frequently do mitotic cells experience DNA damage in normal development and during tumorigenesis?
What is the evolutionary origin of a semi-active DNA damage response during mitosis, at a time when the cell is chiefly preoccupied with the process of faithful chromosome segregation.
How does the DNA damage response differ between mitosis and meiosis?
TRENDS.
The cellular DNA damage response signaling and downstream DNA repair pathways are active in interphase of the cell cycle. Their role in mitosis has long remained unknown.
Emerging evidence suggests that the many proteins involved in DNA repair are also required for the progression of normal mitosis and faithful chromosome segregation.
Mitotic cells can experience DNA damage either exogenously from therapy or endogenously due to unrepaired pre-mitotic damage. Emerging evidence shows that such damage can directly lead to errors in chromosome segregation.
In normal cells a mitotic DNA damage response can serve to either mark damaged chromosomes for subsequent repair or induce p53-mediated cellular senescence to avoid the propagation of damaged genomes.
Cancer cells coopt the mitotic DNA damage response to further propagate chromosomal instability. This offers untapped therapeutic opportunities to target genomic instability in cancer.
Acknowledgments
The authors would like to thank Kristina Godek (Dartmouth) for critical feedback and review of the manuscript. S. Bakhoum is supported by The Department of Defense Breast Cancer Research Breakthrough Award number W81XWH-16-1-0315, the Elsa U. Pardee Foundation, and the NCI MSKCC Cancer Center Core Grant P30 CA008748.
Glossary
- Numerical chromosomal instability
a conspicuous feature of cancer cells whereby they constantly undergo errors in chromosome segregation leading to unstable chromosome copy-numbers (karyotypes) and intratumor heterogeneity.
- Structural chromosomal instability:
the process by which chromosomes frequently undersgo structural alterations in their chromosomes such as breaks and translocation.
- Chromothripsis
A process by which chromosomes (or part of thereof) undergo multiple breaks with segmental loss and random repair leading to clustered complex rearranged regions in whole chromosomes or chromosome arms.
- Kinetochores
Large proteinaceous structures whose functions is to link spindle microtubules to mitotic chromosomes to achieve accurate chromosome segregation. They contain numerous proteins which act in concert to regulate the attachement and detachment rates of microtubules.
- Kinetochore-microtubules
A subset of microtubules that attach to kinetochores. These microtubules are amongst the most stable during mitosis however they nonetheless exhibit a dynamic behavior constantly attaching and detaching from the kinetochores. This dynamic behavior is central to the cellular ability to correct erroneous attachments and prevent chromosome missegregation. Human kinetochores can bind an average of 26 microtubules. Cancer cells have defective kinetochore-microtubule release kinetics often leading to chromosome segregation errors.
- G2/M checkpoint
One of the most important cell-cycle checkpoints; it prevents cells from entering mitosis in the presence of DNA damage.
- Lagging chromosomes
A term used to describe chromosomes, which during anaphase are erroneously attached to microtubules emanating from opposite spindle poles. They typically reside alone separately from the segregating chromosome masses. As a result, these chromosomes can segregate to the wrong daughter cells, become encapsulated into micronuclei, or experience structural damage. Lagging chromosomes are a hallmark of chromosomal instability in cancer and can be observed in both tumor-derived cell lines and fixed tumor samples.
- Chromatin bridges
Denotes continuous chromatin linking the two segregating masses and they can result from any number of causes: telomere fusion, chromosomal translocations causing dicentric chromosomes, broken centromeres, and incompletely replicated chromosomes. They are hallmark of genomic instability and suggest a higher likelihood of pre-mitotic genomic defects.
