Abstract
iNKT cell functional subsets are defined by key transcription factors and output of cytokines such as IL-4, IFNγ, IL-17, and IL-10. To examine how TCR specificity determines iNKT function, we used somatic cell nuclear transfer to generate three lines of mice, cloned from iNKT nuclei. Each line uses the invariant Vα14Jα18 TCRα, paired with unique Vβ7 or Vβ8.2 subunits. We examined tissue homing, expression of PLZF, T-bet, and RORγt, as well as cytokine profiles and found that although monoclonal iNKT cells differentiated into all functional subsets, the NKT17 lineage was reduced or expanded depending on the TCR expressed. We examined iNKT thymic development in limited dilution bone marrow chimeras and show that higher TCR avidity correlates with higher PLZF and reduced T-bet expression. iNKT functional subsets showed distinct tissue distribution patterns. Although each individual monoclonal TCR showed an inherent subset distribution preference that was evident across all tissues examined, the iNKT cytokine profile differed more by tissue of origin than by TCR specificity.
Introduction
NKT cells are a subset of T cells that primarily recognize lipid antigens in a complex with the Class I MHC homolog CD1d 1; 2. Type II NKT cells carry a diverse TCR repertoire, recognize a variety of lipid antigens, such as sulfatides, and will not be discussed further here. In mice, type I NKT cells (iNKT) express an invariant Vα14Jα18 TCRα chain, paired with a limited but diverse set of TCRβ chains. Vβ8.1, 8.2, 7, 8.3, and 2 are preferentially used, but the CDR3 regions vary widely, such that iNKT cells form a polyclonal pool 3. iNKT cells are activated by the ligand α-galactosylceramide (α-GalCer) 1; 4. Other known antigens include both self and microbial lipids 5. Under conditions of infection or inflammation, iNKT cells can skew the ensuing immune response by rapidly producing cytokines such as IFNγ, IL-13, and IL-4 without an obligate need for proliferation 1.
Functional subsets of iNKT cells exist, and they are classified according to the expression of signature transcription factors during thymic development or by the production of signature cytokines. T-bet, PLZF, and RORγt delineate NKT1, NKT2, and NKT17 subsets in the thymus; they produce IFNγ, IL-4, or IL-17, respectively 6; 7; 8. The three major iNKT cell subsets likely differentiate during thymic development, as most convincingly shown by single cell transcriptional profiling of thymic NKT1, NKT2, NKT17, and NKT0 cells 9. Whether interconversions amongst iNKT subsets can occur is not known. Production of NKT17 cells appears to be driven by particular signaling pathways; ThPOK and PTEN expression inversely correlate with acquisition of a RORγt+ IL-17-producing phenotype 10; 11 while mTORC2 is required for NKT17 development 12. Oxidized 5 methylcytosine in DNA suppresses NKT17 development, as revealed by an overabundance of hyperactivated NKT17 cells in mice lacking the epigenetic regulators Tet2 and Tet3 13. Other subsets of iNKT cells including IL-10-producing NKT10 cells 14, follicular helper-like iNKTfh cells 15, IL-9 producing iNKT cells 16, and regulatory iNKT cells 11; 17 have been described, but there is currently no evidence for thymic instruction of these subsets. Certain iNKT subsets are enriched in particular tissues; adipose tissue contains PLZF− E4BP4+ IL-10-producing iNKT cells with a regulatory phenotype 18; 19, while skin-draining lymph nodes are enriched in NK1.1−CD4−CD44+ NKT17 cells 20; 21. NKT2 cells are more frequently found in mesenteric lymph nodes, at least in Balb/c mice 7. In a model of tuberculosis infection, iNKT cells producing GM-CSF were crucial for control of infection in the lung 22. Liver-resident and spleen-resident iNKT cells differ in their ability to reject B16 melanoma lung metastases 23.
Recognition of α-GalCer occurs predominantly through the TCRα chain, with the TCRβ chain forming contacts only with CD1d 24. Nevertheless, Vβ chain usage may influence the spectrum of ligands recognized by iNKT cells 25. Co-crystal structures of TCR, ligand, and CD1d, together with careful measurements of binding kinetics, suggest that ligand merely determines the off-rate of TCR binding 26; 27; 28; 29. Indeed, in contrast to most Class I MHC-restricted TCRs, the iNKT TCR adopts a similar docking mode independent of the identity of the ligand bound 30; 31; 32; 33. On the other hand, a library of recombinant iNKT TCRs with different TCRβ chains showed differential recognition of molecules such as iGb3, GSL-1, and other ligands thought to be more physiologically relevant than α-GalCer 34. Similar effects of TCRβ mutations on ligand recognition were observed for human iNKT TCRs 35, and indeed, selective loss of high affinity iNKT cells has been observed in several human diseases 36; 37. Retrogenic mice expressing several discrete, natural or engineered iNKT TCRs showed that positive selection of iNKT cells correlated with TCR affinity, while lineage choice between NKT1 versus NKT2 was more strongly correlated with the half-life of TCR association 38. iNKT cell TCR fine specificity may play a role in recognition of self-lipids, as Vβ7+ iNKT cells have a higher affinity for self-lipids and are preferentially selected in the thymus 39 despite having a lower affinity for α-GalCer than Vβ8+ iNKT cells 40.
To examine the role of TCR specificity in iNKT cell effector differentiation, we performed somatic cell nuclear transfer using the nuclei of individual iNKT cells to generate three independent lines of transnuclear (TN) mice, all of which use the identical Vα14Jα18 TCRα chain, but with three distinct TCRβ rearrangements. We show by differential staining with OCH, α-GalCer, and α-glucosylceramide tetramers that these iNKT cells recognize different ligands. No comparable set of mice producing distinct iNKT TCRs has been reported. These unique strains of mice thus allowed us to definitively address the contribution of TCR fine specificity to formation of functional iNKT cell subsets, and their presence across different tissues. The Vβ7A and Vβ8.2 TCRs predisposed against NKT17 development across every tissue examined, while the Vβ7C TCR displayed increased NKT17 development. However, iNKT cells obtained from different tissues showed strikingly disparate functions irrespective of TCR usage. TCR specificity therefore plays a minor role in iNKT cell lineage choice.
Materials and Methdods
Animal care
Animals were housed at both the Whitehead Institute for Biomedical Research and the Dana-Farber Cancer Institute and were maintained according to the NIH Guide for the Care and Use of Laboratory Animals and protocols approved by the MIT Committee on Animal Care and the DFCI IACUC, respectively. C57BL/6, Balb/c, and RAG2−/− mice were purchased from Jackson Labs. Jα18−/− mice were obtained from Dr. Michael Brenner (Boston, MA).