Footnotes
Competing financial interests. The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gunderson LL, Tepper JE. Clinical Radiation Oncology. Churchill Livingstone; 2011. [Google Scholar]
- 2.Sinclair WK. Cyclic x-ray responses in mammalian cells in vitro. Radiat Res. 1968;33:620–643. [PubMed] [Google Scholar]
- 3.Sinclair WK, Morton RA. X-ray sensitivity during the cell generation cycle of cultured Chinese hamster cells. Radiat Res. 1966;29:450–474. [PubMed] [Google Scholar]
- 4.Giunta S, et al. DNA damage signaling in response to double-strand breaks during mitosis. The Journal of Cell Biology. 2010;190:197–207. doi: 10.1083/jcb.200911156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrell RA, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakhoum SF, et al. DNA-Damage Response during Mitosis Induces Whole-Chromosome Missegregation. Cancer Discovery. 2014;4:1281–1289. doi: 10.1158/2159-8290.CD-14-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orthwein A, et al. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014;344:189–193. doi: 10.1126/science.1248024. [DOI] [PubMed] [Google Scholar]
- 8.Shang Z, et al. DNA-PKcs activates the Chk2-Brca1 pathway during mitosis to ensure chromosomal stability. Oncogenesis. 2014;3:e85. doi: 10.1038/oncsis.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki M, et al. Phosphorylated histone H2AX foci persist on rejoined mitotic chromosomes in normal human diploid cells exposed to ionizing radiation. Radiat Res. 2006;165:269–276. doi: 10.1667/rr3508.1. [DOI] [PubMed] [Google Scholar]
- 10.Gomez-Godinez V, et al. Analysis of DNA double-strand break response and chromatin structure in mitosis using laser microirradiation. Nucleic Acids Research. 2010;38:e202–e202. doi: 10.1093/nar/gkq836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato TA, et al. Comparison of the induction and disappearance of DNA double strand breaks and gamma-H2AX foci after irradiation of chromosomes in G1-phase or in condensed metaphase cells. Mutat Res. 2008;639:108–112. doi: 10.1016/j.mrfmmm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Williams RS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen RT, et al. TopBP1 is required at mitosis to reduce transmission of DNA damage to G1 daughter cells. The Journal of Cell Biology. 2015;210:565–582. doi: 10.1083/jcb.201502107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minocherhomji S, et al. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015;528:286–290. doi: 10.1038/nature16139. [DOI] [PubMed] [Google Scholar]
- 15.Pantelias GE, Terzoudi GI. Functional cell-cycle chromatin conformation changes in the presence of DNA damage result into chromatid breaks: a new insight in the formation of radiation-induced chromosomal aberrations based on the direct observation of interphase chromatin. Mutat Res. 2010;701:27–37. doi: 10.1016/j.mrgentox.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Terzoudi GI, et al. Chromatin dynamics during cell cycle mediate conversion of DNA damage into chromatid breaks and affect formation of chromosomal aberrations: biological and clinical significance. Mutat Res. 2011;711:174–186. doi: 10.1016/j.mrfmmm.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Rogakou EP, et al. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsvetkov L. Polo-like Kinase 1 and Chk2 Interact and Co-localize to Centrosomes and the Midbody. Journal of Biological Chemistry. 2003;278:8468–8475. doi: 10.1074/jbc.M211202200. [DOI] [PubMed] [Google Scholar]
- 19.Wei J-H, et al. TTK/hMps1 participates in the regulation of DNA damage checkpoint response by phosphorylating CHK2 on threonine 68. J Biol Chem. 2005;280:7748–7757. doi: 10.1074/jbc.M410152200. [DOI] [PubMed] [Google Scholar]
- 20.Cesare AJ, et al. The telomere deprotection response is functionally distinct from the genomic DNA damage response. Mol Cell. 2013;51:141–155. doi: 10.1016/j.molcel.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su TT, Jaklevic B. DNA damage leads to a Cyclin A-dependent delay in metaphase-anaphase transition in the Drosophila gastrula. Curr Biol. 2001;11:8–17. doi: 10.1016/s0960-9822(00)00042-7. [DOI] [PubMed] [Google Scholar]
- 22.Mikhailov A, et al. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr Biol. 2002;12:1797–1806. doi: 10.1016/s0960-9822(02)01226-5. [DOI] [PubMed] [Google Scholar]
- 23.Silva BA, et al. DNA damage to a single chromosome end delays anaphase onset. J Biol Chem. 2014;289:22771–22784. doi: 10.1074/jbc.M113.535955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi MT, et al. A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nat Struct Mol Biol. 2012;19:387–394. doi: 10.1038/nsmb.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi MT, et al. Cell death during crisis is mediated by mitotic telomere deprotection. Nature. 2015;522:492–496. doi: 10.1038/nature14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deckbar D, et al. Chromosome breakage after G2 checkpoint release. J Cell Biol. 2007;176:749–755. doi: 10.1083/jcb.200612047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glass WA, Varma MN. Physical and chemical mechanisms in molecular radiation biology. Plenum Pub Corp; 1991. [Google Scholar]