TN mouse generation
TN mice were generated as previously described 41; 42; 43. Briefly, CD1d-PBS57 tetramer+ Vβ7+ cells were sorted by FACS and used as a source of donor nuclei for SCNT. The mitotic spindle was removed from mouse oocytes and replaced with donor nuclei. The nucleus-transplanted oocytes were then activated in medium containing strontium and TSA, and allowed to develop in culture to the blastocyst stage. SCNT blastocysts were used to derive embryonic stem (ES) cell lines. These ES cell lines were then injected into wild type B6xDBA F1 blastocysts and implanted into pseudopregnant females. The resulting chimeric pups were mated to C57BL/6 females to establish the Vβ7A and Vβ7C lines. Vβ8.2 mice were generated by intercrossing the Vβ7C and TRP1high mouse lines 42, and selecting for founder progeny that received a single copy of the Vα14Jα18 TCR rearrangement and a single copy of the Vβ8.2 TCR rearrangement. Mice were backcrossed 7 generations to C57BL/6 (9 generations for RAG2−/− crossed lines). F1 mice were generated by crossing 7-generations C57BL/6 backcrossed males to Balb/c females. Vα14 mice used throughout the study were littermates of the Vβ7A, Vβ7C or Vβ8.2 mice used. Mice were used at 6–12 weeks of age, and both males and females were included. For each experiment, 5 mice were included per group. A mix of ages and both sexes were included per group, with age and sex matching among the groups as closely as possible. No correlation was observed between age and any of the parameters examined or sex and any parameters examined.
Sequencing of the TCR genes
iNKT cells were sorted by FACS for CD1d-PBS57 tetramer+ cells and used as a source of RNA. 5′RACE was performed according to the manufacturer’s protocol (GeneRacer, #L1502-01 Invitrogen). Gene sequences were searched against the IEDB database (iedb.org).
Tissue preparation
Lymphocytes were harvested from spleen, thymus, and lymph nodes by crushing organs through a 40 micron cell strainer. Adipose tissue was minced with a scalpel and digested in 5mg/mL collagenase II for 30 minutes at 37°C with rotation. Lung tissue was placed in a gentleMACS C tube (Milltenyi 130-093-237) and digested using the lung dissociation kit enzymes (Milltenyi 130-095-927) and the gentleMACS Dissociator (Milltenyi 130-093-235), as per the manufacturer’s recommendation. Liver was homogenized through a 70 micron cell strainer and centrifuged at 1400rpm for 5 minutes. The organ pellet was resuspended in 10mL of 35% Percoll (GE Healthcare 17-0891-01) in RPMI. A bottom layer of 70% Percoll in PBS was added before centrifugation at 1700rpm for 15 minutes with no brakes. The middle layer of lymphocytes was harvested into 10mL PBS.
Flow cytometry
Cells were harvested from spleen, thymus, lymph nodes, liver, epididymal fat pads, or lung. Cell preparations were subjected to hypotonic lysis to remove erythrocytes, stained, and analyzed using a Fortessa (BD). CD1d-PBS57 (CD1d-αgal) tetramers were obtained from the NIH Tetramer Core Facility. CD1d α-GlcCer, α-GalCer (24:1) and OCH tetramers were generated as previously reported with minor modifications 44. Briefly, α-glucosylceramide (phytosphingosine base, C26:0 N-acyl chain), α-galacytosylceramide (C24:1 N-acyl chain) or OCH (both lipids kindly provided by Gurdyal Besra) were solubilized in PBS with 0.25% sodium taurocholate (Sigma) then incubated at a 5:1 molar ratio with mouse CD1d (NIH Tetramer Core Facility) for 16 hrs at 37°C before tetramer assembly with fluorophore-lined streptavidin (Life Technologies). The following antibodies were from Biolegend: IFNγ (Clone XMG1.2, Cat 505830), IL-4 (Clone 11B11, Cat 504109), IL-17A (Clone TC11-18H10.1, Cat 506916), Tbet (Clone 4B10, Cat 644816), Vβ7 (Clone TR310, Cat 118306), CD3 (Clone 17A2, Cat 100236), CD3ε (Clone 145-2C11, Cat 100330), and CD4 (Clone RM4-5, Cat 100531). The following antibodies are from eBioscience: RORγt (Clone B2D, Cat 17-6981-80) and PLZF (Clone Mags.21F7, Cat 53-9320-82).
Cell culturing
Cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 2mM L-glutamine, 100 U/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 0.1 mM 2-ME. For stimulation cultures, total cell preparations from the indicated organs were stimulated with PMA and ionomycin for 4 hours in the presence of GolgiStop (Invitrogen). Cells were then fixed, permeablilized, and stained with antibodies to IL-4, IFNγ, and IL-17. Alternatively, total lymph node cells were stimulated with 100ng/mL α-galcer (Avanti Lipids), and production of IFNγ of 24 hour culture supernatants was measured by ELISA as indicated (BD Pharmingen).
Results
Generation of iNKT transnuclear mice
Somatic cell nuclear transfer is a unique tool used to clone mice from antigen-specific lymphocytes 41; 42; 43; 45; 46 including from iNKT cell nuclei 47. A previous report of transnuclear iNKT cell mice only used mice that express the Vα14 rearranged TCR and relied on polyclonal Vβ expression 47. This yields a mouse model similar to the Vα14 TCR transgenic line 4. To address the contribution of Vβ usage to iNKT cell subset specification, we generated three distinct mouse lines, cloned from the nuclei of individual iNKT cells (Figure 1A). The subset identity of the original nuclei donors was not determined; however all epigenetic marks are erased by nuclear reprogramming, such that only the DNA encoded TCR rearrangements are retained in the germline transmitting TN mouse lines. All three lines use the canonical Vα14Jα18 TCRα rearrangement; two lines express Vβ7 rearrangements with unique CDR3 sequences (A and C), while the remaining line uses Vβ8.2 (Table 1). When the Vβ8.2 line was further crossed to a CD1d−/− background, iNKT cells failed to develop, consistent with wild type iNKT cells (data not shown). Since the TCRα and TCRβ loci map to different chromosomes, backcrossing to C57BL/6 mice yielded mice that retained only the TCRα transnuclear locus (Vα14 line). These mice have an increased frequency of polyclonal iNKT cells that use the same restricted Vβ pattern as seen in C57BL/6 mice.
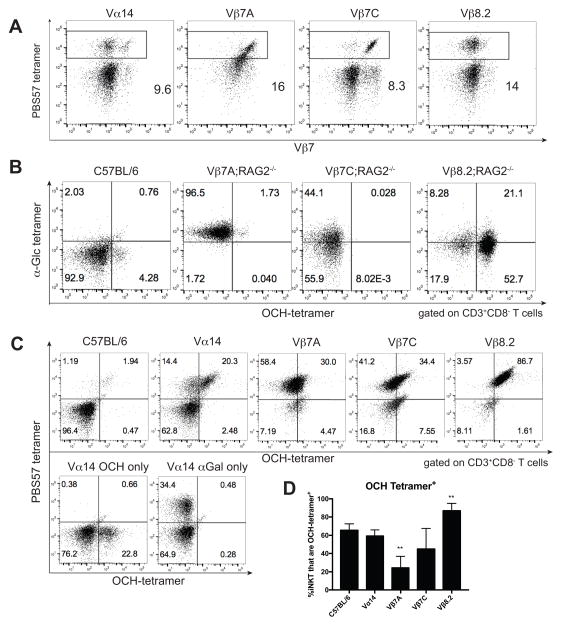
Figure 1. iNKT transnuclear mice have monoclonal iNKT cells with distinct antigenic specificities.