- 28.Yang Z, et al. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat Cell Biol. 2008;10:748–751. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laughney AM, et al. Dynamics of Tumor Heterogeneity Derived from Clonal Karyotypic Evolution. Cell Rep. 2015;12:809–820. doi: 10.1016/j.celrep.2015.06.065. [DOI] [PubMed] [Google Scholar]
- 30.Weaver BAA, et al. Low rates of aneuploidy promote tumorigenesis while high rates of aneuploidy cause cell death and tumor suppression. Cell Oncol. 2008;30:453. doi: 10.3233/CLO-2008-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakhoum SF, Compton DA. Kinetochores and disease: keeping microtubule dynamics in check! Curr Opin Cell Biol. 2012;24:64–70. doi: 10.1016/j.ceb.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakhoum SF, et al. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ertych N, et al. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat Cell Biol. 2014;16:779–791. doi: 10.1038/ncb2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakhoum SF, et al. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabeche L, Compton DA. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature. 2013;502:110–113. doi: 10.1038/nature12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macurek L, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 37.Smits VA, et al. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat Cell Biol. 2000;2:672–676. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- 38.Seki A, et al. Bora and the Kinase Aurora A Cooperatively Activate the Kinase Plk1 and Control Mitotic Entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee DH, et al. Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Mol Cell. 2014;54:512–525. doi: 10.1016/j.molcel.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janssen A, et al. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 41.Maciejowski J, et al. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 2015;163:1641–1654. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duda H, et al. A Mechanism for Controlled Breakage of Under-replicated Chromosomes during Mitosis. Dev Cell. 2016;39:740–755. doi: 10.1016/j.devcel.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 44.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanneste E, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 46.Vogel C, et al. Mechanisms of mitotic cell death induced by chemotherapy-mediated G2 checkpoint abrogation. Cancer Res. 2007;67:339–345. doi: 10.1158/0008-5472.CAN-06-2548. [DOI] [PubMed] [Google Scholar]
- 47.Löbrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 48.Crasta KK, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang CZ, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bakhoum SF, et al. Numerical chromosomal instability mediates susceptibility to radiation treatment. Nat Commun. 2015;6:5990. doi: 10.1038/ncomms6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janssen A, et al. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci USA. 2009;106:19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaki BI, et al. Chromosomal instability portends superior response of rectal adenocarcinoma to chemoradiation therapy. Cancer. 2014;120:1733–1742. doi: 10.1002/cncr.28656. [DOI] [PubMed] [Google Scholar]
- 54.Jamal-Hanjani M, et al. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: a prospective validation cohort study from the TACT trial. Annals of Oncology. 2015;26:1340–1346. doi: 10.1093/annonc/mdv178. [DOI] [PubMed] [Google Scholar]
- 55.Roylance R, et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:2183–2194. doi: 10.1158/1055-9965.EPI-11-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Birkbak NJ, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–3452. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andor N, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hussain M, et al. Targeting DNA repair with combination veliparib (ABT-888) and temozolomide in patients with metastatic castration-resistant prostate cancer. Invest New Drugs. 2014;32:904–912. doi: 10.1007/s10637-014-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bottai G, et al. Targeting the microRNA-regulating DNA damage/repair pathways in cancer. Expert Opin Biol Ther. 2014 doi: 10.1517/14712598.2014.950650. [DOI] [PubMed] [Google Scholar]
- 60.Hosoya N, Miyagawa K. Targeting DNA damage response in cancer therapy. Cancer Sci. 2014;105:370–388. doi: 10.1111/cas.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fokas E, et al. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat Rev. 2014;40:109–117. doi: 10.1016/j.ctrv.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 62.Lee H-S, et al. Effects of Anticancer Drugs on Chromosome Instability and New Clinical Implications for Tumor-Suppressing Therapies. Cancer Res. 2016;76:902–911. doi: 10.1158/0008-5472.CAN-15-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]