(A) Splenocytes were isolated from the indicated TN mouse lines and stained with anti-Vβ7, CD1d (PBS57) tetramer, and anti-CD4. Results are representative of >50 mice per group. (B) Splenocytes were isolated from indicated C57BL/6 or TN mouse lines and stained with anti-CD3, anti-CD8, OCH tetramer, and α-Glc tetramer. Both tetramers were added simultaneously. Plots are gated on CD3+CD8− T cells. (C) Thymocytes from C57BL/6 or TN mouse lines were isolated and stained with anti-CD3 and OCH tetramer. Cells were washed before being stained with CD1d (PBS57) tetramers, to assess displacement. (D) Quantification of OCH staining across multiple, independent experiments. **, p<0.01 compared to C57BL/6.
Table 1.
TCR sequences from iNKT TN mouse lines
| Variable | CDR3 | Joining | |
|---|---|---|---|
| Vβ7A | TRBV29 | CASSPPGQGGRVFF | Trbj 1–7 |
| Vβ7C | TRBV29 | CASSLPGHNERLFF | Trbj 2–4 |
| Vβ8.2 | TRBV13-3 | CASSDRGYEQYF | Trbj 2–7 |
Monoclonal iNKT cells have different ligand specificities
The three iNKT TCRs differ in their ligand specificities. Although all three bind α-GalCer tetramers, Vβ7A iNKT cells stain exclusively with α-Glc tetramers, Vβ7C iNKT cells stain dimly with α-Glc tetramers, and Vβ8.2 iNKT cells stain brightly with OCH tetramers, when the two tetramers are added simultaneously in a competitive binding assay (Figure 1B). Single staining with α–Glc and OCH tetramers revealed that even without competition, Vβ7A and Vβ7C iNKT cells stain dimly with OCH, while Vβ8.2 iNKT cells (like C57BL/6 mice) can bind αGlc and OCH tetramers equally well (Supplemental Figure 1A). OCH tetramers also stained Vβ8.2 cells even in the presence of equimolar αGalCer-CD1d (PBS57) tetramers (Supplemental Figure 1B). In addition, each of the monoclonal lines were stained first with excess OCH tetramer, washed, and then stained with CD1d (PBS57) tetramer; we find very little OCH staining of Vβ7A and Vβ7C iNKT cells compared to Vβ8.2 iNKT cells (Figure 1C). Compared over multiple experiments, Vβ8.2 iNKT cells are consistently more positive for OCH compared to the other monoclonal iNKT cells (Figure 1D). As would be expected from monoclonal lines, the staining patterns of OCH tetramer binding were similar regardless of whether cells were isolated from thymus, spleen, or skin-draining lymph node, although the intensity of tetramer staining varied somewhat by organ (Supplemental Figure 1B). Competition between OCH and a version of αGalCer with a naturally occurring lipid tail length (24.1) shows exclusive staining with αGalCer 24.1 for Vβ7A and Vβ7C iNKT cells, while Vβ8.2 and Vα14 iNKT cells stain both αGalCer 24.1 and OCH (Supplemental Figure 1C). All together, these results confirm differential ligand recognition among our three monoclonal iNKT TN mouse lines, and are similar to those found by Cameron et al. 34.
Transnuclear iNKT cells are similar to C57BL/6 iNKT cells, but with increased abundance
We examined expression of CD4, CD44, and NK1.1, which are commonly used to distinguish subpopulations of iNKT cells. CD44 and NK1.1 expression patterns were similar to polyclonal iNKT cells and varied more by tissue of origin than by TCR (Supplemental Figure 2A–B). Monoclonal iNKT cells expressed CD4 at similar frequencies to iNKT cells from C57BL/6 mice (Supplemental Figure 2C).
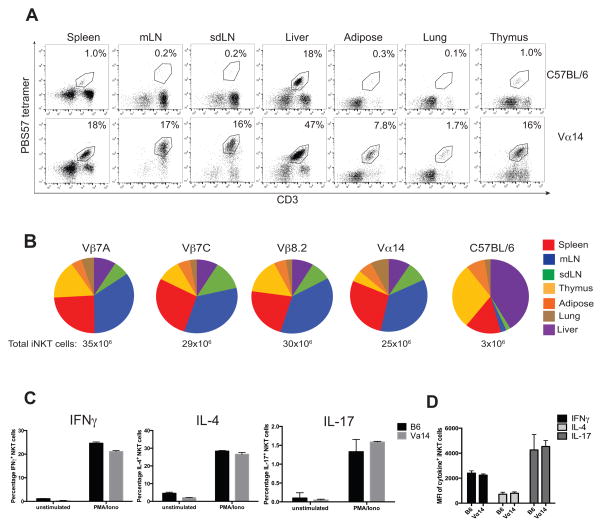
Although C57BL/6 mice have few iNKT cells in most organs except liver, Vα14 TN mice have abundant iNKT cells in all tissues examined, including lung and adipose tissue, making them a useful source of polyclonal iNKT cells (Figure 2A). We next examined iNKT cell tissue distribution in each of the iNKT TN mouse lines. Numbers of iNKT cells were determined for each organ analyzed and added to estimate the total number of iNKT cells per mouse. From each of the monoclonal mouse lines (Vβ7A, Vβ7C, and Vβ8.2) we recovered 29–35 million iNKT cells per animal, as compared to 25 million for the Vα14 line and less than 4 million for C57BL/6 mice (Figure 2B). Compared to C57BL/6 mice, all iNKT TN lines have increased numbers of iNKT cells in lymph nodes, as expected, given that iNKT cells represent the majority of T cells in these lines. None of the monoclonal iNKT lines showed obvious differences in relative tissue distribution of iNKT cells compared to the polyclonal iNKT cells in the Vα14 mice (Figure 2B). When stimulated in vitro with PMA and ionomycin, transnuclear Vα14 iNKT cells produced IL-17, IFNγ, and IL-4 at similar levels and frequencies as those seen from B6 iNKT cells (Figure 2C).
Figure 2. iNKT transnuclear mice have increased frequencies of iNKT cells.
(A) iNKT cells were harvested from the spleen, mesenteric lymph nodes (mLN), skin-draining lymph nodes (sdLN), liver, adipose tissue, lungs, and thymus of C57BL/6 and Vα14 mice and stained with anti-CD3 and CD1d (PBS57) tetramer. Results are representative of 5 mice per group; averaged in (B). (B) Lymphocytes were harvested from the spleen, mesenteric lymph nodes (mLN), skin-draining lymph nodes (sdLN), liver, adipose tissue, lungs, and thymus of the indicated mice and counted. Cells were stained with anti-CD3 and CD1d (PBS57) tetramer and analyzed by flow cytometry. The percentage of iNKT cells from each organ was multiplied by the cell count, and added to obtain an estimated frequency of total iNKT cells in the body. Results shown are average of 5–8 mice per group. (C) Splenocytes from C57BL/6 and Vα14 mice were stimulated in vitro with PMA/Ionomycin for 4 hours in the presence of GolgiStop. Cells were stained with anti-CD3 and CD1d (PBS57) tetramer, before they were fixed, permeabilized, and stained with anti-IFNγ, anti-IL-4, and anti-IL-17. Results shown are gated on CD3+CD1d-tetramer+ cells. Results shown are representative of three independent experiments where n=3 biological replicates.
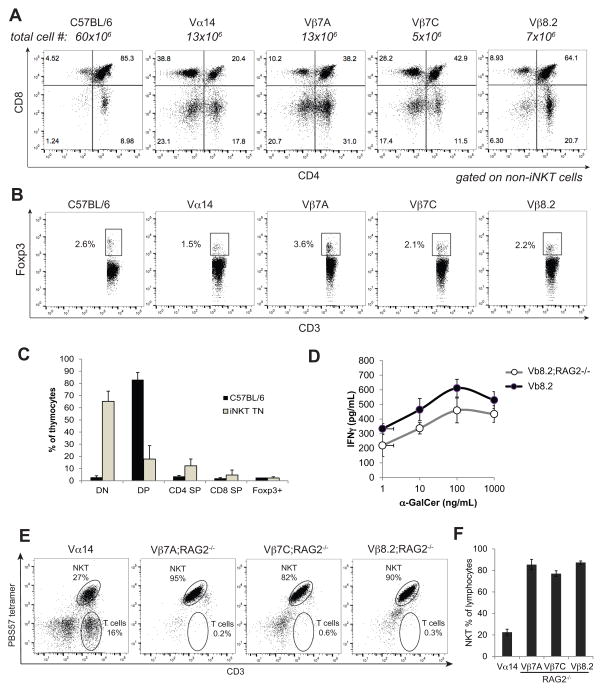
iNKT cells contribute to pathology in several autoimmune diseases, yet all iNKT TN lines are fertile, remain healthy, and showed no evidence of morbidity up to 1 year of age. The TN mice used here were hemizygous for the TN TCR genes to allow for development of conventional CD4+ and CD8+ T cells (Figure 3A), as well as Foxp3+ Tregs (Figure 3B). Thymic development was mostly normal in TN mice, albeit with reduced populations of CD4+CD8+ cells, indicating a more rapid transit of T cells with rearranged TCRα and TCRβ chains (Figure 3A and C), as seen also for conventional TCR transgenic mice and mice expressing a Vα14Jα18 transgenic TCRα 4. To determine whether potentially autoreactive iNKT cells were constrained by Foxp3+ Tregs, we crossed each iNKT TN line to RAG2−/− mice. All iNKT TN;RAG2−/− mice are similarly viable and remain healthy with normal lifespans despite having exclusively CD1d tetramer+ T cells that were not obviously anergic, as evidenced by their capacity to produce IFNγ upon stimulation with α-GalCer (Figure 3D–F). These data suggest that other factors besides the presence of iNKT cells are required for pathologies where iNKT cells have been implicated.
Figure 3. Conventional CD4, CD8 and Foxp3+ Tregs develop in hemizygous RAG-proficient iNKT TN mice.
(A) Thymocytes from C57BL/6 and all TN lines were stained with anti-CD3, CD1d (PBS57) tetramer, anti-CD4, and anti-CD8. Results shown are gated on non-iNKT cells (CD3+ CD1d-tetramer−). (B) Thymocytes from C57BL/6 and all TN lines were stained with anti-CD3 and CD1d (PBS57) tetramer before being fixed, permeabilized, and stained with anti-Foxp3. Results shown are gated on non-iNKT cells. (C) Quantification of A–B where n=3 mice per group. (D) Lymph node cells from Vβ8.2 and Vβ8.2;RAG2−/− mice were stimulated with the indicated concentrations of α-GalCer. IFNγ was measured by ELISA of 24 hour culture supernatants. Error bars are SD. (E) Each monoclonal TN line was crossed to RAG2−/− background. Peripheral blood of mice of the indicated genotypes was stained with CD1d (PBS57) tetramer and anti-CD3. (F) Quantification of (E) averaged over 5 mice per group.
NKT1, NKT2 and NKT17 functional subsets are detectable in all monoclonal TN lines
To address the role of TCR fine specificity in iNKT subset specification, we stimulated iNKT cells from Vα14, Vβ7A, Vβ7C, and Vβ8.2 mice with PMA/ionomycin and measured production of signature cytokines by intracellular staining. NKT1, NKT2, and NKT17 cells were clearly detectable in all monoclonal mouse lines (Supplemental Figure 3); however the frequency of NKT17 cells in both Vβ7A and Vβ8.2 mice trended toward lower abundance (Supplemental Figure 3A). NKT1 cells are the predominant subset in C57BL/6 mice and in our iNKT TN lines which are on a C57BL/6 background. To determine whether TCR specificity might influence iNKT subset differentiation in backgrounds other than C57BL/6, we generated F1 crosses of each iNKT TN line to Balb/c. The resulting F1 mice showed a higher frequency of NKT2 cells, as expected. Vα14 thymii had higher frequencies of IL-4 producing cells than any of the monoclonal lines, although this difference disappeared in the periphery (Figure 4). Again, all three functional subsets were present in monoclonal Vβ7A, Vβ7C and Vβ8.2 mice, with both Vβ7A and Vβ8.2 TCRs skewing slightly away from the NKT17 lineage (Figure 4). These results also argue against potential artifacts induced by insufficient backcrossing of the original founding TN chimeras to C57BL/6.
Figure 4. Monoclonal iNKT cells can produce all major cytokines.
Cells from thymus (A) or spleen (B) of B6xBalb/c F1 mice of all TN lines were stimulated in vitro with PMA/Ionomycin for 4 hours, in the presence of GolgiStop. Cells were stained with anti-CD3 and CD1d (PBS57) tetramer, before being fixed, permeabilized, and stained with antibodies to IFNγ, IL-4, and IL-17. Results shown are gated on CD3+CD1d-tetramer+ cells. n=4 mice per group. Error bars are SD.
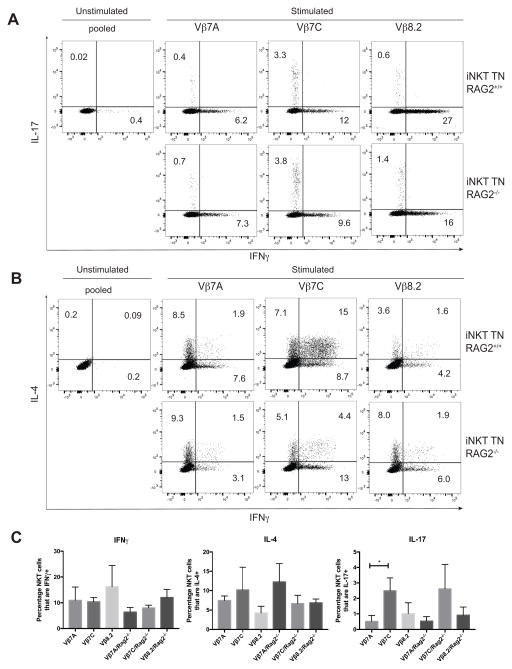
NKT17 cells are a small population, representing only 1–3% of total iNKT cells in C57BL/6 or Balb/c mice. The greatest populations of IL-17 producing iNKT cells are found in the skin draining lymph nodes, as previously reported 21. To better visualize NKT17 cells, we examined skin-draining lymph node cells from each monoclonal TN line on both RAG-sufficient and RAG-deficient backgrounds. Both Vβ7A and Vβ8.2 mice had fewer NKT17 cells, while the Vβ7C TCR favored NKT17 development (Figure 5). These trends were evident even when the TN genes were crossed onto a RAG-deficient background to prevent further V(D)J rearrangements (Figure 5).
Figure 5. NKT1, NKT2 and NKT17 subsets are present in rigorously monoclonal mice.
(A–B) Skin draining lymph node cells from all TN mice were stimulated in vitro with PMA/Ionomycin for 4 hours, in the presence of GolgiStop. Cells were stained with anti-CD3 and CD1d (PBS57) tetramer, before being fixed, permeabilized, and stained with antibodies to IFNγ, IL-4, and IL-17. Results shown are gated on CD3+CD1d-tetramer+ cells. TN mice are either RAG-proficient (top panels) or RAG-deficient (bottom panels). (C) Quantification of panels A–B, n=4 mice per RAG2+/+ group and n=3 mice per RAG2−/− group. Mice were 5–6 weeks of age and both sexes were included in each group.
Lineage development in the thymus correlates with iNKT TCR avidity
iNKT cells are selected in the thymus on CD1d-expressing double positive thymocytes 48. As a surrogate for TCR signaling, we examined Nur77 expression in the thymus of Vα14 TN mice. Similarly to wild-type iNKT cells 49, Vα14 iNKT cells that are stage 0 (CD44lo NK1.1− CD24+ iNKT cells) showed increased levels of TCR stimulation in the thymus, by Nur77 positivity (data not shown). We did not see significant levels of Nur77 in any peripheral tissues; however, iNKT cells in peripheral tissues could be induced to express Nur77 upon intravenous delivery of α-GalCer (data not shown).
Transnuclear mice have greatly increased frequencies of iNKT cells in the thymus, demonstrating that positively selecting ligands are not limiting. However, increased numbers of iNKT cells may affect the development of iNKT cell functional subsets. We therefore generated mixed bone marrow chimeras consisting of 95% Jα18−/− bone marrow mixed with 5% iNKT TN bone marrow. In this setting, iNKT cells represent approximately 1% of total thymocytes, similar to the level seen in C57BL/6 mice (Figure 6A and 2A).
Figure 6. TCR avidity correlates with PLZF expression during thymic development of monoclonal iNKT cells.
Jα18−/− mice were lethally irradiated, reconstituted with 95% Jα18−/− bone marrow and 5% bone marrow of the indicated TN lines, and analyzed 8 weeks later. (A) Thymocytes from C57BL/6 and all TN lines were stained with anti-CD3 and CD1d (PBS57) tetramer, and analyzed by flow cytometry. Mean fluorescence intensities (MFI) of tetramer staining for gated populations are shown on plots. (B) Thymocytes from C57BL/6 and all TN lines were stained with anti-CD3 and CD1d (PBS57) tetramer before being fixed, permeabilized, and stained with antibodies to PLZF, T-bet, and RORγt. Results shown are gated on CD1d (PBS57) tetramer+ iNKT cells. Tetramer bright = top 20% of iNKT cells; tetramer dim = bottom 20% of iNKT cells as defined by intensity of PE signal. (C) Quantification of mice shown in panel A. (D) Quantification of mice shown in panel B. n=4 mice per group. Error bars are SD.
We examined thymic iNKT cells from the 95:5 bone marrow chimeric mice. Among the monoclonal TN lines, the iNKT TCR affinity for CD1d (PBS57) tetramer differs with Vβ7A having the lowest affinity and Vβ8.2 having the highest affinity (Figure 6A and 6C). However, tetramer+ cells appear in a Gaussian distribution with respect to both tetramer staining and CD3 expression (Figure 6A). Since all of the iNKT cells in any given monoclonal line have identical affinities, differences in tetramer staining must be related to avidity. Indeed the positive correlation between CD3 expression and tetramer staining suggests that surface TCR expression is a major contributor, although other factors that affect TCR avidity, such as expression of SLAM family members 50; 51, may be important. We looked at bright versus dim tetramer stained cells with respect to their transcription factor profiles. Within each monoclonal line, we noticed that thymic iNKT cells with higher CD3 expression stained more brightly with tetramer and tended to be PLZFbright NKT2 cells, while lower avidity corresponded with T-bet+ NKT1 development (Figure 6B and 6D). These results are consistent with previous findings showing that Nur77 expression was positively correlated with IL-4 production in thymic iNKT cells 6 and more recent findings showing that TCR half-life of binding to CD1d-lipid complexes correlates with NKT2 development 38.
Limited dilution bone marrow chimeras reveal a role of the iNKT TCR in development of NKT17 cells
The 95:5 Jα18−/− to TN iNKT bone marrow chimera setting represents a more physiologic thymic development, and we examined whether monoclonal iNKT cells would display subset preferences in this setting. Cells were harvested from thymus, spleen, liver, and skin draining lymph nodes, and analyzed by both ICCS (Figure 7) and transcription factor staining (Figure 8 and Supplemental Figure 4). By both methods, we could detect NKT1, NKT2, and NKT17 cells in all organs and among all monoclonal lines. The subset distribution was dramatically different in different tissues, as has been reported by others 7; 19; 21. NKT1 cells were much more abundant in liver, while NKT17 cells were more abundant in skin-draining lymph nodes. However, within each organ, differences in cytokine production based on TCR were observed (Figure 7 and data not shown).
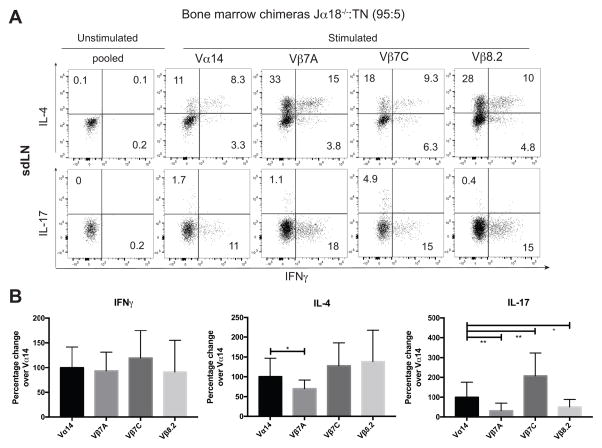
Figure 7. Limited dilution bone marrow chimeras reveal a minor role for TCR specificity in lineage preference of iNKT cells.
Jα18−/− mice were lethally irradiated, reconstituted with 95% Jα18−/− bone marrow and 5% bone marrow of the indicated TN lines, and analyzed 8 weeks later. n=4 mice per group. (A) Skin draining lymph node cells from all TN mice were stimulated in vitro with PMA/Ionomycin for 4 hours, in the presence of GolgiStop. Cells were stained with anti-CD3 and CD1d (PBS57) tetramer, before being fixed, permeabilized, and stained with antibodies to IFNγ, IL-4, and IL-17. Results shown are gated on CD3+CD1d-tetramer+ cells. (B) Quantification of ICCS data from thymus, spleen, skin-draining lymph nodes and liver, normalized to Vα14 values for the same organ. Error bars are SD.
Figure 8. Tissue of origin plays a dominant role in cytokine profiles of monoclonal iNKT cells.
Jα18−/− mice were lethally irradiated, reconstituted with 95% Jα18−/− bone marrow and 5% bone marrow of the indicated TN lines, and analyzed 8 weeks later. n=4 mice per group. (A) Skin draining lymph node cells from all TN mice were stained with anti-CD3 and CD1d (PBS57) tetramer, before being fixed, permeabilized, and stained with antibodies to PLZF, T-bet, and RORγt. Results shown are gated on CD3+CD1d-tetramer+ cells. (B) Quantification of transcription factor expression as shown in (A). n=4 mice per group. ANOVA was used to determine the relative contribution of TCR versus tissue of origin to explain the variation in transcription factor expression.
To better profile the effect of a given TCR independent of tissue of origin, we used intracellular cytokine data and normalized each data point to the average value for Vα14 polyclonal cells from the same organ (Figure 7B). In this meta-analysis, Vβ7A iNKT cells showed significantly less IL-17 and less IL-4 production (Figure 7A and 7B), as well as reduced RORγt expression (Figure 8A and 8B), suggesting a decrease in NKT17 development in this monoclonal line. Vβ8.2 iNKT cells were also significantly less likely to become NKT17 cells (Figure 7 and Figure 8), and trended toward increased IL-4 production and increased PLZF positivity (Figure 7 and Figure 8), suggesting a propensity to become NKT2 cells. Vβ7C iNKT cells were significantly more likely to become NKT17 cells (Figure 7 and Figure 8). All iNKT cells were similarly capable of IFNγ production, demonstrating that the reduction of IL-17 production from Vβ7A and Vβ8.2 iNKT cells is not because the cells are intrinsically refractory to stimulation, but that they are skewed away from the NKT17 lineage.
Tissue of origin in a stronger predictor of iNKT function than TCR specificity
Although TCR specificity can influence NKT effector functions, particularly in settings of low precursor frequency (Figure 6 and ref 38), the largest determinant of iNKT effector function appeared to be tissue of origin. Indeed, when we categorized iNKT cells into NKT1, NKT2, and NKT17 based on their transcription factor profiles, we observed dramatically more NKT17 cells in skin-draining lymph nodes than other tissues, regardless of TCR specificity (Figure 8). Similarly, NKT1 cells were more prevalent in liver (Supplemental Figure 4), and NKT2 cells were found more frequently in spleen and thymus (Supplemental Figure 4). To determine the degree of influence of TCR specificity versus tissue of origin, we performed an ANOVA and revealed that while both TCR and tissue contribute to iNKT cell effector functions, that tissue of origin accounts for a larger fraction of the observed variation (Figure 8B).
Discussion
Here we unequivocally show that monoclonal iNKT cells with different ligand specificities differentiate in vivo into all iNKT subsets, albeit at altered frequencies. As is also the case for CD4+ Tregs 46; 52, in the setting of limited precursor frequency, iNKT cell lineage choice preferences become more apparent. CD1d-presented ligands are not limiting in the thymus, as shown here and by others 38; however particular ligands which might instruct development into NKT1, NKT2, or NKT17 lineages, may be less abundant. Despite the subtle influence of TCR on NKT17 frequencies, tissue of origin had a more dominant role in predicting iNKT cell function, with iNKT cells from liver, skin-draining lymph nodes, and spleen having distinct cytokine and transcription factor profiles.
Whether iNKT lineage choice is determined in the thymus versus the tissues is a subject of some debate. Several reports, including this one, have identified clear populations of NKT1, NKT2, and NKT17 cells in the thymus, which would suggest that lineage commitment occurs in the thymus and that differential chemokine receptor expression explains the differences in accumulation of iNKT subsets in different tissues 7; 9; 53. A recent report by the Mallevaey group correlated CD1d tetramer dissociation rates with NKT1 versus NKT2 thymic development, although no clear relationship between TCR binding kinetics and NKT17 development was determined 38. In the same study, cytokine production by acutely stimulated peripheral iNKT cells was similar across a range of TCR affinities and half-lives 38, suggesting that NKT subset imprinting in the thymus may be altered in the periphery due to differential accumulation of particular subsets or further differentiation upon arrival in tissues. The thymic imprinting model does not account for follicular helper iNKT cells, which arise post-immunization 15, IL-9 producing iNKT cells 16, or NKT10 cells which may undergo further differentiation after seeding the adipose tissue 18; 19. Our observation that a given TCR influences subset choice suggests that iNKT subset formation may occur as early as the positive selection event in the thymus. However, since tissues also contain CD1d+ cells presenting a variety of lipid antigens, the TCR specificity may continue to have an instructive role in subset formation outside of the thymus as well.
Tissue-specific programming of immune cells has been described for both macrophages and Foxp3+ Tregs. Macrophage subsets include alveolar macrophages in the lung, microglia in the brain, and Kupffer cells in the liver, all of which display unique transcriptional profiles. Macrophages are fairly plastic and can adapt to a new environment 54. Likewise, tissue-resident Tregs are present in adipose tissue, muscle, eye, and other sites, where they exert similar regulatory functions, but express transcriptional profiles distinct from lymphoid-resident Tregs 55; 56. In an interesting analogy to iNKT cells, Tregs appear to seed tissues early in life 57. The factors that cause neonatal seeding of iNKT and Tregs into tissues remain to be identified. For iNKT cells, tissue-dependent reprogramming may occur, at least for adipose-resident iNKT that express a transcriptional profile distinct from their spleen or liver counterparts 18; 58.
Here we report four lines of transnuclear mice with individual monoclonal populations of Vβ7 or Vβ8.2 iNKT cells. Although all three lines recognized α-GalCer, they showed differential recognition of α-Glc and OCH, confirming different CD1d-lipid binding properties. These mice may be of great utility in addressing reactivity to particular gut microbial ligands or endogenous ligands. Furthermore, Vα14 transnuclear mice are an excellent source of polyclonal iNKT cells, especially from tissues such as adipose or lung where infiltrating iNKT cells are in low abundance.
Supplementary Material
Acknowledgments
Grant Support: E.C-T. was funded by NIH T32CA207021. P.J.B. was funded by NIH AI102945. S.K.D. was funded by the Bushrod H. Campbell and Adah F. Hall Charity Fund and Harold Whitworth Pierce Charitable Trust, and also by the Melanoma Research Alliance and the Claudia Adams Barr Program in Cancer Research. S.K.D. and H.L.P. were funded by Janssen Pharmaceutical, Inc.
We are grateful to Angelina Bilate and Lydia Lynch for advice and technical assistance, to Patti Wisniewski of the Whitehead Institute Flow Cytometry Core Facility for cell sorting, and to John Jackson for mouse husbandry.
Footnotes
Author contributions: E.C-T. designed and conducted experiments, analyzed data, and helped write the manuscript. G.Z.C., P.M.T., M.M.S., and M.B. conducted experiments. P.J.B. supplied α-Glc and OCH tetramers and contributed to data interpretation. S.K.D. generated the mouse lines. H.L.P. and S.K.D. designed experiments, analyzed data, and wrote the manuscript.
References
- 1.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 2.Macho-Fernandez E, Brigl M. The Extended Family of CD1d-Restricted NKT Cells: Sifting through a Mixed Bag of TCRs, Antigens, and Functions. Front Immunol. 2015;6:362. doi: 10.3389/fimmu.2015.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo T, Chamoto K, Nakatsugawa M, Ochi T, Yamashita Y, Anczurowski M, Butler MO, Hirano N. Mouse and Human CD1d-Self-Lipid Complexes Are Recognized Differently by Murine Invariant Natural Killer T Cell Receptors. PLoS One. 2016;11:e0156114. doi: 10.1371/journal.pone.0156114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendelac A, Hunziker RD, Lantz O. Increased interleukin 4 and immunoglobulin E production in transgenic mice overexpressing NK1 T cells. J Exp Med. 1996;184:1285–93. doi: 10.1084/jem.184.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birkholz AM, Howell AR, Kronenberg M. The Alpha and Omega of Galactosylceramides in T Cell Immune Function. J Biol Chem. 2015;290:15365–70. doi: 10.1074/jbc.R115.647057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–54. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee YJ, Wang H, Starrett GJ, Phuong V, Jameson SC, Hogquist KA. Tissue-Specific Distribution of iNKT Cells Impacts Their Cytokine Response. Immunity. 2015;43:566–78. doi: 10.1016/j.immuni.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watarai H, Sekine-Kondo E, Shigeura T, Motomura Y, Yasuda T, Satoh R, Yoshida H, Kubo M, Kawamoto H, Koseki H, Taniguchi M. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 2012;10:e1001255. doi: 10.1371/journal.pbio.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, Kronenberg M. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol. 2016;17:728–39. doi: 10.1038/ni.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engel I, Zhao M, Kappes D, Taniuchi I, Kronenberg M. The transcription factor Th-POK negatively regulates Th17 differentiation in Valpha14i NKT cells. Blood. 2012;120:4524–32. doi: 10.1182/blood-2012-01-406280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattarollo SR, Yong M, Gosmann C, Choyce A, Chan D, Leggatt GR, Frazer IH. NKT cells inhibit antigen-specific effector CD8 T cell induction to skin viral proteins. J Immunol. 2011;187:1601–8. doi: 10.4049/jimmunol.1100756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei J, Yang K, Chi H. Cutting edge: Discrete functions of mTOR signaling in invariant NKT cell development and NKT17 fate decision. J Immunol. 2014;193:4297–301. doi: 10.4049/jimmunol.1402042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsagaratou A, Gonzalez-Avalos E, Rautio S, Scott-Browne JP, Togher S, Pastor WA, Rothenberg EV, Chavez L, Lahdesmaki H, Rao A. TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells. Nat Immunol. 2016 doi: 10.1038/ni.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest. 2014;124:3725–40. doi: 10.1172/JCI72308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King IL, Fortier A, Tighe M, Dibble J, Watts GF, Veerapen N, Haberman AM, Besra GS, Mohrs M, Brenner MB, Leadbetter EA. Invariant natural killer T cells direct B cell responses to cognate lipid antigen in an IL-21-dependent manner. Nat Immunol. 2012;13:44–50. doi: 10.1038/ni.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monteiro M, Agua-Doce A, Almeida CF, Fonseca-Pereira D, Veiga-Fernandes H, Graca L. IL-9 Expression by Invariant NKT Cells Is Not Imprinted during Thymic Development. J Immunol. 2015;195:3463–71. doi: 10.4049/jimmunol.1403170. [DOI] [PubMed] [Google Scholar]
- 17.Beaudoin L, Laloux V, Novak J, Lucas B, Lehuen A. NKT cells inhibit the onset of diabetes by impairing the development of pathogenic T cells specific for pancreatic beta cells. Immunity. 2002;17:725–36. doi: 10.1016/s1074-7613(02)00473-9. [DOI] [PubMed] [Google Scholar]
- 18.Lynch L, Michelet X, Zhang S, Brennan PJ, Moseman A, Lester C, Besra G, Vomhof-Dekrey EE, Tighe M, Koay HF, Godfrey DI, Leadbetter EA, Sant’Angelo DB, von Andrian U, Brenner MB. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nat Immunol. 2015;16:85–95. doi: 10.1038/ni.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynch L, Nowak M, Varghese B, Clark J, Hogan AE, Toxavidis V, Balk SP, O’Shea D, O’Farrelly C, Exley MA. Adipose tissue invariant NKT cells protect against diet-induced obesity and metabolic disorder through regulatory cytokine production. Immunity. 2012;37:574–87. doi: 10.1016/j.immuni.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doisne JM, Becourt C, Amniai L, Duarte N, Le Luduec JB, Eberl G, Benlagha K. Skin and peripheral lymph node invariant NKT cells are mainly retinoic acid receptor-related orphan receptor (gamma)t+ and respond preferentially under inflammatory conditions. J Immunol. 2009;183:2142–9. doi: 10.4049/jimmunol.0901059. [DOI] [PubMed] [Google Scholar]
- 21.Michel ML, Mendes-da-Cruz D, Keller AC, Lochner M, Schneider E, Dy M, Eberl G, Leite-de-Moraes MC. Critical role of ROR-gammat in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci U S A. 2008;105:19845–50. doi: 10.1073/pnas.0806472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothchild AC, Jayaraman P, Nunes-Alves C, Behar SM. iNKT cell production of GM-CSF controls Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1003805. doi: 10.1371/journal.ppat.1003805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crowe NY, Coquet JM, Berzins SP, Kyparissoudis K, Keating R, Pellicci DG, Hayakawa Y, Godfrey DI, Smyth MJ. Differential antitumor immunity mediated by NKT cell subsets in vivo. J Exp Med. 2005;202:1279–88. doi: 10.1084/jem.20050953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–9. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 25.Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Sullivan LC, Kyparissoudis K, Brooks AG, Reid HH, Gras S, Lucet IS, Koh R, Smyth MJ, Mallevaey T, Matsuda JL, Gapin L, McCluskey J, Godfrey DI, Rossjohn J. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu TN, Lin KH, Chang YJ, Huang JR, Cheng JY, Yu AL, Wong CH. Avidity of CD1d-ligand-receptor ternary complex contributes to T-helper 1 (Th1) polarization and anticancer efficacy. Proc Natl Acad Sci U S A. 2011;108:17275–80. doi: 10.1073/pnas.1114255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel O, Cameron G, Pellicci DG, Liu Z, Byun HS, Beddoe T, McCluskey J, Franck RW, Castano AR, Harrak Y, Llebaria A, Bittman R, Porcelli SA, Godfrey DI, Rossjohn J. NKT TCR recognition of CD1d-alpha-C-galactosylceramide. J Immunol. 2011;187:4705–13. doi: 10.4049/jimmunol.1100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wun KS, Cameron G, Patel O, Pang SS, Pellicci DG, Sullivan LC, Keshipeddy S, Young MH, Uldrich AP, Thakur MS, Richardson SK, Howell AR, Illarionov PA, Brooks AG, Besra GS, McCluskey J, Gapin L, Porcelli SA, Godfrey DI, Rossjohn J. A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity. 2011;34:327–39. doi: 10.1016/j.immuni.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mallevaey T, Clarke AJ, Scott-Browne JP, Young MH, Roisman LC, Pellicci DG, Patel O, Vivian JP, Matsuda JL, McCluskey J, Godfrey DI, Marrack P, Rossjohn J, Gapin L. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity. 2011;34:315–26. doi: 10.1016/j.immuni.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Girardi E, Wang J, Yu ED, Painter GF, Kronenberg M, Zajonc DM. The Valpha14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J Exp Med. 2010;207:2383–93. doi: 10.1084/jem.20101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girardi E, Yu ED, Li Y, Tarumoto N, Pei B, Wang J, Illarionov P, Kinjo Y, Kronenberg M, Zajonc DM. Unique interplay between sugar and lipid in determining the antigenic potency of bacterial antigens for NKT cells. PLoS Biol. 2011;9:e1001189. doi: 10.1371/journal.pbio.1001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellicci DG, Clarke AJ, Patel O, Mallevaey T, Beddoe T, Le Nours J, Uldrich AP, McCluskey J, Besra GS, Porcelli SA, Gapin L, Godfrey DI, Rossjohn J. Recognition of beta-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol. 2011;12:827–33. doi: 10.1038/ni.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu ED, Girardi E, Wang J, Zajonc DM. Cutting edge: structural basis for the recognition of beta-linked glycolipid antigens by invariant NKT cells. J Immunol. 2011;187:2079–83. doi: 10.4049/jimmunol.1101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron G, Pellicci DG, Uldrich AP, Besra GS, Illarionov P, Williams SJ, La Gruta NL, Rossjohn J, Godfrey DI. Antigen Specificity of Type I NKT Cells Is Governed by TCR beta-Chain Diversity. J Immunol. 2015;195:4604–14. doi: 10.4049/jimmunol.1501222. [DOI] [PubMed] [Google Scholar]
- 35.Chamoto K, Guo T, Scally SW, Kagoya Y, Ancruzowski M, Wang CH, Rahman MA, Saso K, Butler MO, Chiu PP, Julien JP, Hirano N. Key Residues at Third CDR3beta Position Impact Structure and Antigen Recognition of Human Invariant NK TCRs. J Immunol. 2016 doi: 10.4049/jimmunol.1601556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tocheva AS, Mansour S, Holt TG, Jones S, Chancellor A, Sanderson JP, Eren E, Elliott TJ, Holt RI, Gadola SD. The Clonal Invariant NKT Cell Repertoire in People with Type 1 Diabetes Is Characterized by a Loss of Clones Expressing High-Affinity TCRs. J Immunol. 2017 doi: 10.4049/jimmunol.1600255. [DOI] [PubMed] [Google Scholar]
- 37.Mansour S, Tocheva AS, Sanderson JP, Goulston LM, Platten H, Serhal L, Parsons C, Edwards MH, Woelk CH, Elkington PT, Elliott T, Cooper C, Edwards CJ, Gadola SD. Structural and Functional Changes of the Invariant NKT Clonal Repertoire in Early Rheumatoid Arthritis. J Immunol. 2015;195:5582–91. doi: 10.4049/jimmunol.1501092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cruz Tleugabulova M, Escalante NK, Deng S, Fieve S, Ereno-Orbea J, Savage PB, Julien JP, Mallevaey T. Discrete TCR Binding Kinetics Control Invariant NKT Cell Selection and Central Priming. J Immunol. 2016;197:3959–3969. doi: 10.4049/jimmunol.1601382. [DOI] [PubMed] [Google Scholar]
- 39.Wei DG, Curran SA, Savage PB, Teyton L, Bendelac A. Mechanisms imposing the Vbeta bias of Valpha14 natural killer T cells and consequences for microbial glycolipid recognition. J Exp Med. 2006;203:1197–207. doi: 10.1084/jem.20060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schumann J, Mycko MP, Dellabona P, Casorati G, MacDonald HR. Cutting edge: influence of the TCR Vbeta domain on the selection of semi-invariant NKT cells by endogenous ligands. J Immunol. 2006;176:2064–8. doi: 10.4049/jimmunol.176.4.2064. [DOI] [PubMed] [Google Scholar]
- 41.Dougan SK, Ashour J, Karssemeijer RA, Popp MW, Avalos AM, Barisa M, Altenburg AF, Ingram JR, Cragnolini JJ, Guo C, Alt FW, Jaenisch R, Ploegh HL. Antigen-specific B-cell receptor sensitizes B cells to infection by influenza virus. Nature. 2013;503:406–9. doi: 10.1038/nature12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dougan SK, Dougan M, Kim J, Turner JA, Ogata S, Cho HI, Jaenisch R, Celis E, Ploegh HL. Transnuclear TRP1-specific CD8 T cells with high or low affinity TCRs show equivalent antitumor activity. Cancer Immunol Res. 2013;1:99–111. doi: 10.1158/2326-6066.CIR-13-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dougan SK, Ogata S, Hu CC, Grotenbreg GM, Guillen E, Jaenisch R, Ploegh HL. IgG1+ ovalbumin-specific B-cell transnuclear mice show class switch recombination in rare allelically included B cells. Proc Natl Acad Sci U S A. 2012;109:13739–44. doi: 10.1073/pnas.1210273109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brennan PJ, Tatituri RV, Brigl M, Kim EY, Tuli A, Sanderson JP, Gadola SD, Hsu FF, Besra GS, Brenner MB. Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol. 2011;12:1202–11. doi: 10.1038/ni.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirak O, Frickel EM, Grotenbreg GM, Suh H, Jaenisch R, Ploegh HL. Transnuclear mice with predefined T cell receptor specificities against Toxoplasma gondii obtained via SCNT. Science. 2010;328:243–8. doi: 10.1126/science.1178590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bilate AMBDML, Agudelo M, Leube J, Kratzert A, Dougan SK, Victora GD, Ploegh HL. Tissue-specific emergence of regulatory and intraepithelial T cells from a clonal T cell precursor. Science Immunology. 2016:1. doi: 10.1126/sciimmunol.aaf7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoue K, Wakao H, Ogonuki N, Miki H, Seino K, Nambu-Wakao R, Noda S, Miyoshi H, Koseki H, Taniguchi M, Ogura A. Generation of cloned mice by direct nuclear transfer from natural killer T cells. Curr Biol. 2005;15:1114–8. doi: 10.1016/j.cub.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 48.Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(−)CD4(+) CD1d-dependent precursor stage. J Exp Med. 2002;195:835–44. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–89. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baev DV, Caielli S, Ronchi F, Coccia M, Facciotti F, Nichols KE, Falcone M. Impaired SLAM-SLAM homotypic interaction between invariant NKT cells and dendritic cells affects differentiation of IL-4/IL-10-secreting NKT2 cells in nonobese diabetic mice. J Immunol. 2008;181:869–77. doi: 10.4049/jimmunol.181.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, Bendelac A. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–62. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bautista JL, Lio CW, Lathrop SK, Forbush K, Liang Y, Luo J, Rudensky AY, Hsieh CS. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol. 2009;10:610–7. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Georgiev H, Ravens I, Benarafa C, Forster R, Bernhardt G. Distinct gene expression patterns correlate with developmental and functional traits of iNKT subsets. Nat Commun. 2016;7:13116. doi: 10.1038/ncomms13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–26. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, Sefik E, Tan TG, Wagers AJ, Benoist C, Mathis D. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–95. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–53. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen NR, Brennan PJ, Shay T, Watts GF, Brigl M, Kang J, Brenner MB ImmGen Project C. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat Immunol. 2013;14:90–9. doi: 10.1038/ni.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.